Abstract
Hepatocellular carcinoma (HCC) is a worldwide malignance, and the underlying mechanisms of this disease are not fully elucidated. In this study, the existence and function of achaete-scute homolog-1 (ASH1)-miR-375-YWHAZ signaling axis in HCC were determined. Our experiments and the Cancer Genome Atlas (TCGA) sequencing data analyses showed that ASH1 and miR-375 were significantly downregulated, whereas YWHAZ was significantly upregulated in HCC. Furthermore, we found that ASH1 positively regulates miR-375, and miR-375 directly downregulates its target YWHAZ. Gain- and loss-of-function study demonstrated ASH1 and miR-375 function as tumor suppressors, whereas YWHAZ acts as an oncogene in HCC. Animal experiment indicated that YWHAZ small interfering RNAs (siRNAs) (si-YWHAZ) delivered by nanoliposomes could suppress the growth of hepatoma xenografts and was well tolerant by nude mice. Further studies revealed that YWHAZ was involved in several protein networks, such as cell autophagy, epithelial-mesenchymal transition (EMT), apoptosis, cell cycle, invasion, and migration. In addition, the patient group with ASH1-high-expression-miR-375-high-expression-YWHAZ-low-expression was correlated with a better clinical prognosis compared with the opposite expression group. In conclusion, we proved the existence of ASH1-miR-375-YWHAZ signaling axis and interpreted its important role in driving HCC tumor progression.
Keywords: liver cancer, microRNA, bioinformatics analysis, YWHAZ, ASH1
Introduction
Hepatocellular carcinoma (HCC) is perhaps a prototypical systems disease, and as such, quantitative systems biology and bioinformatics analysis were extensively used to explore the molecular mechanisms underlying this disease. The most critical component for revitalizing HCC treatment strategies is identification of novel molecular hubs (proteins or genes) in signaling pathways involved in HCC development and progression.1
MicroRNAs (miRNAs) are small noncoding RNAs considered to play fundamental roles in tumor initiation and progression through regulation of gene expression at the level of post-transcription. Increasing evidences have demonstrated that miR-375 is frequently downregulated in multiple types of cancer and acts as an important tumor suppressor by inhibiting malignant properties of cancer cells.2 Previously, we found that miR-375 was one of the core downregulated miRNAs in HCC, and miR-375 inhibits cell proliferation, autophagy, clonogenicity, migration, and invasion and also induces G1 arrest and apoptosis by targeting AEG-1 and ATG7.3, 4, 5 Additionally, Liu et al.6 found that overexpression of miR-375 decreased HCC cell invasion and proliferation by targeting oncogene YAP1. Furthermore, miR-375 was found to strongly inhibit Akt and Ras-induced hepatocarcinogenesis in a primary mouse model of liver cancer.7 Recently, we demonstrated that gold-nanoparticles-delivered miR-375 has potential in HCC treatment,8 and miR-375 and doxorubicin (DOX) co-delivered by nanoparticles had significant synergetic anti-tumor effects and added values in overcoming drug resistance.9, 10 Collectively, these studies suggest that miR-375 is a hub miRNA and an attractive therapeutic target for HCC. However, little is known regarding miR-375’s upstream regulatory mechanisms, and there is a lack of comprehensive understanding of miR-375’s targets in the HCC signaling pathway.
Transcription factors (TFs) are key regulators of gene expression and play critical roles in HCC. Recent studies have revealed that miRNA disorder is often due to the aberrant expression of TF.5, 11 Achaete-scute family basic-helix-loop-helix (bHLH) transcription factor 1 (ASCL1), also well known as Achaete-scute homolog-1 (ASH1), a basic helix-loop-helix TF, is necessary for normal neural or some non-neuronal neuroendocrine (NE) cell type’s differentiation and development.12, 13 It has been reported that miR-375 was identified as a miRNA directly and highly transactivated by ASH1 in lung cancer.14, 15 Our bioinformatics analysis also indicates that ASH1 may be an important upstream regulator of miR-375 in HCC.
As one of miR-375’s predicated targets, YWHAZ aroused our interest. YWHAZ, also named as 14-3-3ζ, acts as a central hub in signaling networks, which promotes cell proliferation and adhesion and inhibits apoptosis in multiple cancers.1 Consistently, upregulation of YWHAZ in gastric cancers, pancreatic, liver, lung, breast, brain, and prostate cancer strengthened its importance in tumorigenesis.16 Several recent studies showed that elevated expression of YWHAZ is significantly associated with reduced survival and increased disease recurrence and chemoresistance in patients.17, 18, 19 Upregulation of YWHAZ increases the potential for invasion and metastasis20, 21 and induces epithelial-mesenchymal transition (EMT) in various tumors.22, 23, 24 Moreover, YWHAZ can promote autophagy by interacting with phosphorylated Atg9.25 These studies strongly suggested an important role of YWHAZ upregulation in cancer; however, little is known about its function and regulatory mechanisms in HCC.
In the present study, we first proposed and proved the ASH1-miR-375-YWHAZ signaling axis existed and further revealed the important role of this signaling axis in HCC.
Results
The ASH1-miR-375-YWHAZ Signaling Axis in HCC
The Expression of ASH1, miR-375, and YWHAZ Is Correlated and Different in HCC and Adjacent Tissues
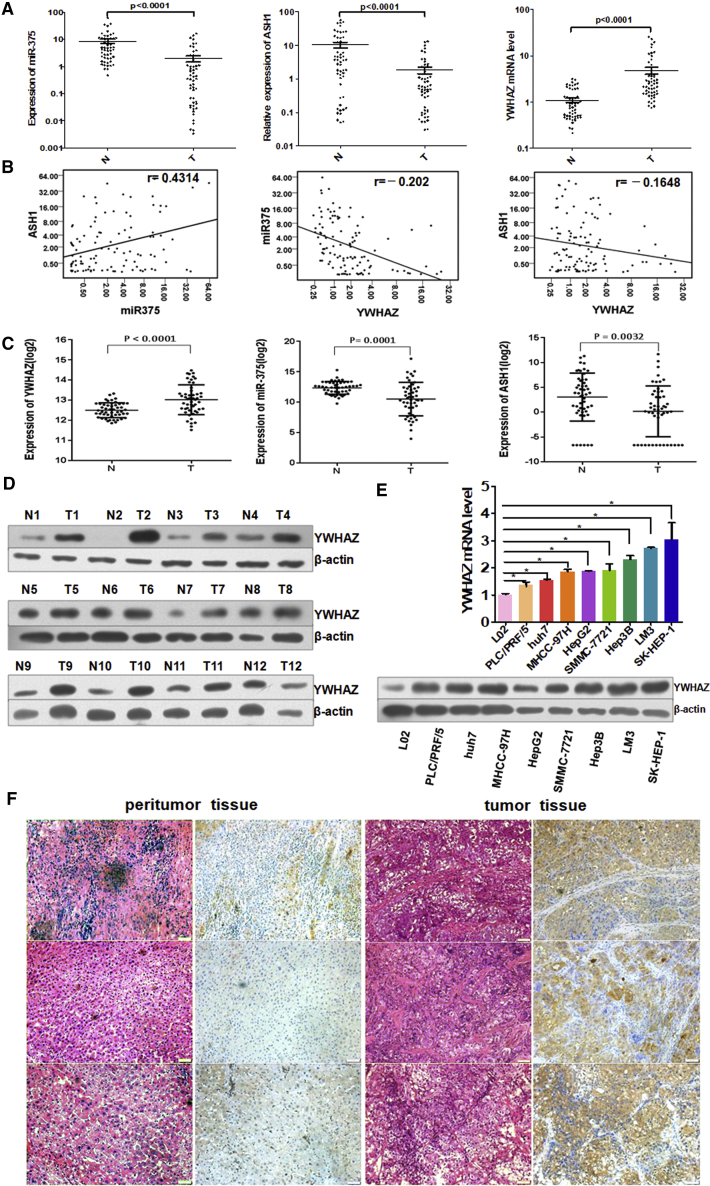
In our previous studies,3, 4, 5 we identified and validated miR-375 as a core miRNA in HCC. We analyzed our previous miR-375 microarray data3 and public TF chromatin immunoprecipitation sequencing (ChIP-seq) data to explore the regulatory signaling axis of miR-375. As a result, we speculated the ASH1-miR-375-YWHAZ signaling axis existed in HCC. To determine this signaling axis, we first examined their RNA expression in 53 paired HCC tissues and adjacent non-tumor tissues. Compared with adjacent non-tumor tissues, YWHAZ was obviously upregulated in HCC tissues. On the contrary, the expression of miR-375 was dramatically decreased in HCC tissues, coincident with the expression of ASH1 (Figure 1A). In addition, there was a strong positive correlation between the expression of ASH1 and miR-375 (p < 0.0001; Pearson’s correlation), whereas the expression of YWHAZ was negatively correlated with miR-375 or ASH1 expression (Figure 1B).
