Abstract
Dynemicin A is a member of a subfamily of enediyne antitumor antibiotics characterized by a 10-membered carbocycle fused to an anthraquinone, both of polyketide origin. Sequencing of the dynemicin biosynthetic gene cluster in Micromonospora chersina previously identified an enediyne polyketide synthase (PKS) but no anthraquinone PKS, suggesting gene(s) for biosynthesis of the latter were distant from the core dynemicin cluster. To identify these gene(s), we sequenced and analyzed the genome of M. chersina. Sequencing produced a short list of putative PKS candidates, yet CRISPR-Cas9 mutants of each locus retained dynemicin production. Subsequently, deletion of two cytochromes P450 in the dynemicin cluster suggested that the dynemicin enediyne PKS, DynE8, may biosynthesize the anthraquinone. Together with 18O-labeling studies, we now present evidence that DynE8 produces the core scaffolds of both the enediyne and anthraquinone, and provide a working model to account for their formation from the programmed octaketide of the enediyne PKS.
The enediyne antitumor antibiotics are a family of cytotoxic natural products distinguished by their 9- or 10-membered carbocyclic core structure containing a double bond flanked by two triple bonds. Early biosynthetic studies of the representative enediyne structural subtypes, exemplified by the 9-membered neocarzinostatin1 and the 10-membered esperamicin2, involved feeding 13C-labeled acetate to fermentations of their bacterial producers. These studies demonstrated that each was formed by the head-to-tail assembly of acetate units, suggesting a polyketide origin for these molecules. More than a decade later, biosynthetic gene cluster (BGC) sequencing efforts revealed a highly-reducing, type I iterative PKS (HR-PKS) in both the 9-membered C-10273 and the 10-membered calicheamicin4 clusters. Inactivation of these PKSs resulted in loss of antibiotic production, confirming their polyketide origin. Several enediyne BGCs have since been sequenced, each having a PKS highly homologous to those found in the C-1027 and calicheamicin clusters. They also share four additional proteins — a thioesterase (TE) and three proteins of unknown function — which together with the enediyne PKS comprise the minimal enediyne PKS cassette5,6.
An analysis of enediyne PKSs with their cognate TEs from five different enediyne producers demonstrated production of heptaene 1 (Fig. 4) as their major product7. Mixed-and-matched PKSs and TEs from 9- and 10-membered systems also produced 1, suggesting a convergent model of biosynthesis for this family of natural products7. Despite the common production of 1 in enediyne systems, it was eliminated as a possible on-pathway precursor to the enediyne core. The calicheamicin enediyne PKS, CalE8, and the C-1027 TE were shown to make heptaene7, but CalE8 was previously demonstrated to not restore antibiotic production in a C-1027 enediyne PKS knockout strain8, leading to the conclusion that 1 was a shunt product of enediyne biosynthesis. Further work has since demonstrated that the programmed product of the enediyne PKS is enzyme-bound β-hydroxyhexaene 2 (Fig. 4)9,10. In an attempt to account for the formation of the principal structural types of enediyne metabolites, a mechanistic framework was advanced for 9- and 10-membered enediyne biosynthesis from this common intermediate9,10. Evidence further in support of this proposal is developed below.
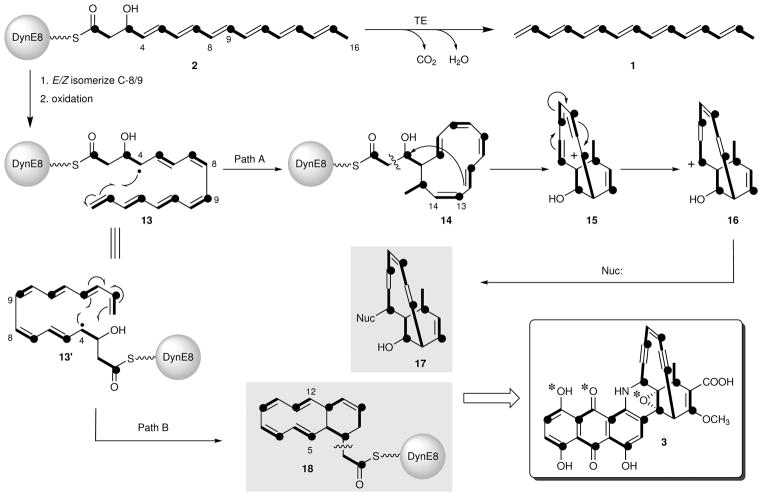
Figure 4. Working model for enediyne and anthraquinone formation by DynE8.
