Abstract
In recent years, CRISPR (clustered regularly interspaced short palindromic repeat)/Cas (CRISPR-associated) genome editing systems have become one of the most robust platforms in basic biomedical research and therapeutic applications. To date, efficient in vivo delivery of the CRISPR/Cas9 system to the targeted cells remains a challenge. Although viral vectors have been widely used in the delivery of the CRISPR/Cas9 system in vitro and in vivo, their fundamental shortcomings, such as the risk of carcinogenesis, limited insertion size, immune responses and difficulty in large-scale production, severely limit their further applications. Alternative non-viral delivery systems for CRISPR/Cas9 are urgently needed. With the rapid development of non-viral vectors, lipid- or polymer-based nanocarriers have shown great potential for CRISPR/Cas9 delivery. In this review, we analyze the pros and cons of delivering CRISPR/Cas9 systems in the form of plasmid, mRNA, or protein and then discuss the limitations and challenges of CRISPR/Cas9-based genome editing. Furthermore, current non-viral vectors that have been applied for CRISPR/Cas9 delivery in vitro and in vivo are outlined in details. Finally, critical obstacles for non-viral delivery of CRISPR/Cas9 system are highlighted and promising strategies to overcome these barriers are proposed.
Keywords: CRISPR/Cas9, non-viral delivery, genetic disorder, cancer, nanomedicine, clinical translation
1. Introduction
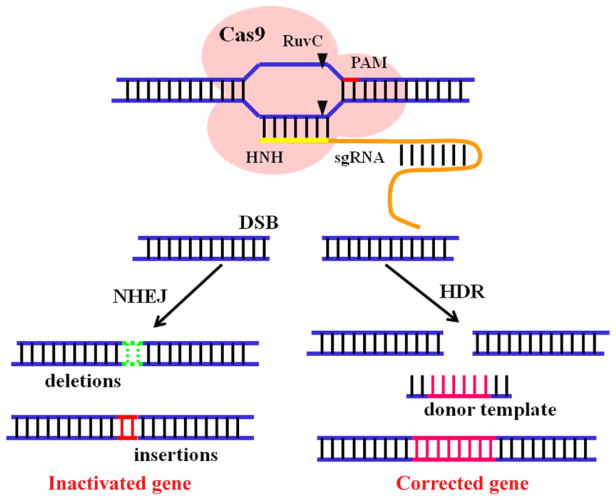
CRISPR (clustered regularly interspaced short palindromic repeat)/Cas (CRISPR-associated) systems are adaptable immune mechanisms of many bacteria and archaea to protect themselves from invading nucleic acids [1–4]. Since first applied in mammalian cells in 2013, CRISPR/Cas system, based on a RNA-guided nuclease, has revolutionized the way that genome editing is performed [5, 6]. To date, CRISPR/Cas systems have been categorized into three main types based on the core element content and sequences [7–9]. In Types I CRISPR/Cas systems, CASCADE (CRISPR-associated complex for antiviral defense) complexes containing multiple Cas protein subunits form complexes with crRNA to trigger the recognition and disruption of the target loci. In type III systems, crRNAs are incorporated into a multi-subunit interference complex called Cmr or Csm to detect and degrade invasive RNA. In sharp contrast, only the Cas9 protein is required for DNA interference in the type II systems [10–13]. Actually, the type II CRISPR/Cas system from streptococcus pyogenes (CRISPR/Cas9) has been widely applied in biomedical applications due to its simplicity, versatility, high specificity, and efficiency [14–18]. CRISPR/Cas9 system contains two critical components: Cas9 nuclease and a guide RNA (gRNA) that is a fusion of a crRNA and a constant tracrRNA. Generally, gRNA can be easily replaced by a synthetic chimeric single guide RNA (sgRNA). In the presence of a protospacer-adjacent motif (PAM) (usually 5′-NGG), the Cas9 nuclease can be directed by a sgRNA to any targeted genomic locus based on base pairing and stimulate site-specific double-stranded DNA breaks (DSBs), where two cellular repair mechanisms-non-homologous end-joining (NHEJ) or homology-directed repair (HDR) pathways-can be exploited to induce error-prone or defined alterations (Fig. 1) [13, 19–21]. Meanwhile, the CRISPR interference (CRISPRi) technology, which uses a catalytically dead Cas9 (dCas9) protein lacking endonuclease activity to regulate genes in an RNA-guided manner, has also been developed in recent years [22–24]. Besides, Cas9 nickases (RuvCD10A or HNHH840A), which cut one strand rather than both strands of the target DNA site have also been shown to be useful for genome editing [25, 26]. This method is applicable for genome editing of any model organism with minimal off-target effects [27, 28]. Recently, an alternative CRISPR-based nuclease Cpf1 has been discovered and share many similar advantages and shortcomings of Cas9 [29, 30].
Fig. 1.
Schematic illustration of the two different repair mechanisms of CRISPR/Cas9-mediated double stranded breaks (DSBs).
With these merits, CRISPR/Cas9 systems have shown great potential in studying the function of genetic elements, disease modeling and therapy [31–38]. For example, CRISPR/Cas9 systems have been applied to recapitulate cancer-associated genes both in vitro and in vivo, creating a convenient and effective platform to investigate cancer-related genetic mutations. Meanwhile, Cas9 nuclease can be easily adjusted to retarget new desired sequences by simply altering the sgRNA sequence and the system can simultaneously target multiple sequences for genome editing [39–41]. In the past decades, generating disease models was an extremely slow and expensive process, which requires complicated embryonic stem cell manipulation, as well as endless mouse husbandry to obtain the desired phenotype and genotype. However, after the emergence of CRISPR/Cas9 systems, novel disease models have been developed with unprecedented speed and precision resulting from the simplicity and flexibility of these systems [42–45]. Furthermore, CRISPR/Cas9 system holds tremendous promise for gene therapy [46–51]. It can correct causal mutations in the original genome and rescue the disease phenotypes of monogenic disorders permanently, which currently represents the most translatable field in CRISPR/Cas9-mediated disease therapy. In addition, CRISPR/Cas9 system has shown potentials in polygenetic diseases, such as cancer, by inhibiting oncogene expression or deactivating oncogenic virus [52–55]. Recently, the first human CRISPR/Cas9-based clinical trial is on the move in China (ClinicalTrials.gov Identifier: NCT02793856) and the first CRISPR clinical trial in the US also received green light from the FDA [56, 57]. Both trials are based on editing ex vivo T cells from patients with CRISPR/Cas9 system and then transplanting these modified cells back into the patients to help augment cancer therapies.
Despite the aforementioned merits, efficient delivery may likely become the main hurdle in the eventual application and clinical translation of CRISPR/Cas9 system. Currently, the strategies of CRISPR/Cas9 delivery are mainly based on physical approaches (microinjection, electroporation, hydrodynamic injection, etc.) and viral vectors (lentivirus, adenovirus (Ad), adeno-associated virus (AAV), etc.) [58–60]. Generally, microinjection, electroporation and nucleofection are limited to use in cultured cells (zygotes, embryonic stem cells, T cells, etc.) in vitro [61–64], while hydrodynamic injection has been employed to deliver CRISPR cassettes to the liver in vivo [46, 60]. Unfortunately, although physical approaches are often successful in the laboratory, these methods are not very amenable for clinical translation. In recent years, viral vectors have been extensively applied to deliver CRISPR cassettes in vitro and in vivo largely owing to their high efficiency in gene delivery and long-term stable transgene expression [34, 58, 65]. Especially, AAV can be flexibly constructed to target specific organs or tissues based on the diverse tissue tropism of AAV serotypes [66, 67]. For instance, Yang et al. developed a dual-AAV8 system delivering CRISPR cassettes to correct the point mutation in newborn liver based on the high liver tropism of AAV8 [68]. Similarly, although highly efficient, the intrinsic drawbacks associated with viral vectors, including the risk of carcinogenesis, limitation of insertion size, immune responses and difficulty in large-scale production, severely limited their applications [69–72]. As an alternative, non-viral vectors may offer tantalizing possibility in CRISPR/Cas9 delivery with respect to their low immunogenicity, absence of endogenous virus recombination, less limitation in delivering larger genetic payloads and ease of large-scale production [73–75]. So far, relatively low gene delivery efficiency and transgene expression have been the major barriers in non-viral vectors-based gene therapy [76, 77]. However, with the rapid development of novel biomaterials in recent years, efficient delivery of gene payloads to pass the multiple barriers under physiological conditions and promote transgene expression can be achieved.
In fact, there have been a number of excellent reviews concerning different delivery systems for CRISPR-based genome editing [78–81]. Thus, in this review, we do not intend to elaborate on the physical approaches and viral vectors in CRISPR/Cas9 delivery. Instead, we will discuss the limitations and challenges in CRISPR/Cas9-based genome editing, with focus on the recent progresses on non-viral vectors for CRISPR/Cas9 delivery, highlight the main extracellular and intracellular barriers of delivery, and finally propose some strategies to overcome these obstacles to fulfill the clinical need of CRISPR/Cas9-based genome editing.