Figure 1.
The Expressions of ASH1, miR-375, and YWHAZ Are Correlated and Different in HCC and Adjacent Tissues
(A) TaqMan qRT-PCR analysis of miR-375 and SYBR Green qRT-PCR analysis of YWHAZ and ASH1 expression in 53 paired HCC tissues (T) and adjacent normal liver tissues (N). (B) miR-375 is consistent with ASH1 expression (p < 0.0001; r = 0.4314; Pearson’s correlation). YWHAZ inversely correlates with miR-375 or ASH1 expression (p = 0.0379, r = −0.202; p = 0.1130, r = −0.1648, respectively; Pearson’s correlation). (C) The expression of YWHAZ, miR-375, and ASH1 in TCGA paired LIHC (liver hepatocellular carcinoma) samples and adjacent tissue samples (paired t test) is shown. (D) The expression of YWHAZ protein was examined in 12 paired HCC tissues and adjacent tissues by western blot. (E) SYBR Green qRT-PCR and western blot analysis YWHAZ in normal hepatocyte and 8 hepatoma cell lines are shown. *p < 0.05. (F) Typical result of the expression of YWHAZ in 3 pairs of adjacent tissues and HCC tissues by H&E staining and immunohistochemistry, respectively, is shown.
Moreover, we analyzed the expressions of ASH1, miR-375, and YWHAZ in the Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) data to confirm the results in our data. We identified the differential expression of ASH1, miR-375, and YWHAZ in 50 pairs of HCC tumor and adjacent non-tumor tissue samples from TCGA. Similarly, YWHAZ was obviously upregulated (paired t test; p < 0.0001), whereas miR-375 and ASH1 were significantly reduced (p = 0.0001; p = 0.0032) in HCC tissues (Figure 1C).
Then, we found that YWHAZ protein was in higher expression in 11/12 HCC tumor tissues compared with corresponding non-tumor tissues by western blot analysis (Figure 1D). Next, we detected YWHAZ mRNA and protein expression in a human normal hepatic cell line L02 and eight liver cancer cell lines (Figure 1E). YWHAZ mRNA and protein was overexpressed in all liver cancer cell lines. Furthermore, we revealed that YWHAZ protein was observed mainly in the cytoplasm and HCC tumor tissues showed stronger positive compared with adjacent non-tumor tissues by immunohistochemistry staining (Figure 1F). Collectively, these data confirmed the overexpression of YWHAZ in HCC tissues and cell lines.
The Existence of Regulations in ASH1-miR-375-YWHAZ Signaling Axis
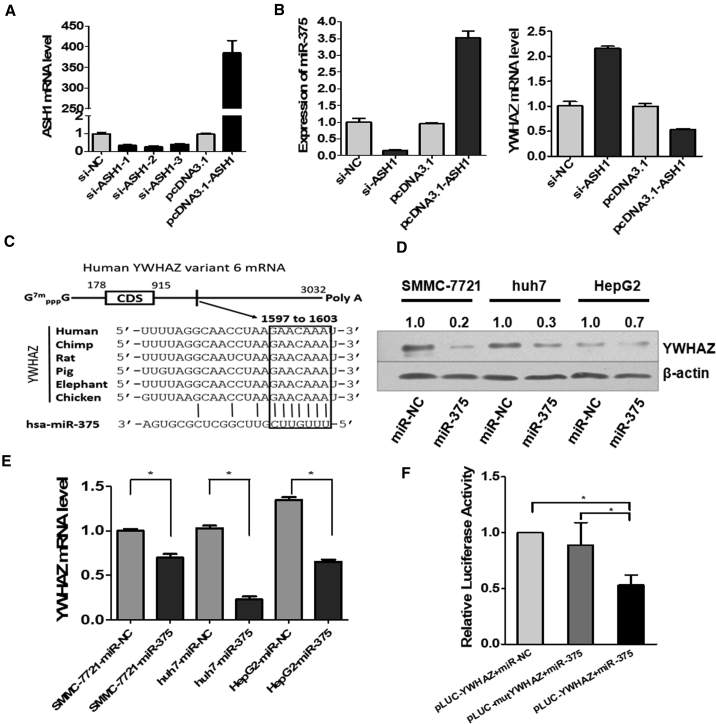
In order to prove the existence of the ASH1-miR-375-YWHAZ signaling axis in HCC, we performed experiments to determine the regulation among the three molecules. We analyzed the expression changes of miR375 and YWHAZ by SYBR Green qRT-PCR when ASH1 expression was knocked down or overexpressed. From Figure 2A, we can see that ASH1 in SMMC-7721 cells was successfully knocked down or overexpressed. As expected, the trend of miR-375 expression was consistent with the expression of ASH1, whereas the expression of YWHAZ was opposite to ASH1 (Figure 2B).
Figure 2.
The Existence of Regulations in the ASH1-miR-375-YWHAZ Signaling Axis
(A) SYBR Green qRT-PCR analysis of ASH1 mRNA after transfected with siRNA or pcDNA3.1 vectors in SMMC-7721 cells. (B) The expression of miR-375 or YWHAZ after knockdown or overexpression of ASH1 in SMMC-7721 cells is shown. (C) Predicted consequential pairing of target region of YWHAZ and miR-375 is shown. (D) The expression of YWHAZ protein was detected by western blot in 3 hepatoma cell lines after the transfection of miR-375. Quantitative analysis of the western blot was performed by ImageJ software, and relative densitometric quantification of YWHAZ proteins was shown. (E) Relative expression of YWHAZ detected by SYBR Green qRT-PCR in three hepatoma cell lines after 48 hr transfected with miR-375 mimics or miR-NC is shown. (F) Luciferase assay in HeLa cell is shown. miR-375 mimics were co-transfected with pLUC-YWHAZ vector or pLUC-mutYWHAZ vector. Relative repression of luciferase expression was standardized to β-gal signal. *p < 0.05.
To verify the miR-375 regulating YWHAZ, we first predicted the potential miRNA binding sites on YWHAZ by the TargetScan tool and identified conserved binding sites of miR-375 on YWHAZ 3′ UTR, which broadly conserved among vertebrates (Figure 2C). To confirm this regulation, we validated the effects of miR-375 on YWHAZ expression in three liver cancer cell lines transfected with miR-375 mimics or control miR-negative control (NC). Western blot and SYBR Green qRT-PCR analysis showed that miR-375 downregulated YWHAZ protein and mRNA levels (Figures 2D and 2E). Further, to determine the direct binding of miR-375 on YWHAZ 3′ UTR, HeLa cells were co-transfected with miR-375 or miR-NC and pLUC-YWHAZ or pLUC-mutYWHAZ vector. As a result, miR-375 showed substantial reduction in luciferase activity in pLUC-YWHAZ+miR-375 group. This effect can be reversed by mutating the miR-375 target site in YWHAZ 3′ UTR fragment (Figure 2F). These results indicate that YWHAZ is a direct target of miR-375.
Thus, we confirm the existence of the ASH1-miR-375-YWHAZ signaling axis.
Functional Effects of the ASH1-miR-375-YWHAZ Axis in HCC
ASH1 Function as a Tumor Suppressor in HCC
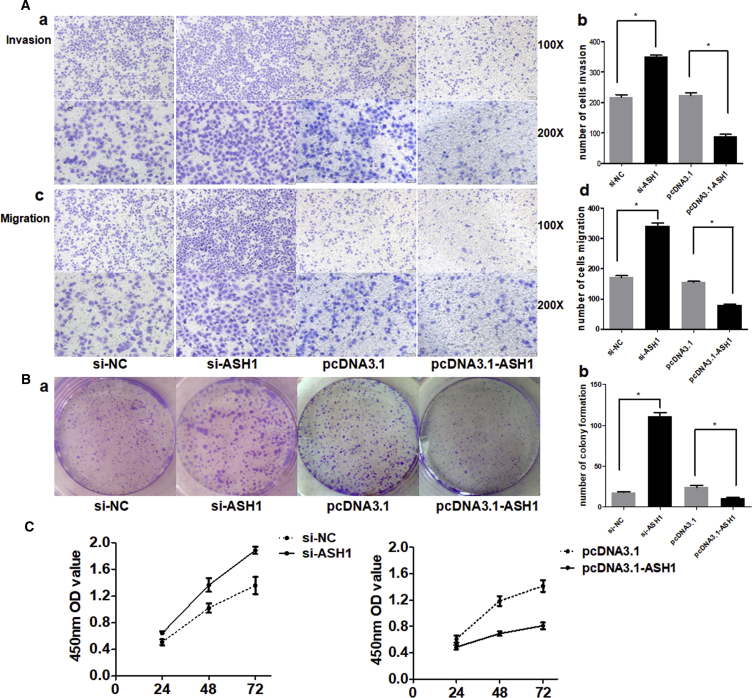
In our previous studies, we proved that miR-375 is a tumor repressor in HCC.3 In the above results, we demonstrated that the expression trend of ASH1 was consistent with that of miR-375. Thus, we further investigated whether the role of ASH1 is similar to miR-375. We tested its function on the malignant phenotypes of SMMC-7721 cells by silencing or overexpressing ASH1. The number of cells migrated through the Matrigel-coated or uncoated membrane into the lower chamber was significantly higher in ASH1 small interfering RNAs (siRNAs) (si-ASH1) transfected cells than in control (Figure 3A). Besides, colony formation and the proliferation of ASH1 deficiency cells were obviously promoted, whereas upregulation of ASH1 showed opposite results (Figures 3B and 3C). These results suggest that, like miR-375, ASH1 also has tumor inhibition potential in HCC cells.
