Abstract
We characterized the dimer of the amyloid-β wild-type (WT) peptide, Aβ, of 42 residues and its disulfide-bond-locked double mutant (S8C) by replica exchange molecular dynamics simulations. Aβ dimers are known to be the smallest toxic species in Alzheimer’s disease, and the S8C mutant has been shown experimentally to form an exclusive homogeneous and neurotoxic dimer. Our 50 µs all-atom simulations reveal similar secondary structures and collision cross-sections but very different intramolecular and intermolecular conformations upon double S8C mutation. Both dimers are very dynamic with hundreds of free-energy minima that differ from the U-shape and S-shape conformations of the peptides in the fibrils. The only common structural feature, shared by both species with a probability of 4% in WT and 12% in S8C–S8C, is a three-stranded β-sheet spanning the 17–23, 29–36, and 39–41 residues, which does not exist in the Aβ40 WT dimers.
Graphical abstract
1. INTRODUCTION
Alzheimer’s disease features extracellular amyloid plaques made of intrinsically disordered protein (IDP) amyloid-β (Aβ) of 39–43 amino acids and intracellular neurofibrillary tangles resulting from the aggregation of the hyperphosphorylated tau protein of 440 amino acids.1 The wild-type (WT) Aβ42 peptide, DAEFRHDSGYEVHHQKLVFFAEDVGSN KGAIIGLMVGGVVIA, has two hydrophilic (residues D1-K16 and E22-G29) and hydrophobic (residues L17-A21 or the central hydrophobic core, CHC and A30–A42) regions. Aβ42 aggregates faster and is more cytotoxic than the most abundant Aβ40 peptide.2 Because toxicity within the amyloid cascade hypothesis is linked to Aβ misfolding, researchers have attempted to determine the structures of fibrils and all other transient aggregates.3,4 The Aβ dimers, known to be structurally heterogeneous among a pool of low-order oligomers, are of high interest as they are the smallest species to lead to tau hyperphosphorylation and neuritic degeneration.5,6
The existence of multiple aggregation pathways producing assemblies with different molecular structures is well established by in vitro and in Alzheimer’s disease brain experimental studies.3,7 Using seeded fibril growth from Alzheimer’s disease brain tissue of two patients and solid-state NMR, Tycko proposed distinct models that differ from in vitro synthetic Aβ40 fibril with U-shape.8 Riek et al. went one step beyond and reduced Aβ42 fibril polymorphism by successive seeding steps and optimal experimental growth conditions. On the basis of solid-state NMR and electron microscopy (EM), they obtained a stable fibrillar model with residues 2–6, 15–18, 26–28, 30–32, and 39–42 forming β-strands and residues 15–42 forming a double-horseshoelike cross-β or S-shape conformation, which is disease-relevant by recognition of monoclonal antibodies.9
Understanding how the dimer and higher aggregate conformations are used during the primary nucleation process and for the propagation of Alzheimer disease with different rates, phenotypes, and clinical declines is of paramount importance. In addition to varying growth conditions and photoinduced cross-linking of the two Y10 residues,10 the heterogeneity of Aβ oligomers can be reduced by engineering bonds at different locations.11–13 By introducing an intermolecular disulphide-bond at positions 8 (S8C) or 35 (M35C) and using biochemical neurophysiological analysis, Sticht designed an exclusive, homogeneous and neurotoxic Aβ dimer for the S8C mutant, in contrast to the AβM35C mutant that assembles into large oligomers.13 On the basis of 100 ns molecular dynamics (MD) simulations of Aβ42 WT and Aβ42 S8C starting from the U-shape conformation of Aβ42 fibrils,14 they proposed that the mutant S8C dimer displays structural properties similar to those of the WT dimer.
In this study, our aim is not to understand why the S8C mutant prevents oligomerization but what makes the S8C dimer neurotoxic. To this end, we compare the equilibrium ensembles of the Aβ42 WT and S8C dimers by all-atom AMBER99sb-ildn replica exchange MD (REMD) simulations using 48 replicas, each for 530 ns. This force field has been shown to fold successfully diverse proteins by MD15,16 and simulated tempering17 in explicit solvent and has led to structural properties for the Aβ42 WT monomer consistent with circular dichroism (CD) and local NMR observables.18 The Aβ42 WT dimer has already been subject to various simulations with coarse-grained (CG) models in implicit solvent,19–21 atomistic MD simulations of 1–4 µs in explicit solvent using OPLS22,23 or AMBER99sb-ildn,24 the TIP3P water model, and OPLS/TIP3P REMD simulations, with replicas of 200 ns25 and 400 ns.26 These simulations show common but different structural features because of variations in the force field, simulation, and analysis protocols; thus, we still have no definitive answer as to which Aβ42 WT dimer conformations are the most correct.
2. MATERIAL AND METHODS
2.1. Simulation Details
To minimize the impact of the choice of the starting structure on the results, the first seven Aβ42 monomer structures predicted by REMD with AMBER parm99sb and the generalized Born implicit solvent and representing 90% of the monomer ensemble were taken as input structures for both sequences (see Figure 8b in ref 27). Among the seven structures, four and three have some β-strand and α-helix contents, respectively. The 48 Aβ42 WT dimer structures were generated by combining randomly the seven monomeric structures and by randomly orienting them with a distance of 1.4–2.4 nm between the peptide centers of mass. Each system was centered in a truncated octahedron box of 225 nm3 with 6868 TIP3P water molecules,28 leading to a peptide concentration of 14.8 mM. The peptides at pH 7 have NH3+ and CO2− termini, deprotonated Glu and Asp, protonated Arg and Lys, and neutral His with a protonated Nε atom. The systems are neutralized by Na+ ions, resulting in 21 858 atoms for WT. Each dimer was then refined by a 50 ns NPT MD simulation with amber99sb-ildn/TIP3P, and the last frame was used for REMD. The same procedure was used for S8C, except that an S8C–S8C intermolecular disulfide bond was enforced. Overall, the secondary structure averaged over the 48 initial REMD structures amounts to 11.7% β-strand, 7% α-helix, and 82% coil-turn for WT and to 16.6% β-strand, 5.9% α-helix, and 77.5% coil-turn for S8C.
The GROMACS program was employed with periodic boundary conditions and a time step of 2 fs using SHAKE and the velocity Verlet integrator.29 Electrostatic interactions were calculated using the particle mesh Ewald method with a cutoff of 1.1 nm30 and van der Waals interactions with a cutoff of 1.2 nm, and a velocity-rescaling thermostat was used.31 REMD simulations were performed with 48 replicas from 300 to 400 K, using van der Spoel’s method.32 Exchanges between two replicas were attempted every 2 ps, leading to a mean acceptance ratio of 25%. Each replica ran for 530 ns.
