Abstract
Introduction
We assessed the molecular epidemiology of multidrug-resistant bacteria colonizing or infecting war-injured patients from Libya and Syria who were treated at the Bundeswehr hospitals Hamburg and Westerstede, Germany.
Methods
Enterobacteriaceae and Gram-negative rod-shaped nonfermentative bacteria with resistance against third-generation methoxyimino cephalosporins or carbapenems as well as methicillin-resistant Staphylococcus aureus (MRSA) from war-injured patients from Libya and Syria were assessed by molecular typing, i.e., spa typing for MRSA strains and rep-PCR and next-generation sequencing (NGS) for Gram-negative isolates.
Results
A total of 66 isolates were assessed – comprising 44 Enterobacteriaceae, 16 nonfermentative rod-shaped bacteria, and 6 MRSA from 22 patients – and 8 strains from an assessment of the patient environment comprising 5 Enterobacteriaceae and 3 nonfermentative rod-shaped bacteria. Although 24 out of 66 patient strains were isolated more than 3 days after hospital admission, molecular typing suggested only 7 likely transmission events in the hospitals. Identified clonal clusters primarily suggested transmission events in the country of origin or during the medical evacuation flights.
Conclusions
Nosocomial transmissions in hospital can be efficiently prevented by hygiene precautions in spite of heavy colonization. Transmission prior to hospital admission like on evacuation flights or in crises zones needs further assessment.
Keywords: multidrug-resistant, typing, next-generation sequencing, rep-PCR, war injuries, Libya, Syria
Introduction
Colonization or infection with multidrug-resistant bacteria is a frequent phenomenon in traumatized patients from war and crisis zones. In a recent assessment of 21 male Ukrainian patients who were treated at 4 Bundeswehr hospitals in Germany, altogether 32 carbapenem-resistant Acinetobacter spp. strains were isolated [1]. Apart from merely colonizing strains, clinically relevant isolates from deep surgical wounds and bioptic material were also detected.
High colonization rates with multidrug-resistant bacteria were also identified in Syrian civilians who were treated at Israeli hospitals after fleeing the Syrian civil war [2]. Similar experience was found with Syrian refugees in German hospitals [3–5]. Various other international surveys confirm high colonization rates of Syrian refugees and patients with multidrug-resistant bacteria [6–14], and the same applies to refugees and patients from Libya [15–28].
Patients from Libya and Syria with war injuries were transferred to the Bundeswehr hospitals Hamburg and Westerstede in 2011 and 2013, respectively. To assess the effectiveness of the hygiene protocols that were enforced, regular swab-based screenings for multidrug-resistant pathogens were performed, and strains collected were subjected to rep-PCR-based and sequence-based typing to identify potential nosocomial transmission events.
Methods
Patients. Criteria for the inclusion of patients in the retrospective assessment comprised the following:
Admission was due to war injuries from the civil war in Libya and Syria in 2011 and 2013, respectively, to the Bundeswehr hospitals Hamburg and Westerstede, Germany.
Multidrug-resistant isolates (as defined in the following) had been isolated from the patients.
There were no specific exclusion criteria.
Anonymous assessment of patient-related data applied to age, sex, and main clinical diagnoses.
Patient Rooms and Wards. For the source tracking of potential nosocomial transmission events, the rooms and wards during the hospital stay of each individual patient were documented, including – if applicable – transfers of patients from one room to another. To ensure the anonymity of assessments, the room and ward numbers were changed for this study.
Environmental Screening. During the treatment of the Libyan patients at the Bundeswehr hospital Hamburg, multidrug-resistant strains from a single environmental screening event in 2011 from the patients' environment were obtained and included in the assessment.
Hygiene-related Information. After their transfer to Germany from Libya or Syria by StratAirMedEvac (strategic air medical evacuation, i.e., transport in specialized medical service airplanes of the German Air Force), strict hygiene procedures were enforced to prevent further nosocomial spread of multidrug-resistant pathogens among the patients, as previously described in detail [29].
In short, the patients were handled by medical staff wearing personal protection equipment that comprised protective gloves, a waterproof one-way coat-apron, a head cover, and a mask protecting the mucous membranes of the nose and the pharynx. The patients were treated in single-room or cohort isolation on especially designated wards as soon as their medical condition allowed for a transfer from the intensive care or intermediate care unit. Special nursing staff supported by interpreters were chosen for their care. On a weekly basis, hygiene screening for multidrug-resistant bacteria involving swabbing with subsequent cultural assessment was performed from the nostrils, the mouth, any wounds, and the perianal region. Local reduction of bacterial load was attempted by disinfection washing with octenidin (Schülke & Mayr, Norderstedt, Germany) based products for the skin and with octenidol (Schülke & Mayr) based products for mucous membranes and wounds. The nostrils and the pharynx were disinfected as often as three times a day. Towels, cloths, and bed linen were changed after each disinfection procedure. The patients' property and medical equipment applied patient-specifically were disinfected several times per day and the surfaces in the patient room twice a day.
Patient Strains and Environmental Strains. Multidrug-resistant strains were defined as Enterobacteriaceae or Gram-negative rod-shaped nonfermentative bacteria with resistance against third-generation methoxyimino cephalosporins or carbapenems as well as methicillin-resistant Staphylococcus aureus (MRSA). Clinical isolates and screening isolates from the Libyan and Syrian patients were provided by the Central Institute of the Bundeswehr Kiel, Department Berlin, for patients from Hamburg as well as by the laboratory “Labor Dr. Enzenauer und Kollegen” for patients from Westerstede after the exclusion of copy strains. Copy strains were defined as repeated isolation events of identical species with identical resistance patterns at any time of treatment from the same patient. Strains from the environmental screening in Hamburg were provided by the Central Institute of the Bundeswehr Kiel.
To ensure that strains were assessed by identical standardized procedures, all strains provided were subjected to repeated identification and resistance testing at the Institute for Medical Microbiology, Virology and Hygiene, University Medicine Rostock. Species identification was performed by matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS) using a Shimadzu/Kratos “AXIMA Assurance” MALDI TOF mass spectrometer (Shimadzu Deutschland GmbH, Duisburg, Germany) using the database “IVD-mode VitekMS-ID” database version 3.2.0.-6 (bioMérieux, Marcy-l'Étoile, France). Resistance was determined by automated resistance testing with a VITEK 2 system (bioMérieux) using the resistance cards AST-N248 (charge 648332310) for Gram-negative nonfermentative rod-shaped bacteria and AST-N263 (charge 663333410) for Enterobacteriaceae. The applied software was the “Vitek 2 systems version 06.01”; automated interpretation of the results was in line with the interpretative guideline EUCAST2014 + CLSI 2014 D using the advanced expert settings (AES) parameter set “EUCAST/CLSI + PHAENOTYPISCH 2014 D.” E-testing (bioMérieux) was added in case of uncertain or noninterpretable VITEK 2 results, i.e., cotrimoxazole testing with Stenotrophomonas maltophilia or any resistance testing with Pseudomonas putida. For Escherichia coli and Klebsiella pneumoniae, ESBL screening was performed via VITEK 2. For all Enterobacteriaceae, the commercial ESBL/ampC disc-based ABCD test kit Mast ID D68C (Mast Diagnostic, Amiens, France) was used to identify ESBL or ampC production [30].
Patient strains that were isolated up to day 3 after admission were considered as nonnosocomial strains, while strains that were isolated after day 3 were defined as potentially nosocomially transmitted.
Spa Typing of MRSA. Spa typing of MRSA isolates was performed exactly as described [31, 32] at the Department of Medical Microbiology, Virology and Hygiene of the University Medicine Rostock using the software RidomStaphType version 2.2.1 (Ridom Ltd., Würzburg, Germany) including allocation of multilocus sequence typing (MLST)-based clonal clusters using the based upon repeat pattern (BURP) algorithm [33].
Rep-PCR (repetitive extragenic palindromic sequence polymerase chain reaction)-based Typing of Gram-negative Rod-shaped Bacteria. Rep-PCR-based typing was performed at the Department of Medical Microbiology, Virology and Hygiene of the University Medicine Rostock for Gram-negative rod-shaped bacteria as described [1] using the bioMérieux DiversiLab system (bioMérieux). Overnight cultures of the isolates from Columbia agar with 5% sheep blood (BD, Heidelberg, Germany) were assessed. After DNA extraction using the DiversiLab MoBio Ultra Clean kit (ref. no. 270 675, bioMérieux), amplification of purified DNA was performed on a T-personal thermal cycler (Biometra, Göttingen, Germany) using the fingerprinting kits DiversiLab Acinetobacter (ref. no. 410 946, bioMérieux), DiversiLab Bacteria (ref. no. 411 007, bioMérieux), DiversiLab Enterobacter (ref. no. 410 968, bioMérieux), DiversiLab Escherichia (410 980), DiversiLab Klebsiella (ref. no. 410 981, bioMérieux), and DiversiLab Pseudomonas (ref. no. 410 946, bioMérieux). Chip-based DNA separation based on a DiversiLab Labchip kit (25 chips, ref. no. 270 670, bioMérieux) was applied for the detection of rep-PCR products on an Agilent 2100 bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA).
Applying the Pearson correlation method and analyses of all entries in duplicate, band-pattern analysis was performed with the help of the DiversiLab software version 3.6.1 (bioMérieux) as described by the manufacturer using an identity cutoff of 95% to suggest clonal identity.
Next-generation Sequencing of Gram-negative Rod-shaped Bacteria. Gram-negative isolates from this study were subjected to next-generation sequencing (NGS)-based assessment of whole genomes in a total of 4 individual 600-cycle sequencing runs. For this, 1 ng of purified chromosomal DNA from all isolates was used to prepare individual libraries employing the Illumina Nextera© XT DNA Library Prep Kit according to the manufacturer's instructions. An Agilent Technology 2100 Bioanalyzer served to verify tagmentation and also final library fragment size distribution on a High Sensitivity DNA Chip. AMPure XP beads were used for DNA library purification. The final pooled libraries were applied to a MiSeq Reagent v3 600-cycle Kit and sequenced on a MiSeq system as 300-cycle paired-end runs. 5% PhiX control library was spiked into the final library pool. On average, a cluster density of 847 (K/mm2) was achieved with 96.46 ± 1.48% of clusters passing filter specifications. In each run, roughly 20.3 million reads (94.7%) of 21.1 million total reads passed filter specifications, leading to 12.52 Gbp (giga base pairs) sequenced. Index reads were evenly distributed across the 20 individual samples routinely used in each run. Generated FASTQ (file containing raw read information) files were subjected to further bioinformatic analysis as outlined below.
Database Storage of Sequence Information. Raw FASTQ files were submitted to the EMBL-EBI (European Molecular Biology Laboratory - European Bioinformatics Institute) (https://www.ebi.ac.uk/ena/submit/sra/#home) European Nucleotide Archive (ENA) and stored in the Short Read Archive (SRA) with the bioproject accession code PRJNA407760 and the study name SRP118558 as unambiguous identifiers. For technical reasons, the sequences of the strains had to be deposited not by their numbers only but by biosample accession codes with the syntax “BW_strain number” (e.g., BW_1 for strain number 1).
Typing Approach Based on the Next-generation Sequencing Data of the Gram-negative Rod-shaped Bacteria. To obtain the sequences used for MLST analyses as well as for average nucleotide identity (ANI)-based comparisons, all reads for the respective sample were assembled using the Newbler assembler, v 2.8 (Roche, Germany). For MLST analyses, the sequences of the gene set(s) used for MLST typing of the respective bacterial species were retrieved from pubmlst.org [34]. For each sample, the set(s) were then compared against a BLAST database containing the contigs larger than 500 bp of the respective assembly using BLASTN. For each gene, the best-matching variant, i.e., the one having 100% nucleotide identity, was identified and checked for completeness, i.e., matching over the full length of the query sequence. Only if they were perfectly identical was the gene variant used for MLST comparisons.
As the coverage obtained for several isolates was insufficient to obtain a complete MLST pattern, an ANI-based approach was used to identify potential clonal isolates. ANI values were calculated using the method described by Goris et al. [35]. By comparison with publicly available genomes of the respective species, it was determined that nonclonal strains differed among each other by thousands to hundreds of thousands of mutations. In contrast, strains differing by only tens to hundreds of SNPs (single nucleotide polymorphisms) are likely to be clonal. Therefore, the cutoffs for potential clonal isolates were set to ANI values greater than or equal to 99.5% (close common ancestor) and 99.9% (likely clonal).
Resistance Testing Approach Based on the Next-generation Sequencing Data of the Gram-negative Rod-shaped Bacteria. Screening of the obtained NGS reads for genetic resistance determinants was performed using the ResFinder version 2.1 (https://cge.cbs.dtu.dk/services/resfinder/, last accessed July 5, 2017) software as described elsewhere [36, 37]. Phenotypic resistance data from the VITEK 2 system were used for comparison.
PCR- and Sanger Sequencing-based Identification of Resistance Genes. Enterobactericeae with a resistance pattern of the ESBL (extended spectrum β-lactamase) type were subjected to PCRs targeting the β-lactamase genes blaTEM and blaSHV as well as four important groups of blaCTX-M as described [38, 39]. The amplicons of the blaTEM and blaSHV PCRs were further analyzed by Sanger sequencing to discriminate ESBL-associated from non-ESBL-associated variants. For the 4 blaCTX-M PCRs, discrimination at group level without sequence-based further differentiation was accepted. In detail, blaCTX-M group I comprised blaCTX-M-1, -3, -10, -11, -12, -15, -22,-23, -28, -29, -30, blaCTX-M group II comprised blaCTX-M-2, -4, -5, -6, -7, -20, blaCTX-M group III comprised blaCTX-M-8, and blaCTX-M group IV comprised blaCTX-M-9, -13, -14, -16 to -19, -21, -27.
PCR-based screening without further differentiation approaches was performed for 11 carbapenemase genes from Gram-negative rod-shaped isolates with reduced sensitivity toward carbapenems as described [1]. In detail, the carbapenemase genes assessed comprised blaIMP, blaVIM, blaNDM, blaSPM, blaAIM, blaDIM, blaGIM, blaSIM, blaKPC, blaBIC, and blaOXA-48.