Figure 3.
ASH1 Functions as a Tumor Suppressor in HCC
(A) (a) and (c) represent the results of cell invasion and migration across an 8-μm-pore-size membrane with or without Matrigel after silencing or overexpressing ASH1. (b) and (d) histograms indicate the number of cells invasion and migration. (B) (a) shows the representative results of colony formation using the indicated clones. (b) histogram indicates the number of colony formation using the indicated clones. (C) Cell proliferation was assessed in SMMC-7721 cells at 24 hr, 48 hr, and 72 hr after transfection with siRNA or pcDNA3.1 vectors by Cell Counting Kit-8 assay. *p < 0.05.
Knockdown or Overexpression of YWHAZ Impacts Cell Malignancy Properties
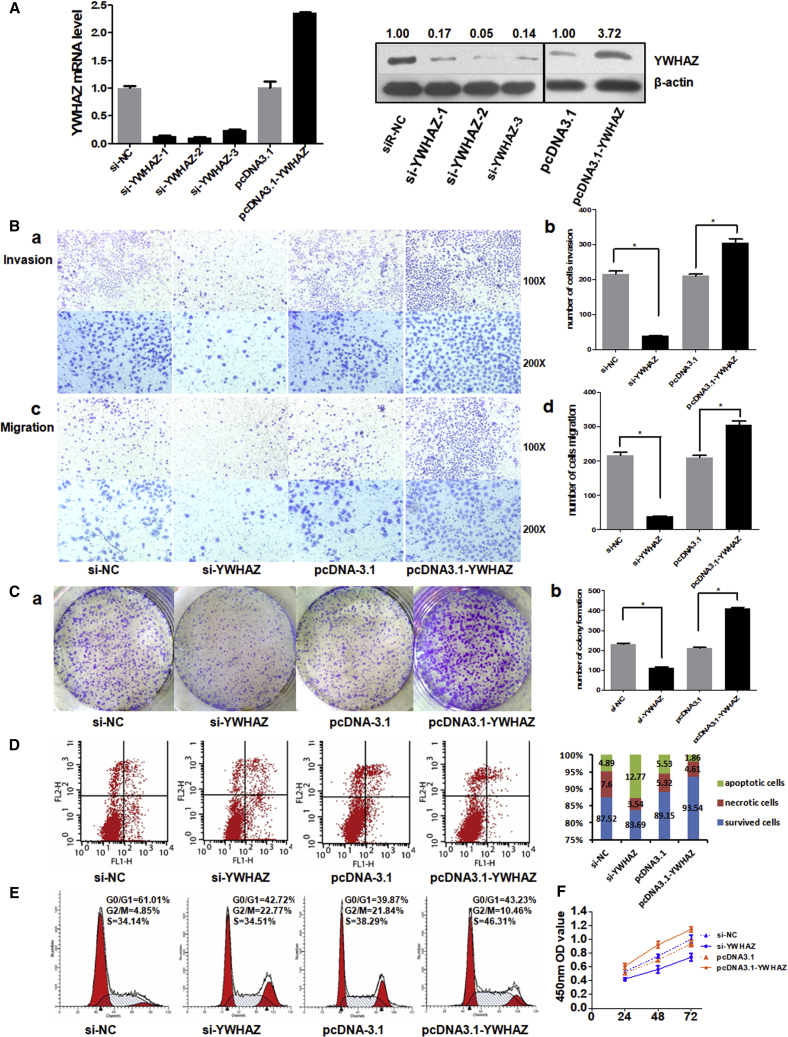
To gain insight into the potential role of YWHAZ in HCC, we used gain- and loss-of-function methods to investigate whether changing YWHAZ expression could affect the malignant phenotypes of liver cancer cells. In SMMC-7721 cells, the expressions of YWHAZ mRNA and protein were efficiently silenced or overexpressed 48 hr after transient introduction of si-YWHAZ or pcDNA3.1-YWHAZ over the control si-NC or pcDNA-3.1 (Figure 4A). The results of Transwell assays with or without Matrigel showed that YWHAZ inhibition effectively suppressed the ability of SMMC-7721 cell invasion and migration, whereas overexpression of YWHAZ showed opposite results (Figure 4B). Besides, colony formation and proliferation of YWHAZ deficiency cells were significantly inhibited, whereas YWHAZ-overexpressed cells showed larger colonies and increased proliferation (Figures 4C and 4F). We further investigated the influence of YWHAZ on apoptosis using Annexin-V and propidium iodide (PI) combined labeling flow cytometry analysis. We found the rates of apoptosis in SMMC-7721 cells infected with si-YWHAZ were higher than those of control cells and the YWHAZ-overexpressed cells (Figure 4D). Next, we investigated whether silencing or overexpressing YWHAZ affected cell cycle. As shown in Figure 4E, the distribution of si-YWHAZ-transfected cells were mainly arrested in G2 phase, whereas the distribution of YWHAZ-overexpressed cells in G2 phase was lower than control. Taken together, these results indicate that YWHAZ augments cell malignancy properties and acts as an oncogene in HCC.
Figure 4.
Knockdown or Overexpression of YWHAZ Impacts Cell Malignancy Properties
(A) SYBR Green qRT-PCR and western blot analysis of YWHAZ expression in SMMC-7721 cell lines 48 hr after transfection. Quantitative analysis of the western blot was performed by ImageJ software, and relative densitometric quantification of YWHAZ proteins was shown. (B) (a) and (c) represent the results of cell invasion and migration across an 8-μm-pore-size membrane with or without Matrigel after silencing or overexpressing YWHAZ. (b) and (d) histograms indicate the number of cell invasions and migrations. (C) (a) shows the representative results of colony formation using the indicated clones. (b) histogram indicates the number of colony formation using the indicated clones. (D) Cell apoptosis was detected by annexin-V and propidium iodide (PI) combined labeling flow cytometry in cells 48 hr after transfection. (E) Cell cycle analysis was determined 48 hr after transfection by PI staining flow cytometry. (F) Cell proliferation was detected at 24 hr, 48 hr, and 72 hr after transfection. *p < 0.05.
miR-375 and YWHAZ Showed Opposite Effects on HCC Cells
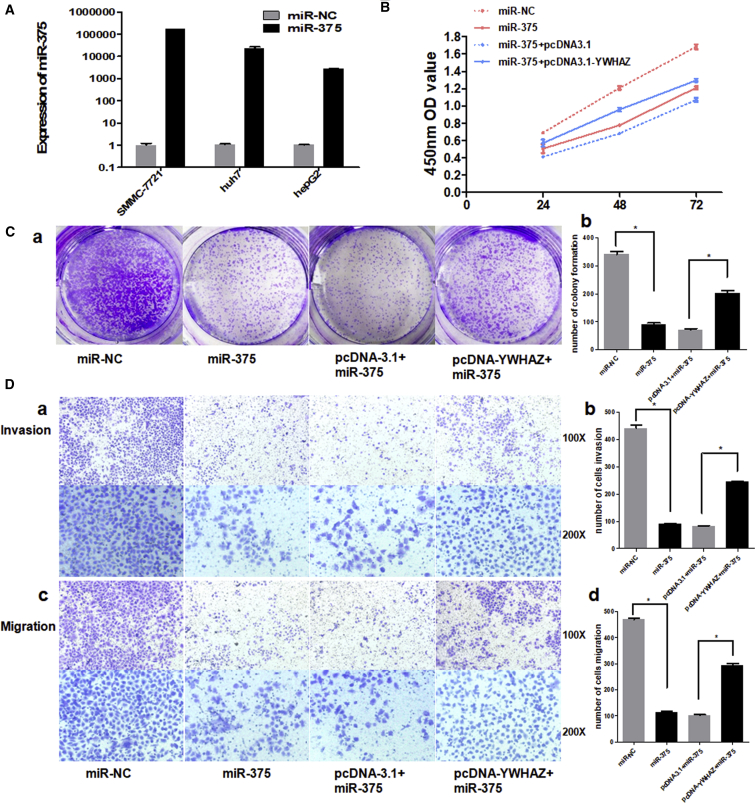
As YWHAZ is a direct target of miR-375, we next investigated the impact of miR-375 on YWHAZ function. We examined the cell proliferation, colony formation, invasion, and migration by co-transfected with miR-375 mimics or miR-NC and pcDNA3.1-YWHAZ or pcDNA-3.1 vector. miR-375 was efficiently overexpressed in three hepatoma cell lines transfected with miR-375 mimics (SMMC-7721, huh7, and HepG2, especially in SMMC-7721 cells; Figure 5A). Thus, we chose SMMC-7721 cells to illustrate the effect of miR-375 on the function of YWHAZ. As shown in Figures 5B and 5C, overexpression of miR-375 resulted in suppression of cell proliferation and colony formation. Also, cells transfected with miR-375 showed reduced invasion and migration ability (Figure 5D). Notably, all these suppressive effects of miR-375 were partially reversed by overexpression of YWHAZ simultaneously (Figures 5B–5D). These results show the important function of YWHAZ in the miR-375 downstream pathway.