2. Limitations and challenges of CRISPR/Cas9-based genome editing
Before CRISPR-based genome editing can be seriously considered for clinical use, several practical issues and technical challenges must be addressed. At molecular level, there are three main issues that need to be overcome. First, setting and reaching the target site with efficiency and accuracy of both cleavage and repair to improve specificity and reduce off-target probability. Second, understanding how to control various repair pathways-NHEJ or HDR- to facilitate switching based on experimental goals. Third, the CRISPR/Cas9 system needs to be more efficient to have therapeutic efficacy in treating diseases. For example, HDR pathway-mediated precise repair dramatically facilitates many areas of biomedical research. However, the desired recombination efficacy often occurs infrequently, which presents enormous challenges for robust applications [61, 82–84]. Although inhibiting the NHEJ pathway with gene silencing or chemical inhibitors can partially increase the efficiency of HDR pathway, the gene repair efficacy still needs to be further improved for disease therapy [82, 85]. Collectively, these issues call for the development of more effective CRISPR systems and more powerful predictable tools. For example, homology-independent targeted integration (HITI) strategy has been developed as an alternative approach for CRISPR/Cas9-mediated gene repair in recent studies [86, 87]. This approach allows for robust DNA knock-in in both dividing and non-dividing cells in vitro and in vivo [87]. There is no doubt that more effective CRISPR systems and tools will be available in the near future with the rapid advances of the scientific research.
Meanwhile, achieving efficient delivery of CRISPR system into particular cell types, tissues or organs for therapeutic benefit is another main ongoing challenge. Development of novel delivery approaches is essential for therapeutic application in vivo. Only when CRISPR/Cas9 systems enter the targeted cells, they can perform more accurately and effectively. Additionally, in this way, the unwanted side effects can be significantly reduced and the systemic safety can be dramatically improved at both extracellular and intracellular levels.
3. Modes of CRISPR/Cas9 delivery
Generally, sgRNA can be incorporated into the plasmids (pX330, pX459 etc.) which contain the sgRNA scaffold or be obtained by in vitro transcription, while the donor template single stranded oligodeoxynucleotide (ssODN) needed for HDR-based gene correction can be synthesized in vitro or constructed in plasmids. Cas9 cassettes can be introduced into the CRISPR systems in three platforms: DNA, mRNA and protein. Each of these delivery strategies has advantages and shortcomings in effectiveness and faces unique challenges (Table 1).
Table 1.
Characteristics of different CRISPR/Cas9 genome editing systems.
| CRISPR/Cas9 platforms | Advantages | Disadvantages | Non-viral vectors | Refs |
|---|---|---|---|---|
| Cas9 protein and sgRNA | swift onset transient duration low off-target effects | high cost endotoxin-contamination | Lipids CPP polymers DNA nanoclew Au nanoparticles zeolitic imidazole frameworks | 69,96–99,102,103,110,113,119 |
| Cas9 mRNA and sgRNA | quick onset transient expression low off-target effects | poor stability | lipids zwitterionic amino-lipid nanoparticles | 104,105,112,116 |
| CRISPR/Cas9 plasmid | low cost great stability | integration risk low efficiency delayed onset | lipids CPP polymers | 100,101,117,118 |
The most straightforward approach is to directly deliver native Cas9 protein with sgRNA. In principle, this format enables the swiftest genome editing as there is no need for transcription and/or translation. It also offers the most transient functionality of genome editing cassette with reduced off-target effects and toxicity. However, due to the large size of Cas9 protein (~ 160 kD), the positive charge of Cas9 protein and the strong negative charge of sgRNA, the effective delivery of Cas9/sgRNA ribonucleoprotein complexes (RNPs) can be difficult [88–91]. Meanwhile, obtaining pure active Cas9 protein is not trivial either. The cost and bacteria endotoxin contamination also need to be carefully addressed.
An alternative option is to use Cas9 mRNA together with sgRNA [27, 92]. Cas9 mRNA can be easily obtained by in vitro transcription. Like RNPs, Cas9 mRNA administration results in quick onset of genome editing, as Cas9 mRNA bypasses the requirement of nuclear entry for transcription. Furthermore, Cas9 mRNA delivery provides transient expression of Cas9 protein, which may be helpful for decreasing the off-target editing events. However, rather short expression duration may also lead to low efficiency. Additionally, the relatively poor stability of Cas9 mRNA is also an important obstacle for this type of delivery strategy. To date, this strategy has been widely applied in editing the genome of zygote, embryo and cultured cells.
The third option is the plasmid-based CRISPR/Cas9 system [93]. This is an appealing delivery approach owing to the simplicity and low cost of manipulation. Both Cas9 and sgRNA cassettes, even HDR template can be facilely packed in the same plasmid, which exhibit greater stability than protein and mRNA. However, the large genetic size of Cas9 (~ 4.5 kb) and the total plasmid size (> 7 kb) significantly increase the difficulty of delivery and expression of CRISPR/Cas9 systems [94]. The second obstacle for plasmid-based expression is the requirement of nuclear entry for DNA transcription, which partially decreases the genome editing efficiency and also induces the delay in therapeutic efficacy. Moreover, the integration of plasmid into the host genome can be another risk. In addition, the plasmid-based expression often leads to a longer duration of Cas9 protein, which may result in higher off-target effects and strong immune responses [95].
4. Current non-viral delivery vectors for CRISPR/Cas9 delivery
4.1 Non-viral vectors in vitro and ex vivo
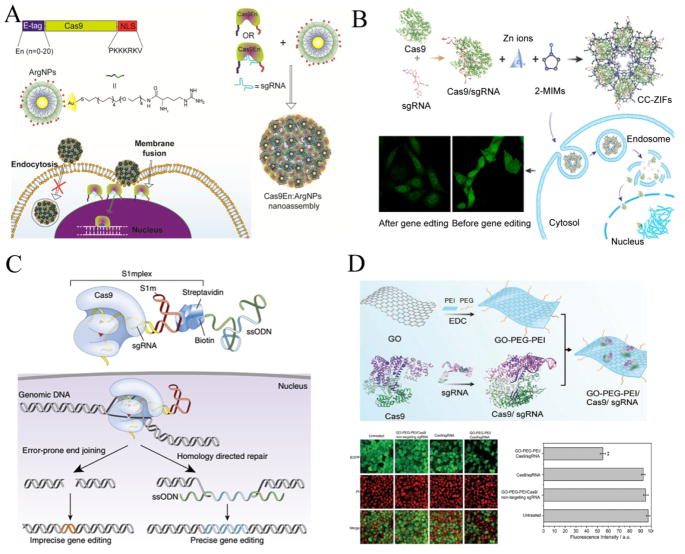
In recent years, CRISPR/Cas9 systems have been widely applied in manipulating mouse zygotes and cultured cells ex vivo and in vitro. Most of these delivery strategies are based on physical approaches (microinjection, electroporation, nucleofection and membrane deformation, etc.) and viral vectors. In the meantime, many non-viral vectors have also been rapidly developed for this purpose. Ramakrishna et al. developed cell-penetrating peptide (CPP)-conjugated recombinant Cas9 protein and CPP-complexed guide RNAs for gene disruptions of CCR5 locus in human cell lines, including HEK293T, HeLa, NCCIT (a human pluripotent embryonal carcinoma cell line), human dermal fibroblasts and human embryonic stem cells. This system led to efficient disruptions of CCR5 gene with minimal off-target mutations [96]. Meanwhile, Mout et al. constructed a nanoplatform based on arginine modified gold nanoparticles to co-assemble with engineered Cas9 protein and sgRNA. This nanoassembly can efficiently enter the targeted cells via membrane fusion, which facilitates direct release of the protein payload into cytoplasm, bypassing endosomes. With this approach, they edited AAVS1 gene and PTEN gene and achieved effective gene editing efficiency up to 30% in HeLa, HEK-293T and Raw 264.7 cell lines (Fig. 2A) [97]. Alsaiari et al. encapsulated negatively charged CRISPR RNPs with positively charged nanoscale zeolitic imidazole frameworks (ZIFs). ZIFs enhanced endosomal escape of RNPs by the protonated imidazole moieties and led to 37% reduction in EGFP gene expression in CHO cells over 4 days (Fig. 2B) [98]. In another study, assembly of CRISPR RNPs with biotinylated oligonucleotides via an RNA aptamer (S1mplex) triggered increased precise gene editing efficacy in hPSC cells (Fig. 2C) [99]. Besides, a flexible dendritic polymer was successfully synthesized to improve the delivery of large plasmid DNA, such as zinc fingers (ZFNs), TALEs and CRISPR/Cas9 systems [100]. Timin et al. reported efficient gene editing with CRISPR/Cas9 system using polymeric and hybrid microcarriers, made of degradable polymers such as polypeptides and polysaccharides [101]. In a recent study, graphene oxide-poly(ethylene glycol)-polyethylenimine (GO-PEG-PEI) nanocarrier mediated efficient CRISPR RNPs delivery via physisorption and π-stacking interaction. This strategy induced efficient genome editing in AGS cells with an efficiency of ~39% (Fig. 2D) [102]. Generally, CRISPR/Cas9 systems delivered by viral or no-viral vectors are more likely to work well in vitro and ex vivo owing to the direct interaction with cells. However, the efficient in vivo delivery of CRISPR/Cas9 systems is nontrivial due to the increased number of barriers under physiological conditions.
Fig. 2.
Non-viral vectors for in vitro CRISPR/Cas9 delivery. (A) Rational design of arginine nanoparticles (ArgNPs) for intracellular delivery of engineering Cas9 protein (Cas9En) or Cas9En/sgRNA ribonucleoprotein complexes (RNPs) via membrane fusion. Engineering Cas9 protein was constructed to carry an N-terminus E-tag and a C-terminus nuclear localization signal (NLS). Reprinted with permission from ref. 97. Copyright 2017 American Chemical Society. (B) The schematic process of preparation, cellular uptake, endosomal eacape and genome editing of zeolitic imidazole frameworks-based CRISPR/Cas9 (CC-ZIFs) delivery. Reprinted with permission from ref. 98. Copyright 2017 American Chemical Society. (C) Design of modular RNA aptamer-streptavidin complexes (S1mplexes) for co-delivery of Cas9/sgRNA ribonucleoprotein complexes and ssODN donor template. Reprinted with permission from ref. 99. Copyright 2015 Nature Publishing Group. (D) The schematic process of preparation and intracellular delivery of GO-PEG-PEI based Cas9/sgRNA delivery. The confocal laser scanning images and the quantification of fluorescence intensity indicated down-regulated expression of EGFP protein after targeted knockout of EGFP genes in AGS.EGFP cells with GO-PEG-PEI based Cas9/sgRNA delivery system. Reprinted with permission from ref. 102. Copyright 2017 John Wiley & Sons. Inc.