Ethics. Ethical clearance was obtained from the ethics committee of the medical association of Hamburg (WF-042/15). The clearance allowed for the retrospective assessment of patient- and strain-related data in the study in an anonymous way without informed consent by the patients or next-of-kin. The respective procedures are in line with German National Laws, in detail, § 9 section 2 of Hamburg's Chamber Law for Medical Professions (“Hamburger Kammergesetz für die Heilberufe”) as well as § 15 section 1 of the Regulations for Hamburg's Doctors (“Berufsordnung für Hamburgs Ärztinnen und Ärzte”).
Results
Patients. Altogether, 8 patients with multidrug-resistant isolates from Libya who were admitted in 2011 and 14 patients from Syria who were admitted in 2013 were included in the assessment. Of these, 4 Libyan patients and 7 Syrian patients were treated at the Bundeswehr hospital of Hamburg and the same numbers at the Bundeswehr hospital Westerstede. All the patients were male. The median age of the Libyan patients was 35.0 years (ranging from 26 to 38 years) and 26.5 years for the Syrian patients (ranging from 4 to 52 years). All patients suffered from war-related injuries, usually complex traumata. Details are shown in Table 1.
Table 1.
Assessed Libyan and Syrian patients and their accommodation in the Bundeswehr Hospitals of Hamburg and Westerstede
| Wards and patient rooms of the Libyan patients in the Bundeswehr Hospital of Hamburg from admission to isolation of the last newly detected multidrug-resistant isolate (22 days) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient ID | Sex | Age (years) | Main diagnosis | Ward HBG-1-ITS, room 1 | Ward HBG-2, room 1 | Ward HBG-2, room 2 | Ward HBG-2, room 3 | Ward HBG-2, room 4 |
| HBG-L1 | Male | 38 | War trauma (multiple) | Day 1–2 | Day 2–18 | Day 18–22 | n.a. | n.a. |
| HBG-L2 | Male | 26 | War trauma (multiple) | Day 1–3 | Day 3–22 | n.a. | n.a. | n.a. |
| HBG-L3 | Male | 35 | War trauma (multiple) | n.a. | Day 1–2 | n.a. | Day 2–22 | n.a. |
| HBG-L4 | Male | 32 | War trauma (multiple) | n.a. | n.a. | Day 13–18 | n.a. | Day 1–13 and 18–22 |
| Wards and patient rooms of the Libyan patients in the Bundeswehr Hospital of Westerstede from admission to isolation of the last newly detected multidrug-resistant isolate (1 day) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient ID | Sex | Age (years) | Main diagnosis | Ward WEST-1, room 1 | Ward WEST-1, room 2 | Ward WEST-1, room 3 | ||
| WEST-L1 | Male | 33 | Thigh bullet | Day 1 | n.a. | n.a. | ||
| WEST-L2 | Male | 35 | Partial traumatic foot amputation | n.a. | Day 1 | n.a. | ||
| WEST-L3 | Male | 37 | Traumatic amputations (multiple) | n.a. | n.a. | Day 1 | ||
| WEST-L4 | Male | 37 | Traumatic amputations (multiple) | n.a. | n.a. | Day 1 | ||
| Wards and patient rooms of the Syrian patients in the Bundeswehr Hospital of Hamburg from admission to isolation of the last newly detected multidrug-resistant isolate (32 days) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient ID | Sex | Age (years) | Main diagnosis | Ward HBG-2, room 1 | Ward HBG-2, room 3 | Ward HBG-2, room 4 | Ward HBG-2, room 5 | Ward HBG-2, room 6 |
| HBG-S1 | Male | 31 | War trauma (multiple) | Day 1–32 | Day 1 | n.a. | n.a. | n.a. |
| HBG-S2 | Male | 23 | Explosion trauma | n.a. | n.a. | Day 1–32 | n.a. | n.a. |
| HBG-S3 | Male | 23 | War trauma (multiple) | n.a. | n.a. | n.a. | Day 11–19, 20–32 | Day 1–11, 19–20 |
| HBG-S4 | Male | 21 | War trauma (multiple) | n.a. | n.a. | Day 11–2a | Day 1–11 | n.a. |
| HBG-S5 | Male | 19 | War trauma (multiple) | Day 19–32 | n.a. | n.a. | n.a. | Day 1–19 |
| HBG-S6 | Male | 39 | War trauma (multiple) | Day 11 | Day 11 | n.a. | Day 1–19a | n.a. |
| HBG-S7 | Male | 25 | War trauma (multiple) | n.a. | Day 1–31a | n.a. | n.a. | n.a. |
| Wards and patient rooms of the Syrian patients in the Bundeswehr Hospital of Westerstede from admission to isolation of the last newly detected multidrug-resistant isolate (69 days) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient ID | Sex | Age (in years) | Main diagnosis | Ward WEST-2, room 1 | Ward WEST-2, room 2 | Ward WEST-2, room 3 | Ward WEST-2, room 4 | Ward WEST-3-ITS, room 1 |
| WEST-S1 | Male | 36 | Jaw injury | Day 1–52b | n.a. | n.a. | n.a. | n.a. |
| WEST-S2 | Male | 28 | Polytrauma | n.a. | n.a. | Day 9–25, 26–69 | Day 1–9 | Day 25–26 |
| WEST-S3 | Male | 4 | Osteomyelitis | n.a. | Day 1–69 | n.a. | n.a. | n.a. |
| WEST-S4 | Male | 31 | Soft tissue injury | n.a. | Day 1–69 | n.a. | n.a. | n.a. |
| WEST-S5 | Male | 21 | Complex bone and soft tissue injury | n.a. | n.a. | n.a. | Day 1–69 | n.a. |
| WEST-S6 | Male | 30 | Severe soft tissue injuries | Day 1–69 | n.a. | n.a. | n.a. | n.a. |
| WEST-S7 | Male | 52 | Pelvic fracture | Day 1 | n.a. | Day 1–69 | n.a. | n.a. |
aPatients were discharged prior to the isolation of the last newly detected multidrug-resistant isolate (32 days).
bPatient was discharged prior to the isolation of the last newly detected multidrug-resistant isolate (69 days).
Patient Rooms and Wards. To allow anonymous tracking of common use of patients' rooms and patient transfers with potential for nosocomial transmission, all patients, rooms, and wards were numbered consecutively. All patients and wards were labeled “HBG” at the Bundeswehr hospital Hamburg and “WEST” at the Bundeswehr hospital Westerstede. The letter “L” indicated origin from Libya and “S” origin from Syria. Intensive care wards were labeled with the acronym “ITS” (Table 1).
Focusing on patients from Libya at the Bundeswehr hospital Hamburg, patients 1 and 2 shared the same room at days 1 and 2 after admission onto the intensive care ward. Afterwards, they shared room 1 on a standard care ward from day 3 to day 18. On day 2 after admission, patient 1 and patient 3 were both in this room 1, too. On day 18 after admission, patient 1 and patient 4 were both in room 2 of the same ward. There were no other instances of common use of patients' rooms. While patient 1 was transferred twice in total, all the other 3 patients were transferred once. Patients' movements were tracked for 22 days altogether until the detection of the last new multidrug-resistant isolate of this assessment.
Focusing on patients from Libya at the Bundeswehr hospital Westerstede, all assessed multidrug-resistant strains had been isolated by the first day after admission. At this day, patients 3 and 4 were in the same room of a standard care ward.
Focusing on patients from Syria at the Bundeswehr hospital Hamburg, the patient movements were tracked until the last new detection of a multidrug-resistant isolate in this assessment at day 32 after admission. Room 1 was shared by patients 1 and 5 from day 19 to day 32 after admission as well as by patients 1 and 6 at day 11. Room 2 was shared on day 1 by patients 1 and 7 as well as on day 11 by patients 6 and 7. Room 4 was shared by patients 2 and 4 on days 11 to 21. Room 5 was shared by patients 3 and 4 on day 11 and by patients 3 and 6 on days 11 to 19. Room 6 was shared by patients 3 and 5 on days 1 to 11 and on day 19. Altogether, patient 6 was transferred twice, patients 1, 3, 4, and 5 were transferred once, and the remaining 2 were not transferred during their hospital stay.
Focusing on patients from Syria at the Bundeswehr hospital Westerstede, a period of 69 days after admission had to be tracked until the last assessed multidrug-resistant strain was isolated. Patients 1, 6, and 7 were together in room 1 at the first day after admission; patients 1 and 6 remained together in this room for altogether 52 days. Patients 3 and 4 were together in the same room 2 from day 1 to day 69, patients 2 and 7 in room 3 from day 9 to day 69, while patients 2 and 5 stayed together in room 4 from day 1 to day 9. Patient 2 was transferred twice, and patient 7 was transferred once, while all other patients remained in their rooms for the whole assessment period.
A clearer overview of patient rooms and patient transfers is provided in Table 1.
Patient Isolates and Environmental Isolates. From 8 Libyan patients, 13 Enterobacteriaceae (comprising 6 Klebsiella pneumoniae, 3 Escherichia coli, 3 Enterobacter cloacae, and 1 Citrobacter freundii) and 5 nonfermentative rod-shaped bacteria (comprising 3 Pseudomonas aeruginosa and 2 Acinetobacter baumannii complex) were isolated. Clinically relevant isolates comprised one Enterobacter cloacae at an external fixator of the patient's left thigh as well 2 Pseudomonas aeruginosa strains, one from an orthopedic wound at the left hand and one from a wound at the dorsal side of the patient's left thigh. The other isolates have to be considered as colonization flora (Supplementary material 1).
Supplementary material 1.
Isolated Enterobacteriaceae with resistance against 3rd generation methoxyimino cephalosporins or carbapenems, nonfermentative Gram-negative rod-shaped bacteria, and methicillin-resistant Staphylococcus aureus (MRSA) from Libyan and Syrian patients.
| Isolates from Libyan patients | ||||||||
|---|---|---|---|---|---|---|---|---|
| Enterobacteriaceae with resistance against 3rd generation methoxyimino cephalosporins or carbapenems | ||||||||
| Patient I.D. | Sample I.D. | Species | Isolation site | Day of isolation | Nosocomial | In rep-PCR 95% identical with | In NGS-based MLSTa/ANIb identical with | Match NGS vs. rep-PCR |
| HBG-L1 | 2 | C. freundii | Rectum | Day 2 | No | |||
| HBG-L1 | 3 | E. cloacae | Rectum | Day 10 | Yes | 14 (WEST-L3), 48 (WEST-S6), 54 (WEST-S6) | 14 (WEST-L3)ab | 14 (WEST-L3) |
| HBG-L1 | 4 | E. coli | Rectum | Day 10 | Yes | |||
| HBG-L1 | 6 | E. coli | Colostomy | Day 22 | Yes | 33 (WEST-S2)(b) | ||
| HBG-L2 | 8 | K. pneumoniae | Inguinal skin | Day 1 | No | 18 (HBG-L4)a,b, 20 (room 4 HBG-2)a,b, 26 (room 4 HBG-2)(a),(b) | ||
| HBG-L2 | 10 | E. cloacae | Inguinal skin | Day 1 | No | 48 (WEST-S6), 54 (WEST-S6), 77 (HBG-S4) | ||
| HBG-L2 | 12 | K. pneumoniae | Inguinal skin | Day 10 | Yes | |||
| HBG-L3 | 16 | K. pneumoniae | Inguinal skin | Day 1 | No | 27 (room 1 HBG-2), 28 (room 3 HBG-2) | 27 (room 1 HBG-2)a,b, 28 (room 3 HBG-2)a,b | 27 (room 1 HBG-2), 28 (room 3 HBG-2) |
| HBG-L3 | 17 | E. coli | Perineal skin | Day 1 | No | None | ||
| HBG-L4 | 18 | K. pneumoniae | Inguinal skin | Day 16 | Yes | 20 (room 4 HBG-2), 26 (room 4 HBG-2) | 8 (HBG-L2)a,b, 20 (room 4 HBG-2)a,b, 26 (room 4 HBG-2)(a),(b) | 20 (room 4 HBG-2), 26 (room 4 HBG-2) |
| WEST-L2 | 13 | K. pneumoniae | Inguinal skin | Day 1 | No | None | ||
| West-L3 | 14 | E. cloacae | External fixator of the left thigh | Day 1 | No | 3 (HBG-L1), 48 (WEST-S6), 54 (WEST-S6) | 3 (HBG-L1)a,b | 3 (HBG-L1) |
| West-L4 | 15 | K. pneumoniae | Inguinal skin | Day 1 | No | |||
| Nonfermentative rod-shaped bacteria | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient I.D. | Sample I.D. | Species | Isolation site | Day of isolation | Nosocomial | In rep-PCR 95% identical with | In NGS-based MLSTa/ANIb identical with | Match NGS vs. rep-PCR |