Figure 5.
miR-375 and YWHAZ Showed Opposite Effects on HCC Cells
(A) TaqMan qRT-PCR was used to evaluate the relative expression of miR-375 in three hepatoma cell lines transfected with miR-375 mimics or miR-NC. The average miRNA expression in miR-NC group was designated as 1. U6 was used as an internal control. (B) Cell proliferation was detected at 24 hr, 48 hr, and 72 hr by Cell Counting Kit-8 assay. (C) (a) shows the representative results of colony formation using the indicated clones. (b) histogram indicates the number of colony formation using the indicated clones. (D) (a) and (c) represent the results of cell invasion and migration across an 8-μm-pore-size membrane with or without Matrigel. (b) and (d) histograms indicate the number of cell invasions and migrations. *p < 0.05.
In Vivo Validation of YWHAZ Function on the Hepatoma Xenografts
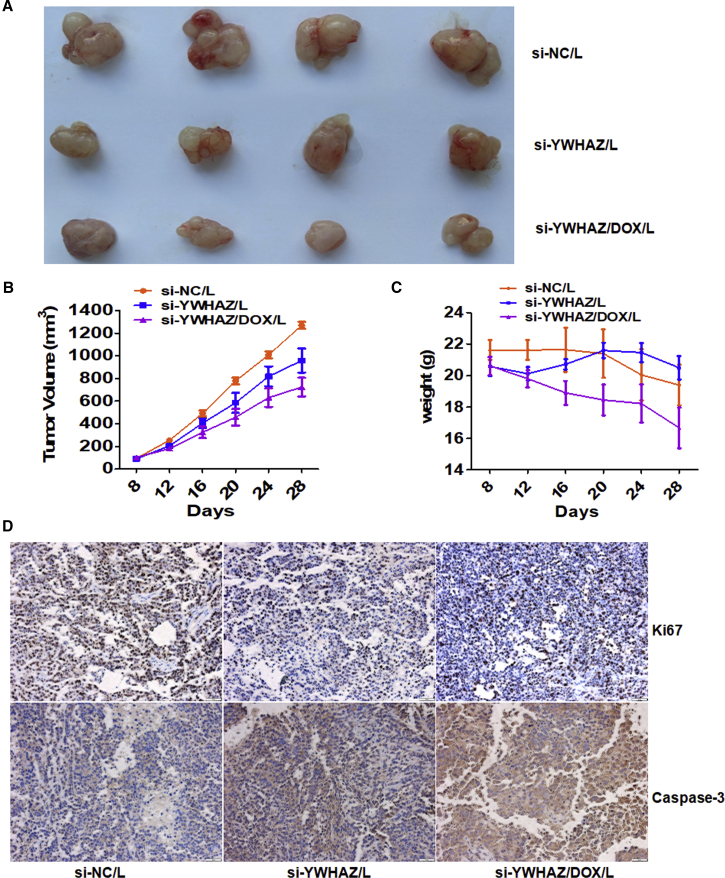
Gain- and loss-of-function-based in vitro experiments demonstrated that YWHAZ augments the migration, invasion, and proliferation abilities of SMMC-7721 cells. We suppose that the nude mouse xenograft model is an efficient system for performing function-oriented therapies of silencing YWHAZ. We successfully constructed HCC xenograft models with SMMC-7721 cells. When the tumor volume is close to 100 mm3, the transplanted nude mice received intratumoral administration with nanoliposomes (L)-delivered si-NC, si-YWHAZ, or si-YWHAZ combined with DOX (n = 4 each group), respectively. Compared with the si-NC/L group, the other two groups showed suppressing effect of tumors growth, which could be presented by gross morphology (Figure 6A). The tumor growth curves showed that si-YWHAZ/L and si-YWHAZ/DOX/L groups showed persistent tumor suppression compared to the control group. Obviously, si-YWHAZ/DOX/L displayed the most visible tumor-suppressor role (Figure 6B). Though si-YWHAZ/DOX/L showed strong combination inhibition of tumor growth, it resulted in a significant reduction in body weight. However, there was no significant loss of body weight in si-YWHAZ/L group (Figure 6C). Immunohistochemistry analysis of YWHAZ, the proliferation marker Ki-67, and apoptosis marker caspase-3 in the tumor tissues indicated that YWHAZ was efficiently silenced in vivo and cell proliferation was inhibited, whereas cell apoptosis increased by si-YWHAZ/L or si-YWHAZ/DOX/L treatment (Figures 7D and 6D). These results show that si-YWHAZ is generally well tolerated by mice and suppressed tumor growth in SMMC-7721 cell xenografts without altering body weight.
Figure 6.
In Vivo Validation of YWHAZ Function on the Hepatoma Xenografts
(A) Effects of YWHAZ on the growth of pre-established SMMC-7721 xenografts at a gross morphology level. After treated with siRNA-NC/L, si-YWHAZ/L, or si-YWHAZ/DOX/L every four days for 5 times, mice were sacrificed and photographed. (B) Tumor growth curve of mice with pre-established SMMC-7721 xenografts treated with siRNA-NC/L, si-YWHAZ/L, or si-YWHAZ/DOX/L is shown. (C) Body weights of mice with xenografts treated with siRNA-NC/L, si-YWHAZ/L, or si-YWHAZ/DOX/L are shown. (D) Immunohistochemistry was assessed for the proliferation marker Ki67 and apoptosis marker caspase-3 in the treated tumor tissues.
Figure 7.
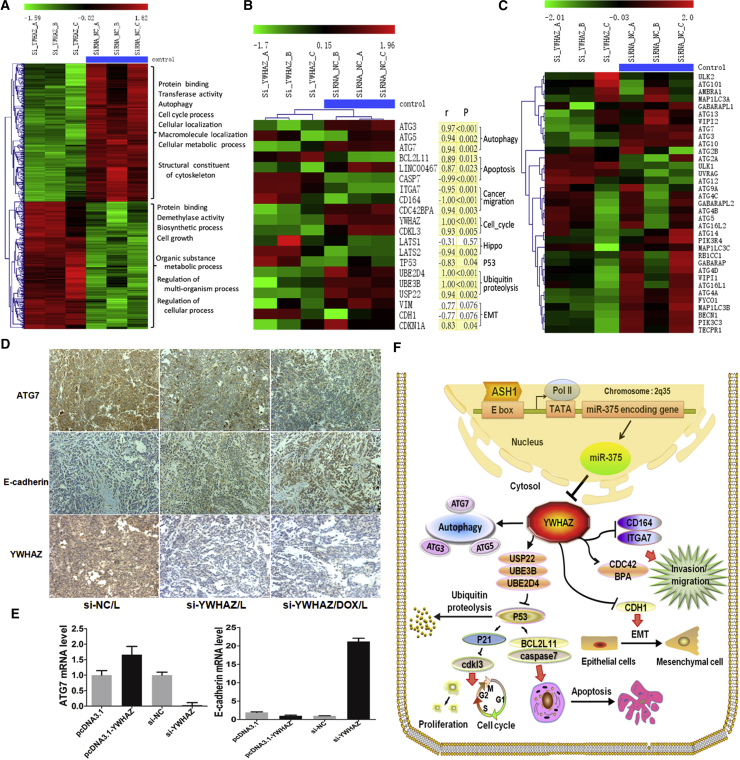
Potential Mechanism and Downstream Pathways of the ASH1-miR-375-YWHAZ Axis in HCC
(A) Heatmap of 645 differentially expressed genes from si-YWHAZ microarray. Each row of gene probes was normalized, and each column sample was clustered by hierarchical clustering. Two different state samples were found to be clustered together (the first three columns were YWHAZ knockout samples, and the last three were control samples [blue scale]). The uppermost scale from green to red indicates that the expression level is low to high. The main function and biological processes of the up and down genes are also shown in the figure. (B) The expression profiles of YWHAZ downstream genes from si-YWHAZ microarray are shown. The heatmap genes were sorted according to different pathways, and most of these genes were significantly differently expressed between si-YWHAZ samples and control. p, significance measure of the correlation; r, Pearson correlation coefficient of each gene and YWHAZ expression. Shading yellow represents the result is significant; otherwise, no color. The results showed that most of these genes were significantly correlated with YWHAZ expression (p < 0.05). (C) Heatmap of 35 autophagy genes in si-YWHAZ microarray data is shown. (D) Immunohistochemistry of ATG7, E-cadherin, and YWHAZ in the three groups of mice tumor tissues treated with siRNA-NC/L, si-YWHAZ/L, or si-YWHAZ/DOX/L is shown. (E) The expressions of ATG7 and E-cadherin were detected in SMMC-7721 cells after transfection by SYBR green qRT-PCR. (F) Schematic illustration of ASH1-miR-375-YWHAZ signaling axis and its possible downstream pathways in hepatocellular carcinoma is shown.