2.2. Simulation Analysis
The Nter spans residues 1–15; CHC, residues 16–22; the loop, residues 23–30; and Cter, residues 31–42. The secondary structure was determined using the STRIDE program33 and the solvent accessible surface area (SASA) per amino acid using GROMACS. A side-chain–side-chain (SC–SC) contact is formed when the minimum distance between their heavy atoms is less than 4.5 Å,34 and the total number of SC–SC contacts is calculated by the Q value (see Methods in ref 35). A H-bond is formed when the acceptor–donor H angle is less than 30° and the acceptor–donor distance is less than 3.5 Å.27 A salt-bridge between charged SCs is formed when the distance between two specific atoms is within 4.6 Å.35 A β-hairpin is formed when there are at least two backbone H-bonds formed between two β-strands, with each strand consisting of at least three consecutive residues belonging to the Ramachandran β-region.35 The single-molecule states, which are the states of a single chain in the presence of another chain,36 were determined using dihedral principal component analysis (dPCA).37 The double-molecule states were determined using the PCA of the inverse distances between Cα atoms, and the overall dimer states are the product of the single- and double-molecule states.36 In the calculations, each system is projected on the eigenvectors obtained from the two REMD trajectories. To identify geometric clusters in these two subspaces, we performed clustering analysis as described in refs 35 and 37. The collision cross-sections (CCSs) were calculated using MOBCAL38 and the trajectory method reported in other simulations.25,35,37
3. RESULTS
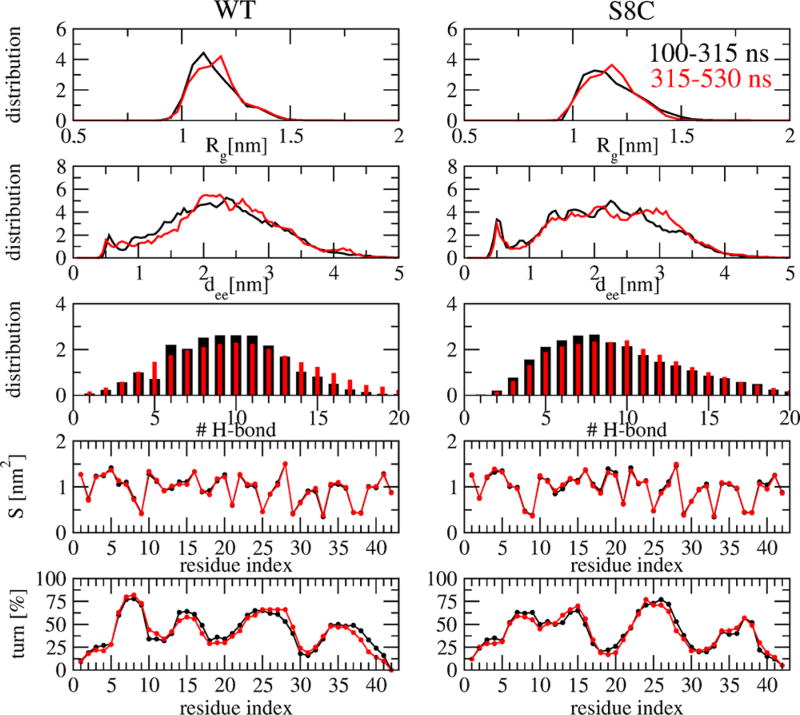
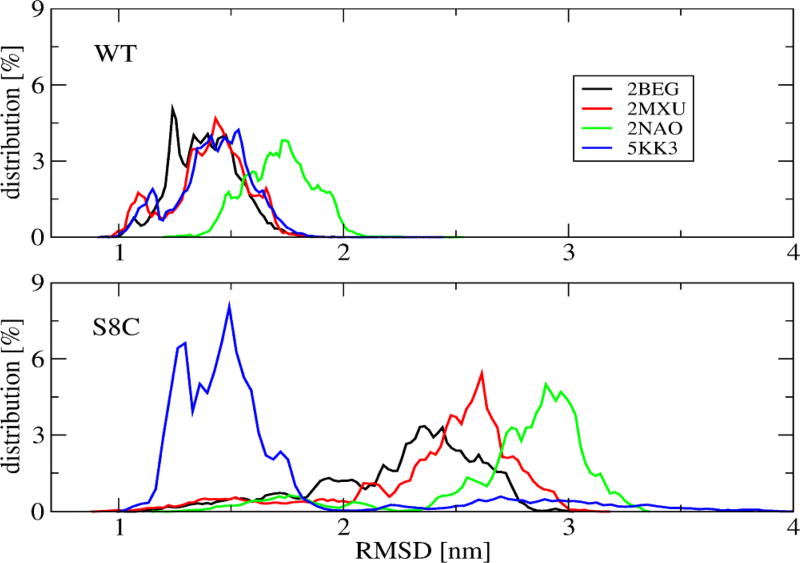
Figure 1 demonstrates the convergence of the REMD simulation at 315 K, near the physiological temperature, by comparing five metrics calculated over 100–315 and 315–530 ns that remain essentially unchanged over both intervals. The same metrics calculated between 100–400 and 100–530 ns are nearly identical, suggesting a simulation time of 400 ns for obtaining equilibrium structures (data not shown). The metrics include the radius of gyration, Rg, of individual chains, the intramolecular Cα end-to-end distance between the first and last residues (dee), the total number of intermolecular H-bonds, the SASA of each side-chain, and the percentage of each residue to be a turn. The very high profile similarity of Rg, dee, H-bond, and SASA distributions between WT and S8C seems to suggest that both dimers behave similarly, but this is not confirmed by the turn profile, where significant differences are observed in the 5–25 region (Figure 1), and the distributions of the root-mean-square deviation (RMSD) using residues 1–42 with respect to four Aβ42 fibril models (PDB entries 2BEG,14 2MXU,39 2NA0,9 and 5KK340), where the values lie within 1–2 nm for WT and 1–3.5 nm for S8C (Figure 2). Note that 2BEG has a U-shape and 2NA0 and 5KK3 have an S-shape, whereas 2MXU displays triple parallel β-sheet segments spanning residues 12–20, 27–32, and 36–41. In what follows, analysis was performed at 315 K within the time interval of 100–535 ns, and statistical deviations were determined by using block averages over different time intervals.
Figure 1.
Convergence of REMD simulations at 315 K. Shown are the distributions of various quantities including the radius of gyration (Rg), the Ca end-to-end distance (dee), the total number of intermolecular hydrogen bonds (H-bond), the SASA (S), and the turn secondary propensity of each residue. The results are calculated for two time intervals 100–315 ns (black curves) and 315–530 ns (red curves). The panels on the left and right display the results of the WT and S8C, respectively.
Figure 2.
Distribution of the RMSD of the single chains at 315 K of WT (upper) and S8C (lower). Shown are the results calculated with respect to different PDB structures (2BEG, 2MXU, 2NAO, 5KK3) using residues 1–42 in all calculations.
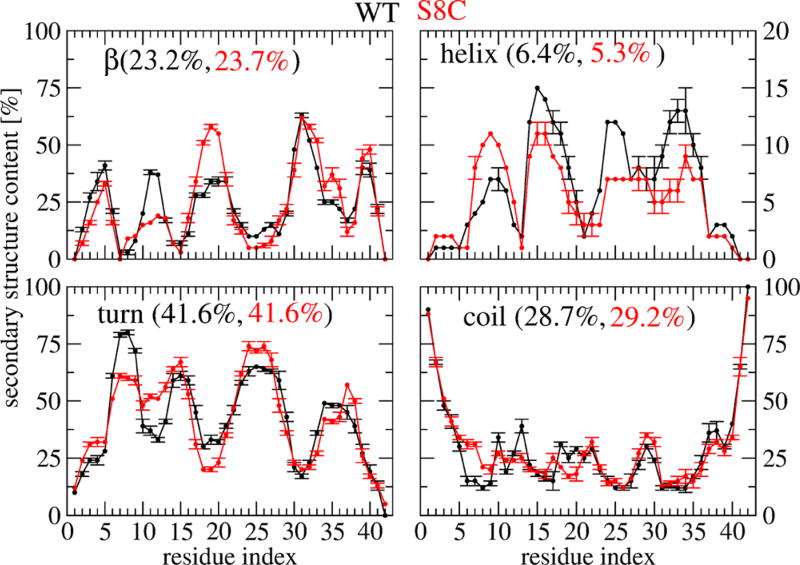
The percentage of the secondary structure averaged over all residues is identical between WT and S8C within statistical deviations: 23 ± 3.9% for β-strand, 6 ± 2% for α-helix, 41.6 ± 4% for turn, and 29 ± 4% for random coil. Figure 3 shows the secondary structure along each sequence. Whereas the coil profile is identical between WT and S8C, we note (a) for β-strand content a decrease from 37 to 12% for residues 11–12 and an increase from 34 to 60% for residues 17–18–19 upon S8C mutation, (b) for α-helix content a reduction for residues 15–35 of few percents compensated by an increase in the 7–10 region upon S8C mutation, and (c) for the turn region four distinct peaks centered at residues 8, 15, 24, and 35 in WT versus a broad distribution in the 6–15 region and two peaks centered at 24 and 37 in S8C.