| HBG-L1 | 1 | A. baumannii complex | Inguinal skin | 2 | no | |||
| HBG-L1 | 5 | P. aeruginosa | Perineal skin | 16 | yes | |||
| HBG-L2 | 9 | A. baumannii complex | Inguinal skin | 1 | no | |||
| HBG-L2 | 11 | P. aeruginosa | Orthopedic wound at the left hand | 1 | no | 7 (WEST-L1)a,(b) | ||
| WEST-L1 | 7 | P. aeruginosa | Wound at the dorsal side of the left thigh | 1 | no | 11 (HBG-L2)a,(b) | ||
| Isolates from Syrian patients | ||||||||
|---|---|---|---|---|---|---|---|---|
| Enterobacteriaceae with resistance against 3rd generation methoxyimino cephalosporins or carbapenems | ||||||||
| Patient I.D. | Sample I.D. | Species | Isolation site | Day of isolation | Nosocomial | In rep-PCR 95% identical with | In NGS-based MLSTa/ANIb identical with | Match NGS vs. rep-PCR |
| HBG-S1 | 57 | E. cloacae | Deep wound at the right heel | Day 2 | No | |||
| HBG-S1 | 75 | E. coli | Perineal skin | Day 9 | Yes | |||
| HBG-S2 | 59 | K. pneumoniae | Perineal skin | Day 2 | No | 78 (HBG-S4)a,b | ||
| HBG-S2 | 60 | E. coli | Perineal skin | Day 2 | No | 37 (WEST-S4), 61 (HBG-S3), 66 (HBG-S4), 72 (HBG-S6) | 37 (WEST-S4)a,(b), 61 (HBG-S3)a,(b), 72 (HBG-S6)(a),(b) | 37 (WEST-S4), 61 (HBG-S3), 72 (HBG-S6) |
| HBG-S3 | 61 | E. coli | Perineal skin | Day 2 | No | 37 (WEST-S4), 60 (HBG-S2), 66 (HBG-S4), 72 (HBG-S6) | 37 (WEST-S4)a,(b) 60 (HBG-S2)a,(b) 72 (HBG-S6)(a),(b) | 37 (WEST-S4), 60 (HBG-S2), 72 (HBG-S6) |
| HBG-S4 | 63 | E. coli | Perineal skin | Day 2 | No | |||
| HBG-S4 | 65 | C. freundii | Perineal skin | Day 2 | No | |||
| HBG-S4 | 66 | E. coli | Deep wound at the right gluteal region | Day 2 | No | 37 (WEST-S4), 60 (HBG-S2), 61 (HBG-S3), 72 (HBG-S6) | ||
| HBG-S4 | 76 | E. coli | Catheter urine | Day 30 | yes | |||
| HBG-S4 | 77 | E. cloacae | Perineal skin | Day 16 | Yes | 10 (HBG-L2), 54 (WEST-S6) | 54 (WEST-S6)a,(b) | 54 (WEST-S6) |
| HBG-S4 | 78 | K. pneumoniae | Superficial wound at the groin | Day 32 | Yes | 59 (HBG-S2)ab | ||
| HBG-S5 | 68 | E. coli | Perineal skin | Day 2 | no | 51 (WEST-S6)a,(b) | ||
| HBG-S5 | 69 | Proteus mirabilis | Perineal skin | Day 2 | no | |||
| HBG-S5 | 70 | E. cloacae | Deep wound at the upper side of a fixator | Day 2 | no | |||
| HBG-S5 | 71 | K. pneumoniae | Perineal skin | Day 9 | Yes | |||
| HBG-S6 | 72 | E. coli | Perineal skin | Day 2 | No | 37 (WEST-S4), 60 (HBG-S2), 61 (HBG-S3), 66 (HBG-S4) | 37 (WEST-S4)(a),b, 60 (HBG-S2)(a),b, 61 (HBG-S3)(a),b | 37 (WEST-S4), 60 (HBG-S2), 61 (HBG-S3) |
| HBG-S7 | 74 | E. coli | Perineal skin | Day 9 | Yes | |||
| WEST-S1 | 32 | R. planticola | Anus | Day 15 | yes | 52 (WEST-S6) | 52 (West-S6)b | 52 (WEST-S6) |
| WEST-S2 | 33 | E. coli | Wound | Day 51 | yes | 6 (HBG-L1)(b) | ||
| WEST-S2 | 38 | E. coli | Anus | Day 1 | no | |||
| WEST-S2 | 39 | P. mirabilis | Wound | Day 3 | no | |||
| WEST-S2 | 41 | M. morganii | Wound | Day 69 | yes | |||
| WEST-S3 | 36 | M. morganii | Wound | Day 1 | no | |||
| WEST-S4 | 37 | E. coli | Wound | Day 1 | No | 60 (HBG-S2), 61 (HBG-S3), 66 (HBG-S4), 72 (HBG-S6) | 60 (HBG-S2)a,(b) 61 (HBG-S3)a,(b), 72 (HBG-S6)(a),(b) | 60 (HBG-S2), 61 (HBG-S3), 72 (HBG-S6) |
| WEST-S5 | 42 | K. pneumoniae | Wound | Day 8 | Yes | |||
| WEST-S5 | 45 | C. freundii | Wound | Day 1 | no | 55 (West-S7) | 55 (West-S7)b | 55 (West-S7) |
| WEST-S6 | 48 | E. cloacae | Wound | Day 4 | Yes | 3 (HBG-L1), 10 (HBG-L2), 14 (WEST-L3), 54 (WEST-S6) | ||
| WEST-S6 | 51 | E. coli | Wound | Day 1 | No | 68 (HBG-S5)(b) | ||
| WEST-S6 | 52 | R. planticola | Wound | Day 1 | No | 32 (West-S1) | 32 (West-S1)b | 32 (West-S1) |
| WEST-S6 | 54 | E. cloacae | Skin | Day 1 | no | 3 (HBG-L1), 10 (HBG-L2), 14 (WEST-L3), 48 (WEST-S6), 77 (HBG-S4) | 77 (HBG-S4)a,(b) | 77 (HBG-S4) |
| WEST-S7 | 55 | C. freundii | Wound | Day 1 | No | 45 (WEST-S5) | 45 (WEST-S5)b | 45 (WEST-S5) |
| Nonfermentative rod-shaped bacteria | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient I.D. | Sample I.D. | Species | Isolation site | Day of isolation | Nosocomial | In rep-PCR 95% identical with | In NGS-based MLSTa/ANIb identical with | Match NGS vs. rep-PCR |
| HBG-S1 | 56 | A. baumannii complex | Deep wound at the right heel | Day 2 | no | 31 (WEST-S1)b, 50 (West-S6)b, 62 (HBG-S2)(b), 64 (HBG-S4)b, 73 (HBG-S7)b | ||
| HBG-S2 | 62 | A. baumannii complex | Deep wound at the upper pin of the fixator at the thigh | Day 2 | no | 31 (WEST-S1), 50 (WEST-S6), 64 (HBG-S4), 73 (HBG-S7) | 31 (WEST-S1)(b), 50 (West-S6)(b), 56 (HBG-S1)(b), 64 (HBG-S4)(b), 73 (HBG-S7)(b) | 31 (WEST-S1), 50 (WEST-S6), 64 (HBG-S4), 73 (HBG-S7) |
| HBG-S4 | 64 | A. baumannii complex | Perineal skin | Day 2 | no | 31 (WEST-S1), 50 (WEST-S6), 62 (HBG-S2), 73 (HBG-S7) | 31 (WEST-S1)a,b, 50 (West-S6)a,b, 56 (HBG-S1)b, 62 (HBG-S2)(b),73 (HBG-S7)a,b | 31 (WEST-S1), 50 (West-S6), 62 (HBG-S2), 73 (HBG-S7) |
| HBG-S7 | 73 | A. baumannii complex | Pharynx | Day 9 | yes | 31 (WEST-S1), 50 (WEST-S6), 62 (HBG-S2), 64 (HBG-S4) | 31 (WEST-S1)a,b, 50 (West-S6)a,b, 56 (HBG-S1)b, 62 (HBG-S2)(b), 64 (HBG-S4)ab | 31 (WEST-S1), 50 (West-S6), 62 (HBG-S2), 64 (HBG-S4) |
| WEST-S1 | 31 | A. baumannii complex | Anus | Day 21 | yes | 50 (WEST-S6), 62 (HBG-S2), 64 (HBG-S4), 73 (HBG-S7) | 50 (West-S6)a,b, 56 (HBG-S1)b, 62 (HBG-S2)(b), 64 (HBG-S4)a,b, 73 (HBG-S7)a,b | 50 (West-S6), 62 (HBG-S2), 64 (HBG-S4), 73 (HBG-S7) |
| WEST-S2 | 40 | P. aeruginosa | Wound | Day 3 | no | |||
| WEST-S5 | 44 | A. radioresistens | Wound | Day 2 | no | |||
| WEST-S5 | 46 | S. maltophilia | Wound | Day 11 | yes | |||
| WEST-S6 | 47 | A. baumannii complex | Wound | Day 4 | yes | |||
| WEST-S6 | 49 | A. baumannii complex | Groin | Day 1 | no | |||
| WEST-S6 | 50 | A. baumannii complex | Wound | Day 1 | no | 31 (WEST-S1), 62 (HBG-S2), 64 (HBG-S4), 73 (HBG-S7) | 31 (WEST-S1)a,b, 56 (HBG-S1)b, 62 (HBG-S2)(b) 64 (HBG-S4)a,b, 73 (HBG-S7)a,b | 31 (WEST-S1), 62 (HBG-S2), 64 (HBG-S4), 73 (HBG-S7) |
| Methicillin-resistant Staphylococcus aureus (MRSA) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient I.D. | Sample I.D. | Species | Isolation site | Day of isolation | Nosocomial | In spa-typing identical with | spa type | MLST type (according to BURP algorithm) |
| HBG-S1 | 58 | Staphylococcus aureus (MRSA) | Deep wound at the left elbow | Day 2 | no | t274 | n.a. | |
| HBG-S5 | 67 | Staphylococcus aureus (MRSA) | Nasal vestibulum | Day 2 | no | 53 (WEST-S6) | t223 | ST-22 |
| WEST-S2 | 34 | Staphylococcus aureus (MRSA) | Groin | Day 23 | yes | t026 | ST-45, ST-47 | |
| WEST-S2 | 35 | Staphylococcus aureus (MRSA) | Wound | Day 18 | yes | 43 (WEST-S5) | t376 | n.a. |
| WEST-S5 | 43 | Staphylococcus aureus (MRSA) | Groin | Day 64 | yes | 35 (WEST-S2) | t376 | n.a. |
| WEST-S6 | 53 | Staphylococcus aureus (MRSA) | Skin | Day 1 | no | 67 (HBG-S5) | t223 | ST-22 |
a Completely identical based on MLST
(a) identical based on MLST but not all markers complete due to lack of coverage.
b Identical based on ANI (>= 99.9%)
(b) highly similar based on ANI (>= 99.5%, < 99.9%).
From one assessed environmental screening of the surroundings of the Libyan patients at the Bundeswehr hospital Hamburg at day 11 after admission, 5 Enterobacteriaceae (Klebsiella pneumoniae without exemption) and 3 nonfermentative rod-shaped bacteria (2 Pseudomonas aeruginosa, 1 P. putida) were isolated (Supplementary material 2).
Supplementary material 2.
Isolated Enterobacteriaceae with resistance against 3rd generation methoxyimino cephalosporins or carbapenems, and nonfermentative Gram-negative rod-shaped bacteria, from the environment of Libyan patients at the Bundeswehr Hospital of Hamburg.
| Isolates from the environment of Libyan patients at the Bundeswehr Hospital of Hamburg | ||||||||
|---|---|---|---|---|---|---|---|---|
| Enterobacteriaceae with resistance against 3rd generation methoxyimino cephalosporins or carbapenems | ||||||||
| Ward | Room number | Sample I.D. | Species | Isolation site | Day of isolation | In rep-PCR 95% identical with | In NGS-based MLSTa/ANIb identical with | NGS-based MLST type |
| HBG-2 | Room 4 | 20 | K. pneumoniae | hands | Day 11 | 18 (HBG-L4), 26 (room 4 HBG-2) | 8 (HBG-L2)a,b, 18 (HBG-L4)a,b, 26 (room 4 HBG-2)(a),(b) | 18 (HBG-L4), 26 (room 4 HBG-2)(a),(b) |
| HBG-2 | Room 3 | 21 | K. pneumoniae | hands | Day 11 | None | ||
| HBG-2 | Room 4 | 26 | K. pneumoniae | handle of a bag | Day 11 | 18 (HBG-L4), 20 (room 4 HBG-2) | 8 (HBG-L2)(a),b, 18 (HBG-L4)(a),b, 20 (room 4 HBG-2)(a),b | 18 (HBG-L4), 20 (room 4 HBG-2) |
| HBG-2 | Room 1 | 27 | K. pneumoniae | hands and beard | Day 11 | 16 (HBG-L3), 28 (room 3 HBG-2) | 16 (HBG-L3)a,b, 28 (room 3 HBG-2)a,b | 16 (HBG-L3), 28 (room 3 HBG-2) |
| HBG-2 | Room 3 | 28 | K. pneumoniae | walking frame | Day 11 | 16 (HBG-L3), 27 (room 1 HBG-2), | 16 (HBG-L3)a,b, 27 (room 1 HBG-2)a,b | 16 (HBG-L3), 27 (room 1 HBG-2), |
| Nonfermentative rod-shaped bacteria | ||||||||
|---|---|---|---|---|---|---|---|---|
| Ward | Room number | Sample I.D. | Species | Isolation site | Day of isolation | In rep-PCR 95% identical with | In NGS-based MLSTa/ANIb identical with | NGS-based MLST type |
| HBG-2 | Room 3 | 19 | P. aeruginosa | walker | Day 11 | 29 (room 3 HBG-2)b | ||
| HBG-2 | Room 3 | 29 | P. aeruginosa | hands | Day 11 | 19 (room 3 HBG-2)b | ||
| HBG-1-ITS | Room 1 | 30 | P. putida | sink (spout) | Day 11 | None | ||
a Completely identical based on MLST
(a) identical based on MLST but not all markers found/complete due to lack of coverage.
b Identical based on ANI (>= 99.9%)
(b) highly similar based ANI (>= 99.5%, < 99.9%).
From 14 Syrian patients, 31 Enterobacteriaceae (comprising 13 E. coli, 5 E. cloacae, 4 K. pneumoniae, 3 C. freundii, 2 Morganella morganii, 2 Proteus mirabilis, and 2 Raoultella planticola), 11 nonfermentative rod-shaped bacteria (8 A. baumannii complex, 1 A. radioresistens, 1 P. aeruginosa, and 1 S. maltophilia), and 6 MRSA were isolated. Clinically relevant isolates comprised 5 E. coli from a deep wound at the right gluteal region, 3 not further specified wounds, and a catheter urine sample; 3 E. cloacae from a not-further specified wound, a deep wound at the right heel, and a deep wound at the upper side of a fixator, respectively; 2 C. freundii, 1 K. pneumoniae, 2 Morganella morganii, 1 P. mirabilis, and 1 R. planticola from not further specified wounds; 4 A. baumannii complex from a deep wound at the right heel, a deep wound at the upper pin of the fixator at the thigh, and 2 not further specified wounds; 1 A. radioresistens, 1 P. aeruginosa, and 1 S. maltophilia each from not further specified wounds; and 2 MRSA from a deep wound at the left elbow and a not further specified wound. Isolates from other sites, including a K. pneumoniae isolate from a superficial wound, were considered as colonization flora (Supplementary material 1).