DynE8 makes 2, which is proposed to form the dynemicin enediyne and anthraquinone in the presence of accessory enzymes, and heptaene 1 in the presence of its native TE, DynE7. Intact acetate units are indicated in bold, and the carbonyl carbons of acetate are indicated with black dots. Asterisks in 3 indicate atoms originating from molecular oxygen. Numbering for 3 is presented in Fig. 1a.
Beyond the enediyne core, several structural features of the enediyne antitumor antibiotics are polyketide-derived. Examples include the naphthoate of neocarzinostatin and the hexa-substituted orsellinate of calicheamicin, and their respective BGCs have additional PKSs and accessory proteins to biosynthesize these moieties4,11. Dynemicin A (3; Fig. 1a) is a third example of an enediyne with two components of polyketide origin12. Produced by Micromonospora chersina, dynemicin A was first discovered in 1989,13 and features a 10-membered enediyne coupled to a highly-oxidized anthraquinone. Studies with 13C-labeled acetate suggested that the enediyne and anthraquinone portions of the molecule were biosynthesized separately as heptaketides, and then joined to form the final natural product12. It came as a surprise, then, that sequencing of the dynemicin BGC only revealed one PKS — the enediyne PKS, DynE814. Physical separation of biosynthetic genes has been described previously15,16, so until now, it has been assumed that the anthraquinone-producing PKS was located elsewhere on the M. chersina genome14.
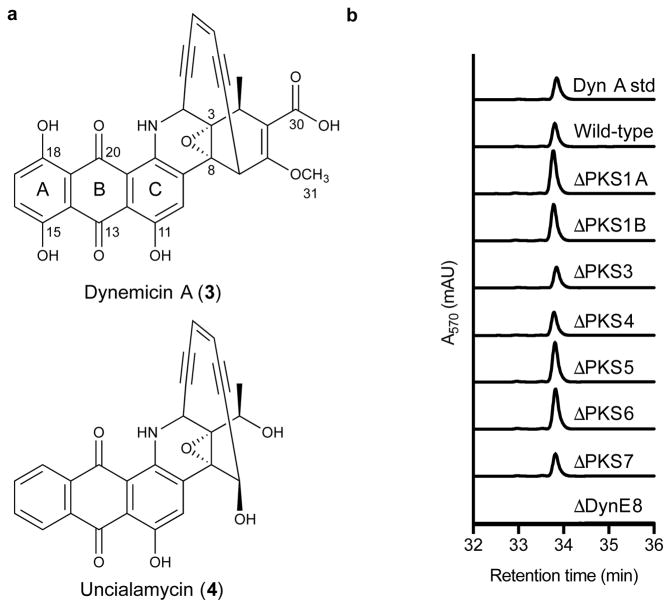
Figure 1. Structures of anthraquinone-containing enediynes and comparison of dynemicin A production by wild-type M. chersina and PKS gene deletion strains.
(a) Structures of dynemicin A (3) and uncialamycin (4). (b) HPLC chromatograms illustrating production of dynemicin A by all PKS mutants except ΔDynE8.
To identify this second PKS, we sequenced the genome of M. chersina NRRL B-24756 and individually inactivated all potential PKS-encoding genes. Against our expectations, the knockouts failed to identify the anthraquinone PKS. These negative results combined with a comparative bioinformatics analysis (Supplementary Fig. 1) with the BGC of the anthraquinone-containing 10-membered enediyne, uncialamycin (4; Fig. 1a)17, suggested that all the genes necessary to produce the dynemicin anthraquinone were within the bounds of the dynemicin BGC. Here, through a series of gene deletions in the dynemicin BGC and oxygen-labeling experiments of dynemicin A, we present evidence that the dynemicin enediyne PKS, DynE8, plays a dual role in the biosynthesis of both the enediyne and the anthraquinone halves of the molecule.
Results
The evident lack of anthraquinone PKS genes in the dynemicin BGC prompted us to sequence the M. chersina genome in search of candidate PKSs. After the sequencing reads were assembled, a draft genome of M. chersina was released by the Joint Genome Institute (JGI Project ID 1059218), providing a reference against which to evaluate our data. Both datasets were analyzed for the presence of BGCs using antiSMASH v 3.0.518, which resulted in identical lists of nine potential PKSs, including genes predicted to more closely resemble fatty acid synthases (FASs) (Supplementary Tables 1 and 2). We hypothesized that one of the nine was involved in anthraquinone biosynthesis, and accordingly planned gene deletion experiments for all (including DynE8 as a control) to test every possibility. We chose to carry out our gene deletions using CRISPR-Cas9 owing to its recent development for use in Actinomycetes19. CRISPR-Cas9 required the design of a single guide RNA (sgRNA) and a homologous recombination template (HRT) for each knockout, which were cloned into the pCRISPR-Cas9 plasmid. Plasmids were delivered to M. chersina by conjugation with Escherichia coli. After induction of CRISPR-Cas9, mutants were screened by colony PCR, and gene excision confirmed by isolation, PCR, and sequencing of the genomic DNA (gDNA).