Potential Mechanism and Downstream of the ASH1-miR-375-YWHAZ Axis in HCC
As we demonstrated the function of ASH1-miR-375-YWHAZ axis in HCC, we tried to explore the mechanism and downstream pathways of this axis. We performed microarray assay in SMMC-7721 cells transfected with si-YWHAZ or si-NC to identify the downstream genes of YWHAZ. The microarray data were deposited in NCBI’s GEO public database (https://www.ncbi.nlm.nih.gov/geo/; GEO: GSE98026). As a result, we obtained 507 (245 upregulation and 262 downregulation) differentially expressed genes (DEGs) significantly affected by YWHAZ knockdown (Figure 7A). Function enrichment analysis showed that there are many genes located in nucleus and cytosol, as well as 185 protein binding proteins. These genes were also enriched in the “regulation of actin cytoskeleton” pathway (Figure S1A), which is related to cell migration and tumor metastasis.26 We further analyzed the regulation of TFs in the DEGs and found 9 TFs potentially regulated 356 DEGs (Figure S1B). TFs SPIB, KLF11, ZNF148, and RUNX1 were the hub nodes in the network and all upregulated by si-YWHAZ. ZNF148 and KLF11 were reported as tumor repressors to inhibit cell growth and induce cell death in HCC.27, 28 These TFs may play important roles in the YWHAZ downstream signaling, as miRNAs and TFs can regulate the same targets to form feedforward loop regulatory modules in a refined regulation.29 Next, we analyzed the co-regulation of miR-375 together with these hub TFs and found that 3 TFs (KLF11, SPIB, and ZNF148) can collaborate with miR-375 to regulate dozens of genes, among which 25% were located in lysosome (Figures S1C and S1D). We also analyzed the co-expression genes of YWHAZ in TCGA HCC samples and identified 78 genes with high correlation (r > 0.5 or r < −0.5). Interestingly, although these 78 correlated genes had only few overlaps with the above DEGs, they were also enriched in cytosol and protein binding GO (gene ontology) terms (Table S4; Figure S1E), similar to our si-YWHAZ DEGs (Figure S1A).
Based on our analysis and literature review, we knew that YWHAZ involves in autophagy, epithelial-mesenchymal transition (EMT), cell cycle, apoptosis, etc. We further analyzed the expressions of reported YWHAZ downstream genes and found that the expressions of most genes were highly correlated with YWHAZ and changed by silencing YWHAZ (Figure 7B). Especially most of the autophagy-associated genes, such as ATG3, ATG5, and ATG7, and EMT-related gene CDH1 (also known as E-cadherin) were changed by si-YWHAZ (Figures 7C, S1F, S2A, and S2B). Further, we validated these gene expression alterations. The mice tumor tissues from our animal experiment were used to analyze the expression of core genes in autophagy and EMT by immunohistochemistry. As shown in Figure 7D, the expression of autophagy-associated gene ATG7 was decreased, and EMT-associated gene E-cadherin was increased in both si-YWHAZ/L and si-YWHAZ/DOX/L groups. We also tested the expression of ATG7 in SMMC-7721 cells after silencing or overexpression YWHAZ and found that ATG7 expression was markedly promoted by YWHAZ (Figure 7E). The above results reinforce our inference that YWHAZ promotes autophagy by inducing ATG7. Next, we used SYBR Green PCR to test the expression of E-cadherin after silencing or overexpression of YWHAZ in SMMC-7721 cells. Consistently, E-cadherin was upregulated after transfected with si-YWHAZ whereas downregulated after transfected with pcDNA3.1-YWHAZ (Figure 7E). However, SYBR Green PCR results found that another EMT-associated gene VIM expression was not altered with YWHAZ (Figure S2C). The EMT-mediated increase in invasion and metastasis is largely contributed by loss of E-cadherin function.30, 31 This provides us favorable evidence that YWHAZ induced HCC cells migration and invasion by downregulating E-cadherin. These results support that YWHAZ plays important roles in autophagy and EMT.
Our bioinformatics analysis found that the expressions of apoptosis- and cell-cycle-related genes, such as p53, p21, BCL2L11, CASP7, and CDKL3, were also highly correlated with YWHAZ (Figure 7B). Besides, our SYBR Green PCR results found that the expression of p21, p53, BCL2L11, and CASP7 were increased and the expression of CDKL3 was decreased after silencing YWHAZ (Figures S2D–S2F), which was consistent with our microarray results. Moreover, the expression of p53 protein was also increased after downregulating YWHAZ and decreased after overexpressing YWHAZ in SMMC-7721 cells (Figure S2E). The dysregulation of apoptosis and cell cycle genes after silencing YWHAZ interprets the potential mechanisms of YWHAZ knockdown, accelerates cell apoptosis, and slows down cell division, thus inhibiting tumor growth. Our results indicated that YWHAZ involves in various biological processes associated with cancer progression through interaction with different downstream genes. Potential mechanism and downstream pathways of the ASH1-miR-375-YWHAZ axis were summarized in Figure 7F.
Overall Survival Significance of the ASH1-miR-375-YWHAZ Axis in HCC
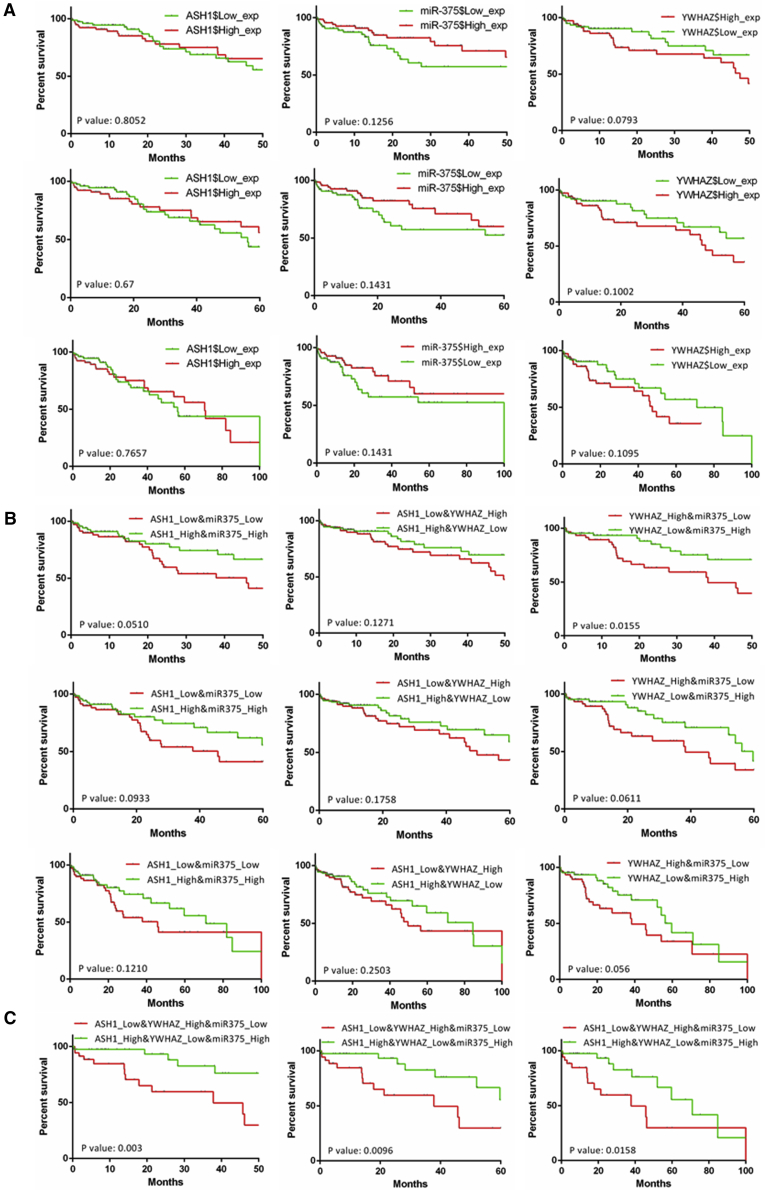
To understand their clinical significance in human HCC, Kaplan-Meier (KM) survival analysis was used to analyze the relationship between their expressions and the overall survival (OS) months in TCGA HCC samples. As shown in Figure 8A, single gene expression of the ASH1-miR-375-YWHAZ axis didn’t show obvious correlation with OS at 50 months, 60 months, or 100 months. However, the combination of miR-375 and YWHAZ has significance in the OS analysis at 50 months (p = 0.0155; Figure 8B). When combining expressions of the three genes, we obtained significant difference on OS at 50 months (p = 0.003), 60 months (p = 0.0096), and 100 months (p = 0.0158; Figure 8C). The high ASH1-high miR-375-low YWHAZ group has significantly longer OS months than the opposite expression group. Overall, these results indicate that the expression of ASH1-miR-375-YWHAZ axis could be a significant marker for prognosis.