Resistance as Determined by Phenotypic Assessment and PCR. Among the Enterobacteriaceae from Libyan patients, an ESBL mechanism was phenotypcially identified in 12/13 instances (92.3%). When the PCR-based screening was added, the percentage of ESBL-positive strains increased to 100%. In three isolates (all of them E. cloacae), the ESBL mechanism was associated with ampC production. PCR-based screening showed blaTEM β-lactamases in 10/13 instances (76.9%), blaSHV β-lactamases in 6/13 instances (46.2%), blaCTX-M group I β-lactamases in 10/13 instances (76.9%), and blaCTX-M group IV β-lactamases in 1/13 instances (7.7%) (Supplementary material 7). PCR-based carbapenemase screening detected blaOxa-48 in 2 carbapenem-resistant Klebsiella pneumoniae strains (Supplementary material 8). Phenotypical resistance testing indicated high resistance rates for fluoroquinolones (10 out of 13), aminoglycosides (9 out of 13), and cotrimaxazole (9 out of 13) (Supplementary material 3) among Enterobacteriaceae. Focusing on the nonfermentative rod-shaped bacteria, the number of isolates was too small for any general conclusions to be drawn (Supplementary materials 4 and 5). While the 2 isolated A. baumannii complex tested sensitive for colistin only (Supplementary material 4), the 3 P. aeruginosa strains showed a differentiated resistance pattern without carbapenem-resistance (Supplementary material 5).
Supplementary material 7.
Detection of ESBL genes in Enterobacteriaceae that tested positive or not determined for ESBL by ABCD testing.
| blaTEM | blaSHV | blaCTX-M group I | blaCTX-M group II | blaCTX-M group III | blaCTX-M group IV | |
|---|---|---|---|---|---|---|
| Isolates from Libyan patients (n=13) | 10a (76.9%) | 6d (46.2%) | 10 (76.9%) | 1 (7.7%) | ||
| Isolates from the environment of Libyan patients at the Bundeswehr Hospital of Hamburg (n=5) | 5b (100%) | 4e (80%) | 5 (100%) | |||
| Isolates from Syrian patients (n=29) | 15c (51,7%) | 5f (17.2%) | 26 (89.7%) | 1 (3.4%) |
aDistribution of sequences: blaTEM-1 without exception.
bDistribution of sequences: blaTEM-1 without exception.
cDistribution of sequences: blaTEM-1 in 14 instances, not resolved due to different types with identical matching in 1 instance.
dDistribution of sequences: blaSHV-12, blaSHV-1/blaSHV-28 (no further sequence discrimination possible), and blaSHV-33 in one instance each, insufficient discriminatory power in additional 3 instances.
eDistribution of sequences: blaSHV-1/blaSHV-1a (no further sequence discrimination possible), blaSHV-11, and blaSHV-33 in one instance each, insufficient discriminatory power in another instance
fDistribution of sequences: blaSHV-1 in two instances, blaSHV-12/blaSHV-5 (no further sequence discrimination possible) and blaSHV-121/blaSHV-136 (no further sequence discrimination possible) in one instance each, insufficient discriminatory power in another instance.
Supplementary material 8.
Detection of carbapenemase genes in Enterobacteriaceae and nonfermentative, Gram-negative rod-shaped bacteria that did not test fully sensitive against carbapenems.
| blaNDM | blaKPC | blaBIC | blaOXA48 | blaAIM | blaGIM | blaSIM | blaDIM | blaIMP | blaVIM | blaSPM | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Enterobacteriaceae from Libyan patients (n=2) | 2x* | ||||||||||
| A. baumannii complex from Libyan patients (n=2) | |||||||||||
| Enterobacteriaceae from Syrian patients (n=2) | 1° | 1° | |||||||||
| A. baumannii complex from Syrian patients (n=8) |
*In 2 K. pneumoniae isolates.
°In the same E. cloacae isolate.
Supplementary material 3.
Resistance of isolated Enterobacteriaceae with resistance against 3rd generation methoxyimino cephalosporins (n=49) and/or carbapenems (n=6/49) as identified by VITEK-II.
| Enterobacteriaceae from Libyan patients at the Bundeswehr Hospitals of Hamburg and Westerstede (n=13) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Piperacillin / Tazobactam | Ceftazidime | Imipenem | Meropenem | Gentamicin | Ciprofloxacin | Levofloxacin | Tigecyclin | Fosfomycin | Nitrofurantoin | Cotrimoxazole | |
| R | 8 | 10 | 1 | 9 | 10 | 10 | 1 | 5 | 9 | ||
| I | 5 | 3 | 2 | 1 | 1 | 7 | 4 | ||||
| S | 0 | 11 | 11 | 4 | 2 | 3 | 5 | 13 | 4 | 4 | |
| Enterobacteriaceae from the environment of Libyan patients at the Bundeswehr Hospital of Hamburg (n=5) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Piperacillin / Tazobactam | Ceftazidime | Imipenem | Meropenem | Gentamicin | Ciprofloxacin | Levofloxacin | Tigecyclin | Fosfomycin | Nitrofurantoin | Cotrimoxazole | |
| R | 4 | 5 | 1 | 5 | 5 | 5 | 2 | 1 | 2 | 2 | |
| I | 1 | 1 | 2 | 2 | 2 | ||||||
| S | 0 | 3 | 3 | 1 | 4 | 1 | 3 | ||||
| Enterobacteriaceae from Syrian patients at the Bundeswehr Hospitals of Hamburg and Westerstede (n=31) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Piperacillin / Tazobactam | Ceftazidime | Imipenem | Meropenem | Gentamicin | Ciprofloxacin | Levofloxacin | Tigecyclin | Fosfomycin | Nitrofurantoin | Cotrimoxazole | |
| R | 9 | 25 | 2 | 2 | 16 | 18 | 17 | 4 | 7 | 7 | 18 |
| I | 21 | 6 | 6 | 5 | |||||||
| S | 1 | 29 | 29 | 15 | 7 | 14 | 22 | 24 | 24 | 13 | |
Supplementary material 4.
Resistance of isolated Acinetobacter baumannii complex as identified by VITEK-II.
| Acinetobacter baumannii complex from Libyan patients at the Bundeswehr Hospitals of Hamburg and Westerstede (n=2) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Piperacillin | Ceftazidime | Cefepime | Aztreonam | Imipenem | Meropenem | Gentamicin | Tobramycin | Ciprofloxacin | Colistin | Cotrimoxazole | |
| R | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | |
| I | 1 | 1 | |||||||||
| S | 2 | ||||||||||
| Acinetobacter baumannii complex from Syr an patients at the Bundeswehr Hospitals of Hamburg and Westerstede (n=8) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Piperacillin | Ceftazidime | Cefepime | Aztreonam | Imipenem | Meropenem | Gentamicin | Tobramycin | Ciprofloxacin | Colistin | Cotrimoxazole | |
| R | 8 | 8 | 8 | 8 | 6 | 6 | 7 | 7 | 8 | 0 | 5 |
| I | 2 | 2 | |||||||||
| S | 1 | 1 | 8 | 3 | |||||||
Supplementary material 5.
Resistance of isolated Pseudomonas aeruginosa as identified by VITEK-II.
| Pseudomonas aeruginosa from Libyan patients at the Bundeswehr Hospitals of Hamburg and Westerstede (n=3) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Piperacilli n | Ceftazidim e | Cefepim e | Aztreona m | Imipene m | Meropene m | Amikaci n | Gentamici n | Tobramyci n | Ciprofloxaci n | Colisti n | Cotrimoxazol e | |
| R | 3 | 3 | 2 | 1 | 2 | 2 | 2 | 2 | ||||
| I | 2 | 1 | ||||||||||
| S | 1 | 3 | 3 | 1 | 1 | 1 | 3 | 3 | ||||
| Pseudomonas aeruginosa from the environment of Libyan patients at the Bundeswehr Hospital of Hamburg (n=2) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Piperacilli n | Ceftazidim e | Cefepim e | Aztreona m | Imipene m | Meropene m | Amikaci n | Gentamici n | Tobramyci n | Ciprofloxaci n | Colisti n | Cotrimoxazol e | |
| R | ||||||||||||
| I | 2 | |||||||||||
| S | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Pseudomonas aeruginosa from Syrian patients at the Bundeswehr Hospitals of Hamburg and Westerstede (n=1) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Piperacilli n | Ceftazidim e | Cefepim e | Aztreona m | Imipene m | Meropene m | Amikaci n | Gentamici n | Tobramyci n | Ciprofloxaci n | Colisti n | Cotrimoxazol e | |
| R | 1 | |||||||||||
| I | 1 | |||||||||||
| S | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
Among the Enterobacteriaceae from Syrian patients, an ESBL mechanism was detected in 27 out of 31 strains (87.1%) by phenotypical approaches. In two instances (6.5%) (1 M. morganii and 1 E. coli), phenotypical screening allowed the detection of ampC mechanisms. In additional two instances (6.5%) (1 Proteus mirabilis, 1 E. cloacae), phenotypical screening for ESBL and ampC mechanisms led to nonconclusive results. In the carbapenem-resistant E. cloacae strain, PCR screening for carbapenemases indicated blaBIC and blaVIM (Supplementary material 8). Within the 27 ESBL-positive strains and the two strains with uncertain phenotypical screening results, PCR indicated blaTEM β-lactamases in 15/29 instances (51.7%), blaSHV β-lactamases in 5/29 instances (17.2%), blaCTX-M group I β-lactamases in 26/29 instances (89.7%), and blaCTX-M group IV β-lactamases in 1/29 instances (3.4%) (Supplementary material 7). In contrast to the Libyan Enterobacteriaceae, the Enterobacteriaceae from Syria showed an almost equal distribution of sensitive and resistant strains with respect to fluoroquinolones (18 out of 31 resistant), amino-glycosides (16 out of 31 resistant), and cotrimaxazole (18 out of 31 resistant) (Supplementary material 3). Of the 8 Acinetobacter baumannii complex strains from Syria, all showed sensitivity toward colistin, 3 toward cotrimoxazole, and 1 toward aminoglycosides (Supplementary material 4). The single P. aeruginosa strain assessed could not be compared with other isolates from the Libyan patients in any useful way (Supplementary material 5). The Syrian MRSA isolates showed broad sensitivity with only 2 out of 6 strains showing resistance to erythromycin and clindamycin and 3 out of 6 strains being resistant against fusidic acid (Supplementary material 6).
Supplementary material 6.
Resistance of isolated MRSA as identified by VITEK-II.
| MRSA isolates from Syria | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Levoflo xacin | Moxifl oxacin | Genta micin | Clinda mycin | Erythro mycin | Cotrimo xazole | Tetrac ycline | Tigecy cline | Teicop lanin | Vanco mycin | Linez olid | Dapto mycin | Fosfo mycin | Fusi dic acid | Rifam picin | Mupir ocin | Nitrofur antoin | |
| R | 2 | 2 | 1 | 3 | |||||||||||||
| I | |||||||||||||||||
| S | 6 | 6 | 6 | 4 | 4 | 5 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 3 | 6 | 6 | 6 |
Focusing on the 5 K. pneumoniae strains that were isolated from the environment of the patients from Libya at the Bundeswehr hospital Hamburg at day 11 after admission, phenotypical assessment indicated 80% ESBL and PCR-based assessment 100% ESBL. Detected β-lactamases comprised blaTEM and blaCTX-M group 1 in 5 instances (100%) each and blaSHV in 4 instances (80%) (Supplementary material 7). In all instances, resistance to aminoglycosides and fluoroquinolones was observed (Supplementary material 3). In contrast, resistance patterns without any hint for multidrug resistance were identified in the P. aeruginosa strains isolated from the patient environment (Supplementary material 5).
NGS Coverage. In total, 66 Gram-negative isolates were assessed by NGS, using Nextera XT libraries sequenced on the MiSeq desktop sequencing platform. Coverages varied widely, ranging from 13-fold to 206-fold. Due to variations in the amounts of data and the use of Nextera XT libraries, the assemblies using Newbler v.2.8 resulted in quite high numbers of contigs, ranging between 86 and 17,427 contigs larger than 100 bp. This problem was especially pronounced for assemblies with less than 30-fold input data. Thus, the genome assemblies for the samples 71 (HBG-S5) and 74 (HBG-S7) were not suitable for MLST analysis, while the assemblies for the samples 1 (HBG-L1), 4 (HBG-L1), 6 (HBG-L1), 14 (West-L3), 26 (HBG-L1), 57 (HBG-S1), 70 (HBG-S5), 72 (HBG-S6), and 75 (HBG-S1) yielded incomplete MLST patterns. Therefore, any presentation MLST types was skipped and another typing approach was chosen as follows. In order to classify all strains for which NGS data were obtained, an approach based on ANI using the method described by Goris et al. [35] was applied. Using a cutoff value of 99.9% allowed identification of potential clonal isolates even for extremely fragmented and incomplete datasets.
Nosocomial Transmission and Clonal Identity of Isolates. Based on a 3-day cutoff after admission to hospital, detection of nosocomial strains from Libyan patients had to be postulated for 5 Enterobacteriaceae (2 E. coli, 2 K. pneumonia, 1 E. cloacae) and in the case of 1 P. aeruginosa isolate, all from the Bundeswehr hospital Hamburg (Supplementary material 1).
Among the Syrian patients, 11 nosocomial Enterobacteriaceae strains (3 E. coli, 2 K. pneumoniae, 1 E. cloacae from the Bundeswehr hospital Hamburg; 1 E. cloacae, 1 E. coli, 1 K. pneumoniae, 1 M. morganii, 1 R. planticola from the Bundeswehr hospital Westerstede), 4 nosocomial nonfermentative rod-shaped bacteria (1 A. baumannii complex from the Bundeswehr hospital Hamburg; 2 A. baumannii complex and 1 S. maltophilia from the Bundeswehr hospital Westerstede), and 3 nosocomial MRSA (all from the Bundeswehr hospital Westerstede) were identified (Supplementary material 1).