Figure 3.
Secondary structure propensities of each amino acid of Aβ42 WT (black) and S8C dimers at 315 K with error bars obtained using three independent time windows.
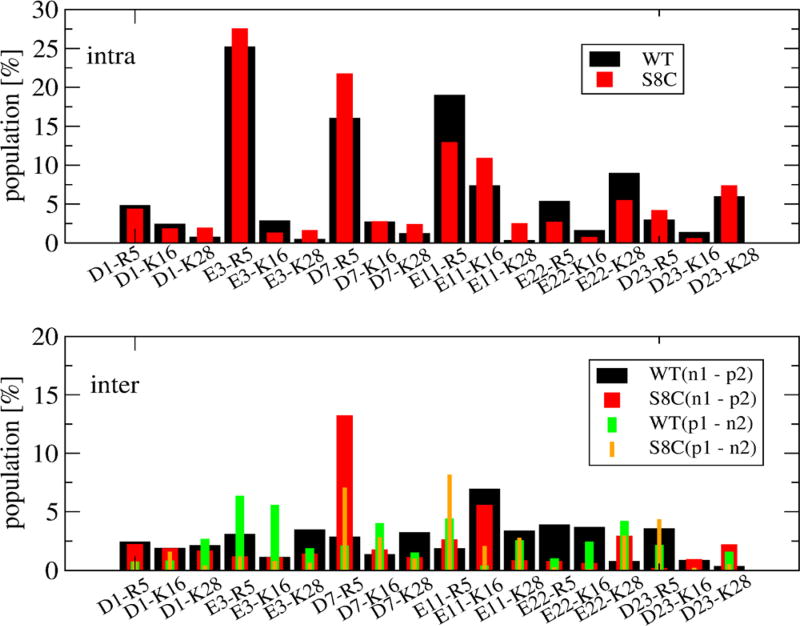
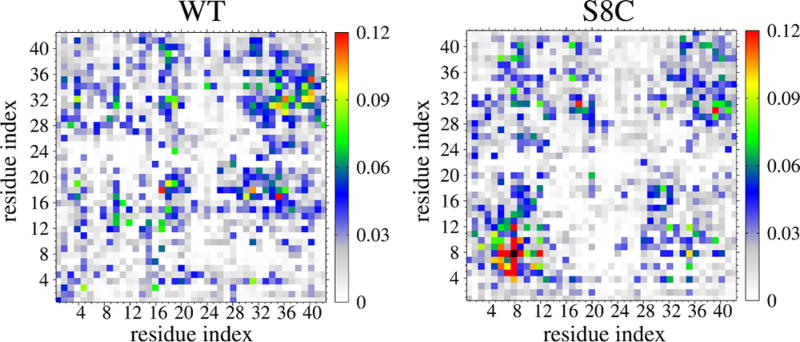
Although the populations of all intramolecular and intermolecular salt bridges formed in WT and S8C (in Figure 4) are very similar (with the exception of the long-range intramolecular E11-R5 contact varying from 19 to 12% and the intermolecular D7-R5 contact varying from 2 to 10% upon S8C mutation), there are substantial differences in the interpeptide SC–SC contact maps shown in Figure 5. Both contact maps are rather sparse, indicating a small number of interactions with probabilities between 9 and 12%. Looking at the WT map, we identify three dominant spots involving the two CHC regions (residues 17–20), the CHC region and the Cter (residues 32–40), and the two C-terminus regions (four maxima among residues 31–35, 32–36, 35–40, and 33–40). The S8C species reveals a completely different pattern with one clearly identified region involving residues 4–12, two spots involving Nter and Cter, and the two C-termini. As can be seen, the contacts are reduced between CHC and Cter and between the two N-termini upon S8C mutation.
Figure 4.
Population (in %) of intramolecular (upper) and intermolecular salt bridges formed in Aβ42 WT and S8C dimers. The salt-bridge distances are calculated between atoms CG (in Asp1, Asp7, or Asp23) or CD (in Glu3, Glu11, or Glu22) and atoms NH2 (in Arg5) or NZ (in Lys16 or Lys28). The notation (ni − pj) or (pi − nj) (i,j = 1,2) stands for the salt bridges between negative charged residues (Asp1, Glu3, Asp7, Glu11, Glu22, and Asp23) and positive charged residues (Arg5, Lys16, and Lys28) or vice versa. The error bars are at most of 3% using three independent time windows.
Figure 5.
Interpeptide side-chain–side-chain contact probabilities at 315 K. The black box in the right panel indicates the 100% probability of the CYS8–CYS8 contact.
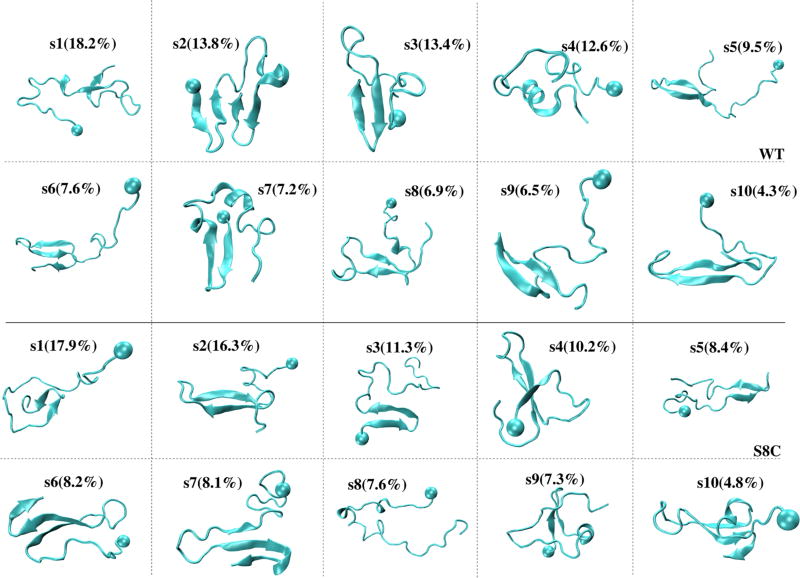
As a first step to characterize the similarities and differences between the equilibrium ensembles of WT and S8C, we determined the single-molecule states using dPCA and the k-means clustering leads to 10 free-energy minima denoted s1–s10 for WT and S8C. The conformations closest to the center of each minimum are shown in Figure 6. For each free-energy minimum and using all conformations belonging to each state, we give in Table 1 the population, the (β-strand, α-helix) locations, and the averaged values of the Q number of intramolecular SC–SC contacts between Nter and CHC, CHC and Cter, and Nter and Cter. For both sequences, the populations vary between 18% (s1) and 4% (s10). The 10 single-molecule states of WT (S8C) have Q = 3–5 (2–5) Nter–CHC contacts, Q = 4–9 (3–10) CHC–Cter contacts, and Q = 2–6 (2–6) Nter–Cter contacts. The first five states (s1–s5) represent 67 and 53% of the ensemble in WT and S8C, respectively.
Figure 6.
Representative structures of the 10 single-molecule states of Aβ42 WT (upper panels) and S8C (lower panels) dimers. The ball indicates the first residue.
Table 1.