4.2 Non-viral vectors in vivo
CRISPR/Cas9-based genome editing systems are of great interest in biomedical research. They have shown great potential in different fields, including studying the function of genetic elements, disease modeling and correcting genetic disorders. The delivery vectors of CRISPR/Cas9 system are vital for in vivo application. A large fraction of viral vectors and some physical methods have been investigated for their potential to facilitate in vivo delivery of CRISPR/Cas9 system. Nevertheless, with the rapid development of various synthetic vectors, non-viral vectors are very likely to provide unique advantages over viral vectors in these fields. Here, we review recent advances of non-viral vectors for CRISPR/Cas9 delivery in vivo and focus on those non-viral delivery systems applied for CRISPR/Cas9-based gene therapy.
Self-assembled DNA nanoclews were constructed for CRISPR/Cas9-based genome editing system delivery. After being loaded with CRISPR RNPs, these nanoclews enabled efficient target disruption of EGFP gene in vitro and in U2OS.EGFP xenograft tumor models in vivo after intratumoral injection [103]. Jiang et al. developed lipid-like nanoparticles for targeted delivery of Cas9 mRNA and sgRNA to the liver. This system achieved disruption of HBV DNA and the proprotein convertase subtilisin/kexin type 9 (pcsk9) gene in vivo [104]. In another study, Miller et al. reported a non-viral vector based on zwitterionic amino lipids to deliver Cas9 mRNA and sgRNAs. In contrast to transient therapies, they showed permanent editing of LoxP gene in genetically engineered mice containing a homozygous Rosa26 promoter Lox-Stop-Lox tdTomato (tdTO) cassette after intravenous administration [105]. Additionally, cationic lipid nucleic acid transfection reagents were also applied for topical delivery of modified Cas9 protein for genome editing in vivo. Zuris et al. injected Cas9:sgRNA complexes targeting GFP gene into the cochlea of transgenic Atoh1-GFP mice. This system resulted in ~20% Cas9-mediated genome modification in hair cells [69]. These are exploratory studies on the in vivo delivery of CRISPR/Cas9 systems with non-viral vectors. These preliminary studies give hopes for in vivo delivery of CRISPR/Cas9 systems with non-viral vectors to treat genetic diseases.
4.2.1 Curing monogenic disorders
Monogenic disorders are diseases arising from a single gene defect in the genome [106, 107]. Gene editing exhibits significant potential in curing monogenic disorders due to the possibility of permanently modifying a false genomic sequence through targeted disruption or HDR-mediated correction [108]. Recently, the use of non-viral vectors to deliver CRISPR/Cas9 system for correcting monogenic disorders has been explored.
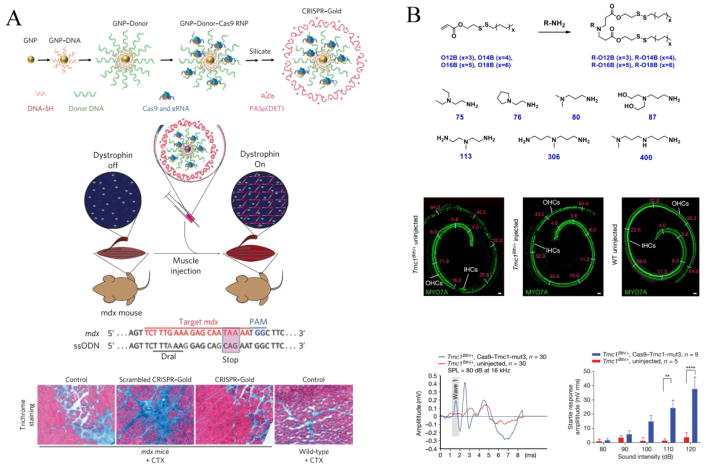
Duchenne muscular dystrophy (DMD) is an inherited X-linked disease caused by the mutations in the genomic sequence encoding dystrophin, a protein crucial for stabilization of muscle sarcolemma and signaling [109]. In a recent study, Lee et al. developed a delivery vehicle (CRISPR–Gold) composed of gold nanoparticles conjugated to DNA and complexed with cationic endosomal disruptive polymers to deliver CRISPR RNPs and donor template. After being injected into the hindlimb muscle of mdx mice simultaneously with cardiotoxin (CTX), this system efficiently corrected the DNA mutation that causes DMD in mice with reduced off-target effect and reduced muscle fibrosis in mdx mice (Fig. 3A) [110].
Fig. 3.
Non-viral vectors for in vivo CRISPR/Cas9 delivery to treat monogenic disorders. (A) Rational design of CRISPR–gold nanoparticles to deliver Cas9/sgRNA RNPs and donor DNA template to correct dystrophin gene in the mouse model of Duchenne muscular dystrophy (DMD). CRISPR–Gold was injected into the hind leg muscle of mdx mice simultaneously with cardiotoxin (CTX), which activated the proliferation of muscle stem cells by muscle damage. Two weeks later, Trichrome staining was performed on the tibialis anterior muscle to determine the levels of muscle fibrosis after different treatment. Reprinted with permission from ref. 110. Copyright 2015 Nature Publishing Group. (B) Rational synthesis of cationic lipids for Cas9/sgRNA RNPs delivery to ameliorate hearing loss in Beethoven (Bth) mouse model of human genetic deafness. Cas9/sgRNA–lipid complexes targeting the Tmc1Bth allele were locally injected into the cochlea of neonatal Tmc1Bth/+ mice. The Effects of Cas9–Tmc1-mut3 sgRNA–lipid injection on hair-cell function and hearing rescue in mice was assessed by confocal microscopy imaging and acoustic startle responses. Reprinted with permission from ref. 113. Copyright 2015 Nature Publishing Group.
Hereditary tyrosinemia type 1 (HT1) is an autosomal recessive disorder resulted from genetic defects of fumarylacetoacetate hydrolase (FAH), the last enzyme in the degradation of tyrosine. HT1 is characterized by severe liver failure, impaired coagulation, renal tubular dysfunction and a risk of hepatocellular carcinoma [111]. Yin et al. combined lipid nanoparticle-mediated delivery of Cas9 mRNA with AAV encoding sgRNAs and donor templates to repair FAH gene in a mouse model of human hereditary tyrosinemia. The efficiency of correction was >6% of hepatocytes after a single application and the treatment rescued disease symptoms such as weight loss and liver damage [112]. Most recently, the same group also applied cationic lipid to deliver Cas9-guide RNA complexes for the treatment of autosomal dominant hearing loss. Local injection of Cas9-guide RNA (Cas9-Tmc1-mut3 sgRNA)-lipid complexes targeting the Tmc1Bth allele into the cochlea of neonatal Tmc1Bth/+ mice significantly enhanced the survival of inner hair cells (IHCs) and outer hair cells (OHCs). Greater auditory brainstem responses (ABRs) wave 1 amplitudes and a more normal ABR waveform pattern in injected ears were observed than that in uninjected controls. Besides, significant startle responses were detected in Cas9-Tmc1-mut3-lipid-injected Tmc1Bth/+ mice following stimulus at 110 and 120 dB. All these results demonstrated that this system substantially reduced progressive hearing loss in Beethoven mouse model (Fig. 3B) [113].
Transthyretin (TTR)-mediated amyloidosis (ATTR) is an inherited, progressive and fatal disease resulted from mutations in the TTR gene. Mutations in TTR cause abnormal amyloid proteins accumulation and result in intractable peripheral sensory neuropathy, autonomic neuropathy, and cardiomyopathy [114, 115]. In a recent report, a lipid nanoparticle (LNP-INT01) mediated effective delivery of both Cas9 mRNA and chemically modified sgRNA in vivo [116]. LNP-INT01 was a biodegradable non-viral delivery system and can be cleared from the body, which minimized the undesired prolonged duration of CRISPR/Cas9 in cells after gene editing and reduced the off-target effect associated with CRISPR/Cas9-based genome editing. With a single intravenous administration, this system enabled significant editing of the TTR gene in the liver of mice or rats, and resulted in a dramatic reduction (>97% for mice, >90% for rats) in serum TTR protein levels. More importantly, the editing levels can persist for at least 12 months, despite the transient nature of the delivery system and the editing components. Additionally, unlike viral delivery systems, the synthetic nature of LNP-INT01 allowed for multi-dosing regimens to obtain cumulative editing efficacy without significant changes in cytokine levels or body weight.
Collectively, using non-viral vectors for in vivo delivery of CRISPR/Cas9 system has shown great potential for monogenic disorders treatment. These preliminary promising results open an avenue to understanding the impact of the chemical structure and components of synthetic materials on the in vivo duration, gene editing efficacy, off-target effect and other safety concern of CRISPR/Cas9 systems. These precious experiences are bound to greatly promote the development of novel approaches based on synthetic non-viral vectors-mediated in vivo CRISPR/Cas9 delivery to cure other monogenic diseases, such as sickle cell disease, cystic fibrosis, polycystic kidney disease, etc., which were treated with viral vectors-mediated gene therapy before.