The spa typing for MRSA suggested clonal identity of two nonnosocomial MRSA strains from Syrian patient 5 from the Bundeswehr hospital Hamburg (HBG-S5) and Syrian patient 6 from the Bundeswehr hospital Westerstede (WEST-S6) (both t223), and also of two nosocomial MRSA strains from Syrian patients 2 and 5 from the Bundeswehr hospital Westerstede (WEST-S2, WEST-S5) (both t376). For HBG-S5 and WEST-S6, hospital transmission is excluded. In the case of WEST-S2 and WEST-S5, the same spa type was identified on day 18 after admission from the wound of the patient WEST-S2 and on day 64 after admission from the wound of the patients WEST-S5 (Supplementary material 6). Nosocomial transmission from WEST-S2 to WEST-S5 is likely here. Although the patients were in the same room at the beginning of their hospital stay until day 9 after admission, they were in different rooms during the period of the likely transmission event (Table 1). In addition, 2 strains with the spa types t026 and t274 were isolated from Syrian patients (Supplementary material 6).
The Gram-negative isolates were assessed by rep-PCR and NGS (Supplementary materials 1 and 2) and associated with the patients and patient rooms (Table 1) to identify potential nosocomial transmission events. The clonal complexes identified were as follows.
Typing suggested complex interlinks between 6 E. cloacae isolates from 5 patients. In detail, these 6 strains comprised a nonnosocomial isolate from the skin and a nosocomial isolate from a wound of Syrian patient 6 at the Bundeswehr hospital Westerstede (WEST-S6), a nosocomial isolate from the perineal skin of Syrian patient 4 at the Bundeswehr hospital Hamburg (HBG-L4), a nosocomial isolate from the rectum of Libyan patient 1 at the Bundeswehr hospital Hamburg (HBG-L1), a nonnosocomial isolate from the inguinal skin of Libyan patient 2 at the Bundeswehr hospital Hamburg (HBG-L2), and a nonnosocomial isolate from an external fixator of the left thigh of Libyan patient 3 at the Bundeswehr hospital Westerstede (WEST-L3). Only the association between the isolates of HBG-L1 and WEST-L3 was confirmed by both typing approaches. A questionable confirmation of the rep-PCR-based association by NGS was also available for the nonnosocomial isolate from the skin of WEST-S6 and the isolate of HBG-S4. In both instances, hospital transmission was excluded because of the spatial distance. Additional possible associations that were based on rep-PCR results comprised only associations of the nonnosocomial isolate of WEST-S6 with all other isolates, of the nosocomial isolate of WEST-S6 with all other isolates with the exception of the isolate of HBG-S4, and the additional association of the isolate of HBG-L2 with the isolate of HBG-S4. Given the spatial and temporal distribution of the isolation events, the only plausible transmission in hospital was by auto-inoculation from the skin into the wound in case of the patient WEST-S6 (Figure 1).
Figure 1.
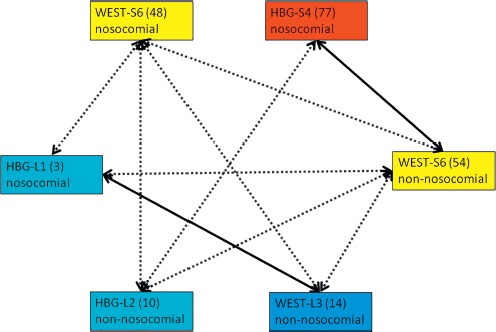
Clonal identities of E. cloacae strains. Light blue boxes = isolates from anonymously numbered Libyan (L) patients at the Bundeswehr Hospital Hamburg (HBG) with anonymous strain numbers in brackets. Dark blue box = isolate from an anonymously numbered Libyan (L) patient at the Bundeswehr Hospital Westerstede (WEST) with an anonymous strain number in brackets. Red box = isolate from an anonymously numbered Syrian (S) patient at the Bundeswehr Hospital Hamburg (HBG) with an anonymous strain number in brackets. Yellow boxes = isolates from anonymously numbered Syrian (S) patients at the Bundeswehr Hospital Westerstede (WEST) with anonymous strain numbers in brackets. Dotted arrows = clonally identical by rep-PCR but not by NGS typing. Continuous arrows = clonally identical by rep-PCR and by NGS typing
Regarding A. baumannii complex strains, there was a cluster of 4 strains from Syrian patients for which clonal identity was confirmed by rep-PCR and NGS. These strains comprised a nonnosocomial isolate from the perineal skin of Syrian patient 4 at the Bundeswehr hospital Hamburg (HBG-S4), a nosocomial isolate from the pharynx of Syrian patient 7 at the Bundeswehr hospital Hamburg (HBG-S7), a nosocomial isolate from the anus of Syrian patient 1 at the Bundeswehr hospital Westerstede (WEST-S1), and a nonnosocomial isolate from a wound of Syrian patient 6 at the Bundeswehr hospital Westerstede (WEST-S6). Two further strains were potentially part of this cluster. Clonal identity with a nonnosocomial isolate of a deep wound at the right heel of the Syrian patient 1 at the Bundeswehr hospital Hamburg (HBG-S1) was suggested by NGS but not by rep-PCR. In contrast, clonal identity with a nonnosocomial isolate from a deep wound at the upper pin of the fixator at the thigh of Syrian patient 2 at the Bundeswehr hospital Hamburg (HBG-S2) was suggested by rep-PCR but was clearly excluded by NGS as similar but not identical. Neither rep-PCR nor NGS suggested identity between these two latter strains. The origin of the nosocomial isolate that was isolated from the pharynx of HBG-S7 9 days after admission remains unresolved. From the Syrian patients at the Bundeswehr hospital Hamburg, only patient HBG-S1 was in the same patient room on the first day after admission. In contrast, the patients WEST-S1 and WEST-S6 were in the same room at the Bundeswehr hospital Westerstede, so nosocomial transmission from patient WEST-S6 to patient WEST-S1 is likely to explain the detection of the nosocomial isolate at the anus of patient WEST-S1 at day 21 after admission (Figure 2).
Figure 2.
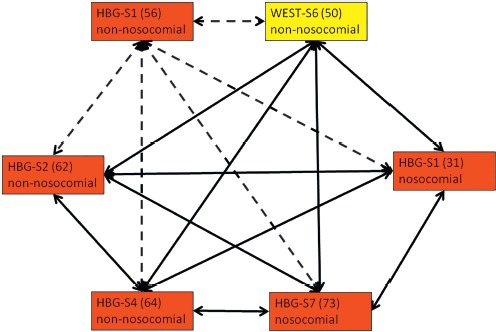
Clonal identities of A. baumannii strains. Red boxes = isolates from anonymously numbered Syrian (S) patients at the Bundeswehr Hospital Hamburg (HBG) with anonymous strain numbers in brackets. Yellow box = isolate from an anonymously numbered Syrian (S) patient at the Bundeswehr Hospital Westerstede (WEST) with an anonymous strain number in brackets. Dashed arrows = clonally identical by NGS typing but not by rep-PCR. Continuous arrows = clonally identical by rep-PCR and by NGS typing
Focusing on E. coli, the typing approaches suggested three confirmed or potential clonal clusters. Both rep-PCR and NGS confirmed clonal identity of 4 nonnosocomial strains isolated from the perineal skin of Syrian patients 2, 3, and 6 at the Bundeswehr hospital Hamburg (HBG-S2, HBG-S3, HBG-S6), and from a wound of Syrian patient 4 at the Bundeswehr hospital Westerstede (WEST-S4). Potential clonal identity with this cluster was further suggested by rep-PCR but again was clearly excluded by NGS as similar but not identical to a nonnosocomial isolate from a deep wound at the right gluteal region of Syrian patient 4 at the Bundeswehr hospital Hamburg (HBG-S4). The second potential cluster was suggested by rep-PCR but not by NGS and comprised a nonnosocomial isolate from a wound from Syrian patient 6 at the Bundeswehr hospital Westerstede (WEST-S6) and a nonnosocomial isolate from the perineal skin of the Syrian patient 5 at the Bundeswehr hospital Hamburg (HBG-S5). A high degree of similarity but questionable clonal identity was suggested by NGS but not by rep-PCR for a nosocomial strain isolated 22 days after admission from the colostomy of Libyan patient 1 at the Bundeswehr hospital Hamburg (HBG-L1) and for a nosocomial strain isolated 51 days after admission from a wound of Syrian patient 2 at the Bundeswehr hospital Westerstede (WEST-S2). The temporal and spatial distribution of the isolation events suggests that the sequence similarity occurred by chance (Figure 3).
Figure 3.
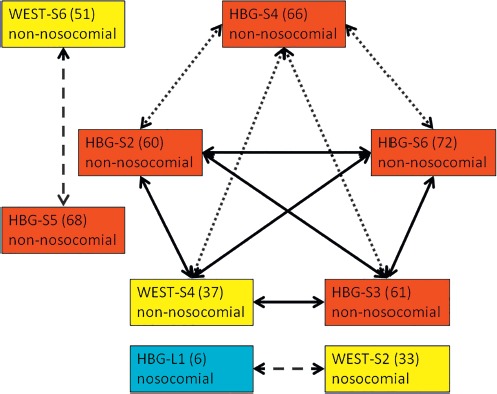
Clonal identities of E. coli strains. Red boxes = isolates from anonymously numbered Syrian (S) patients at the Bundeswehr Hospital Hamburg (HBG) with anonymous strain numbers in brackets. Light blue box = isolate from an anonymously numbered Libyan (L) patient at the Bundeswehr Hospital Hamburg (HBG) with an anonymous strain number in brackets. Yellow boxes = isolates from anonymously numbered Syrian (S) patients at the Bundeswehr Hospital Westerstede (WEST) with anonymous strain numbers in brackets. Dotted arrows = clonally identical by rep-PCR but not by NGS typing. Dashed arrows = clonally identical by NGS typing but not by rep-PCR. Continuous arrows = clonally identical by rep-PCR and by NGS typing
Focusing on K. pneumoniae, there were three distinct confirmed or potential clonal clusters. The first cluster was confirmed by rep-PCR and NGS and comprised three isolates, i.e., a nonnosocomial isolate from the inguinal skin of Libyan patient 3 at the Bundeswehr hospital Hamburg (HBG-L3) and two isolates from the environmental screening at day 11 after admission of the Libyan patients at the Bundeswehr hospital Hamburg from hand and beard of a patient in room 1 of the ward HGB-2 and from a walker in room 3 of the ward HBG-2. The patient HBG-L3 had been in room 1 during the first two days after admission, potentially leading to the introduction of the strain in this room during this period. At the time of the environmental screening, patient HBG-L3 had been in room 3 where the other environment strain was found. In the second cluster, clonal identity of a nosocomial strain isolated at day 16 from the inguinal skin of Libyan patient 4 at the Bundeswehr hospital Hamburg (HBG-L4) and a strain isolated during the environmental screening at day 11 after admission from the hands of this patient in room 4 of the ward HBG-2 suggests auto-inoculation. Potential clonal identity of these strains with a further environmental strain on the handle of a bag in room 4 of the ward HBG-2 that was isolated during the environmental screening as well was suggested by rep-PCR and confirmed by NGS. Rep-PCR but not NGS also suggested clonal identity of a nonnosocomial isolate from the inguinal skin of Libyan patient 2 at the Bundeswehr hospital Hamburg (HBG-L2) with the nosocomial isolates from the inguinal skin and the hands of the patient HBG-L4 but not with the isolate from the handle of the bag. The patients HBG-L2 and HBG-L4 had never occupied the same room together. However, nosocomial transmission via the hands of the medical staff is not excluded here. The third cluster was suggested by rep-PCR and confirmed by NGS and comprised a nonnosocomial isolate from the perineal skin of Syrian patient 2 at the Bundeswehr hospital Hamburg (HBG-S2) and a nosocomial strain isolated 32 days after admission from a superficial wound at the groin of Syrian patient 4 at the Bundeswehr hospital Hamburg (HBG-S4). Of note, this latter strain was collected during an out-patient control assessment even after discharge of patient HBG-S4. Nevertheless, the fact that the patients HBG-S2 and HBG-S4 stayed in the same room until the discharge of patient HBG-S4 makes nosocomial transmission likely (Figure 4).
Figure 4.
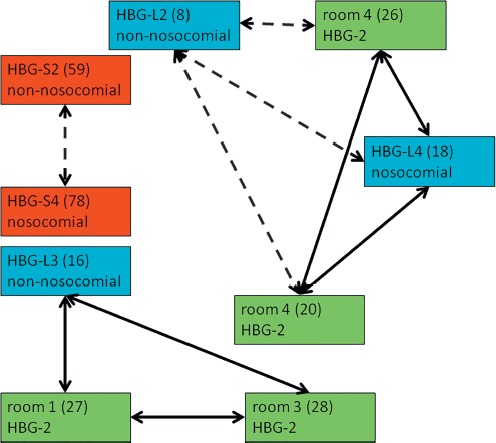
Clonal identities of K. pneumoniae strains. Light blue boxes = isolates from anonymously numbered Libyan (L) patients at the Bundeswehr Hospital Hamburg (HBG) with anonymous strain numbers in brackets. Red boxes = isolates from anonymously numbered Syrian (S) patients at the Bundeswehr Hospital Hamburg (HBG) with anonymous strain numbers in brackets. Green boxes = isolates from an environmental screening of the rooms of Libyan patients at the Bundeswehr Hospital Hamburg (HBG, anonymous ward 2) with anonymous strain numbers in brackets. Dashed arrows = clonally identical by NGS typing but not by rep-PCR. Continuous arrows = clonally identical by rep-PCR and by NGS typing
Regarding the less frequently isolated species, rep-PCR suggested clonal identity of two nonnosocomial C. freundii strains isolated from wounds of Syrian patients 5 and 7 at the Bundeswehr hospital Westerstede (WEST-S5, WEST-S7), which was clearly confirmed by NGS (Figure 5). Furthermore, the nosocomial R. planticola isolate from the anus of the Syrian patient 1 (WEST-S1) was clonally identical with the nonnosocomial isolate from a wound of the Syrian patient 6 (WEST-S6) in rep-PCR and highly similar in NGS-based typing (Supplementary material 1). Nosocomial transmission is likely, because both patients had been in the same room (Figure 6).