The desired mutant was obtained for all but one of the nine CRISPR-Cas9 experiments; the failed deletion repeatedly returned a negative result by colony PCR despite many CRISPR-Cas9 attempts. Closer examination of these genes revealed a genomic organization identical to that of type II FASs in Streptomyces20, and our inability to knock them out suggested their essential nature, consistent with a FAS identity. These genes were eliminated, therefore, as possible anthraquinone PKS candidates, and their inactivation no longer pursued. The remaining eight mutants were fermented and their dynemicin production compared to that of the wild-type strain by HPLC. The ΔDynE8 strain was the only mutant that resulted in loss of dynemicin production (Fig. 1b).
While the mutants were being processed, BGC sequencing data was released for uncialamycin 4 on the NCBI database (KT762610). At the time, uncialamycin was the only other structurally characterized enediyne with an anthraquinone similar to dynemicin, but with one obvious difference that the two hydroxy groups on the A-ring of the dynemicin anthraquinone are absent17. The availability of the uncialamycin BGC sequence allowed us to pursue a comparative bioinformatics analysis (Supplementary Fig. 1) with the dynemicin BGC with the hope of gaining insight into anthraquinone biosynthesis. This analysis brought to our attention two putative cytochrome P450 genes in the dynemicin BGC (E10; ACB47071 and orf19; ACB47070) that are absent in the uncialamycin BGC. It was speculated previously that these genes were involved in joining the enediyne and anthraquinone halves of dynemicin14. However, we envisioned a different biosynthetic role for them: hydroxylation of the A-ring of the anthraquinone. We hypothesized that knocking out both P450s would produce an uncialamycin-like intermediate in M. chersina, whose occurrence by other means had been suggested previously in the literature14. Importantly, our comparative bioinformatics analysis also revealed that the uncialamycin BGC lacks an anthraquinone PKS. This observation, together with the thought that a more highly-reduced uncialamycin-like anthraquinone is an intermediate in dynemicin biosynthesis, led us to consider whether DynE8 biosynthesizes the carbon skeletons of both the enediyne and the anthraquinone. An uncialamycin-like anthraquinone is closer in oxidation state to the characterized polyene products of the enediyne PKSs than the dynemicin anthraquinone. Additionally, a role for DynE8 in anthraquinone biosynthesis would explain why all CRISPR-Cas9 gene inactivations except DynE8 failed to abolish dynemicin production.
E10 and orf19 were knocked out in tandem in M. chersina using CRISPR-Cas9. The resulting blocked strain produced compounds 5 and 6, both with chromophores blue-shifted by 34 nm in comparison to dynemicin A (Fig. 2, Supplementary Fig. 3). UPLC-ESI-MS analysis of the major peak, 5, was consistent with the loss of two oxygen atoms from dynemicin A, while the minor peak, 6, had a mass corresponding to the loss of CH2O3 (Supplementary Fig. 3). To structurally characterize these intermediates by 1H and 13C NMR spectroscopy, the P450 double mutant was fermented and extracted, and efforts made to purify the enediynes. Following reports that per-acetylation of dynemicin A increased its solubility21, purification of the enediynes included derivatization with acetic anhydride and pyridine. Low production of 6 hampered its isolation, but the 1H NMR spectrum of 7 (acetylated 5; Fig. 2a and Supplementary Fig. 8) revealed five aromatic hydrogens, two more than dynemicin A. The mutual coupling pattern indicated four neighboring hydrogens on the A-ring of the anthraquinone, confirming the production of an uncialamycin-like intermediate en route to the more highly-oxidized dynemicin anthraquinone.
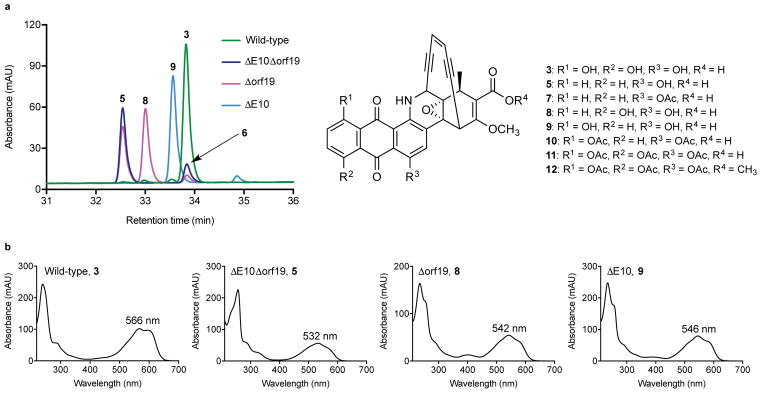
Figure 2. Comparison of enediynes produced by wild-type M. chersina and P450 gene deletion strains.