4.2.2 Manipulating cancer genome
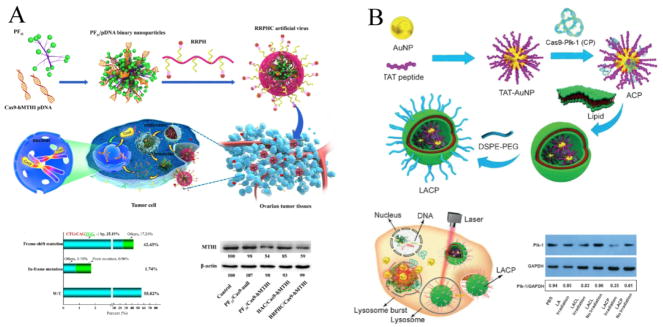
Given that cancer is a genetic disease and CRISPR/Cas9 system is a versatile genome editing tool, it is tempting to apply this technology to cancer treatment. Generally, CRISPR/Cas9 systems were used to induce loss-of-function of oncogenes in these studies. In recent years, many instances of CRISPR/Cas9-mediated therapeutic genome manipulations in cancer have been explored. In one study, Li et al. reported a multifunctional nucleus-targeting “core-shell” artificial virus for CRISPR/Cas9-based plasmid delivery. This system induced higher targeted gene disruption efficacy than that of Lipofectamine 3000 in vitro. After intraperitoneal administration, they also showed effective disruption of targeted MTH1 gene and significantly inhibited the progression of ovarian tumor in vivo (Fig. 4A) [117]. Meanwhile, Wang et al. condensed Cas9-sgPlk-1 plasmids on TAT peptide-modified Au nanoparticles (LACP) via electrostatic interactions, and coated lipids on them. LACP can release plasmid into the cytosol by laser-triggered thermo-effects of the Au nanoparticles and the plasmid can enter nuclei with the assistance of TAT peptide. This delivery system enabled effective knock-outs of target gene (Plk-1) and inhibited the growth of melanoma both in vitro and in vivo (Fig. 4B) [118]. In another report, core-shell liposome-templated hydrogel nanoparticles (LHNPs) were constructed to deliver CRISPR RNPs for cancer gene therapy. After loading with CRISPR/Cas9 targeting polo-like kinase 1 (plk1) gene, LHNPs effectively inhibited U87MG tumor growth [119].
Fig. 4.
Non-viral vectors for in vivo CRISPR/Cas9 delivery to treat cancer. (A) Schematic process of RRPHC artificial virus-mediated in vivo CRISPR/Cas9 delivery for gene editing-based cancer therapy. RRPHC/Cas9-hMTH1 treatment induced ~ 42.43% frame-shift mutation of hMTH1-target loci and down-regulated the expression of MTH1 protein in SKOV3 cells. Reprinted with permission from ref. 117. Copyright 2017 American Chemical Society. (B) Preparation process of LACP and the schematic process of laser-enhanced knock-outs of targeted genes by LACP in A375 cells. LACP encapsulating Cas9-Plk-1 plasmid down-regulated the expression of Plk-1 protein in A375 cells with laser irradiation. Reprinted with permission from ref. 118. Copyright 2017 John Wiley & Sons. Inc.
Owing to the polygenic and heterogeneous properties of cancer, genomic aberration profiles are different in tumors and even in different stages of tumor progression. Disruption of one or two over-expressed genes in tumors with CRISPR/Cas9 system may not be very effective in cancer treatment. Even more problematic is that the genome editing efficiency of CRISPR/Cas9 system is not high enough under the current conditions. Monogenic diseases can tolerate a low initial gene editing efficiency due to the selective advantages of the corrected cells, while cancer treatment requires relatively high editing efficiency to ensure effective therapy, because the unedited cancer cells possess selective advantages and proliferate quickly. Additionally, due to the instability of the genome of cancer cells, they have evolved many mechanisms to escape CRISPR/Cas9-mediated genome editing, such as mutation of the PAM sequence and the recognition site etc. [120–122]. However, with the rapid development of CRISPR/Cas9 technology and materials science, we believe these barriers in CRISPR/Cas9-mediated cancer gene therapy would certainly be conquered by enhancing activity and delivery of the CRISPR/Cas9 system in the near future.
In recent years, epigenetics has attracted much attention with a growing understanding of tumorigenesis. Epigenetics are heritable changes in gene expression which do not involve any alteration in the DNA sequence. Epigenetics is on top of the traditional genetic basis for inheritance and donated changes in chromosomes that affect activity and expression of a series of genes. Many reports have demonstrated that epigenetic alterations play important roles in driving cancer initiation and progression [123, 124]. Three systems, including DNA methylation, RNA-associated silencing and histone modification, are used to initiate and sustain epigenetic silencing. Generally, these processes require the catalysis of some specific enzymes, such as DNA methyltransferase (DNMT), histone deacetylase (HDAC) and histone acetyltransferase (HAT), etc. Therefore, we conceive that manipulating epigenome or the genes encoding the associated enzymes would be alternative strategies for CRISPR/Cas9-based cancer gene therapy. Most recently, dCas9 fusion proteins-mediated CRISPRi, such as dCas9-KRAB (Krüppel-associated box) and dCas9-LSD1 (lysine-specific histone demethylase 1) system, has been introduced [125–127]. dCas9-KRAB system, recruiting a diverse array of histone modifiers that reversibly suppresses gene expression through the formation of heterochromatin, significantly reduced the expression of a series of endogenous genes in the genome [125, 128]. However, due to the multiple functions of epigenetics, safety is a big concern that needs special attention when applying these strategies. Non-viral vectors-mediated transient expression of CRISPR/Cas9 system may be an effective approach to alleviate this problem.
In addition to epigenetics-based therapy, combination strategy of gene editing with other treatment, such as chemotherapy, may be another effective way for cancer treatment. Acquired drug resistance is the major cause of chemotherapy-based cancer treatment failure [129, 130]. Overexpression of drug resistance-associated genes, such as P-gp and Bcl-2, is one of the most consistent alterations in multidrug resistant cells [131–134]. The conventional strategy is to inhibit the expression or activity of these genes through small interfering RNA (siRNA) or chemical inhibitors [135–137]. However, the effects of either siRNA or chemical inhibitor are transient and are limited to the mother cell that takes the drug. If the cell divides, these drugs will lose activity in the daughter cells. Due to the permanent gene editing effect of CRISPR/Cas9 systems in chromosome, the daughter cells will inherit the edited gene sequences of the mother cells. Manipulating drug resistance-associated genes using CRISPR/Cas9 systems together with chemotherapy may be a preferable approach to cancer treatment over conventional combination therapy. Besides, in contrast with viral vectors, cumulative editing efficacy can be obtained after multiple administrations of non-viral vectors-mediated CRISPR/Cas9 delivery owing to limited immune responses [73, 74, 116, 138].
As a rapid developing field for CRISPR/Cas9 delivery, non-viral vectors also face some hurdles especially in systemic delivery in vivo. There are two main limitations of non-viral vectors-mediated CRISPR/Cas9 delivery in vivo. The first one is the serum stability of polyplexes after systemic administration [73]. The second one is the relatively low transfection efficiency in the targeted tissues after administration in vivo [76, 77]. These issues are not just for non-viral vectors-mediated CRISPR/Cas9 delivery, but for non-viral vectors-mediated gene delivery in general. In the following section, we will systematically discuss the critical obstacles for non-viral vectors-mediated CRISPR/Cas9 delivery in order to provide some basis for designing better vectors in the future.
5. Critical obstacles for non-viral delivery of CRISPR/Cas9 system
Similar to other common gene therapy strategies, CRISPR/Cas9 system delivery faces critical challenges for in vivo applications. Following systemic administration, the elements of CRISPR/Cas9 system should remain stable before reaching its target site. Then the elements need to be effectively taken up by the targeted cell types and escape from endo-lysosome system to avoid degradation. After being released from endosome or lysosome, they also need to enter the nucleus to initiate gene editing in the genome. In addition, in case of CRISPR/Cas9-based plasmids, they must traffic through the cytosol to the nucleus for transcription.
The first obstacle is effective encapsulation of the editing tools in delivery vectors due to the large size and different charge properties of Cas9 protein, mRNA and donor DNA. The molecular weight of the commonly used Cas9 protein from streptococcus pyogenes is about 160 KD, larger than that of the most proteins [89]. Meanwhile, native Cas9 protein is positively charged, while the conventional proteins are negatively charged [88, 97, 103, 139]. Thus, the cationic lipids or polymers widely used to encapsulate negatively charged proteins based on electrostatic interaction cannot be applied to deliver the native form of Cas9 protein. Similar to the protein form, Cas9 mRNA and DNA (~ 4.5 kb) also exhibit large size, which increases the difficulty of efficient encapsulation. Although the newly discovered Cas9 form from Staphylococcus aureus strain (SaCas9) is about 1 kb shorter than SpCas9, the overall genetic size is still too large for application [94, 140]. All of these defects need to be taken into consideration in future non-viral vectors designing.
The second obstacle is the stability of complex under physiological conditions. On the one hand, protein, plasmid and RNA are vulnerable to proteases and nucleases in the blood. They can degrade quickly in naked form in systemic circulation. On the other hand, cationic non-viral vectors used to condense negatively charged CRISPR RNPs and plasmids are prone to interact with plasma proteins and the physiological salt in physiological conditions, resulting in the recognition and clearance by the reticuloendothelial system (RES) [141–143]. PEGylation, the most commonly used strategy to reduce the opsonization and minimize the clearance by RES, may also be applicable for non-viral vectors-mediated CRISPR/Cas9 delivery [144, 145]. However, the accompanying decreased cellular uptake after PEGylation cannot be neglected. Like other gene therapy strategies, environmental stimuli-triggered detachment may be a feasible solution to this problem [146].