Figure 5.

Clonal identities of C. freundii strains. Yellow boxes = isolates from anonymously numbered Syrian (S) patients at the Bundeswehr Hospital Westerstede (WEST) with anonymous strain numbers in brackets. Continuous arrow = clonally identical by rep-PCR and by NGS typing
Figure 6.

Clonal identities of R. planticola strains. Yellow boxes = isolates from anonymously numbered Syrian (S) patients at the Bundeswehr Hospital Westerstede (WEST) with anonymous strain numbers in brackets. Continuous arrow = clonally identical by rep-PCR and by NGS typing
Furthermore, NGS but not rep-PCR suggested clonal identity between two P. aeruginosa isolates without unusual resistance patterns that were isolated during the environmental screening at the Bundeswehr hospital Hamburg 11 days after admission of the Libyan patients from a walking frame and the hands of a patient in room 3 of the ward HBG-2. No clonal identity with any other patient isolates was confirmed for these environmental strains. In addition, NGS but not rep-PCR indicated clonal identity between two P. aeruginosa isolates from an orthopedic wound at the left hand of a Libyan patient at the Bundeswehr hospital Hamburg (HBG-L2) and a wound at the dorsal side of the left thigh of a Libyan patient at the Bundeswehr hospital Westerstede (WEST-L1), both without hints of nosocomial transmission (Figure 7).
Figure 7.

Clonal identities of P. aeruginosa strains. Light blue box = isolate from an anonymously numbered Libyan (L) patient at the Bundeswehr Hospital Hamburg (HBG) with an anonymous strain number in brackets. Dark blue box = isolate from an anonymously numbered Libyan (L) patient at the Bundeswehr Hospital Westerstede (WEST) with an anonymous strain number in brackets. Green boxes = isolates from an environmental screening of the rooms of Libyan patients at the Bundeswehr Hospital Hamburg (HBG, anonymous ward 2) with anonymous strain numbers in brackets. Dashed arrows = clonally identical by NGS typing but not by rep-PCR
Of note, the environmental screening led to a detection of an ESBL-positive K. pneumoniae from the hands of a patient in room 3 of the ward HBG-2 that did not show any clonal identity with other isolates from the Libyan patients.
Rep-PCR-based typing patterns are shown in Supplementary materials 10–18.
Supplementary material 10.
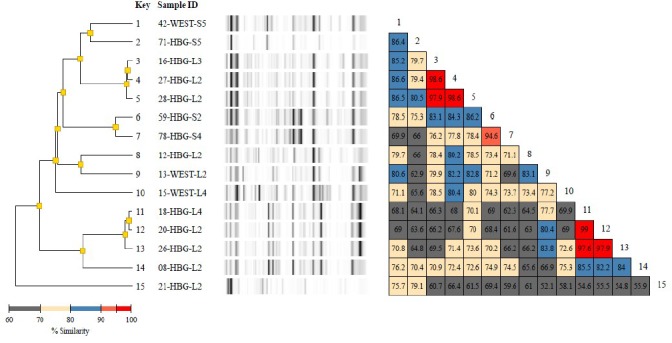
Diversilab typing report of 16 Escherichia coli isolates including a dendrogram, band patterns of the virtual gels, and a similarity matrix for ease of data interpretation. The dendrogram shows fingerprint similarities as a treelike structure. The virtual band patterns give a short overview of the experimental data. The similarity matrix provides percentages of similarity between every pair of samples. Data interpretation criteria—similarity matrix: lower than 95% means different, 95%–97% means similar, above 97% means indistinguishable; color code of % similarity: bright red: 95% to 100%, dull red: 90% to 95%, blue: 80% to 90%, yellow: 70% to 80%, grey: less than 70%.
Supplementary material 11.
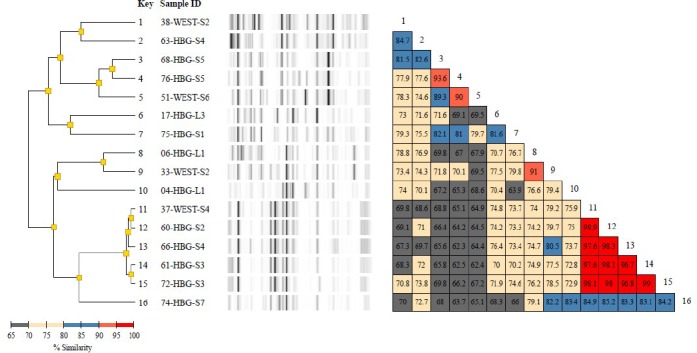
Diversilab typing report of 15 Klebsiella pneumoniae isolates including a dendrogram, band patterns of the virtual gels, and a similarity matrix for ease of data interpretation. The dendrogram shows fingerprint similarities as a treelike structure. The virtual band patterns give a short overview of the experimental data. The similarity matrix provides percentages of similarity between every pair of samples. Data interpretation criteria—similarity matrix: lower than 95% means different, 95%-97% means similar, above 97% means indistinguishable; color code of % similarity: bright red: 95% to 100%, dull red: 90% to 95%, blue: 80% to 90%, yellow: 70% to 80%, grey: less than 70%
Supplementary material 12.

Diversilab typing report of 8 Enterobacter cloacae isolates including a dendrogram, band patterns of the virtual gels, and a similarity matrix for ease of data interpretation. The dendrogram shows fingerprint similarities as a treelike structure. The virtual band patterns give a short overview of the experimental data. The similarity matrix provides percentages of similarity between every pair of samples. Data interpretation criteria—similarity matrix: lower than 95% means different, 95%-97% means similar, above 97% means indistinguishable; color code of % similarity: bright red: 95% to 100%, dull red: 90% to 95%, blue: 80% to 90%, yellow: 70% to 80%, grey: less than 70%.
Supplementary material 13.

Diversilab typing report of 4 Citrobacter freundii isolates including a dendrogram, band patterns of the virtual gels, and a similarity matrix for ease of data interpretation. The dendrogram shows fingerprint similarities as a treelike structure. The virtual band patterns give a short overview of the experimental data. The similarity matrix provides percentages of similarity between every pair of samples. Data interpretation criteria—similarity matrix: lower than 95% means different, 95%–97% means similar, above 97% means indistinguishable; color code of % similarity: bright red: 95% to 100%, dull red: 90% to 95%, blue: 80% to 90%, yellow: 70% to 80%, grey: less than 70%.
Supplementary material 14.

Diversilab typing report of 2 Morganella morganii isolates including a dendrogram, band patterns of the virtual gels, and a similarity matrix for ease of data interpretation. The dendrogram shows fingerprint similarities as a treelike structure. The virtual band patterns give a short overview of the experimental data. The similarity matrix provides percentages of similarity between every pair of samples. Data interpretation criteria—similarity matrix: lower than 95% means different, 95%–97% means similar, above 97% means indistinguishable; color code of % similarity: bright red: 95% to 100%, dull red: 90% to 95%, blue: 80% to 90%, yellow: 70% to 80%, grey: less than 70%
Supplementary material 15.

Diversilab typing report of 2 Proteus mirabilis isolates including a dendrogram, band patterns of the virtual gels, and a similarity matrix for ease of data interpretation. The dendrogram shows fingerprint similarities as a treelike structure. The virtual band patterns give a short overview of the experimental data. The similarity matrix provides percentages of similarity between every pair of samples. Data interpretation criteria—similarity matrix: lower than 95% means different, 95%–97% means similar, above 97% means indistinguishable; color code of % similarity: bright red: 95% to 100%, dull red: 90% to 95%, blue: 80% to 90%, yellow: 70% to 80%, grey: less than 70%
Supplementary material 16.

Diversilab typing report of 2 Raoultella planticola isolates including a dendrogram, band patterns of the virtual gels, and a similarity matrix for ease of data interpretation. The dendrogram shows fingerprint similarities as a treelike structure. The virtual band patterns give a short overview of the experimental data. The similarity matrix provides percentages of similarity between every pair of samples. Data interpretation criteria—similarity matrix: lower than 95% means different, 95%–97% means similar, above 97% means indistinguishable; color code of % similarity: bright red: 95% to 100%, dull red: 90% to 95%, blue: 80% to 90%, yellow: 70% to 80%, grey: less than 70%
Supplementary material 17.
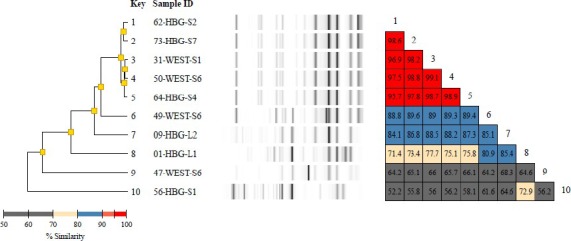
Diversilab typing report of 10 Acinetobacter baumanii isolates including a dendrogram, band patterns of the virtual gels, and a similarity matrix for easy data interpretation. The dendrogram shows fingerprint similarities as a treelike structure. The virtual band patterns give a short overview of the experimental data. The similarity matrix provides percentages of similarity between every pair of samples. Data interpretation criteria—similarity matrix: lower than 95% means different, 95%–97% means similar, above 97% means indistinguishable; color code of % similarity: bright red: 95% to 100%, dull red: 90% to 95%, blue: 80% to 90%, yellow: 70% to 80%, grey: less than 70%
Supplementary material 18.

Diversilab typing report of 6 Pseudomonas aeruginosa isolates including a dendrogram, band patterns of the virtual gels, and a similarity matrix for ease of data interpretation. The dendrogram shows fingerprint similarities as a treelike structure. The virtual band patterns give a short overview of the experimental data. The similarity matrix provides percentages of similarity between every pair of samples. Data interpretation criteria—similarity matrix: lower than 95% means different, 95%–97% means similar, above 97% means indistinguishable; color code of % similarity: bright red: 95% to 100%, dull red: 90% to 95%, blue: 80% to 90%, yellow: 70% to 80%, grey: less than 70%.
NGS-based Detection of Resistance Genes. In spite of the low NGS coverage, the NGS-based approach led to the detection of resistance genes in 23 of the 66 NGS-sequenced Gram-negative strains in the ResFinder assessment (Supplementary material 9). In comparison, no interpretative standards were deposited in the VITEK 2 system for the definition of phenotypic resistance or susceptibility of one assessed S. maltophilia strain and one A. radioresistens strain.
Supplementary material 9.
Resistance testing as determined by NGS with the ResFinder Software.