Characterization of the 10 Single-Molecule States of the Aβ42 WT and S8C Dimersa
| system | state | P | (β, α)a | (β, α)b | (β, α)c | N1 | N2 | N3 | resα | resβ |
|---|---|---|---|---|---|---|---|---|---|---|
| WT | 1 | 18.21 | (21, 2) | (21, 4) | (34, 5) | 5 | 5 | 3 | 15–17 | 20–22, 37–39 |
| 2 | 13.78 | (28, 2) | (10, 7) | (35, 12) | 4 | 5 | 4 | 25–29 | 2–5, 11–13, 31–33, 38–40 | |
| 3 | 13.43 | (17, 2) | (22, 1) | (30, 5) | 5 | 7 | 5 | 30–33, 37–40 | ||
| 4 | 12.56 | (17, 6) | (10, 16) | (27, 10) | 4 | 4 | 3 | 28–36 | ||
| 5 | 9.52 | (24, 3) | (66, 0) | (54, 0) | 6 | 9 | 3 | 17–23, 28–33 | ||
| 6 | 7.64 | (11, 4) | (69, 0) | (51, 1) | 3 | 9 | 2 | 19–22, 29–32, 36–37 | ||
| 7 | 7.18 | (31, 9) | (3, 28) | (15, 29) | 4 | 4 | 6 | 14–16, 24–29 | 3–6, 10–13 | |
| 8 | 6.87 | (6, 8) | (24, 3) | (29, 2) | 4 | 6 | 2 | 9–11 | 18–20, 33–35 | |
| 9 | 6.49 | (6, 10) | (17, 34) | (54, 3) | 5 | 7 | 3 | 21–27, 30–36 | ||
| 10 | 4.33 | (1, 9) | (68, 0) | (60, 9) | 3 | 9 | 4 | 17–21, 30–36, 39–40 | ||
| S8C | 1 | 17.92 | (14, 3) | (28, 3) | (35, 4) | 5 | 6 | 3 | 30–31, 35–36 | |
| 2 | 16.28 | (8, 11) | (51, 0) | (47, 3) | 4 | 9 | 4 | 18–22, 29–33 | ||
| 3 | 11.25 | (19, 3) | (8, 12) | (28, 7) | 4 | 3 | 3 | 3–6, 9–13 | ||
| 4 | 10.22 | (7, 4) | (66, 1) | (51, 2) | 3 | 9 | 3 | 3–5, 17–21, 30–34 | ||
| 5 | 8.42 | (17, 1) | (61, 0) | (40, 2) | 2 | 9 | 4 | 19–21, 28–30 | ||
| 6 | 8.23 | (6, 8) | (73, 0) | (72, 0) | 2 | 10 | 5 | 16–20, 31–36, 39–40 | ||
| 7 | 8.06 | (15, 1) | (33, 2) | (39, 6) | 4 | 6 | 2 | 19–21, 30–33, 38–40 | ||
| 8 | 7.63 | (13, 10) | (4, 19) | (19, 5) | 5 | 5 | 3 | 7–9, 14–16 | ||
| 9 | 7.25 | (30, 6) | (1, 36) | (20, 22) | 5 | 5 | 5 | 15–19 | 4–5, 11–12 | |
| 10 | 4.75 | (4, 10) | (72, 0) | (76, 0) | 3 | 10 | 6 | 9–11 | 16–20, 31–36, 39–40 |
Shown are the state population P (in %), the population of β-strand and helix (β, α)a,b,c spanning the Nter (a), CHC (b), and Cter (c) regions, the average Q value of intramolecular contacts between Nter–CHC (N1), Cter–CHC (N2), Nter–Cter (N3), and the positions of the residues forming helical (Resa) and β (Resb) segments. Using two independent time intervals, the mean error bar of the populations for both species is ±2.5%.
In both species, the equilibrium ensemble consists of highly disordered structures with short β-hairpins (six states in WT vs five states in S8C) spanning various locations (CHC–Cter, Nter, Cter, and loop–Cter) and amino acid positions. For instance, within the CHC–Cter region, there are β-hairpins spanning 20–22 and 37–39 (s1) or 18–20 and 33–35 (s8) in WT and spanning 18–22 and 29–33 (s2) in S8C. Within the Nter region, there is a β-hairpin spanning 3–6 and 10–13 (s7) in WT and spanning 3–6 and 9–13 (s3) or 4–5 and 11–12 (s9) in S8C. Within the Cter region, we find a β-hairpin at positions 30–31 and 35–36 (s1) in S8C and at positions 30–33 and 37–40 (s3) in WT. Of interest is the presence of a β-hairpin spanning the Cter (residues 30–36) and loop (23–37) regions in WT (s9), which is absent in S8C.
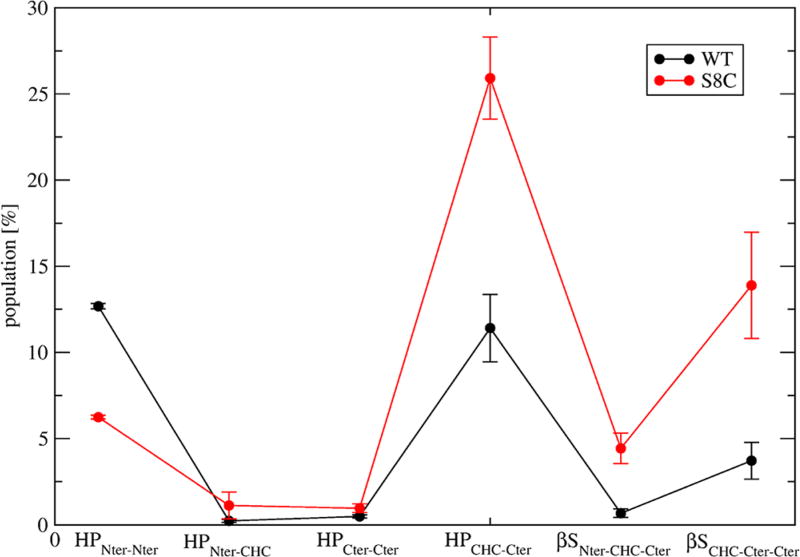
Then, the two ensembles contain transient three-stranded β-sheets: two states in WT spanning CHC–Cter–Cter: s6 (residues 19–22, 29–32, and 36–37) and s10 (residues 17–21, 30–36, and 39–40) versus four states in S8C: s4 spanning Nter–CHC–Cter and the s6, s7, and s10 states (21%) spanning CHC–Cter–Cter. Finally, the equilibrium structures also contain transient four-stranded β-sheets in WT (s2) with two strands in Nter (residues 2–5 and 11–13) and two strands in Cter (residues 31–33 and 38–40), which are absent in S8C, and more transient 3–10 and α-helical conformations in WT (s4, Figure 6) than in S8C. Overall, as seen in Figure 7, which reports the populations of all types of β-sheets, upon S8C mutation, the population of β-hairpin drops from 12.5 to 6% in the Nter region and increases from 11 to 26% in the CHC–Cter region and the population of intramolecular three-stranded β-sheets increases from 1 to 4% in the Nter–CHC–Cter region and from 3.8 to 14% in the CHC–Cter–Cter region.
Figure 7.
Populations (in %) of Aβ42 WT and S8C dimers forming β-hairpins only at Nter (HPNter–Nter), between Nter and CHC (HPNter–CHC), at Cter (HPCter–CHC), and between CHC and Cter (HPCHC–Cter). The populations of intramolecular three-stranded β-sheets at Nter–CHC–Cter (βSNter–CH–Cter) and CHC–Cter–Cter (βSCHC–Cter–Cter) are also shown. Error bars based on three independent time windows.