(a) HPLC chromatograms demonstrating loss of 3 production in P450 mutant strains and accumulation of major intermediates 5, 8, and 9 with indicated structures. P450 mutant and wild-type HPLC traces are shown at 540 and 570 nm, respectively. (b) UV-visible spectrum of 3 compared to the UV-visible spectra of 5, 8, and 9.
Although we presumed that each P450 installed a single hydroxyl group, our results did not preclude the possibility that one P450 carried out both hydroxylations. We therefore excised E10 and orf19 individually from the genome of M. chersina using CRISPR-Cas9 and compared the HPLC product profiles of the mutants (Fig. 2a). HPLC analysis of the Δorf19 strain revealed a ~1:1 mixture of two products, while the ΔE10 strain produced one major product. The earlier-eluting Δorf19 product matched the UV-visible spectrum and retention time of the unhydroxylated A-ring intermediate 5 (Supplementary Fig. 4), which was later confirmed by UPLC-ESI-MS analysis. The remaining Δorf19 product, 8, and the major ΔE10 product, 9, had UV-visible spectra that were blue-shifted in comparison to dynemicin A but less so than that of 5 (24 and 20 nm, respectively, vs 34 nm; Fig. 2b). UPLC-ESI-MS analysis of HPLC-purified 8 and 9 revealed identical masses consistent with the loss of a single oxygen from dynemicin A (Supplementary Fig. 4 and 5), establishing that each P450 performs a single hydroxylation. To distinguish the regiochemistry of –OH installation by each P450, the ΔE10 mutant was cultured and 9 isolated from its fermentation as the diacetate. Taking advantage of the known carbon-labeling pattern of the dynemicin anthraquinone (Fig. 4)12, the fermentation included [1-13C]-sodium acetate to create carbon label asymmetry in the anthraquinone (2–3% enrichment per site). Direct inspection of the 13C NMR spectrum of the intermediate led to the assignment of the A-ring acetate to an unlabeled carbon, and HSQC analysis showed that two of the three A-ring hydrogens were on labeled carbons (Supplementary Fig. 12–13,15–16), identifying its structure as 10 (Fig. 2a). Together these results establish that E10 and orf19 install their hydroxyl groups at C-15 and C-18 of the dynemicin anthraquinone, respectively. Furthermore, based on the accumulation of 5 in the Δorf19 mutant but not the ΔE10 mutant, there appears to be a preferred order to the P450 hydroxylations: orf19 first, followed by E10.
With evidence from our P450 mutants that two of the dynemicin anthraquinone hydroxyl groups arise by aryl oxidation and that DynE8 gives rise to both the enediyne and anthraquinone halves of dynemicin, we looked to the established chemistry of the enediyne PKS for clues as to how it could produce two such chemically dissimilar structures. We previously demonstrated that β-hydroxyhexaene 2 is the programmed product of the enediyne PKS10. We reasoned that if DynE8 makes the anthraquinone from this polyene or a polyene like heptaene 1, then the source of the oxygens on the anthraquinone would be molecular oxygen, not acetate. To investigate the origin of the remaining oxygens, the M. chersina ΔPKS5 mutant was fermented in a closed system under a ~1:1 16O2:18O2 atmosphere. ΔPKS5 was used for the fermentation as it consistently afforded greater yields of dynemicin A than the wild-type strain. Dynemicin A was purified from the closed-system fermentation as its triacetate 11 (Fig. 2a), and the isotope incorporation analyzed by UPLC-ESI-MS and 13C NMR (18O produces a small, but diagnostic upfield shift in the directly attached 13C NMR resonances22). The 13C NMR spectrum revealed that six of nine oxygens in dynemicin A — all five on the anthraquinone portion and the epoxide — clearly originate from molecular oxygen (Fig. 3a).
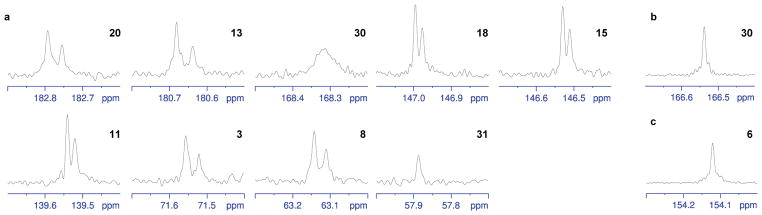
Figure 3. 13C NMR resonances of oxygen-attached carbons in dynemicin A derivatives.