| ID and species | Phenotypic resistance by Vitek II (acronyms of antibiotics that were not tested sensitive) | Genotypic detection of resistance genes by NGS with low coverage* | Phenotypic resistance without detected genotypic correlate | Genotypic resistance without detected phenotypic correlate |
|---|---|---|---|---|
| 1 Acinetobacter baumannii | PRL, TZP, CTX, CAZ, FEP, ATM, IPM, MEM, GC, NN, CIP, COT | execution failed | ||
| 2 Citrobacter freundii | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, TGC | execution failed | ||
| 3 Enterobacter cloacae ssp. cloacae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, TGC, NF, COT | strA, aadB, aph(3’)-Ic, blaOXA-23, blaOXA-64, blaADC-25, sul2 | fluoroquinolone, tetracycline, trimethoprim | none |
| 4 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP, LEV, COT | execution failed | ||
| 5 Pseudomonas aeruginosa | PRL, TZP, CAZ, ATM, CIP, COT | execution failed | ||
| 6. Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, COT | aadA2, aadA1, blaSHV-12, blaTEM-135, sul3, sul2, dfrA12 | none | aminoglycoside |
| 7 Pseudomonas aeruginosa | PRL, TZP, CAZ, FEP, ATM, AK, GC, NN, CIP, COT | aph(3’)-XV, strA, aph(3’)-Ic, aacA4, strB, aadA6, aph(3’)-IIb, blaPAO, blaGES-1, blaOXA-50, aac(6’)lb-cr, fosA, catB7, sul1, tet(G) | none | fosfomycin (not tested phenotypically) |
| 8 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, NF | execution failed | ||
| 9 Acinetobacter baumannii | PRL, TZP, CTX, CAZ, FEP, ATM, IPM, MEM, GC, NN, CIP, COT | execution failed | ||
| 10 Enterobacter cloacae ssp. cloacae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, TGC, NF, COT | execution failed | ||
| 11 Pseudomonas aeruginosa | PRL, TZP, CAZ, FEP, ATM, AK, GC, NN, CIP, COT | execution failed | ||
| 12 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, IPM, MEM, GC, CIP, LEV, NF, COT | execution failed | ||
| 13 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, IPM, MEM, GC, CIP, LEV, TGC, NF | execution failed | ||
| 14 Enterobacter cloacae ssp. cloacae | AMP, SAM, TZP, CXM, CXMAX, CPD. CTX, CAZ, GC, CIP, LEV, TGC, NF, COT | execution failed | ||
| 15 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP, TGC, NF, COT | execution failed | ||
| 16 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, TGC, NF | execution failed | ||
| 17 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, COT | execution failed | ||
| 18 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CN, CIP, LEV, TGC, NF, COT | execution failed | ||
| 19 Pseudomonas aeruginosa | ATM, COT | execution failed | ||
| 20 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, IMP, MEM, GC, CIP, LEV, TGC, NF, COT | execution failed | ||
| 21 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV | no sequence data | ||
| 26 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, IMP, MEM, GC, CIP, LEV, TGC, FOS, NF, COT | execution failed | ||
| 27 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, TGC, NF | no sequence data | ||
| 28 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, TGC, NF | execution failed | ||
| 29 Pseudomonas aeruginosa | ATM, COT | aadB, aac(6’)lb-cr, aac(3)-lla, strA, strB, aadA2, blaTEM-1B, blaCMY-4, blaCTX-M-15, blaSHV-33, blaOXA-1, oqxA, aac(6’)lb-cr, oqxB, QnrA1, fosA, mph(E), msr(E), catB3, floR, sul1, sul2, tet(A) | trimethoprim | aminoglycoside, beta-lactam, fluoroquinolone, fosfomycin (not tested phenotypically) |
| 30 Pseudomonas putida | PRL, MEM, CIP, LEV, | execution failed | ||
| 31 Acinetobacter baumannii | PRL, TZP, CTX, CAZ, FEP, ATM, IPM, MEM, GC, NN, CIP, COT | execution failed | ||
| 32 Enterobacter cloacae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, FOS, NF, COT | execution failed | ||
| 33 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ | execution failed | ||
| 36 Morganella morganii ssp. morganii | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, TGC, FOS, NF, COT | aac(3)-lld, aadA5, blaDHA-1, mph(A), catA2, sul1, tet(B), dfrA17 | fosfomycin, fluoroquinolone, nitrofurantoin | none |
| 37 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV | execution failed | ||
| 38 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, COT | execution failed | ||
| 39 Proteus mirabilis | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, TGC, NF | execution failed | ||
| 40 Pseudomonas aeruginosa | PRL, PZP, ATM, COT | aph(3’)-llb, blaPAO, blaOXA-50, fosA, catB7 | sulfonamide, trimethoprim | aminoglycoside, fosfomycin |
| 41 Morganella morganii | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP, LEV, TGC, FOS, NF, COT | strA, aac(6’)lb-cr, aph(3’)-la, aadA5, strB, aadA1, blaTEM-1B, blaDHA-1, blaCTX-M-15, blaOXA-1, aac(6’)lb-cr, catB3, catA2, catA1, sul1, sul2, tet(B), dfrA17, dfrA1 | fosfomycin | aminoglycoside |
| 42 Klebsiella pneumoniae ssp. pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP, LEV, TGC, NF, COT | aac(6’)lb-cr, strB, strA, blaSHV-83, blaCTX-M-15, blaTEM-1B, blaOXA-1, aac(6’)lb-cr, oqxB, oqxA, QnrB66, fosA, catB3, sul2, dfrA14 | tetracycline | aminoglycoside, fosfomycin |
| 44 Acinetobacter radioresistens | no EUCAST breakpoints for VITEK available | blaOXA-133 | ||
| 45 Citrobacter freundii | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP, LEV, COT | execution failed | ||
| 46 Stenotrophomonas maltophila | no EUCAST breakpoints for VITEK available | sph, blaL1 | ||
| 47 Acinetobacter baumannii | PRL, CTX, CAZ, FEP, ATM, IPM, MEM, GC, NN, CIP, COT | aacA4, strB, strA, aadA2, aadB, aph(3’)-Vla, blaADC-25, blaGES-11, blaOXA-89, aac(6’)lb-cr, cmlA1, dfrA7 | sulphonamide | none |
| 48 Enterobacter cloacae ssp. cloacae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, FOS, COT | aac(6’)lb-cr, strA, aadA1, strB, blaTEM-1B, blaACT-16, blaCTX-M-15, blaOXA-1, aac(6’)lb-cr, QnrB1, fosA, catB3, catA1, sul2, tet(A), dfrA14 | none | tetracycline |
| 49 Acinetobacter baumannii | PRL, CTX, CAZ, FEP, ATM, IPM, MEM, GC, NN, CIP, COT | aph(3’)-Vla, aadB, blaADC-25, blaOXA-71, tet(A) | fluoroquinolone, sulphonamide, trimethoprim | tetracycline (not tested phenotypically) |
| 50 Acinetobacter baumannii | PRL, CTX, CAZ, FEP, ATM, IPM, MEM, GC, NN, CIP, COT | strB, aph(3’)-lc, strA, armA, blaADC-25, blaTEM-1D, blaOXA-23, blaOXA-66, msr(E), mph(E), tet(B) | sulphonamide, trimethoprim, fluoroquinolone | tetracycline (not tested phenotypically) |
| 51 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, COT | aac(3)-lld, strA, strB, blaTEM-1B, blaCTX-M-15, QnrS1, sul2, dfrA14 | ||
| 52 Raoultella planticola | AMP, SAM, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, FOS | aac(3)-lla, aac(6’)lb-cr, blaPLA1a, blaCTX-M-15, blaOXA-1, fosA, catB3, tet(B) | none | tetracycline |
| 54 Enterobacter cloacae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, COT | execution failed | ||
| 55 Citrobacter freundii | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP, LEV, COT | aac(6’)-lb-cr, aadA2, blaTEM, blaCTX-M-15, blaCMY-84, blaOXA-1, QnrS2, mph(A), catB3, ARR-3, sul1, dfrA12 | ||
| 56 Acinetobacter baumannii | PRL, CTX, CAZ, FEP, ATM, IPM, MEM, CIP | aph(3’)-lc, strA, strB, blaOXA-23, blaOXA-66, blaADC-25, blaTEM-1D, tet(B) | fluoroquinolone | aminoglycoside, tetracycline (not tested phenotypically) |
| 57 Enterobacter cloacae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, TGC, FOS, NF, COT | execution failed | ||
| 59 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, TGC, NF | no resistance genes found | ||
| 60 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP, LEV, COT | no resistance genes found | ||
| 61 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP, LEV, COT | aadA5, strB, strA, blaCTX-M-15, blaTEM-1B, mph(A), sul1, sul2, tet(A), dfrA17 | none | aminoglycoside, tetracycline |
| 62 Acinetobacter baumannii | PRL, CTX, CAZ, FEP, ATM, IPM, MEM, GC, NN, CIP, COT | aacA4, aadA2, aadB, strB, aph(3’)-Vla, strA, blaOXA-23, blaGES-11, blaOXA-66, blaOXA-117, blaTEM-1D, aac(6’)lb-cr, cmlA1, sul1, sul2, dfrA7 | ||
| 63 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP | no resistance genes found | ||
| 64 Acinetobacter baumannii | PRL, CTX, CAZ, FEP, ATM, IPM, MEM, GC, NN, CIP | execution failed | ||
| 65 Citrobacter freundii | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ | qnrS1 | beta-lactam | fluoroquinolone |
| 66 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC | aac(3’)-lla, aadA1, aadB, blaCTX-M-15, mph(A), cmlA1, tet(B), dfrA17 | none | tetracycline |
| 68 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP, LEV | strA, strB, blaTEM-1B | fluoroquinolone | aminoglycoside |
| 69 Proteus mirabilis | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, TGC, NF, COT | execution failed | ||
| 70 Enterobacter cloacae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, IPM, MEM, CIP, LEV, TGC, FOS, COT | no resistance genes found | ||
| 71 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, CIP, LEV, NF | execution failed | ||
| 72 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, COT | execution failed | ||
| 73 Acinetobacter baumannii | PRL, CTX, CAZ, FEP, ATM, IPM, MEM, GC, NN, CIP | execution failed | ||
| 74 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ | execution failed | ||
| 75 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, COT | execution failed | ||
| 76 Escherichia coli | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV | execution failed | ||
| 77 Enterobacter cloacae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, NF, COT | execution failed | ||
| 78 Klebsiella pneumoniae | AMP, SAM, TZP, CXM, CXMAX, CPD, CTX, CAZ, GC, CIP, LEV, TGC, NF | execution failed |
Matching between detected genotypic and phenotypic resistance was observed in 3 instances. In 15 instances, genotypic resistance determinants were identified without a detected or assessed phenotypic correlate. In 12 instances, the genotypic correlate for a phenotypically identified resistance was not found using the approach.
Details are provided in Supplementary material 9.
Discussion
The study assessed the molecular epidemiology of multidrug-resistant bacteria isolated from war-injured Libyan and Syrian patients who were treated at two Bundeswehr hospitals in Germany. With multidrug-resistant bacteria being rapidly on the rise in Europe [40], patients with history of hospitalization abroad provide potential sources of influx of resistant bacteria, as also suggested by others [41]. As expected from previous results with war-injured patients from the Ukraine [1], high rates of colonization and infection with such strains were observed, with 66 clinical and screening isolates being obtained from 22 patients and 8 suspicious isolates being obtained from an environmental screening during the Libyan patients' hospital stay at the Bundeswehr hospital Hamburg. With 29 out of 66 strains having potential etio-logical relevance, colonizing bacteria dominated quantitatively.
From 24 isolates that were formally considered as nosocomially transmitted from consideration of the time of isolation, rep-PCR and NGS-based typing suggested potential transmission in the hospitals in only 7 instances, comprising 3 K. pneumoniae, 2 A. baumannii complex, 1 MRSA, and 1 R. planticola. Of these likely nosocomial transmission events, only 2 isolates (i.e., one MRSA and one K. pneumoniae, from wounds of the respective patients) were considered as potentially etiologically relevant, while the other transmissions led to harmless colonization of the mouth of a patient with A. baumannii complex and the anus of two patients with A. baumannii complex and R. planticola, respectively, or identification of K. pneumoniae during the environmental screening from the hands and beard of two Libyan patients. In addition, the definitions when potential nosocomial transmission was considered are very conservative and thus even likely to overestimate the true transmission rate. As shown by others [20], many formally nosocomial isolates are simply missed by initial patient screening, suggesting insufficiency of single screening sets for a reliable inventory of a patients' colonization status with resistant bacteria.
These results suggest that the hygiene procedures enforced were basically effective and that healthcare-associated infections due to multidrug-resistant pathogens, which impose a considerable burden on the European healthcare systems [42], were widely prevented. Decolonizing washing procedures which were applied during the care for the patients were shown to reduce nosocomial transmission [43, 44] and even to be effective in preventing colonization-derived bloodstream infections [45]. In contrast, purely screening-based prevention is usually too late and thus lacks efficacy [46, 47].
Obtaining both clinical and screening isolates, as was done in this assessment, was nevertheless useful for performing typing analyses in order to identify transmission chains. Real-time NGS-based typing was recently demonstrated to be cost-efficient by providing a precise infection control instrument for hospital hygiene approaches [48]. High reproducibility of NGS-based typing in ring trials recently confirmed the suitability of the approaches for the diagnostic routine laboratory [49].
In this study, the retrospective typing approaches that were applied provided interesting insights into the epidemiology of the isolates. Although the suggested transmission events in the Bundeswehr hospital were few, several clonal clusters comprised nonnosocomial isolates that were isolated both at the Bundeswehr hospital Westerstede and at the Bundeswehr hospital Hamburg. A similar phenomenon had already been described for carbapenem-resistant A. baumannii complex isolates that were derived from Ukrainian war-injured patients who were treated in 4 Bundeswehr hospitals [1]. Two explanations are plausible: The underlying nosocomial transmission events may have occurred within the medical infrastructure in the home countries of the patients or during the medical evacuation flights. The latter option should be systematically assessed in future studies.
Molecular resistance testing was performed to assess the resistance mechanisms of the isolates. The PCR-based analyses for ESBL enzymes showed a similar distribution pattern as recently observed for European soldiers on deployment in West African Mali [38] and for German soldiers on various subtropical and tropical deployments [39] with a dominance of blaTEM and blaCTX-M group I. Among the carbapenemases assessed, blaOxa-48 was repeatedly detected, as also described for other European countries [40]. Of note, blaOxa-48-positive K. pneumoniae strains have been described by several study groups for Libyan patients [15, 17, 19, 21], suggesting a frequent regional occurrence.
In spite of a low coverage, NGS-based screening for molecular resistance mechanisms was attempted [36, 37]. Note that at least a 30-fold coverage is recommended by EUCAST (European Committee on Antimicrobial Susceptibility Testing) if whole-genome sequencing is used for molecular resistance testing at strain level [50]. This coverage was missed by a wide margin in this assessment. However, the approach at least allowed a first insight into underlying molecular mechanisms of phenotypically observed resistance, although the ResFinder-based assessment of several strains failed due to poor data quality and no genotypic correlate was identified for a considerable proportion of phenotypically identified resistance. In fact some resistance genes were identified for which no phenotypical correlate had been observed. Although the approach confirms that adherence to the EUCAST recommendations is advisable [50], NGS-based resistance detection despite low coverage has been attempted – partially successfully – in various resistome approaches using samples from blood [51, 52], urine [53], stool [54], and even from sewers [55], toilets [56], and environmental samples [57]. However, as shown for the stool samples [54], high proportions of resistant bacteria are necessary for a reliable identification of resistance genes using such procedures.
In summary, the data confirm a considerable influx of multidrug-resistant bacteria from international war and crisis zones [6-28, 58] with the potential of nosocomial transmission and further spreading. Intermediate-term suppression of resistant strains due to fitness costs because of the expression of resistance determinants cannot be reliably expected, because several strains with low or even no reduced fitness have been described [59]. Although the development of new antimicrobial drugs continues [60-62], such procedures require considerable time investment. Accordingly, thorough prevention of the spread of multidrug-resistant strains in the hospital environment remains a major hygiene issue.
Future studies should address whether hygiene conditions on medical evacuation flights require optimization in order to prevent nosocomial transmission during airborne evacuation procedures. The available data cannot answer this question because no screening analyses were performed prior to the evacuation flights, which is an undeniable limitation of this study.
Conclusions
Considerable influx of multidrug-resistant bacteria with patients from international war and crisis zones has to be expected. However, uncontrolled nosocomial transmission in the hospital setting can be widely prevented if strict hygiene precautions are enforced. Likely settings of nosocomial transmissions prior to hospital admission, e.g., during evacuation flights or in medical camps in war and crisis zones, should be further addressed in future studies.
Acknowledgments
The authors are grateful to Annett Michel, Simone Priesnitz, and Steffen Lohr for excellent technical assistance.
Footnotes
Funding sources
The molecular typing was funded by grant 12K2-S-451315 “Pathogen typing on deployment” (German original: “Erregertypisierung im Einsatz”) of the German Ministry of Defense (MoD).