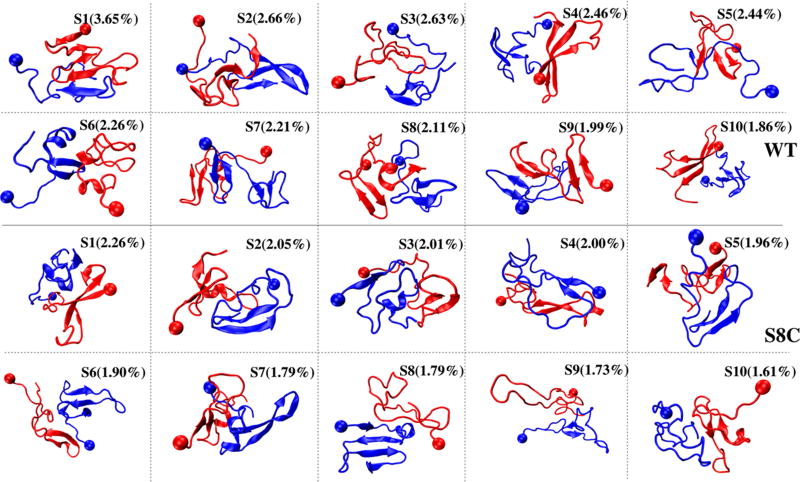
The 10 dominant clusters of Aβ42 WT and S8C dimers denoted S1–S10 of populations between 3.6 and 1.8% for WT and between 2.3 and 1.6% for S8C, representing therefore 24 and 18% of all conformations, are displayed in Figure 8 and analyzed in Table 2. In total, there are 400 clusters in both WT and S8C. Each structure refers to the center of a cluster. Each cluster is described by its two single-molecule s states (Figure 8), α-helix, turn and β-strand contents, and CCS values. Also, we give the Q number of interpeptide SC–SC contacts between the Nter and CHC region (N1), the Cter and CHC region (N2), Nters (N3), the CHC and CHC region (N4), and Cters (N5). All values are obtained using all conformations belonging to each cluster. The first 10 states of each species are highly disordered structures with various topologies, interfaces, and compositions of single-molecule states. The 20 states have a β-strand, varying between 14 and 38%, but four states in WT (S1, S3, S6, and S9) have between 3 and 13% of α-helix, whereas seven states in S8C have between 3 and 15% of α-helix.
Figure 8.
Representative structures of the first 10 overall states of the Aβ42 WT and S8C dimers. The overall state Si of the WT dimer is formed by two single-molecule states (sk, sl) with S1 = (s2, s1), S2 = (s6, s5), S3 = (s5, s4), S4 = (s6, s10), S5 = (s7, s1), S6 = (s8, s4), S7 = (s3, s2), S8 = (s7, s5), S9 = (s7, s2), and S10 = (s9, s6). The overall states of the S8C dimer consist of S1 = (s4, s1), S2 = (s2, s9), S3 = (s1, s9), S4 = (s2, s3), S5 = (s3, s2), S6 = (s10, s5), S7 = (s9, s2), S8 = (s8, s6), S9 = (s8, s5), and S10 = (s2, s8). The representative structures of the single-molecule states are shown in Figure 6. The ball indicates the first residue.
Table 2.
Characterization of the First 10 Overall States of the Aβ42 WT and S8C Dimersa
| system | state | P | β | α | turn | N1 | N2 | N3 | N4 | N5 | CCS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | 1 | 3.65 | 22 | 6 | 51 | 2 | 7 | 1 | 6 | 18 | 1229 |
| 2 | 2.66 | 31 | 1 | 37 | 2 | 5 | 3 | 1 | 8 | 1296 | |
| 3 | 2.63 | 27 | 4 | 40 | 3 | 6 | 4 | 4 | 0 | 1232 | |
| 4 | 2.46 | 38 | 1 | 41 | 4 | 9 | 5 | 0 | 5 | 1312 | |
| 5 | 2.44 | 35 | 1 | 37 | 2 | 7 | 5 | 18 | 2 | 1374 | |
| 6 | 2.26 | 18 | 13 | 40 | 3 | 0 | 1 | 19 | 4 | 1285 | |
| 7 | 2.21 | 37 | 0 | 35 | 2 | 6 | 4 | 2 | 1 | 1315 | |
| 8 | 2.11 | 34 | 1 | 41 | 3 | 3 | 2 | 1 | 4 | 1359 | |
| 9 | 1.99 | 22 | 4 | 41 | 5 | 6 | 1 | 5 | 9 | 1301 | |
| 10 | 1.86 | 32 | 1 | 45 | 6 | 2 | 2 | 0 | 3 | 1352 | |
| S8C | 1 | 2.26 | 24 | 3 | 45 | 1 | 2 | 7 | 0 | 8 | 1313 |
| 2 | 2.05 | 31 | 5 | 42 | 0 | 2 | 11 | 0 | 4 | 1212 | |
| 3 | 2.01 | 18 | 12 | 43 | 0 | 7 | 4 | 1 | 11 | 1234 | |
| 4 | 2.00 | 27 | 3 | 36 | 2 | 4 | 6 | 3 | 11 | 1269 | |
| 5 | 1.96 | 29 | 1 | 38 | 1 | 2 | 10 | 0 | 3 | 1264 | |
| 6 | 1.90 | 37 | 4 | 38 | 1 | 1 | 6 | 0 | 1 | 1389 | |
| 7 | 1.79 | 26 | 15 | 42 | 4 | 1 | 10 | 0 | 9 | 1321 | |
| 8 | 1.79 | 21 | 7 | 41 | 2 | 0 | 8 | 0 | 1 | 1281 | |
| 9 | 1.73 | 22 | 2 | 48 | 2 | 4 | 8 | 5 | 5 | 1454 | |
| 10 | 1.61 | 14 | 5 | 58 | 1 | 4 | 5 | 3 | 7 | 1240 |
Shown are the state population P (in %); the average values of various secondary structure contents (in %); the average Q value of the intermolecular contacts between Nter and CHC (N1), Cter and CHC (N2), Nter and Nter (N3), CHC and CHC (N4), and Cter and Cter (N5); and the CCS (in Å2) of each representative structure. Using two independent time intervals, the mean error bar of the populations for both species is ±1.5%.
In the first WT state, S1, with a population of 3.6% and consisting of the s1 and s2 states, the dimer forms a five-stranded β-sheet with an intermolecular parallel β-sheet at the Cter spanning residues 37–40 and 31–33. This state, displaying one chain with an extended Nter and the second chain with a β-hairpin spanning Cter, is dominated by intermolecular Cter–Cter contacts (N5 = 18) followed by intermolecular Cter–CHC (N2 = 7) and CHC–CHC (N4 = 6) contacts. The S2 WT state, with a population of 2.6% and consisting of the s6 and s5 states with two rather extended Nters, displays an intermolecular antiparallel β-sheet spanning residues 16–22 (CHC) and 38–40 (Cter), with a small number of intermolecular CHC–CHC (N4 = 1) contacts.
For the S8C dimer, S1, of population 2.3% and with one chain with three β-strands spanning Nter–CHC–Cter, displays the same number of intermolecular Nter–Nter (N3 = 7) and Cter–Cter (N5 = 8) contacts and an interpeptide antiparallel β-sheet spanning residues 39–40. The S8C S2 state, of population 2.1%, with one chain displaying an α-helix at residues 15–19 and a β-hairpin spanning the Cter and the second chain with a β-hairpin spanning residues 18–22 and 29–33, is characterized by more intermolecular Nter–Nter (N3 = 11) contacts than Cter–Cter (N5 = 4) contacts.
The high dissimilarity between the first 10 states of WT and S8C is revealed by the values of the N1–N5 intermolecular contacts. As seen in Table 2, the WT states are characterized by a high number of Cter–CHC interactions (N2 on the order of 5.1 for WT vs 2.7 for S8C) and CHC–CHC interactions (N4 varying between 0 and 19 for WT). On the other hand, the S8C states are characterized by a higher number of Nter–Nter interactions (N3 varying between 1 and 5 in WT vs 4 and 11 in S8C, as expected from the formation of the disulfide bridge) and a lower number of CHC–CHC interactions (N4 varying between 0 and 5 for S8C). Considering the first 100 states, which represent 79 and 72% of the full WT and SC equilibrium ensembles, does not change the previous pictures.