Figure 8.
Overall Survival Analysis of the ASH1-miR375-YWHAZ Axis in HCC
The x axis is the overall survival (OS) month, and the y axis represents the survival rate. (A) Kaplan-Meier survival analysis of ASH1, miR-375, and YWHAZ at 50, 60, and 100 months is shown. (B) Kaplan-Meier survival analysis of two gene combinations in ASH1-miR375-YWHAZ axis at 50, 60, and 100 months is shown. (C) Kaplan-Meier survival analysis of three gene combinations in ASH1-miR375-YWHAZ axis at 50, 60, and 100 months is shown.
Discussion
Given a goal of tailoring treatment to the individual tumor, we face a need to integrate diverse molecular measurements and interpret these in terms of pathway changes driving tumor growth and gene or protein aberrations that drive these pathways.32 miRNAs are endogenously produced small size regulators of global gene expression and gaining recognition as essential regulators involved in cancers. Previously, we reviewed the literature about the emerging role of miR-375 in cancer and found that miR-375 is consistently downregulated in multiple types of cancers and acts as a core miRNA in HCC.2, 3, 4, 6, 8, 9, 10 However, little is known regarding phenotype-dependent mechanism circuitry involving TFs, miR-375, and its targets in HCC pathogenesis and progression.
In this study, we found the existence of ASH1-miR-375-YWHAZ signaling axis in HCC. Our experiment and TCGA sequencing data showed that YWHAZ was obviously upregulated, whereas ASH1 and miR-375 were significantly downregulated in HCC. In addition, there was a strong positive correlation between the expression of ASH1 and miR-375, whereas YWHAZ expression was negatively correlated with miR-375 or ASH1 expression. Further studies revealed that ASH1 positively regulates miR-375 and miR-375 directly downregulates its target YWHAZ. Some similar results have been reported in lung cancer and gastric cancer.15, 33 Gain- and loss-of-function study demonstrated that ASH1 decreased HCC cell proliferation, clonogenicity, and migration, and invasion and functioned as a tumor suppressor in HCC-like miR-375, as we reported before.3 We also demonstrated that inhibiting the expression of YWHAZ in liver cancer cells decreased cell proliferation, clonogenicity, migration, and invasion and also induced G2 arrest and apoptosis, whereas upregulation of YWHAZ showed opposite results. Co-transfection of pcDNA3.1-YWHAZ and miR-375 mimics in SMMC-7721 cells partially reverse the anti-tumor effects induced by miR-375. Importantly, our animal experiment indicated that si-YWHAZ delivered by nanoliposomes could suppress the growth of hepatoma xenografts and were well tolerant by nude mice. It has been reported that reduced expression of miR-375 indicates a poor prognosis in a majority of cancers.34, 35, 36 Overexpression of YWHAZ was also found to be related to a bad prognosis in various cancers.17, 33, 37 Similarly, our analysis showed that high expression of miR-375 or low expression of YWHAZ were correlated with a better overall survival in HCC, especially the group miR-375 and ASH1 high expression and YWHAZ low expression; this means that the expression of ASH1-miR375-YWHAZ axis may become a good prognostic biomarker for HCC.
Given the importance of ASH1-miR-375-YWHAZ axis in HCC, further research was conducted to explore the possible downstream pathways of this axis. Through bioinformatics analysis (differential expression analysis, GO, and Kyoto Encyclopedia of Genes and Genomes [KEGG] pathway enrichment analysis) of our microarray data, we revealed that YWHAZ was involved in several protein networks and acts as a central hub regulating cell autophagy, EMT, apoptosis, cell cycle, migration, and p53 ubiquitination. It was reported that suppression of Ube2d family members (Ube2d1, Ube2d2, Ube2d3, and Ube2d4) could induce accumulation and phosphorylation of p53, followed by p53-dependent apoptotic cell death.38, 39 Ubiquitin-specific protease 22 (USP22) stabilizes substrate proteins by inhibiting their ubiquitin-dependent degradation. Several studies demonstrated that USP22 silencing could activate p53.40, 41 Because USP22, Ube2d4, and Ube3b were important downstream genes of YWHAZ, we infer that YWHAZ downregulates p53 by activating USP22, Ube2d4, and Ube3b, which make P53 ubiquitination and lead to its proteasomal degradation. From our microarray data, YWHAZ is directly or indirectly involved in many important pathways related to tumor progression. Moreover, we confirmed several key results from the microarray data using SYBR Green qRT-PCR and provided primary explanation of the physiological mechanisms of YWHAZ involved in cell autophagy, EMT, cell cycle, apoptosis, invasion and migration, and p53 ubiquitination. Therefore, more experiment studies are needed to clarify the mechanism of ASH1-miR-375-YWHAZ axis in HCC.
In conclusion, our data revealed that ASH1-miR-375-YWHAZ signaling axis existed and played an important role in hepatocarcinogenesis, and it may be a significant biomarker for HCC prognosis. It provides us a better understanding of the molecular mechanisms and novel molecular hubs of HCC, which may benefit the treatment strategies for patients with unresectable HCC.
Materials and Methods
Cell Lines and Patient Samples
The human normal hepatic cell line L02 was cultured in 1640 medium supplemented with 10% fetal bovine serum and the hepatoma cell lines PLC/PRF/5, huh7, MHCC-97H, HepG2, SMMC-7721, Hep3B, and LM3 SK-HEP-1, and the human HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum. All cells were incubated in a humidified atmosphere at 37°C with 5% CO2. Following ethical and institutional guidelines and after informed consent of the tissue donors, pair-matched tumorous and adjacent nontumorous hepatic tissues from 53 patients undergoing resection for liver cancer were obtained from Tongji Hospital, Huazhong University of Science and Technology.
miRNA Mimics, Plasmid, siRNA, and Cell Transfection
miR-375 mimics, si-YWHAZ, and si-ASH1 and their matched negative controls (miR-NC and si-NC) were purchased from Guangzhou Ribobio, China. Sequences of siRNAs were listed in Table S1. YWHAZ- or ASH1-overexpressing vectors were purchased from Shanghai Genechem, China. Cell transfections were performed using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions.
SYBR Green RT-PCR and TaqMan qRT-PCR
These assays were detected as we have described before.3 Primers used in SYBR Green qRT-PCR were listed in Table S2.
Cell Proliferation, Cell Cycle and Apoptosis, Cell Migration and Invasion, and Clonogenicity Assay
Cell proliferation assay was implemented according to the cell counting kit-8 (CCK-8) instructions (Promoter, Wuhan, China). Cell cycle and apoptosis, cell migration and invasion, and clonogenicity assay were detected as described.3
Western Blotting
Western blot analysis was detected as we have described before.3 The following primary antibodies were used: anti-YWHAZ (Proteintech); anti-p53 (Cell Signaling Technologies); and anti-β-actin (Promotor).
Immunohistochemistry
Immunohistochemical (IHC) protocols have been described before.42 Briefly, human and mice tumor tissue sections (4 μm) were cut and then placed on glass slides. Deparaffinize slides in 2 changes of xylene, each for 10 min. Then, rehydrate sections by sequentially incubating with 100%, 95%, 80%, and 60% ethanol for 5 min each. For antigen retrieval, the sections were boiled in citrate buffer (pH 6.0) for 15 min in a microwave oven. Endogenous peroxidase activity was blocked by incubation with 3% hydrogen peroxide solution for 10 min. Subsequently, the primary antibodies, anti-YWHAZ (1:400; Proteintech), anti-ATG7 (1:200; Cell Signaling Technology), anti-E-cadherin (1:200; Promotor), anti-Ki-67 (1:200; Abcam), and anti-caspase-3 (1:100; Proteintech), were applied to the tissue sections and allowed to incubate overnight at 4°C. Then, samples were further processed as per manufacturer’s instructions and a series of procedures, including incubating with peroxidase-conjugated secondary antibody, staining, mild re-dyeing with hematoxylin, dehydration, coverslipping, and microscope observation, were performed in sequence.