(a) Triacetyldynemicin A from the closed-system fermentation supplied with ~1:1 16O2:18O2 and (b) its methyl ester derivative. (c) Triacetyldynemicin A methyl ester from the fermentation supplied with [1-13C,18On]-sodium acetate (n = 0–2). Numbering corresponds to the carbon labeling scheme illustrated in Fig. 1a.
Since 13C-labeling studies had demonstrated that the carboxylic acid carbon of dynemicin is derived from the methyl carbon of acetate12, we reasoned that one aerobic oxidation must occur at minimum to produce the free acid. The number of oxidations that occurred at the carboxylic acid of 18O-labeled 11 was obscured, however, by the broadness of its carbon resonance (C-30; Fig. 3a). To enhance resolution at this position, the acid was converted to the methyl ester 12 (Fig. 2a) with diazomethane. Repeat 13C NMR analysis showed sharpening of the desired carbon signal, but unexpectedly, the complete absence of an 18O label (Fig. 3b). This observation finally brought us to the methoxyl oxygen, which was the only other oxygen in the molecule that was not labeled during the 16O2:18O2 fermentation. A potential alternative source of oxygen at this position is acetate; to investigate this possibility, [1-13C,18On]-sodium acetate (n = 0–2; Supplementary Fig. 25–26) was synthesized and administered to a fermentation of the M. chersina ΔPKS5 mutant to achieve a ratio of ~60:40 18O:16O during polyketide extension. Dynemicin A was purified from the fermentation as 12, and 18O-isotope incorporation analyzed by 13C NMR spectroscopy. The vinyl carbon (C-6) attached to the methoxyl in 12 again showed no 18O label (Fig. 3c).
Discussion
The serial gene deletion and 18O-labeling experiments reveal an unanticipated intricacy in the story of enediyne biosynthesis by uncovering a role for DynE8 in the production of both the enediyne and anthraquinone halves of dynemicin. Although it was originally postulated that the genes responsible for anthraquinone biosynthesis were located on the M. chersina genome removed from the dynemicin BGC, our results exclude this hypothesis. Inactivations of all potential PKSs in M. chersina, with the exception of DynE8, do not derail dynemicin production. Additionally, the key finding that all oxygens on the dynemicin anthraquinone originate from O2 and not acetate is fully consistent with a HR-PKS being involved in anthraquinone biosynthesis. Analysis of the M. chersina genome by antiSMASH reveals that DynE8 is the only HR-PKS in the organism. These observations mutually reinforce our confidence in the participation of DynE8 in enediyne and anthraquinone biosynthesis, and have prompted us to consider how DynE8 could carry out this chemistry.
Figure 4 depicts a working model for enediyne and anthraquinone biosynthesis from octaketide 2 that is in accord with the acetate-labeling pattern of dynemicin12. Notably, this model extends hypotheses previously proposed for enediyne biosynthesis to formation of the dynemicin anthraquinone: (1) accessory proteins in the enediyne BGC initiate and control the cyclization chemistry9,23, and (2) ring formation begins at C-4, which is the carbon of 2 uniquely incorporated into the cyclizations of all known enediyne families9,10. Biosynthesis begins with the production of 2 by DynE8 with its double bonds all-trans following conventional PKS logic. Due to the nature of the conjugated polyene, these double bonds are anticipated to isomerize readily in light, heat, or acid. Reversible addition of dioxygen resulting in isomerization is also known to occur with polyenes, and is invoked in Fig. 4, as was done previously to account for 9- and 10-membered enediyne biosynthesis from 139,10,24. E/Z-Isomerization at C-8/9 with subsequent hydrogen abstraction at the terminal methyl group, C-16, would result in the production of the highly delocalized radical (or cation) 13. Following Path A with C-13/14 in the cis-configuration, C-4/15 bond formation and radical quenching would produce hypothetical intermediate 14. This intermediate could undergo retro-aldol cleavage to achieve the required heptaketide chain length of the enediyne, and the resulting aldehyde could further close in a Prins reaction to the bicyclic cation 15. Resonance of this triply allylic cation would produce 16, with capture by a – possibly nitrogen – nucleophile affording enediyne precursor 17. Importantly, this intermediate is correctly functionalized for joining the enediyne and anthraquinone halves of dynemicin, and secures the positions of unsaturation (C=C geometries not specified) in the nascent enediyne bridge.