4. DISCUSSION
We have only low-resolution-structure information on the Aβ42 WT dimers. The Aβ42 dimer size was investigated by an IM-MS study, leading to a mean CCS of 1252 Å2.41 The CCS values of the first 10 WT clusters range between 1229 and 1374 Å2. Using the first 200 clusters, the calculated CCS is 1269 Å2, in perfect agreement with the experiment but very different from those reported by a recent OPLS/TIP3P REMD simulation, with CCS values of the first four clusters ranging between 923 and 1053 Å2.26 This discrepancy may arise from the choice of the force fields but also from the method used to identify the most populated clusters. We estimate the clusters by a very complex procedure that considers the product basis of single-molecule and double-molecule states and identified a total of 400 clusters, whereas the previous REMD simulations calculated the potential of mean force using the average number of intrachain CHC–Cter contacts and the total number of interchain contacts between CHC and Cter and identified four dominant clusters with a population of 74%.26
The secondary structure composition was investigated by two CD experiments on the Aβ42 WT peptide at pH 7 and led to the α-helix content of 3% and β-strand content of 13% at time zero and thus consisting of monomers and small oligomers.2,42 Using the same sample preparation, identical results were obtained for the Aβ40 WT peptide, but another study based on another separation procedure for the Aβ40 WT dimer led to 9% of helix and 38.6% of β-sheet.43 Our AMBER99sb-ildn/TIP3P simulation gave 6 ± 2% of α-helix and 23 ± 3.9% of β-strand, in agreement with the first set of CD experiments and the recent REMD simulation using OPLS/TIP3P (<3%, 15–19%).26 For comparison, multiple short OPLS/TIP3P MD simulations starting from CG predictions22 and a 4 µs MD trajectory followed by 500 ns of accelerated MD simulations using AMBER99sb-ildn/TIP3P24 gave α-helix and β-strand of 1 and 6% and 4 and 5%, respectively, suggesting limited sampling of the conformational space. In contrast, atomistic Monte Carlo simulations with an implicit solvent predicted a too high β-strand population.44
Looking at the β-strand and turn profiles along the WT sequence, we find five regions with high strand propensity (>20%): residues 2–6 and 11–13, the CHC region, and residues 30–35 and 39–41, that are connected by turns at positions 7–8, 14–16, 24–29, and 34–37. Similar profiles with different contents have been observed by OPLS/TIP3P REMD simulations for the Aβ42 dimer26 and AMBER99sb-ildn/TIP4P-Ew REMD simulations for the Aβ42 monomer.45 For instance, the former predicts turn propensity of 90% in the loop region versus 67% in this study and β-strand propensity of >20% for CHC and residues 31–40 and 10–15% for residues 2–3 and 5–7.26
The WT simulations confirm some three-dimensional (3D) features that have already discussed by previous Aβ42 monomer45–48 and dimer21,26 simulations using various protein force fields (CG/atomistic) and solvent representations (implicit/explicit): the high propensity of CHC and the N-terminus of Cter (residues 30–36) for transient β-hairpins that also form in Aβ40 and the high probability of residues 17–21, 30–36, and 39–41 to adopt transient intramolecular three-stranded β-sheets, which are absent in Aβ40.37,46
However, our extensive simulations reveal new features. First, there are no well-defined interfaces in the dimer. Whereas previous MD and REMD simulations reported that the Aβ42 dimers are stabilized primarily by intermolecular hydrophobic interactions of the CHC regions,24 of the CHC and Cter regions,25,26 or between the Cter and Cter regions with some CHC–CHC contributions,21,26 we find a diverse ensemble of interfaces stabilized by intermolecular interactions between any pairs of segments, that is, Nter–Nter, Nter–CHC, CHC–CHC, CHC–Cter, Cter–Cter, and Nter–Cter, indicating that almost all orientations and positions of the peptides are populated in the dimers. Second, the dimer cannot be described by a few free-energy minima24,26 but by a plethora of minima that can act as seeds for further polymerization. Although there is a high propensity for β-strands at the CHC and Cter regions compatible with the fibril of the Aβ42 synthetic peptide,4,14 some dimers contain β-strands in the loop region (residues 25–28) and the N-terminus (residues 2–6) compatible with the disease-relevant Aβ42 fibril model.9 It is of interest that two-dimensional IR experiments on amylin suggested the initial stages of aggregation involving β-sheet formation in the loop region.49 Whether the formation of a short β-strand at positions 2–6, already discussed in the simulation of the Aβ42 monomer with AMBERs99-ildn/TIP4P but not with OPLS,45 is an artifact of the force field remains to be determined.
We do not have any CD and IM-MS data on Aβ42 S8C dimers. Our simulations reveal that the CCS value (1276 Å2) and the secondary structures are rather invariant. Upon S8C–S8C mutation, the β-strand content varies however with a decrease of 75% for residues 11–12 and an increase of 50% for residues 17–18–19, and this is accompanied by the presence of three peaks in the turn profile versus four in WT, with a change in the region 6–15 and an increase in the turn content of residues 36–37, the latter residues in turn are considered as the sine qua non of A42β WT with respect to Aβ40 WT.50 Upon mutation, the lifetime of β-hairpins in CHC–Cter and that of a double β-hairpin spanning residues 17–21, 30–36, and 39–41 increase by a factor of 2 and 3, respectively. A comparison of the single-molecule and overall states between the WT and S8C dimers reveals substantial differences in terms of intramolecular and intermolecular conformations. However, detailed analysis reveals one common structural feature shared by both species, that is, the double β-hairpin discussed above, in the single-molecule states (state s10 in WT and states s6 and S10 in S8C in Figure 6) and in the overall dimer conformations (e.g., state S4 in WT and states S6 and S8 in S8C in Figure 8). Application of our clustering procedure and the standard RMSD clustering (using all main-chain and side-chain atoms of residues 17–42 or 9–42 and a cutoff of 3 Å) leads to the populations of 4 and 12% of double hairpin in WT and S8C, respectively. Therefore, it is reasonable to speculate that the toxicity in the WT and S8C species is at least mediated by this 3D structure. This result is consistent with recent experimental evidence that the Aβ39–42 peptide can modulate Aβ oligomerization and toxicity.51,52
5. CONCLUSIONS
We have characterized the structure of the Aβ1–42 dimer and its disulfide-bond-locked double mutant (S8C) using extensive REMD simulations at pH 7. Our goal was not to understand why the intermolecular disulfide bond prevents Aβ oligomerization upon S8C mutation. REMD simulations in explicit solvent are computationally demanding, and only CG simulations of the Aβ1–42 S8C tetramers could provide physical insights explaining this fact.53–55 Instead, in this study, we reveal that the intermolecular disulfide bond at position 8 significantly impacts the equilibrium structures and propose one possible structural scenario for preserving the toxicity of the S8C mutant that cannot form large aggregates. This study, in concert with recent simulations and experiments on new variants,56–59 may help design new inhibitors for early-stage Alzheimer disease.60–62
Acknowledgments
This work was supported by grants from the GRAL ANR SIMI 12-BS07-0017-01, the National Institute of Health (NIH-R01GM118508), the National Science Foundation (NSF, grant SI2-SEE-1534941), and computer facilities by the NC state HPC, and the IDRIS, CINES, and TGCC centers (grant x2016077604).
Footnotes
The authors declare no competing financial interest.