The third obstacle is the host immune responses triggered by the elements of CRISPR/Cas9 system, which is derived from bacteria. Generally, after being sheltered by non-viral vectors, the recognition by the host immune system would be reduced to some extent. Encapsulating these elements in the interior of the vectors would significantly prevent this recognition and subsequent clearance. However, it is also worth noting that partial shielding via simple electrostatic interaction may not be effective enough to eliminate the recognition by host immune cells owing to the exposure of neutral domains of these elements. Additionally, the properties such as size, surface charge, hydrophobicity/hydrophilicity, and the steric effects of particle coating have influence on the compatibility of nanoparticles with the immune system [147–150]. Attaching PEG or other hydrophilic polymers to the surface of the nanoparticles can partially shield them from immune recognition [151, 152]. However, some reports also demonstrated the generation of PEG-specific antibodies after administration of PEG-coated liposomes [153, 154]. It is worth noting that formation of specific antibodies against nanoparticles may affect the efficacy and the safety of nanoparticle-based therapeutics [149, 150]. Despite all these, the immunogenicity of non-viral vectors is still considered to be much lower than that of viral vectors [74].
The fourth obstacle is the insufficient accumulation of non-viral vector-mediated CRISPR/Cas9 systems in the targeted tissues. This issue is very important for targeted gene editing efficacy as well as the off-target probability in other tissues. In the past decades, there have been extensive efforts on facilitating particles accumulation in the targeted tissues for non-viral delivery vectors-mediated gene therapy [155, 156]. We assume that these strategies can also be applied to non-viral vectors-mediated CRISPR/Cas9 genome editing. For example, as a unique feature of solid tumors, the enhanced permeability and retention (EPR) effect can be exploited to promote particles accumulation into tumors. Generally, the particles should be nano-sized and the junction of endothelial cells has a great influence on the extravasation of these particles. Some reports have demonstrated that EPR effect can provide 20–30% increase in delivery compared with critical normal organs [157]. Some auxiliary means, such as topical hyperthermia, radiotherapy and high intensity focused ultrasound, etc., can also be used to enhance this effect [158–160]. Besides, for the fibrotic lesion, degradation of the extracellular matrix using collagenase or hyaluronidase can be effective strategies to enhance accumulation [161, 162]. Additionally, surface decorating active targeting ligands on the vectors should be another effective method to promote targeted accumulation via receptor-mediated recognition. For example, due to the overexpression of αvβ3 integrin receptors on tumor vasculature, the nanoparticles decorated with arginine-glycine-glutamic acid (RGD) peptide, a ligand for αvβ3 integrin receptors, have more access to the targeted tumor tissues [163–165].
The fifth encountered obstacle is the phagocytic clearance, blockage of RES in targeted tissues especially in liver and spleen, and the negatively charged components of extracellular matrix. Smaller-sized nanoparticles may be preferred to enhance permeability and penetration over large-sized ones [166, 167]. However, smaller-sized nanoparticles can also be rapidly cleared from the systemic circulation through the kidneys. Due to the threshold for rapid renal excretion being about 5.5 nm, the hydrodynamic diameters of the prepared nanoparticles need to be larger than this size [168–170]. To balance the effect of penetration and renal excretion, environmental stimuli-responsive size changeable tactics would be a possible compromising solution. The particles remain stable in systemic circulation, while exhibiting size conversion or directly broken into smaller parts at the target site triggered by specific stimulation to boost deep penetration.
Cellular uptake is the next obstacle owing to the selective transmission of the hydrophobic and negatively charged plasma membrane. In principle, naked CRISPR plasmid and sgRNA cannot directly transport through the lipid bilayer of the cell membrane due to the hydrophilic and anionic properties. Generally, after complexation with non-viral vectors to form nanoparticles, the cellular uptake would be promoted. For cationic complexes, cellular uptake is partly induced through electrostatic interaction between the positive charge of the complexes and the negative charge of the cell membrane. Meanwhile, many types of ligands, such as folate, transferrin, antibody and aptamer, etc., can also be incorporated into the non-viral vectors to boost cellular uptake via receptor-mediated endocytosis [171–173]. Besides, cell penetrating peptides (CPP), such as TAT and R8 peptide, have also been extensively used to facilitate cell entry based on the positive charge and amphipathic in nature [174, 175]. Some studies have shown that hyperthermia can increase the membrane permeability of compounds and enhance cellular uptake. But the temperature and duration of this approach need to be carefully controlled. Additionally, ultrasound has also been implicated as a potential approach to promote cell entry owing to ultrasound-induced mild hyperthermia and cell membrane deformation [176, 177].
In addition to cellular uptake, endosomal and lysosomal degradation is also a critical challenge for efficient CRISPR/Cas9 delivery due to the acidic environment (pH 5.0–6.2) and hydrolases. The “proton-sponge effect” of polymers with proton buffer capacity can be exploited to promote endo-lysosomal escape. Briefly, 2° and 3° amines on polymer chains can be protonated inside endo-lysosomes and then counterbalance chloride ions flow into the vesicles. This process creates an osmotic gradient between the exterior and interior of the endo-lysosomes, leading to swelling and bursting of these vesicles and releasing the polyplex into the cytosol [178]. A variety of non-viral vectors, including but not limited to polyethylenimine (PEI), poly[2-(dimethylamino) ethyl methacrylate] (PDMAEMA), have “proton-sponge effect”. Meanwhile, some compounds, such as chloroquine, imidazole, and zinc chloride, have also been combined with cationic vectors to further enhance endosomal escape [179–182]. Recently, gas-generating nanoparticles have also been designed to boost endo-lysosomal escape in some studies. Liu et al. fabricated pH-responsive poly(D,L-lactic-co-glycolic acid) (PLGA) nanoparticles encapsulating ammonium bicarbonate (NH4HCO3) to trigger antigen release from endo-lysosomes. NH4HCO3 interacted with protons in endo-lysosomes, then produced CO2 and NH3 to disrupt the endo-lysosomes and promote antigen release into the cytosol [183].
In addition, inspired by the mechanism of viral infection, some membrane fusion peptides, such as GALA peptides and acid-activatable melittin, have been introduced into non-viral vectors to facilitate endosomal escape [184–186]. One underlying mechanism is these peptides undergo a structural transformation from a random coil to an α-helical conformation in lower pH environments, thus promoting endosomal fusion. In fact, direct transmembrane delivery of the particles into the cytosol bypassing the endo-lysosomes may be an alternative way [187, 188]. Ding et al. described a nanoparticle formulation based on reconstituted high-density lipoprotein (rHDL) can efficiently transfer siRNA across the cell membrane directly into the cytosol via scavenger receptor BI (SR-BI)-mediated non-endocytotic mechanism. This approach markedly promoted RNA interference (RNAi)-mediated degradation of the targeted mRNA and resulted in down-regulated expression of the corresponding protein [189].
Cytoplasmic mobility and nuclear import is another big obstacle in the intracellular environment. CRISPR/Cas9 system must be delivered to the nucleus to initiate genome editing effect. Once released into the cytosol, non-viral vectors loaded with CRISPR/Cas9 system need to overcome additional barriers in the cytosol, including the mesh-like cytoskeleton and nucleolytic enzymes within the cytosolic milieu [190]. Disruption or reorganization of the actin cytoskeleton, or depletion of keratin and vimentin in cells, significantly increases the diffusion of gene payloads [190–192]. Virus such as herpes simplex virus and some serotypes of adenovirus could travel through the cytoplasm via microtubule-mediated transport [76, 193, 194]. Cationic polyplexes could move along microtubules by non-specifically interacting with anionic microtubules or motor proteins [76]. For CRISPR/Cas9-based plasmids, they need to overcome more barriers than that of CRISPR RNPs and mRNA. They have to enter the nucleus for transcription and traffic back to the cytosol for translation. Non-viral vectors that compact plasmid into small particles may aid the movement to the nucleus. Nanoparticles smaller than 9 nm can passively diffuse through nuclear pore complexes (NPCs), while larger ones requiring the assistance of import proteins (e.g., importins) to actively shuttle them into the nucleus [73]. Inspired by the active transportation process of proteins to nucleus, nuclear localization sequences (NLSs) have been introduced into the vectors to facilitate transportation to nucleus [195–197]. Most NLSs are characterized by short basic and cationic peptide sequences. NLSs can be recognized by importin α, the nuclear import receptor, and trigger the subsequent transportation from the cytosol to the nucleus [198].
6. Perspectives
RNA-guided CRISPR/Cas9 system is a powerful genome editing tool, which greatly simplifies the previous cumbersome genetic manipulations. In recent years, CRISPR/Cas9 system has been broadly applied for disease modeling to therapeutic applications. Undoubtedly, with further development of this technology, its application will certainly be more extensive in the near future.
However, the specificity of this technology will need to be further improved to decrease the potential off-target probability for increasing safety. The term “off-target” generally has dual meanings, one for manipulating at the targeted genes but in the non-targeted tissues or cells, while the other for gene editing at the non-targeted site in the genome of the targeted cells at the intracellular level. For the former off-target effects, tissue-specific promoters in CRISPR/Cas9-based plasmid have been introduced into the system to partially solve this problem. Meanwhile, targeted delivery of CRISPR/Cas9 system by tissue-specific AAV type or non-viral vectors could be another effective solution to this issue. In addition, proper selection of the targeting site, rational design of sgRNA and using paired Cas9 nickases could be effective approaches to alleviate the latter off-target effects.