A similar cyclization to form the anthraquinone could also be envisioned from the common radical 13. Generation of 13′ and its cyclization to form 18 (for example, by radical closure to the α,β-unsaturated thioester through Path B) would possibly be controlled by an auxiliary protein in the dynemicin BGC distinct from that used to cyclize the enediyne precursor. The intermediate 18 that would result from the cyclization has a double bond that is positioned correctly in the C-ring to couple to the developing enediyne partner 17. One could imagine oxygen addition across C-5/12 of 18 to aromatize the A-ring and form the B-ring quinone. Oxidative scission as indicated would finally introduce oxygen at C-3, as dictated by the 18O-incorporation experiment. The remaining oxidative tailoring reactions to form the final natural product are anticipated to occur post-fusion of the enediyne and anthraquinone halves, but their timing cannot be determined at present. The methoxyl oxygen and at least one of the carboxylic acid oxygens are proposed to derive from aerial oxidation (the latter from C-C bond cleavage of acetate), but the labeled oxygens must be lost by exchange with water during the course of biosynthesis.
Recently, the structure and BGC sequence of new anthraquinone-containing enediynes, the tiancimycins, was reported25. Further strengthening our assertion that the enediyne PKS biosynthesizes both the enediyne and anthraquinone moieties, the tiancimycin BGC contains only a single PKS with high sequence homology to DynE8. Sequencing of the tiancimycin BGC also revealed the presence of one putative cytochrome P450 gene that is homologous to both E10 and orf19. Like dynemicin, the A-ring of the tiancimycin anthraquinone is doubly hydroxylated, suggesting that this one P450 carries out both hydroxylations. The assignment of E10 and orf19 in the dynemicin BGC as cytochromes P450 involved in A-ring hydroxylation simplifies the list of genes in the cluster that may be involved in cyclization of the enediyne and anthraquinone carbon skeletons. However, since E10 and orf19 were originally proposed to be involved in joining the enediyne and anthraquinone halves of dynemicin, the enzymes responsible for this chemistry remain to be identified.
The chemistry of enediyne biosynthesis has long perplexed natural product biochemists. Identification of the programmed product of the enediyne PKS was a practical and conceptual advance toward elucidating the inner-workings of these systems. Now, the demonstration of a role for this HR-PKS family in anthraquinone as well as enediyne biosynthesis adds a new layer of complexity, yet clarity, to the problem. Understanding the differentiation of enzyme-bound β-hydroxyhexaene 2 to these structurally complex and elaborately functionalized products remains an estimable challenge. The conceptual mechanistic framework developed in Fig. 4 is intended to stimulate further research, while the results presented here give fresh insights into these processes and the dramatic elevations of oxidation state that occur from a simple, common precursor.
Methods
Gene deletions using CRISPR-Cas9
CRISPR-Cas9 plasmids with HRTs and sgRNAs were delivered to M. chersina by conjugation26, as follows. Plasmids were electroporated into E. coli GM2929 hsdS::Tn10 (pUB307::Tn7)27, and transformants selected on LB agar with streptomycin and apramycin (50 and 100 μg/mL, respectively). The resulting colonies were grown overnight in LB at 37 °C with streptomycin and apramycin, and 500 μL each culture then used to inoculate 50 mL LB with the same drug selection. When OD600 ~ 0.4 was reached at 37 °C, cells were pelleted by centrifugation at 6500 x g and 4 °C, washed twice with ~40 mL LB, and finally resuspended in ~2.5 mL LB.
Concentrated M. chersina spores (50 μL) were diluted with 500 μL 2xYT and germinated at 50°C for 10 min. Germinated spores were combined with 500 μL pCRISPR-Cas9-bearing E. coli. Spore-E.coli mixtures were concentrated into 100 μL LB and plated on medium 5328 with 2% agar and supplemented with 10 mM MgCl2. After overnight growth at 28 °C, plates were overlaid with nalidixic acid and apramycin in sterile ddH2O to achieve 20 and 50 μg/mL in the plates, respectively. Incubation proceeded at 28 °C for 4–7 days until colonies appeared.
Ex-conjugants were re-streaked on medium 53 agar with nalidixic acid and apramycin and grown at 28 °C for 3–4 days. The resulting colonies were then re-streaked on medium 53 agar with nalidixic acid, apramycin, and thiostrepton (1 μg/mL) to induce CRISPR-Cas919. After 6–7 days at 28 °C, colonies were re-streaked on medium 53 agar without drug and incubated at 37 °C for 3 days. Mutants were finally screened by colony PCR, and gene deletions confirmed by isolation, PCR, and sequencing of the gDNA.