References
- 1.Selkoe DJ, Hardy J. The Amyloid Hypothesis of Alzheimer’s Disease at 25 Years. EMBO Mol. Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid-b Protein (Ab) Assembly: Ab40 and Ab42 Oligomerize Through Different Pathways. Proc. Natl. Acad. Sci. U.S.A. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nasica-Labouze J, Nguyen PH, Sterpone F, Berthoumieu O, Buchete NV, Coté S, De Simone A, Doig AJ, Faller P, Garcia A, et al. Amyloid beta Protein and Alzheimer’s Disease: When Computer Simulations Complement Experimental Studies. Chem. Rev. 2015;115:3518–3563. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riek R, Eisenberg DS. The Activities of Amyloids From a Structural Perspective. Nature. 2016;539:227–235. doi: 10.1038/nature20416. [DOI] [PubMed] [Google Scholar]
- 5.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Natural Oligomers of the Amyloid-beta Protein Specifically Disrupt Cognitive Function. Nat. Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 6.Jin M, Shepardson N, Yang T, Chen G, Walsh D, Selkoe DJ. Soluble Amyloid beta-protein Dimers Isolated from Alzheimer Cortex directly Induce Tau Hyperphosphorylation and Neuritic Degeneration. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5819–5824. doi: 10.1073/pnas.1017033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tycko R. Amyloid Polymorphism: Structural Basis and Neurobiological Relevance. Neuron. 2015;86:632–45. doi: 10.1016/j.neuron.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular Structure of β-amyloid Fibrils in Alzheimer’s Disease Brain Tissue. Cell. 2013;154:1257–68. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wälti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Böckmann A, Güntert P, Meier BH, Riek R. Atomic-resolution Structure of a Disease-relevant Aβ(1–42) Amyloid Fibril. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahimi F, Maiti P, Bitan GJ. Photo-induced Cross-linking of Unmodified Proteins (PICUP) Applied to Amyloidogenic Peptides. J. Visualized Exp. 2009;23 doi: 10.3791/1071. No. e1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandberg A, Luheshi LM, Söllvander S, Pereira de Barros T, Macao B, Knowles TP, Biverstål H, Lendel C, Ekholm-Petterson F, Dubnovitsky A, et al. Stabilization of Neurotoxic Alzheimer Amyloid-beta Oligomers by Protein Engineering. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15595–15600. doi: 10.1073/pnas.1001740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi T, Yagi H, Goto Y, Matsuzaki K, Hoshino M. A Disulfide-linked Amyloid-beta Peptide Dimer Forms a Protofibril-like Oligomer Through a Distinct Pathway from Amyloid Fibril formation. Biochemistry. 2010;49:7100–7107. doi: 10.1021/bi100583x. [DOI] [PubMed] [Google Scholar]
- 13.Müller-Schiffmann A, Andreyeva A, Horn AH, Gottmann K, Korth C, Sticht H. Molecular Engineering of a Secreted, Highly Homogeneous, and Neurotoxic Aβ Dimer. ACS Chem. Neurosci. 2011;2:242–248. doi: 10.1021/cn200011h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. 3D Structure of Alzheimer’s Amyloid-beta(1–42) Fibrils. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, et al. Atomic-level Characterization of the Structural Dynamics of Proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 16.Saladino G, Marenchino M, Gervasio FL. Bridging the Gap between Folding Simulations and Experiments: The Case of the Villin Headpiece. J. Chem. Theory Comput. 2011;7:2675–2680. doi: 10.1021/ct2002489. [DOI] [PubMed] [Google Scholar]
- 17.Man HV, Derreumaux P, Nguyen PH. Communication: Multiple Atomistic Force Fields in a Single Enhanced Sampling Simulation. J. Chem. Phys. 2015;143 doi: 10.1063/1.4926535. No. 021101. [DOI] [PubMed] [Google Scholar]
- 18.Carballo-Pacheco M, Strodel B. Comparison of Force Fields for Alzheimer’s Aβ42: A Case Study for Intrinsically Disordered Proteins. Protein Sci. 2017;26:174–185. doi: 10.1002/pro.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Côté S, Laghaei R, Derreumaux P, Mousseau N. Distinct Dimerization for Various Alloforms of the amyloid-beta Protein: Abeta(1–40), Abeta(1–42), and Abeta(1–40)(D23N) J. Phys. Chem. B. 2012;116:4043–4055. doi: 10.1021/jp2126366. [DOI] [PubMed] [Google Scholar]
- 20.Sterpone F, Melchionna S, Tuffery P, Pasquali S, Mousseau N, Cragnolini T, Chebaro Y, St-Pierre J, Kalimeri M, Barducci A, et al. The OPEP Protein Model: From Single Molecules, Amyloid Formation, Crowding and Hydrodynamics to DNA/RNA Systems. Chem. Soc. Rev. 2014;43:4871–4893. doi: 10.1039/c4cs00048j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urbanc B, Betnel M, Cruz L, Bitan G, Teplow DB. Elucidation of Amyloid β-protein Oligomerization Mechanisms: Discrete Molecular Dynamics Study. J. Am. Chem. Soc. 2010;132:4266–4280. doi: 10.1021/ja9096303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barz B, Urbanc B. Dimer Formation Enhances Structural Differences between Amyloid β-protein (1–40) and (1–42): an Explicit-solvent Molecular Dynamics Study. PloS One. 2012;7 doi: 10.1371/journal.pone.0034345. No. e34345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Man VH, Nguyen PH, Ngo ST, Li MS, Derreumaux P. Effect of the Tottori Familial Disease Mutation (D7N) on the Monomers and Dimers of Aβ40 and Aβ42. ACS Chem. Neurosci. 2013;4:1446–1457. doi: 10.1021/cn400110d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Hashemi M, Lv Z, Lyubchenko YL. Self-assembly of the Full-length Amyloid Aβ42 Protein in Dimers. Nanoscale. 2016;8:18928–18937. doi: 10.1039/c6nr06850b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T, Zhang J, Derreumaux P, Mu Y. Molecular Mechanism of the Inhibition of EGCG on the Alzheimer A β(1–42) Dimer. J Phys Chem B. 2013;117:3993–4002. doi: 10.1021/jp312573y. [DOI] [PubMed] [Google Scholar]
- 26.Das P, Chacko AR, Belfort G. Alzheimer’s Protective Cross-interaction between wild-type and A2T Variants Alters Abeta42 Dimer Structure. ACS Chem. Neurosci. 2016 doi: 10.1021/acschemneuro.6b00357. in press. [DOI] [PubMed] [Google Scholar]
- 27.Yang M, Teplow DB. Amyloid beta-protein Monomer Folding: Free-energy Surfaces Reveal Alloform-specific Differences. J. Mol. Biol. 2008;384:450–464. doi: 10.1016/j.jmb.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 29.van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 30.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 31.Bussi G, Donadio D, Parrinello M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007;126 doi: 10.1063/1.2408420. No. 014101. [DOI] [PubMed] [Google Scholar]
- 32.Patriksson A, van der Spoel DA. Temperature Predictor for Parallel Tempering Simulations. Phys. Chem. Chem. Phys. 2008;10:2073–2077. doi: 10.1039/b716554d. [DOI] [PubMed] [Google Scholar]