Next, the efficiency will also need to be improved for in vivo application, especially in correcting majority of genetic disorders. Since the corrected cells have no selection advantages in most genetic disorders, the correction efficiency will undoubtedly need to improve. Meanwhile, different cell types or targeted genes can also affect the efficiency of the CRISPR/Cas9 system [93, 199]. The mitotic state of the targeted cells is another major concern. Non-mitotic cells are very inefficient for HDR-based repair, which is the main pathway to correct genetic disorders to date [87, 200]. Besides, due to the recognition preference of some specific bases of CRISPR/Cas9 system and the diverse epigenetic landscape, different targeted gene loci may exhibit distinctive different gene editing efficiencies [140, 201–203]. These issues may be resolved by enhancing activity and delivery efficiency of the CRISPR-Cas9 system. The recently developed HITI strategy exhibits much higher knock-in efficiency than HDR both in vitro and in vivo [86, 87]. More importantly, HITI keeps its high efficiency in the non-mitotic cells, which are present in most adult tissues [87]. It is likely that this approach will show great potential in treating genetic disorders in the near future.
In addition, delivery alternatives are another main challenge for the application of CRISPR/Cas9 gene editing system both in vitro and in vivo. The latter would particularly benefit from non-viral vectors. Ideally, effective non-viral delivery vectors should induce efficient targeted gene editing with lower off-target effect. Although numerous non-viral vectors have been developed and exploited in the field of gene therapy over the past decades, the non-viral vectors applied in CRISPR/Cas9 delivery are still very few so far. One possible reason is that the large-sized Cas9 protein or plasmid is difficult to be efficiently encapsulated and delivered. Moreover, unlike commonly delivery approaches for siRNA or protein, whose action site is in the cytosol, CRISPR/Cas9 systems must be delivered to the nucleus to initiate genome editing in the chromosome. It seems like that the cationic lipid-based vectors were one of the most effective delivery approaches for CRISPR/Cas9 system to date. But it is mainly limited to local administration. Novel effective non-viral vector for CRISPR/Cas9 delivery is still one of the bottlenecks for the successful clinical translation of CRISPR/Cas9-based genome editing to date. In conclusion, despite these challenges, there is no doubt that the rapid advances in the field of genome editing and biomaterials science will certainly pave the way for translating these promising results to clinical disease treatment in the near future.
Acknowledgments
This research was supported in part, by the Intramural Research Program, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, and the National Science Foundation of China (81771873).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 3.Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–209. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 5.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasiunas G, Siksnys V. RNA-dependent DNA endonuclease Cas9 of the CRISPR system: Holy Grail of genome editing? Trends Microbiol. 2013;21:562–567. doi: 10.1016/j.tim.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y, Zhou H, Fan X, Zhang Y, Zhang M, Wang Y, et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25:67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 16.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 19.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 28.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dow LE, Fisher J, O’Rourke KP, Muley A, Kastenhuber ER, Livshits G, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33:390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 33.Ye L, Wang J, Tan Y, Beyer AI, Xie F, Muench MO, et al. Genome editing using CRISPR- Cas9 to create the HPFH genotype in HSPCs: An approach for treating sickle cell disease and beta-thalassemia. Proc Natl Acad Sci U S A. 2016;113:10661–10665. doi: 10.1073/pnas.1612075113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 36.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimi K, Kaneko T, Voigt B, Mashimo T. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat Commun. 2014;5:4240. doi: 10.1038/ncomms5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 40.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koike-Yusa H, Li Y, Tan EP, del Velasco-Herrera MC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 42.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 43.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maresch R, Mueller S, Veltkamp C, Ollinger R, Friedrich M, Heid I, et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun. 2016;7:10770. doi: 10.1038/ncomms10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansen AK, Molenaar B, Versteeg D, Leitoguinho AR, Demkes C, Spanjaard B, et al. Postnatal Cardiac Gene Editing Using CRISPR/Cas9 With AAV9-Mediated Delivery of Short Guide RNAs Results in Mosaic Gene Disruption. Circ Res. 2017;121:1168–1181. doi: 10.1161/CIRCRESAHA.116.310370. [DOI] [PubMed] [Google Scholar]
- 46.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 48.Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaj T, Ojala DS, Ekman FK, Byrne LC, Limsirichai P, Schaffer DV. In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci Adv. 2017;3:eaar3952. doi: 10.1126/sciadv.aar3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao-Jie L, Hui-Ying X, Zun-Ping K, Jin-Lian C, Li-Juan J. CRISPR-Cas9: a new and promising player in gene therapy. J Med Genet. 2015;52:289–296. doi: 10.1136/jmedgenet-2014-102968. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castillo A. Gene editing using CRISPR-Cas9 for the treatment of lung cancer. Colomb Med (Cali) 2016;47:178–180. [PMC free article] [PubMed] [Google Scholar]
- 55.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539:479. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 57.Reardon S. First CRISPR clinical trial gets green light from US panel. Nature. 2016 [Google Scholar]
- 58.Yu W, Mookherjee S, Chaitankar V, Hiriyanna S, Kim JW, Brooks M, et al. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun. 2017;8:14716. doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 60.Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen S, Lee B, Lee AY, Modzelewski AJ, He L. Highly Efficient Mouse Genome Editing by CRISPR Ribonucleoprotein Electroporation of Zygotes. J Biol Chem. 2016;291:14457–14467. doi: 10.1074/jbc.M116.733154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 67.Gao G, Vandenberghe LH, Wilson JM. New recombinant serotypes of AAV vectors. Curr Gene Ther. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- 68.Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lehrman S. Virus treatment questioned after gene therapy death. Nature. 1999;401:517–518. doi: 10.1038/43977. [DOI] [PubMed] [Google Scholar]
- 70.Gore ME. Adverse effects of gene therapy: gene therapy can cause leukaemia: no shock, mild horror but a probe. Gene Ther. 2003;10:4–4. [Google Scholar]
- 71.Donahue RE, Kessler SW, Bodine D, McDonagh K, Dunbar C, Goodman S, et al. Helper virus induced T cell lymphoma in nonhuman primates after retroviral mediated gene transfer. J Exp Med. 1992;176:1125–1135. doi: 10.1084/jem.176.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun JY, Anand-Jawa V, Chatterjee S, Wong KK. Immune responses to adeno-associated virus and its recombinant vectors. Gene Ther. 2003;10:964–976. doi: 10.1038/sj.gt.3302039. [DOI] [PubMed] [Google Scholar]
- 73.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 74.Li SD, Huang L. Non-viral is superior to viral gene delivery. J Control Release. 2007;123:181–183. doi: 10.1016/j.jconrel.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 75.Li L, He ZY, Wei XW, Gao GP, Wei YQ. Challenges in CRISPR/CAS9 delivery: potential roles of nonviral vectors. Hum Gene Ther. 2015;26:452–462. doi: 10.1089/hum.2015.069. [DOI] [PubMed] [Google Scholar]
- 76.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 77.Davis ME. Non-viral gene delivery systems. Curr Opin Biotechnol. 2002;13:128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- 78.Hao Y, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nat Rev Drug Discov. 2017;16:387–399. doi: 10.1038/nrd.2016.280. [DOI] [PubMed] [Google Scholar]
- 79.Wang HX, Li M, Lee CM, Chakraborty S, Kim HW, Bao G, et al. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem Rev. 2017;117:9874–9906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- 80.Glass Z, Lee M, Li Y, Xu Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 2018;36:173–185. doi: 10.1016/j.tibtech.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26. doi: 10.1016/j.jconrel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 83.Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He X, Tan C, Wang F, Wang Y, Zhou R, Cui D, et al. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016;44:e85. doi: 10.1093/nar/gkw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Subburaj S, Chung SJ, Lee C, Ryu SM, Kim DH, Kim JS, et al. Site-directed mutagenesis in Petunia x hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016;35:1535–1544. doi: 10.1007/s00299-016-1937-7. [DOI] [PubMed] [Google Scholar]