Luciferase Assay
To construct a luciferase reporter vector, YWHAZ 3′ UTR fragment containing putative binding sites for miR-375 or a mutant YWHAZ 3′ UTR sequence (Table S3) was cloned into the pMIR-REPORT luciferase plasmid. HeLa cells were co-transfected with 100 ng of pLUC-YWHAZ or pLUC-mut YWHAZ-1 and 50 nM pre-miR-375 or pre-miR-NC; 10 ng of pMIR-REPORT β-gal plasmid served as an internal transfection efficiency control. 48 hr after transfection, luciferase and β-galactosidase activities were measured using the Dual-Light System (Applied Biosystems).
Microarray Expression Profiling
SMMC-7721 cells transfected with si-YWHAZ_1 or siRNA-NC were used to synthesize double-stranded cDNA, which was labeled and hybridized on the SurePrint G3 Human Gene Expression Microarray 8 × 60K v2 (Agilent Technologies, Santa Clara, CA, USA). Processed slides were scanned with the Agilent G2565CA Microarray Scanner (Agilent Technologies) after hybridization and washing. Data were extracted with Feature Extraction software 10.7 (Agilent). Quantile normalization and subsequent data processing were performed using the R software limma package. Differentially expressed long non-coding RNAs (lncRNAs) and mRNAs between two samples were identified through fold change filtering and Student’s t test. The normalized intensity for each lncRNA and mRNA was presented as a log2-transformed pattern; fold changes >2 and p values < 0.05 were selected as significantly differentially expressed. The microarray profiling was conducted by the Shanghai Biotechnology.
Animal Experiments
BALB/c athymic nude mice (male; 4–6 weeks old and 16–18 g) were purchased from Beijing HFK Bioscience and bred at pathogen-free conditions in Animal Center of Tongji Medical College. All animal experiments were carried out in accordance with the Guide for the Care and Use of Laboratory Animals of Tongji Medical College. SMMC-7721 cells (4 × 106) were suspended in 100 μL fetal bovine serum (FBS)-free DMEM medium and inoculated subcutaneously into the flanks of nude mice to establish hepatoma xenograft model. When the tumor volume is close to 100 mm3, the transplanted nude mice were randomly divided into three groups (n = 4 each). 1.0 mg/kg doxorubicin combined with 2.5 nmol si-YWHAZ, 2.5 nmol si-YWHAZ, or 2.5 nmol si-NC (RiboBio, Guangzhou, China), respectively, was delivered by L into the implanted tumor per mouse every 4 days for 5 times. Tumor volume (V) was monitored by measuring the length (L) and width (W) with digital caliper and calculated with the formula V = (L × W2) × 0.5.
Liver Cancer Data from TCGA
We have downloaded RNA-seq (V2) and miRNA-seq level3 data of liver cancer (374 samples) from The Cancer Genome Atlas (TCGA) data portal (https://portal.gdc.cancer.gov/). The paired cancer and adjacent tissue samples were used to analyze gene differential expression. The expressions of YWHAZ, ASH1, and miR375 were significantly different (p value < 0.01; paired t test) between tumor and adjacent tissue.
Survival Analysis
We used TCGA tumor data to perform Kaplan-Meier survival analysis. For survival analysis for a single gene, the low expression was defined by expression levels in the lowest quartile (25th percentile) and high expression by levels in the upper quartile (75th percentile). The numbers of high-expression and low-expression samples of YWHAZ and miR375 are both 83. p value was calculated by log rank (Mantel-Cox) test. For multi-gene survival analysis, we divided the high and low expression based on the median of their gene expression levels. We compared the best prognostic subgroups with the worst prognostic subgroups (39 samples in each group) and found that the combined survival results were more significant than the single genes.
Identification of miRNA and TF Targets
We have constructed interaction networks for all 553 si-YWHAZ DEGs and miRNA. The experimental validated or predicted target genes of TFs and miRNAs were summarized in the previous project.29 We selected all genes and TFs in DEGs and constructed a TF-target network. Then, the miRNA target gene data were added to construct a co-regulatory network of miR-375 and TFs.
Enrichment and Co-expression Analysis
The gene enrichment analysis for GO and KEGG was carried out by the DAVID online tool (https://david.ncifcrf.gov/) and FunRich,43 and the significant results (p < 0.01; Fisher’s test) visualization was through R package ggplot2. The co-expression data were derived from TCGA liver cancer data in the cBioPortal database (http://www.cbioportal.org/). The Pearson and Spearman correlation analyses were performed for all genes with YWHAZ. The genes with high correlation (| r | > 0.5) between the two algorithms and p < 0.01 were selected as co-expression genes of YWHAZ.
Statistic Analyses
All experiments were performed in triplicate unless specified. Results are represented as the mean ± SD. Statistical analysis was performed using unpaired Student’s t test by Graphpad Prism 5.0 software. p < 0.05 was considered significant.
Author Contributions
X.-X.H., A.-Y.G., and Q.Z. designed the study. J.-F.Z., H.H., and C.X. performed the experiments. Y.C. and J.L. contributed reagents and materials. J.-F.Z., X.-X.H., A.-Y.G., and Q.Z. analyzed the data and wrote the paper. J.-Z.L., J.-S.L., and X.-X.H. contributed to data interpretation and provided intellectual input. All authors reviewed, participated in revision, and approved the final version of the manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (nos. 81772969, 81472832, and 31471247) and the Applied Basic Research Program of Wuhan City (no. 2017060201010155).
Footnotes
Supplemental information includes two figures and four tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.04.007.
Contributor Information
An-Yuan Guo, Email: guoay@hust.edu.cn.
Xing-Xing He, Email: xxhe@tjh.tjmu.edu.cn.
Supplemental Information
References
- 1.Matta A., Siu K.W., Ralhan R. 14-3-3 zeta as novel molecular target for cancer therapy. Expert Opin. Ther. Targets. 2012;16:515–523. doi: 10.1517/14728222.2012.668185. [DOI] [PubMed] [Google Scholar]
- 2.Yan J.W., Lin J.S., He X.X. The emerging role of miR-375 in cancer. Int. J. Cancer. 2014;135:1011–1018. doi: 10.1002/ijc.28563. [DOI] [PubMed] [Google Scholar]
- 3.He X.X., Chang Y., Meng F.Y., Wang M.Y., Xie Q.H., Tang F., Li P.Y., Song Y.H., Lin J.S. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 4.Chang Y., Yan W., He X., Zhang L., Li C., Huang H., Nace G., Geller D.A., Lin J., Tsung A. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143 doi: 10.1053/j.gastro.2012.04.009. 177–87.e8, e178. [DOI] [PubMed] [Google Scholar]
- 5.He X.X., Guo A.Y., Xu C.R., Chang Y., Xiang G.Y., Gong J., Dan Z.L., Tian D.A., Liao J.Z., Lin J.S. Bioinformatics analysis identifies miR-221 as a core regulator in hepatocellular carcinoma and its silencing suppresses tumor properties. Oncol. Rep. 2014;32:1200–1210. doi: 10.3892/or.2014.3306. [DOI] [PubMed] [Google Scholar]
- 6.Liu A.M., Poon R.T., Luk J.M. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem. Biophys. Res. Commun. 2010;394:623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 7.Tao J., Ji J., Li X., Ding N., Wu H., Liu Y., Wang X.W., Calvisi D.F., Song G., Chen X. Distinct anti-oncogenic effect of various microRNAs in different mouse models of liver cancer. Oncotarget. 2015;6:6977–6988. doi: 10.18632/oncotarget.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue H.Y., Liu Y., Liao J.Z., Lin J.S., Li B., Yuan W.G., Lee R.J., Li L., Xu C.R., He X.X. Gold nanoparticles delivered miR-375 for treatment of hepatocellular carcinoma. Oncotarget. 2016;7:86675–86686. doi: 10.18632/oncotarget.13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Y.P., Liao J.Z., Lu Y.Q., Tian D.A., Ye F., Zhao P.X., Xiang G.Y., Tang W.X., He X.X. MiR-375 and doxorubicin co-delivered by liposomes for combination therapy of hepatocellular carcinoma. Mol. Ther. Nucleic Acids. 2017;7:181–189. doi: 10.1016/j.omtn.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao P., Wu S., Cheng Y., You J., Chen Y., Li M., He C., Zhang X., Yang T., Lu Y. MiR-375 delivered by lipid-coated doxorubicin-calcium carbonate nanoparticles overcomes chemoresistance in hepatocellular carcinoma. Nanomedicine (Lond.) 2017;13:2507–2516. doi: 10.1016/j.nano.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Ruffalo M., Bar-Joseph Z. Genome wide predictions of miRNA regulation by transcription factors. Bioinformatics. 2016;32:i746–i754. doi: 10.1093/bioinformatics/btw452. [DOI] [PubMed] [Google Scholar]