Analysis of enediyne production in CRISPR-Cas9 mutants
CRISPR-Cas9 mutants were fermented as described in the Supplementary Methods. On the seventh day of fermentation, 5 mL each mutant was extracted with 5 mL ethyl acetate (EtOAc) by vortexing for 1 min. Extracts were centrifuged at 4000 x g and 4 °C, and 4 mL each EtOAc layer dried by SpeedVac without heating. Samples were resuspended in 200 μL DMSO, filtered through 0.2 μm PTFE filters, and analyzed on an Agilent 1200 HPLC equipped with a Prodigy ODS3 100 Å, 5 μm, 250 x 4.6 mm column (Phenomenex). Injection volumes were 20 μL. Samples were separated using a gradient method of 5–95% ACN + 0.1% (v/v) formic acid over 40 min, followed by a 10 min hold at 95% ACN before column re-equilibration (1 mL/min flow rate). Production of dynemicin and related biosynthetic intermediates was monitored at 540 and 570 nm.
Closed-system fermentation of M. chersina
Spores of the M. chersina ΔPKS5 mutant (10 μL) were plated on medium 53 agar and incubated at 28 °C. After 5 days, the resulting mycelia on agar were inoculated into 2 x 50 mL medium 53 in 125 mL flasks. Cultures were shaken at 250–300 rpm and 37 °C for 5 days, following which they were combined and 14 x 4.4 mL inoculated into 14 x 110 mL H88128 in 500 mL flasks. Flasks were shaken again at 250–300 rpm and 37 °C. After 24 h, 2 mM [1-13C]- and [2-13C]-sodium acetate (4 mM total; stocks were adjusted to pH 7.0 with 1 M HCl and sterilized by filtration through 0.2 μm PES filters) and 1% (w/v) SupeliteTM DAX-828 (sterilized by autoclave) were added to each flask. The flasks were connected in series as illustrated in Supplementary Fig. 18, and shaking resumed at 37 °C with the system closed. Over the next 24 h, ~1300 mL 18O2 (97% isotopic purity) were added to the system to achieve an approximately equal ratio of 16O2 and 18O2. Subsequently, ~1:1 16O2:18O2 was supplied as needed as oxygen was consumed. The closed-system fermentation was complete after 72 h (96 h from the start of the experiment).
Purification of dynemicin and related biosynthetic intermediates
Fermentations were extracted with EtOAc and the products purified by flash chromatography on silica gel. Crude dynemicin A or related intermediates were then derivatized with acetic anhydride and pyridine, and purified again as above. The acetylated material was finally purified twice on Sephadex LH-20 (GE Healthcare Life Sciences). Detailed protocols for purification of 7, 10, 11, and 12 are provided in the Supplementary Methods.
Data availability
All data supporting the findings herein are available within the Article or the Supplementary Information, or upon request from the corresponding author.
Supplementary Material
Acknowledgments
We thank the Experimental and Computational Genomics Core at The Johns Hopkins University for sequencing and assembling the M. chersina genome, Dr. A. Majumdar, Dr. I. P. Mortimer, B. Steinberg, F. B. d’Andrea, and C. L. Malbon for NMR, UPLC-ESI-MS, mechanical, synthesis, and coding help, respectively, and A. L. Eller for performing preliminary experiments. We are particularly grateful for generous donations made by the following: Prof. H. Ikeda for E. coli GM2929 hsdS::Tn10 (pUB307::Tn7), Y. Tong, Prof. T. Weber, and Prof. S. Y. Lee for pCRISPR-Cas9, and Dr. R. F. Li for obtaining these items; finally, the Agricultural Research Service for M. chersina NRRL B-24756, J. Stuart at Omega Protein Corporation for fish meal, and Dr. C. Fischer and Prof. J. C. Vederas for use of the O2 fermentation system. This work was supported by NIH Grant R01 ES001670 and T32 GM080189.