- 90.Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33:1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 91.Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hughes TS, Langer SJ, Virtanen SI, Chavez RA, Watkins LR, Milligan ED, et al. Immunogenicity of intrathecal plasmid gene delivery: cytokine release and effects on transgene expression. J Gene Med. 2009;11:782–790. doi: 10.1002/jgm.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mout R, Ray M, Yesilbag Tonga G, Lee YW, Tay T, Sasaki K, et al. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017;11:2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alsaiari SK, Patil S, Alyami M, Alamoudi KO, Aleisa FA, Merzaban JS, et al. Endosomal escape and delivery of CRISPR/Cas9 genome editing machinery enabled by nanoscale zeolitic imidazolate framework. J Am Chem Soc. 2018;140:143–146. doi: 10.1021/jacs.7b11754. [DOI] [PubMed] [Google Scholar]
- 99.Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, et al. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat Commun. 2017;8:1711. doi: 10.1038/s41467-017-01875-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kretzmann JA, Ho D, Evans CW, Plani-Lam JHC, Garcia-Bloj B, Mohamed AE, et al. Synthetically controlling dendrimer flexibility improves delivery of large plasmid DNA. Chem Sci. 2017;8:2923–2930. doi: 10.1039/c7sc00097a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Timin AS, Muslimov AR, Lepik KV, Epifanovskaya OS, Shakirova AI, Mock U, et al. Efficient gene editing via non-viral delivery of CRISPR-Cas9 system using polymeric and hybrid microcarriers. Nanomedicine. 2018;14:97–108. doi: 10.1016/j.nano.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 102.Yue H, Zhou X, Cheng M, Xing D. Graphene oxide-mediated Cas9/sgRNA delivery for efficient genome editing. Nanoscale. 2018;10:1063–1071. doi: 10.1039/c7nr07999k. [DOI] [PubMed] [Google Scholar]
- 103.Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54:12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang C, Mei M, Li B, Zhu X, Zu W, Tian Y, et al. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017;27:440–443. doi: 10.1038/cr.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, et al. Non-Viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew Chem Int Ed Engl. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jullien D. Pathogenesis of the metabolic syndrome. Ann Dermatol Venereol. 2008;135(Suppl 4):S243–248. doi: 10.1016/S0151-9638(08)70542-8. [DOI] [PubMed] [Google Scholar]
- 107.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 108.Prakash V, Moore M, Yanez-Munoz RJ. Current Progress in Therapeutic Gene Editing for Monogenic Diseases. Mol Ther. 2016;24:465–474. doi: 10.1038/mt.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L, et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 110.Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nature Biomedical Engineering. 2017:889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aponte JL, Sega GA, Hauser LJ, Dhar MS, Withrow CM, Carpenter DA, et al. Point mutations in the murine fumarylacetoacetate hydrolase gene: Animal models for the human genetic disorder hereditary tyrosinemia type 1. Proc Natl Acad Sci U S A. 2001;98:641–645. doi: 10.1073/pnas.98.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao X, Tao Y, Lamas V, Huang M, Yeh WH, Pan B, et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hund E. Familial amyloidotic polyneuropathy: current and emerging treatment options for transthyretin-mediated amyloidosis. Appl Clin Genet. 2012;5:37–41. doi: 10.2147/TACG.S19903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sekijima Y. Transthyretin (ATTR) amyloidosis: clinical spectrum, molecular pathogenesis and disease-modifying treatments. J Neurol Neurosurg Psychiatry. 2015;86:1036–1043. doi: 10.1136/jnnp-2014-308724. [DOI] [PubMed] [Google Scholar]
- 116.Jonathan DF, Amy RS, Mihir CP, Lucinda S, Madeleine RY, Jane VH. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 117.Li L, Song L, Liu X, Yang X, Li X, He T, et al. Artificial vrus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano. 2017;11:95–111. doi: 10.1021/acsnano.6b04261. [DOI] [PubMed] [Google Scholar]
- 118.Wang P, Zhang L, Zheng W, Cong L, Guo Z, Xie Y, et al. Thermo-triggered Release of CRISPR-Cas9 System by Lipid-Encapsulated Gold Nanoparticles for Tumor Therapy. Angew Chem Int Ed Engl. 2018;57:1491–1496. doi: 10.1002/anie.201708689. [DOI] [PubMed] [Google Scholar]
- 119.Chen Z, Liu F, Chen Y, Liu J, Wang X, Chen AT. Targeted delivery of CRISPR/Cas9-mediated cancer gene therapy via liposome-templated hydrogel nanoparticlesaptation repertoires. Adv Funct Mater. 2017;27:1703036. doi: 10.1002/adfm.201703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 121.Shen Z. Genomic instability and cancer: an introduction. J Mol Cell Biol. 2011;3:1–3. doi: 10.1093/jmcb/mjq057. [DOI] [PubMed] [Google Scholar]
- 122.Tubbs A, Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell. 2017;168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 124.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 125.Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13:127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kitano H. Cancer as a robust system: implications for anticancer therapy. Nat Rev Cancer. 2004;4:227–235. doi: 10.1038/nrc1300. [DOI] [PubMed] [Google Scholar]
- 130.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 131.Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance- associated proteins. J Natl Cancer Inst. 2000;92:1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- 132.Thomas H, Coley HM. Overcoming multidrug resistance in cancer: an update on the clinical strategy of inhibiting p-glycoprotein. Cancer Control. 2003;10:159–165. doi: 10.1177/107327480301000207. [DOI] [PubMed] [Google Scholar]
- 133.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 134.Delbridge AR, Grabow S, Strasser A, Vaux DL. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat Rev Cancer. 2016;16:99–109. doi: 10.1038/nrc.2015.17. [DOI] [PubMed] [Google Scholar]
- 135.He C, Lu K, Liu D, Lin W. Nanoscale metal-organic frameworks for the co-delivery of cisplatin and pooled siRNAs to enhance therapeutic efficacy in drug-resistant ovarian cancer cells. J Am Chem Soc. 2014;136:5181–5184. doi: 10.1021/ja4098862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li L, Gu W, Chen J, Chen W, Xu ZP. Co-delivery of siRNAs and anti-cancer drugs using layered double hydroxide nanoparticles. Biomaterials. 2014;35:3331–3339. doi: 10.1016/j.biomaterials.2013.12.095. [DOI] [PubMed] [Google Scholar]
- 137.Huang W, Chen L, Kang L, Jin M, Sun P, Xin X, et al. Nanomedicine-based combination anticancer therapy between nucleic acids and small-molecular drugs. Adv Drug Deliv Rev. 2017;115:82–97. doi: 10.1016/j.addr.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 138.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 139.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li L, Wei Y, Gong C. Polymeric nanocarriers for non-viral gene delivery. J Biomed Nanotechnol. 2015;11:739–770. doi: 10.1166/jbn.2015.2069. [DOI] [PubMed] [Google Scholar]
- 142.Dash PR, Read ML, Barrett LB, Wolfert MA, Seymour LW. Factors affecting blood clearance and in vivo distribution of polyelectrolyte complexes for gene delivery. Gene Ther. 1999;6:643–650. doi: 10.1038/sj.gt.3300843. [DOI] [PubMed] [Google Scholar]
- 143.Xu L, Anchordoquy T. Drug delivery trends in clinical trials and translational medicine: challenges and opportunities in the delivery of nucleic acid-based therapeutics. J Pharm Sci. 2011;100:38–52. doi: 10.1002/jps.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6:595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 145.Malam Y, Lim EJ, Seifalian AM. Current trends in the application of nanoparticles in drug delivery. Curr Med Chem. 2011;18:1067–1078. doi: 10.2174/092986711794940860. [DOI] [PubMed] [Google Scholar]
- 146.Kuai R, Yuan W, Li W, Qin Y, Tang J, Yuan M, et al. Targeted delivery of cargoes into a murine solid tumor by a cell-penetrating peptide and cleavable poly(ethylene glycol) comodified liposomal delivery system via systemic administration. Mol Pharm. 2011;8:2151–2161. doi: 10.1021/mp200100f. [DOI] [PubMed] [Google Scholar]
- 147.Zolnik BS, Gonzalez-Fernandez A, Sadrieh N, Dobrovolskaia MA. Nanoparticles and the immune system. Endocrinology. 2010;151:458–465. doi: 10.1210/en.2009-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol Pharm. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dobrovolskaia MA, McNeil SE. Immunological properties of engineered nanomaterials. Nat Nanotechnol. 2007;2:469–478. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 150.Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv Drug Deliv Rev. 2009;61:428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Moghimi SM. Chemical camouflage of nanospheres with a poorly reactive surface: towards development of stealth and target-specific nanocarriers. Biochim Biophys Acta. 2002;1590:131–139. doi: 10.1016/s0167-4889(02)00204-5. [DOI] [PubMed] [Google Scholar]
- 152.Haque MR, Jeong JH, Byun Y. Combination strategy of multi-layered surface camouflage using hyperbranched polyethylene glycol and immunosuppressive drugs for the prevention of immune reactions against transplanted porcine islets. Biomaterials. 2016;84:144–156. doi: 10.1016/j.biomaterials.2016.01.039. [DOI] [PubMed] [Google Scholar]
- 153.Ishida T, Wang X, Shimizu T, Nawata K, Kiwada H. PEGylated liposomes elicit an anti-PEG IgM response in a T cell-independent manner. J Control Release. 2007;122:349–355. doi: 10.1016/j.jconrel.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 154.Wang X, Ishida T, Kiwada H. Anti-PEG IgM elicited by injection of liposomes is involved in the enhanced blood clearance of a subsequent dose of PEGylated liposomes. J Control Release. 2007;119:236–244. doi: 10.1016/j.jconrel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 155.Liu L, Yi H, He H, Pan H, Cai L, Ma Y. Tumor associated macrophage-targeted microRNA delivery with dual-responsive polypeptide nanovectors for anti-cancer therapy. Biomaterials. 2017;134:166–179. doi: 10.1016/j.biomaterials.2017.04.043. [DOI] [PubMed] [Google Scholar]