- 12.Guillemot F., Lo L.C., Johnson J.E., Auerbach A., Anderson D.J., Joyner A.L. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 13.Ito T., Udaka N., Okudela K., Yazawa T., Kitamura H. Mechanisms of neuroendocrine differentiation in pulmonary neuroendocrine cells and small cell carcinoma. Endocr. Pathol. 2003;14:133–139. doi: 10.1385/ep:14:2:133. [DOI] [PubMed] [Google Scholar]
- 14.Zhao H., Zhu L., Jin Y., Ji H., Yan X., Zhu X. miR-375 is highly expressed and possibly transactivated by achaete-scute complex homolog 1 in small-cell lung cancer cells. Acta Biochim. Biophys. Sin. (Shanghai) 2012;44:177–182. doi: 10.1093/abbs/gmr110. [DOI] [PubMed] [Google Scholar]
- 15.Nishikawa E., Osada H., Okazaki Y., Arima C., Tomida S., Tatematsu Y., Taguchi A., Shimada Y., Yanagisawa K., Yatabe Y. miR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Res. 2011;71:6165–6173. doi: 10.1158/0008-5472.CAN-11-1020. [DOI] [PubMed] [Google Scholar]
- 16.Neal C.L., Yu D. 14-3-3ζ as a prognostic marker and therapeutic target for cancer. Expert Opin. Ther. Targets. 2010;14:1343–1354. doi: 10.1517/14728222.2010.531011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neal C.L., Yao J., Yang W., Zhou X., Nguyen N.T., Lu J., Danes C.G., Guo H., Lan K.H., Ensor J. 14-3-3zeta overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res. 2009;69:3425–3432. doi: 10.1158/0008-5472.CAN-08-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maxwell S.A., Li Z., Jaye D., Ballard S., Ferrell J., Fu H. 14-3-3zeta mediates resistance of diffuse large B cell lymphoma to an anthracycline-based chemotherapeutic regimen. J. Biol. Chem. 2009;284:22379–22389. doi: 10.1074/jbc.M109.022418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Zou L., Li Q., Haibe-Kains B., Tian R., Li Y., Desmedt C., Sotiriou C., Szallasi Z., Iglehart J.D. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat. Med. 2010;16:214–218. doi: 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M., Liu T., Xu L., Gao X., Liu X., Wang C., He Q., Zhang G., Liu L. Direct interaction of 14-3-3ζ with ezrin promotes cell migration by regulating the formation of membrane ruffle. J. Mol. Biol. 2014;426:3118–3133. doi: 10.1016/j.jmb.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 21.Xu J., Acharya S., Sahin O., Zhang Q., Saito Y., Yao J., Wang H., Li P., Zhang L., Lowery F.J. 14-3-3ζ turns TGF-β’s function from tumor suppressor to metastasis promoter in breast cancer by contextual changes of Smad partners from p53 to Gli2. Cancer Cell. 2015;27:177–192. doi: 10.1016/j.ccell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C.H., Chuang S.M., Yang M.F., Liao J.W., Yu S.L., Chen J.J.W. A novel function of YWHAZ/β-catenin axis in promoting epithelial-mesenchymal transition and lung cancer metastasis. Mol. Cancer Res. 2012;10:1319–1331. doi: 10.1158/1541-7786.MCR-12-0189. [DOI] [PubMed] [Google Scholar]
- 23.Lu J., Guo H., Treekitkarnmongkol W., Li P., Zhang J., Shi B., Ling C., Zhou X., Chen T., Chiao P.J. 14-3-3zeta cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial-mesenchymal transition. Cancer Cell. 2009;16:195–207. doi: 10.1016/j.ccr.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi J.E., Hur W., Jung C.K., Piao L.S., Lyoo K., Hong S.W., Kim S.W., Yoon H.Y., Yoon S.K. Silencing of 14-3-3ζ over-expression in hepatocellular carcinoma inhibits tumor growth and enhances chemosensitivity to cis-diammined dichloridoplatium. Cancer Lett. 2011;303:99–107. doi: 10.1016/j.canlet.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Weerasekara V.K., Panek D.J., Broadbent D.G., Mortenson J.B., Mathis A.D., Logan G.N., Prince J.T., Thomson D.M., Thompson J.W., Andersen J.L. Metabolic-stress-induced rearrangement of the 14-3-3ζ interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3ζ interaction with phosphorylated Atg9. Mol. Cell. Biol. 2014;34:4379–4388. doi: 10.1128/MCB.00740-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman A.K., Morrison D.K. 14-3-3 proteins: diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011;22:681–687. doi: 10.1016/j.semcdb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye C.G., Chen G.G., Ho R.L.K., Merchant J.L., He M.L., Lai P.B.S. Epigenetic upregulation of Bak by ZBP-89 inhibits the growth of hepatocellular carcinoma. Biochim. Biophys. Acta. 2013;1833:2970–2979. doi: 10.1016/j.bbamcr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buck A., Buchholz M., Wagner M., Adler G., Gress T., Ellenrieder V. The tumor suppressor KLF11 mediates a novel mechanism in transforming growth factor beta-induced growth inhibition that is inactivated in pancreatic cancer. Mol. Cancer Res. 2006;4:861–872. doi: 10.1158/1541-7786.MCR-06-0081. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H.M., Kuang S., Xiong X., Gao T., Liu C., Guo A.Y. Transcription factor and microRNA co-regulatory loops: important regulatory motifs in biological processes and diseases. Brief. Bioinform. 2015;16:45–58. doi: 10.1093/bib/bbt085. [DOI] [PubMed] [Google Scholar]
- 30.Beavon I.R. The E-cadherin-catenin complex in tumour metastasis: structure, function and regulation. Eur. J. Cancer. 2000;36:1607–1620. doi: 10.1016/s0959-8049(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 31.Guarino M. Epithelial-mesenchymal transition and tumour invasion. Int. J. Biochem. Cell Biol. 2007;39:2153–2160. doi: 10.1016/j.biocel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Geman D., Ochs M., Price N.D., Tomasetti C., Younes L. An argument for mechanism-based statistical inference in cancer. Hum. Genet. 2015;134:479–495. doi: 10.1007/s00439-014-1501-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimura Y., Komatsu S., Ichikawa D., Nagata H., Hirajima S., Takeshita H., Kawaguchi T., Arita T., Konishi H., Kashimoto K. Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma. Br. J. Cancer. 2013;108:1324–1331. doi: 10.1038/bjc.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathé E.A., Nguyen G.H., Bowman E.D., Zhao Y., Budhu A., Schetter A.J., Braun R., Reimers M., Kumamoto K., Hughes D. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin. Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang C., Shi H., Wang C., Wang J., Geng N., Jiang X., Wang X. Correlation of microRNA-375 downregulation with unfavorable clinical outcome of patients with glioma. Neurosci. Lett. 2012;531:204–208. doi: 10.1016/j.neulet.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 37.Fan T., Li R., Todd N.W., Qiu Q., Fang H.B., Wang H., Shen J., Zhao R.Y., Caraway N.P., Katz R.L. Up-regulation of 14-3-3zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 2007;67:7901–7906. doi: 10.1158/0008-5472.CAN-07-0090. [DOI] [PubMed] [Google Scholar]
- 38.Tokumoto M., Fujiwara Y., Shimada A., Hasegawa T., Seko Y., Nagase H., Satoh M. Cadmium toxicity is caused by accumulation of p53 through the down-regulation of Ube2d family genes in vitro and in vivo. J. Toxicol. Sci. 2011;36:191–200. doi: 10.2131/jts.36.191. [DOI] [PubMed] [Google Scholar]
- 39.Tokumoto M., Satoh M. [Cadmium induces p53-dependent apoptosis through the inhibition of Ube2d family gene expression] Nippon Eiseigaku Zasshi. 2012;67:472–477. doi: 10.1265/jjh.67.472. [DOI] [PubMed] [Google Scholar]
- 40.Lin Z., Yang H., Kong Q., Li J., Lee S.M., Gao B., Dong H., Wei J., Song J., Zhang D.D., Fang D. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell. 2012;46:484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 41.Ding F., Bao C., Tian Y., Xiao H., Wang M., Xie X., Hu F., Mei J. USP22 promotes NSCLC tumorigenesis via MDMX up-regulation and subsequent p53 inhibition. Int. J. Mol. Sci. 2014;16:307–320. doi: 10.3390/ijms16010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan J.J., Zhang Y.N., Liao J.Z., Ke K.P., Chang Y., Li P.Y., Wang M., Lin J.S., He X.X. MiR-497 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting VEGFA and AEG-1. Oncotarget. 2015;6:29527–29542. doi: 10.18632/oncotarget.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.