Footnotes
Author contributions
D.R.C. and C.A.T. designed experiments, analyzed data, and wrote the manuscript, and D.R.C. performed experiments.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Hensens OD, Giner JL, Goldberg IH. Biosynthesis of NCS chrom A, the chromophore of the antitumor antibiotic neocarzinostatin. J Am Chem Soc. 1989;111:3295–3299. [Google Scholar]
- 2.Lam KS, et al. Biosynthesis of esperamicin A1, an enediyne antitumor antibiotic. J Am Chem Soc. 1993;115:12340–12345. [Google Scholar]
- 3.Liu W, Christenson SD, Standage S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 4.Ahlert J, et al. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 5.Zazopoulos E, et al. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- 6.Liu W, et al. Rapid PCR amplification of minimal enediyne polyketide synthase cassettes leads to a predictive familial classification model. Proc Natl Acad Sci USA. 2003;100:11959–11963. doi: 10.1073/pnas.2034291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horsman GP, Yihua C, Thorson JS, Shen B. Polyketide synthase chemistry does not direct biosynthetic divergence between 9- and 10-membered enediynes. Proc Natl Acad Sci USA. 2010;107:11331–11335. doi: 10.1073/pnas.1003442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, et al. A phosphopantetheinylating polyketide synthase producing a linear polyene to initiate enediyne antitumor antibiotic biosynthesis. Proc Natl Acad Sci USA. 2008;105:1460–1465. doi: 10.1073/pnas.0711625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belecki K, Townsend CA. Environmental control of the calicheamicin polyketide synthase leads to detection of a programmed octaketide and a proposal for enediyne biosynthesis. Angew Chem Int Ed. 2012;51:11316–11319. doi: 10.1002/anie.201206462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belecki K, Townsend CA. Biochemical determination of enzyme-bound metabolites: preferential accumulation of a programmed octaketide on the enediyne polyketide synthase CalE8. J Am Chem Soc. 2013;135:14339–14348. doi: 10.1021/ja406697t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W, et al. The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chem Biol. 2005;12:293–302. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Tokiwa Y, et al. Biosynthesis of dynemicin A, a 3-ene-1,5-diyne antitumor antibiotic. J Am Chem Soc. 1992;114:4107–4110. [Google Scholar]
- 13.Konishi M, et al. Dynemicin A, a novel antibiotic with the anthraquinone and 1,5-diyn-3-ene subunit. J Antibiot. 1989;42:1449–1452. doi: 10.7164/antibiotics.42.1449. [DOI] [PubMed] [Google Scholar]
- 14.Gao Q, Thorson JS. The biosynthetic genes encoding for the production of the dynemicin enediyne core in Micromonospora chersina ATCC53710. FEMS Microbiol Lett. 2008;282:105–114. doi: 10.1111/j.1574-6968.2008.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahlan K, Park HU, Jensen SE. Three unlinked gene clusters are involved in clavam metabolite biosynthesis in Streptomyces clavuligerus. Can J Microbiol. 2004;50:803–810. doi: 10.1139/w04-070. [DOI] [PubMed] [Google Scholar]
- 16.Lazos O, et al. Biosynthesis of the putative siderophore erythrochelin requires unprecedented crosstalk between separate nonribosomal peptide gene clusters. Chem Biol. 2010;17:160–173. doi: 10.1016/j.chembiol.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Davies J, et al. Uncialamycin, a new enediyne antibiotic. Org Lett. 2005;7:5233–5236. doi: 10.1021/ol052081f. [DOI] [PubMed] [Google Scholar]
- 18.Weber T, et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong Y, Charusanti P, Zhang L, Weber T, Lee SY. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol. 2015;4:1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 20.Summers RG, Ali A, Shen B, Wessel WA, Hutchinson CR. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: a possible link between fatty acid and polyketide biosynthesis. Biochemistry. 1995;34:9389–9402. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]
- 21.Konishi M, et al. Crystal and molecular structure of dynemicin A: a novel 1,5-diyn-3-ene antitumor antibiotic. J Am Chem Soc. 1990;112:3715–3716. [Google Scholar]
- 22.Vederas JC. The use of stable isotopes in biosynthetic studies. Nat Prod Rep. 1987;4:277–337. doi: 10.1039/np9870400277. [DOI] [PubMed] [Google Scholar]
- 23.Belecki K, Crawford JM, Townsend CA. Production of octaketide polyenes by the calicheamicin polyketide synthase CalE8: implications for the biosynthesis of enediyne core structures. J Am Chem Soc. 2009;131:12564–12566. doi: 10.1021/ja904391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin H, Xu L, Porter NA. Free radical lipid peroxidation: mechanisms and analysis. Chem Rev. 2011;111:5944–5972. doi: 10.1021/cr200084z. [DOI] [PubMed] [Google Scholar]
- 25.Yan X, et al. Strain prioritization and genome mining for enediyne natural products. mBio. 2016;7:1–12. doi: 10.1128/mBio.02104-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich: 2000. [Google Scholar]
- 27.Komatsu M, Tsuda M, Ōmura S, Oikawa H, Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci USA. 2008;105:7422–7427. doi: 10.1073/pnas.0802312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam KS, Veitch JA, Lowe SE, Forenza S. Effect of neutral resins on the production of dynemicins by Micromonospora chersina. J Ind Microbiol. 1995;15:453–456. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings herein are available within the Article or the Supplementary Information, or upon request from the corresponding author.