- 156.Li Y, Wang H, Wang K, Hu Q, Yao Q, Shen Y, et al. Targeted Co-delivery of PTX and TR3 siRNA by PTP Peptide Modified Dendrimer for the Treatment of Pancreatic Cancer. Small. 2017;13 doi: 10.1002/smll.201602697. [DOI] [PubMed] [Google Scholar]
- 157.Kobayashi H, Watanabe R, Choyke PL. Improving conventional enhanced permeability and retention (EPR) effects; what is the appropriate target? Theranostics. 2013;4:81–89. doi: 10.7150/thno.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kong G, Braun RD, Dewhirst MW. Hyperthermia enables tumor-specific nanoparticle delivery: effect of particle size. Cancer Res. 2000;60:4440–4445. [PubMed] [Google Scholar]
- 159.Lammers T, Peschke P, Kuhnlein R, Subr V, Ulbrich K, Debus J, et al. Effect of radiotherapy and hyperthermia on the tumor accumulation of HPMA copolymer-based drug delivery systems. J Control Release. 2007;117:333–341. doi: 10.1016/j.jconrel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 160.Ranjan A, Jacobs GC, Woods DL, Negussie AH, Partanen A, Yarmolenko PS, et al. Image-guided drug delivery with magnetic resonance guided high intensity focused ultrasound and temperature sensitive liposomes in a rabbit Vx2 tumor model. J Control Release. 2012;158:487–494. doi: 10.1016/j.jconrel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Eikenes L, Bruland OS, Brekken C, de Davies CL. Collagenase increases the transcapillary pressure gradient and improves the uptake and distribution of monoclonal antibodies in human osteosarcoma xenografts. Cancer Res. 2004;64:4768–4773. doi: 10.1158/0008-5472.CAN-03-1472. [DOI] [PubMed] [Google Scholar]
- 162.Eikenes L, Tari M, Tufto I, Bruland OS, de Lange Davies C. Hyaluronidase induces a transcapillary pressure gradient and improves the distribution and uptake of liposomal doxorubicin (Caelyx) in human osteosarcoma xenografts. Br J Cancer. 2005;93:81–88. doi: 10.1038/sj.bjc.6602626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang J, Mao F, Niu G, Peng L, Lang L, Li F. 68Ga-BBN-RGD PET/CT for GRPR and Integrin αvβ3 Imaging in Patients with Breast Cancer. Theranostics. 2018;8:1121–1130. doi: 10.7150/thno.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Li L, Song L, Yang X, Li X, Wu Y, He T, et al. Multifunctional “core-shell” nanoparticles-based gene delivery for treatment of aggressive melanoma. Biomaterials. 2016;111:124–137. doi: 10.1016/j.biomaterials.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 165.Amin M, Mansourian M, Koning GA, Badiee A, Jaafari MR, Ten Hagen TLM. Development of a novel cyclic RGD peptide for multiple targeting approaches of liposomes to tumor region. J Control Release. 2015;220:308–315. doi: 10.1016/j.jconrel.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 166.Dong YD, Tchung E, Nowell C, Kaga S, Leong N, Mehta D, et al. Microfluidic preparation of drug-loaded PEGylated liposomes, and the impact of liposome size on tumour retention and penetration. J Liposome Res. 2017:1–9. doi: 10.1080/08982104.2017.1391285. [DOI] [PubMed] [Google Scholar]
- 167.Sun Q, Ojha T, Kiessling F, Lammers T, Shi Y. Enhancing Tumor Penetration of Nanomedicines. Biomacromolecules. 2017;18:1449–1459. doi: 10.1021/acs.biomac.7b00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, et al. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Longmire M, Choyke PL, Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine (Lond) 2008;3:703–717. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Choi HS, Liu W, Liu F, Nasr K, Misra P, Bawendi MG, et al. Design considerations for tumour-targeted nanoparticles. Nat Nanotechnol. 2010;5:42–47. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Molas M, Gomez-Valades AG, Vidal-Alabro A, Miguel-Turu M, Bermudez J, Bartrons R, et al. Receptor-mediated gene transfer vectors: progress towards genetic pharmaceuticals. Curr Gene Ther. 2003;3:468–485. doi: 10.2174/1566523034578195. [DOI] [PubMed] [Google Scholar]
- 172.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 173.Steichen SD, Caldorera-Moore M, Peppas NA. A review of current nanoparticle and targeting moieties for the delivery of cancer therapeutics. Eur J Pharm Sci. 2013;48:416–427. doi: 10.1016/j.ejps.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Torchilin VP. Tat peptide-mediated intracellular delivery of pharmaceutical nanocarriers. Adv Drug Deliv Rev. 2008;60:548–558. doi: 10.1016/j.addr.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 175.Torchilin VP. Cell penetrating peptide-modified pharmaceutical nanocarriers for intracellular drug and gene delivery. Biopolymers. 2008;90:604–610. doi: 10.1002/bip.20989. [DOI] [PubMed] [Google Scholar]
- 176.Cho CW, Liu Y, Cobb WN, Henthorn TK, Lillehei K, Christians U, et al. Ultrasound-induced mild hyperthermia as a novel approach to increase drug uptake in brain microvessel endothelial cells. Pharm Res. 2002;19:1123–1129. doi: 10.1023/a:1019837923906. [DOI] [PubMed] [Google Scholar]
- 177.Wang YX, Leung KC, Cheung WH, Wang HH, Shi L, Wang DF, et al. Low-intensity pulsed ultrasound increases cellular uptake of superparamagnetic iron oxide nanomaterial: results from human osteosarcoma cell line U2OS. J Magn Reson Imaging. 2010;31:1508–1513. doi: 10.1002/jmri.22173. [DOI] [PubMed] [Google Scholar]
- 178.Behr JP. The proton sponge: A trick to enter cells the viruses did not exploit. Chimia International Journal for Chemistry. 1997;51:34–36. [Google Scholar]
- 179.Hughes JA, Aronsohn AI, Avrutskaya AV, Juliano RL. Evaluation of adjuvants that enhance the effectiveness of antisense oligodeoxynucleotides. Pharm Res. 1996;13:404–410. doi: 10.1023/a:1016044609972. [DOI] [PubMed] [Google Scholar]
- 180.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 181.Pichon C, Guerin B, Refregiers M, Goncalves C, Vigny P, Midoux P. Zinc improves gene transfer mediated by DNA/cationic polymer complexes. J Gene Med. 2002;4:548–559. doi: 10.1002/jgm.303. [DOI] [PubMed] [Google Scholar]
- 182.Cho YW, Kim JD, Park K. Polycation gene delivery systems: escape from endosomes to cytosol. J Pharm Pharmacol. 2003;55:721–734. doi: 10.1211/002235703765951311. [DOI] [PubMed] [Google Scholar]
- 183.Liu Q, Chen X, Jia J, Zhang W, Yang T, Wang L, et al. pH-responsive poly(D,L-lactic-co-glycolic acid) nanoparticles with rapid antigen release behavior promote immune response. ACS Nano. 2015;9:4925–4938. doi: 10.1021/nn5066793. [DOI] [PubMed] [Google Scholar]
- 184.Li W, Nicol F, Szoka FC., Jr GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv Drug Deliv Rev. 2004;56:967–985. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 185.Hou KK, Pan H, Ratner L, Schlesinger PH, Wickline SA. Mechanisms of nanoparticle-mediated siRNA transfection by melittin-derived peptides. ACS Nano. 2013;7:8605–8615. doi: 10.1021/nn403311c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Moore NM, Sheppard CL, Barbour TR, Sakiyama-Elbert SE. The effect of endosomal escape peptides on in vitro gene delivery of polyethylene glycol-based vehicles. J Gene Med. 2008;10:1134–1149. doi: 10.1002/jgm.1234. [DOI] [PubMed] [Google Scholar]
- 187.Deng H, Zhao X, He D, Guo W, Yang K, Dong A, et al. One-step gene delivery into the cytoplasm in a fusion-dependent manner based on a new membrane fusogenic lipid. Chem Commun (Camb) 2016;52:7406–7408. doi: 10.1039/c6cc01996j. [DOI] [PubMed] [Google Scholar]
- 188.Tang R, Kim CS, Solfiell DJ, Rana S, Mout R, Velazquez-Delgado EM, et al. Direct delivery of functional proteins and enzymes to the cytosol using nanoparticle-stabilized nanocapsules. ACS Nano. 2013;7:6667–6673. doi: 10.1021/nn402753y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ding Y, Wang Y, Zhou J, Gu X, Wang W, Liu C, et al. Direct cytosolic siRNA delivery by reconstituted high density lipoprotein for target-specific therapy of tumor angiogenesis. Biomaterials. 2014;35:7214–7227. doi: 10.1016/j.biomaterials.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 190.Lechardeur D, Lukacs GL. Nucleocytoplasmic transport of plasmid DNA: a perilous journey from the cytoplasm to the nucleus. Hum Gene Ther. 2006;17:882–889. doi: 10.1089/hum.2006.17.882. [DOI] [PubMed] [Google Scholar]
- 191.Dauty E, Verkman AS. Actin cytoskeleton as the principal determinant of size-dependent DNA mobility in cytoplasm: a new barrier for non-viral gene delivery. J Biol Chem. 2005;280:7823–7828. doi: 10.1074/jbc.M412374200. [DOI] [PubMed] [Google Scholar]
- 192.Geiger RC, Taylor W, Glucksberg MR, Dean DA. Cyclic stretch-induced reorganization of the cytoskeleton and its role in enhanced gene transfer. Gene Ther. 2006;13:725–731. doi: 10.1038/sj.gt.3302693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez-Boulan E, et al. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum Gene Ther. 2000;11:151–165. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- 195.Haberland A, Bottger M. Nuclear proteins as gene-transfer vectors. Biotechnol Appl Biochem. 2005;42:97–106. doi: 10.1042/BA20050063. [DOI] [PubMed] [Google Scholar]
- 196.Chen K, Guo L, Zhang J, Chen Q, Wang K, Li C, et al. A gene delivery system containing nuclear localization signal: Increased nucleus import and transfection efficiency with the assistance of RanGAP1. Acta Biomater. 2017;48:215–226. doi: 10.1016/j.actbio.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 197.Maity AR, Stepensky D. Efficient subcellular targeting to the cell nucleus of Quantum Dots densely decorated with a nuclear localization sequence peptide. ACS Appl Mater Interfaces. 2016;8:2001–2009. doi: 10.1021/acsami.5b10295. [DOI] [PubMed] [Google Scholar]
- 198.Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J Mol Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 199.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 201.Nishimasu H, Yamano T, Gao L, Zhang F, Ishitani R, Nureki O. Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1. Mol Cell. 2017;67:139–147. e132. doi: 10.1016/j.molcel.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]