- 33.Frishman D, Argos P. Knowledge-based Protein Secondary Structure Assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 34.Zhang T, Xu W, Mu Y, Derreumaux P. Atomic and Dynamic Insights into the Beneficial Effect of the 1,4-naphthoquinon-2-yl-L-tryptophan Inhibitor on Alzheimer’s Aβ1–42 Dimer in terms of Aggregation and Toxicity. ACS Chem. Neurosci. 2014;5:148–59. doi: 10.1021/cn400197x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen PH, Sterpone F, Campanera JM, Nasica-Labouze J, Derreumaux P. Impact of the A2V Mutation on the Heterozygous and Homozygous Aβ1–40 Dimer Structures from Atomistic Simulations. ACS Chem. Neurosci. 2016;7:823–832. doi: 10.1021/acschemneuro.6b00053. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen PH, Li MS, Derreumaux P. Amyloid Oligomer Structure Characterization from Simulations: a General Method. J. Chem. Phys. 2014;140 doi: 10.1063/1.4866902. No. 094105. [DOI] [PubMed] [Google Scholar]
- 37.Tarus B, Tran TT, Nasica-Labouze J, Sterpone F, Nguyen PH, Derreumaux P. Structures of the Alzheimer’s Wild-Type Aβ1–40 Dimer from Atomistic Simulations. J. Phys. Chem. B. 2015;119:10478–10487. doi: 10.1021/acs.jpcb.5b05593. [DOI] [PubMed] [Google Scholar]
- 38.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. Structural Information from Ion Mobility Measurements: Effects of the Long-Range Potential. J. Phys. Chem. 1996;100:16082–16086. [Google Scholar]
- 39.Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y. Aβ (1–42) Fibril Structure Illuminates Self-recognition and Replication of Amyloid in Alzheimer’s Disease. Nat. Struct. Mol. Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, et al. Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils. J. Am. Chem. Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea JE, Ruotolo BT, Robinson CV, et al. Amyloid-β Protein Oligomerization and the Importance of Tetramers and Dodecamers in the Aetiology of Alzheimer’s Disease. Nat. Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirkitadze MD, Condron MM, Teplow DB. Identification and Characterization of Key Kinetic Intermediates in Amyloid beta-protein Fibrillogenesis. J. Mol. Biol. 2001;312:1103–1119. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- 43.Ono K, Condron MM, Teplow DB. Structure-neurotoxicity Relationships of Amyloid beta-protein Oligomers. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14745–14750. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitternacht S, Staneva I, Härd T, Irbäck A. Monte Carlo Study of the Formation and Conformational Properties of Dimers of Aβ42 Variants. J. Mol. Biol. 2011;410:357–367. doi: 10.1016/j.jmb.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 45.Rosenman DJ, Wang C, García AE. Characterization of Aβ Monomers through the Convergence of Ensemble Properties among Simulations with Multiple Force Fields. J. Phys. Chem. B. 2016;120:259–277. doi: 10.1021/acs.jpcb.5b09379. [DOI] [PubMed] [Google Scholar]
- 46.Rosenman DJ, Connors CR, Chen W, Wang C, García AE. Aβ Monomers Transiently Sample Oligomer and Fibril-like Configurations: Ensemble Characterization Using a Combined MD/NMR Approach. J. Mol. Biol. 2013;425:3338–3359. doi: 10.1016/j.jmb.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das P, Murray B, Belfort G. Alzheimer’s Protective A2T Mutation Changes the Conformational Landscape of the Aβ1–42 Monomer Differently than Does the A2V Mutation. Biophys. J. 2015;108:738–747. doi: 10.1016/j.bpj.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Côté S, Derreumaux P, Mousseau N. Distinct Morphologies for Amyloid Beta Protein Monomer: Aβ1–40, Aβ1–42, and Aβ1–40(D23N) J. Chem. Theory Comput. 2011;7:2584–92. doi: 10.1021/ct1006967. [DOI] [PubMed] [Google Scholar]
- 49.Buchanan LE, Dunkelberger EB, Tran HQ, Cheng PN, Chiu CC, Cao P, Raleigh DP, de Pablo JJ, Nowick JS, Zanni MT. Mechanism of IAPP Amyloid Fibril Formation Involves an Intermediate with a Transient β-sheet. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19285–19290. doi: 10.1073/pnas.1314481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roychaudhuri R, Yang M, Deshpande A, Cole GM, Frautschy S, Lomakin A, Benedek GB, Teplow DB. C-terminal Turn Stability Determines Assembly Differences between Aβ40 and Aβ42. J. Mol. Biol. 2013;425:292–308. doi: 10.1016/j.jmb.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fradinger EA, Monien BH, Urbanc B, Lomakin A, Tan M, Li H, Spring SM, Condron MM, Cruz L, Xie C-W, et al. C-Terminal Peptides Coassemble into Aβ42 Oligomers and Protect Neurons against Aβ42-Induced Neurotoxicity. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14175–14180. doi: 10.1073/pnas.0807163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng X, Wu C, Liu D, Li H, Bitan G, Shea JE, Bowers MT. Mechanism of C-Terminal Fragments of Amyloid β-Protein as Aβ Inhibitors: Do C-Terminal Interactions Play a Key Role in Their Inhibitory Activity? J. Phys. Chem. B. 2016;120:1615–1623. doi: 10.1021/acs.jpcb.5b08177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chebaro Y, Pasquali S, Derreumaux P. The Coarse-grained OPEP Force Field for Non-amyloid and Amyloid Proteins. J. Phys. Chem. B. 2012;116:8741–8752. doi: 10.1021/jp301665f. [DOI] [PubMed] [Google Scholar]
- 54.Morriss-Andrews A, Shea JE. Simulations of Protein Aggregation: Insights from Atomistic and Coarse-Grained Models. J. Phys. Chem. Lett. 2014;5:1899–1908. doi: 10.1021/jz5006847. [DOI] [PubMed] [Google Scholar]
- 55.Emperador A, Orozco M. Discrete Molecular Dynamics Approach to the Study of Disordered and Aggregating Proteins. J. Chem. Theory Comput. 2017 doi: 10.1021/acs.jctc.6b01153. in press. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen PH, Sterpone F, Pouplana R, Derreumaux P, Campanera JM. Dimerization Mechanism of Alzheimer Aβ40 Peptides: The High Content of Intrapeptide-Stabilized Conformations in A2V and A2T Heterozygous Dimers Retards Amyloid Fibril Formation. J. Phys. Chem. B. 2016;120:12111–12126. doi: 10.1021/acs.jpcb.6b10722. [DOI] [PubMed] [Google Scholar]
- 57.Xu L, Nussinov R, Ma B. Allosteric Stabilization of the Amyloid-β Peptide Hairpin by the Fluctuating N-terminal. Chem. Commun. 2016;52:1733–1736. doi: 10.1039/c5cc08107f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adler J, Baumann M, Voigt B, Scheidt HA, Bhowmik D, Häupl T, Abel B, Madhu PK, Balbach J, Maiti S, et al. A Detailed Analysis of the Morphology of Fibrils of Selectively Mutated Amyloid β (1–40) Chem Phys Chem. 2016;17:2744–2753. doi: 10.1002/cphc.201600413. [DOI] [PubMed] [Google Scholar]
- 59.Marshall KE, Vadukul DM, Dahal L, Theisen A, Fowler MW, Al-Hilaly Y, Ford L, Kemenes G, Day IJ, Staras K, et al. A Critical Role for The Self-assembly of Amyloid-β1–42 in Neurodegeneration. Sci. Rep. 2016;6 doi: 10.1038/srep30182. No. 30182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doig AJ, Derreumaux P. Inhibition of Protein Aggregation and Amyloid Formation by Small Molecules. Curr. Opin. Struct. Biol. 2015;30:50–56. doi: 10.1016/j.sbi.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen P, Derreumaux P. Understanding Amyloid Fibril Nucleation and Aβ Oligomer/Drug Interactions from Computer Simulations. Acc. Chem. Res. 2014;47:603–611. doi: 10.1021/ar4002075. [DOI] [PubMed] [Google Scholar]
- 62.Habchi J, Chia S, Limbocker R, Mannini B, Ahn M, Perni M, Hansson O, Arosio P, Kumita JR, Challa PK, et al. Systematic Development of Small Molecules to Inhibit Specific Microscopic Steps of Aβ42 Aggregation in Alzheimer’s Disease. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E200–E208. doi: 10.1073/pnas.1615613114. [DOI] [PMC free article] [PubMed] [Google Scholar]