Authors' contributions
HF, RMH, AP, and BK planned the study. TK, MM, WW, RG, US, AB, and PW participated in performing the microbiological analyses. KPE assessed and provided epidemiological and hygiene-related data. HF, TK, RMH, RS, and CR performed the assessment and evaluation of the data. All authors jointly wrote the paper, performed the revision, had full access to all data in the study, and take responsibility for the integrity of the data and the accuracy of the data analysis.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Granzer H, Hagen RM, Warnke P, Bock W, Baumann T, Schwarz NG, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii complex isolates from patients that were injured during the eastern Ukrainian conflict. Eur J Microbiol Immunol (Bp). 2016;6:109-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peretz A, Labay K, Zonis Z, Glikman D. Disengagement does not apply to bacteria: a high carriage rate of antibiotic-resistant pathogens among Syrian civilians treated in Israeli hospitals. Clin Infect Dis. 2014;59:753–4. [DOI] [PubMed] [Google Scholar]
- 3.Heudorf U, Albert-Braun S, Hunfeld KP, Birne FU, Schulze J, Strobel K, et al. Multidrug-resistant organisms in refugees: prevalences and impact on infection control in hospitals. GMS Hyg Infect Control. 2016;11: Doc16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinheimer C, Kempf VA, Göttig S, Hogardt M, Wichelhaus TA, O'Rourke F, et al. Multidrug-resistant organisms detected in refugee patients admitted to a University Hospital, Germany June-December 2015. Euro Surveill. 2016;21:2. [DOI] [PubMed] [Google Scholar]
- 5.Reinheimer C, Kempf VA, Jozsa K, Wichelhaus TA, Hogardt M, O'Rourke F, et al. Prevalence of multidrug-resistant organisms in refugee patients, medical tourists and domestic patients admitted to a German university hospital. BMC Infect Dis. 2017;17:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Assil B, Mahfoud M, Hamzeh AR. First report on class 1 integrons and Trimethoprim-resistance genes from dfrA group in uropathogenic E. coli (UPEC) from the Aleppo area in Syria. Mob Genet Elements. 2013;3:e25204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Assil B, Mahfoud M, Hamzeh AR. Resistance trends and risk factors of extended spectrum β-lactamases in Escherichia coli infections in Aleppo Syria. Am J Infect Control. 2013;41:597-600. [DOI] [PubMed] [Google Scholar]
- 8.Teicher CL, Ronat JB, Fakhri RM, Basel M, Labar AS, Herard P, Murphy RA. Antimicrobial drug-resistant bacteria isolated from Syrian war-injured patients, August 2011-March 2013. Emerg Infect Dis. 2014;20:1949-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doganay M, Demiraslan H. Refugees of the Syrian civil war: impact on reemerging infections, health services, and biosecurity in Turkey. Health Secur. 2016;14:220-5. [DOI] [PubMed] [Google Scholar]
- 10.Sahli ZT, Bizri AR, Abu-Sittah GS. Microbiology and risk factors associated with war-related wound infections in the Middle East. Epidemiol Infect. 2016;144:2848-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbara A, Al-Harbat N, Karah N, Abo-Yahya B, El-Amin W, Hatcher J, et al. Antimicrobial drug resistance among refugees from Syria. Jordan. Emerg Infect Dis. 2017;23:885-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rafei R, Dabboussi F, Hamze M, Eveillard M, Lemarié C, Mallat H, et al. First report of blaNDM-1-producing Acinetobacter baumannii isolated in Lebanon from civilians wounded during the Syrian war. Int J Infect Dis. 2014;21:21-3. [DOI] [PubMed] [Google Scholar]
- 13.Kassem DF, Hoffmann Y, Shahar N, Ocampo S, Salomon L, Zonis Z, et al. Multidrug-resistant pathogens in hospitalized Syrian children. Emerg Infect Dis. 2017;23:166-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piso RJ, Käch R, Pop R, Zillig D, Schibli U, Bassetti S, et al. A cross-sectional study of colonization rates with methicillin-resistant Staphylococcus aureus (MRSA) and extended-spectrum beta-lactamase (ESBL) and carbapenemase-producing Enterobacteriaceae in four Swiss refugee centres. PLoS One. 2017;12:e0170251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pirš M, Andlovic A, Cerar T, Žohar-Čretnik T, Kobola L, Kolman J, et al. A case of OXA-48 carbapenemase-producing Klebsiella pneumoniae in a patient transferred to Slovenia from Libya, November 2011. Euro Surveill. 2011;16:20042. [PubMed] [Google Scholar]
- 16.Franka EA, Shembesh MK, Zaied AA, El-Turki E, Zorgani A, Elahmer OR, et al. Multidrug resistant bacteria in wounds of combatants of the Libyan uprising. J Infect. 2012;65:279-81. [DOI] [PubMed] [Google Scholar]
- 17.Hammerum AM, Larsen AR, Hansen F, Justesen US, Friis-Møller A, Lemming LE, et al. Patients transferred from Libya to Denmark carried OXA-48-producing Klebsiella pneumoniae, NDM-1-producing Acinetobacter baumannii and meticillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2012;40:191-2. [DOI] [PubMed] [Google Scholar]
- 18.Ghenghesh KS, Rahouma A, Tawil K, Zorgani A, Franka E. Antimicrobial resistance in Libya: 1970-2011. Libyan J Med. 2013;8:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kocsis E, Savio C, Piccoli M, Cornaglia G, Mazzariol A. Klebsiella pneumoniae harbouring OXA-48 carbapenemase in a Libyan refugee in Italy. Clin Microbiol Infect. 2013;19:E409-11. [DOI] [PubMed] [Google Scholar]
- 20.Koole K, Ellerbroek PM, Lagendijk R, Leenen LP, Ekkelenkamp MB. Colonization of Libyan civil war casualties with multidrug-resistant bacteria. Clin Microbiol Infect. 2013;19:E285-7. [DOI] [PubMed] [Google Scholar]
- 21.Lafeuille E, Decré D, Mahjoub-Messai F, Bidet P, Arlet G, Bingen E. OXA-48 carbapenemase-producing Klebsiella pneumoniae isolated from Libyan patients. Microb Drug Resist. 2013;19:491-7. [DOI] [PubMed] [Google Scholar]
- 22.Zorgani A, Ziglam H. Injured Libyan combatant patients: both vectors and victims of multiresistance bacteria? Libyan J Med 2013;8 DOI: 10.3402/ljm.v8i0.20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djahmi N, Dunyach-Remy C, Pantel A, Dekhil M, Sotto A, Lavigne JP. Epidemiology of carbapenemase-producing Enterobacteriaceae and Acinetobacter baumannii in Mediterranean countries. Biomed Res Int. 2014;2014:305784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seiffert SN, Perreten V, Johannes S, Droz S, Bodmer T, Endimiani A. OXA-48 carbapenemase-producing Salmonella enterica serovar Kentucky isolate of sequence type 198 in a patient transferred from Libya to Switzerland. Antimicrob Agents Chemother. 2014;58:2446–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leistner R, Denkel LA, Gastmeier P, Werner G, Layer F, Pfeifer Y. Prevalence of MRSA and Gram-negative bacteria with ESBLs and carbapenemases in patients from Northern Africa at a German hospital. J Antimicrob Chemother. 2015;70:3161–4. [DOI] [PubMed] [Google Scholar]
- 26.Manenzhe RI, Zar HJ, Nicol MP, Kaba M. The spread of carbapenemase-producing bacteria in Africa: a systematic review. J Antimicrob Chemother. 2015;70:23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathlouthi N, Al-Bayssari C, El Salabi A, Bakour S, Ben Gwierif S, Zorgani AA, et al. Carbapenemases and extended-spectrum β-lactamases producing Enterobacteriaceae isolated from Tunisian and Libyan hospitals. J Infect Dev Ctries. 2016;10:718–27. [DOI] [PubMed] [Google Scholar]
- 28.Mathlouthi N, El Salabi AA, Ben Jomàa-Jemili M, Bakour S, Al-Bayssari C, Zorgani AA, et al. Early detection of metallo-β-lactamase NDM-1- and OXA-23 carbapenemase-producing Acinetobacter baumannii in Libyan hospitals. Int J Antimicrob Agents. 2016;48:46–50. [DOI] [PubMed] [Google Scholar]
- 29.Klein B, Becker D, Urbach M, Mügge G. Versorgung libyscher Kriegsverletzter.am Bundeswehrkrankenhaus Hamburg unter spezieller Berücksichtigung der Hygienemaßnahmen. Wehrmed & Wehrpharm. 2013;1:52–4. [Google Scholar]
- 30.Ingram PR, Inglis TJ, Vanzetti TR, Henderson BA, Harnett GB, Murray RJ. Comparison of methods for AmpC betalactamase detection in Enterobacteriaceae. J Med Microbiol. 2011;60:715–21. [DOI] [PubMed] [Google Scholar]
- 31.Micheel V, Hogan B, Köller T, Warnke P, Crusius S, Hinz R, et al. Screening agars for MRSA: evaluation of a stepwise diagnostic approach with two different selective agars for the screening for methicillin-resistant Staphylococcus aureus (MRSA). Mil Med Res. 2015;2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hogan B, Rakotozandrindrainy R, Al-Emran H, Dekker D, Hahn A, Jaeger A, et al. Prevalence of nasal colonisation by methicillin-sensitive and methicillin-resistant Staphylococcus aureus among healthcare workers and students in Madagascar. BMC Infect Dis. 2016;16:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellmann A, Weniger T, Berssenbrügge C, Rothgänger J, Sammeth M, Stoye J, et al. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol. 2007;7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. [DOI] [PubMed] [Google Scholar]
- 36.Kleinheinz KA, Joensen KG, Larsen MV. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage. 2014;4:e27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zankari E. Comparison of the web tools ARG-ANNOT and ResFinder for detection of resistance genes in bacteria. Antimicrob Agents Chemother. 2014;58:4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagen RM, Hinz R. Frickmann H: β-Lactamases encoded by blaCTX-M group i genes as determinants of resistance of ESBL-positive Enterobacteriaceae in European soldiers in tropical Mali. Eur J Microbiol Immunol (Bp). 2015;5:281–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frickmann H, Wiemer D, Frey C, Hagen RM, Hinz R, Podbielski A, et al. Low enteric colonization with multidrug-resistant pathogens in soldiers returning from deployments — experience from the Years 2007–2015. PLoS One. 2016;11:e0162129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cantón R, Akóva M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, et al. European Network on Carbapenemases: Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18:413–31. [DOI] [PubMed] [Google Scholar]
- 41.Khawaja T, Kirveskari J, Johansson S, Väisänen J, Djupsjöbacka A, Nevalainen A, et al. Patients hospitalized abroad as importers of multiresistant bacteria — a cross-sectional study. Clin Microbiol Infect. 2017. DOI: 10.1016/j.cmi.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Cassini A, Plachouras D, Eckmanns T, Abu Sin M, Blank HP, Ducomble T, et al. Burden of six healthcare-associated infections on European population health: estimating incidence-based disability-adjusted life years through a population prevalence-based modelling study. PLoS Med. 2016;13: e1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Climo MW, Yokoe DS, Warren DK, Perl TM, Bolon M, Herwaldt LA, et al. Effect of daily chlorhexidine bathing on hospital-acquired infection. N Engl J Med. 2013;368:533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boonyasiri A, Thaisiam P, Permpikul C, Judaeng T, Suiwongsa B, Apiradeewajeset N, et al. Effectiveness of chlorhexidine wipes for the prevention of multidrug-resistant bacterial colonization and hospital-acquired infections in intensive care unit patients: a randomized trial in Thailand. Infect Control Hosp Epidemiol. 2016;37:245–53. [DOI] [PubMed] [Google Scholar]
- 45.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, et al. CDC Prevention Epicenters Program; AHRQ DECIDE Network and Healthcare-Associated Infections Program: Targeted versus universal decolonization to prevent ICU infection. N Engl J Med. 2013;368:2255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huskins WC, Huckabee CM, O'Grady NP, Murray P, Kopetskie H, Zimmer L, et al. STAR*ICU Trial Investigators: Intervention to reduce transmission of resistant bacteria in intensive care. N Engl J Med. 2011;364:1407–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derde LP, Cooper BS, Goossens H, Malhotra-Kumar S, Willems RJ, Gniadkowski M, et al. MOSAR WP3 Study Team: Interventions to reduce colonisation and transmission of antimicrobial-resistant bacteria in intensive care units: an interrupted time series study and cluster randomized trial. Lancet Infect Dis 2014;14:31–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mellmann A, Bletz S, Böking T, Kipp F, Becker K, Schultes A, et al. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol 2016;54:2874–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mellmann A, Andersen PS, Bletz S, Friedrich AW, Kohl TA, Lilje B, et al. High interlaboratory reproducibility and accuracy of next-generation-sequencing-based bacterial genotyping in a ring trial. J Clin Microbiol. 2017;55:908–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect. 2017;23:2–22. [DOI] [PubMed] [Google Scholar]
- 51.Grumaz S, Stevens P, Grumaz C, Decker SO, Weigand MA, Hofer S, et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gyarmati P, Kjellander C, Aust C, Song Y, Öhrmalm L, Giske CG. Metagenomic analysis of bloodstream infections in patients with acute leukemia and therapy-induced neutropenia. Sci Rep. 2016;6:23532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt K, Mwaigwisya S, Crossman LC, Doumith M, Munroe D, Pires C, et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother. 2017;72:104–14. [DOI] [PubMed] [Google Scholar]
- 54.Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, et al. The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrob Agents Chemother. 2015;59, 6551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pehrsson EC, Tsukayama P, Patel S, Mejía-Bautista M, Sosa-Soto G, Navarrete KM, et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533:212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nordahl Petersen T, Rasmussen S, Hasman H, Carøe C, Bælum J, Schultz AC, et al. Meta-genomic analysis of toilet waste from long distance flights; a step towards global surveillance of infectious diseases and antimicrobial resistance. Sci Rep. 2015;5:11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DG. The structure and diversity of human, animal and environmental resistomes. Microbiome. 2016;4:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lohr B, Pfeifer Y, Heudorf U, Rangger C, Norris DE, Hunfeld KP. High prevalence of multidrug-resistant bacteria in Libyan war casualties admitted to a tertiary care hospital, Germany. Microb Drug Resist. 2017;2017 DOI: 10.1089/mdr.2017.0141. [DOI] [PubMed] [Google Scholar]
- 59.Schaufler K, Semmler T, Pickard DJ, de Toro M, de la Cruz F, Wieler LH, et al. Carriage of extended-spectrum beta-lactamase-plasmids does not reduce fitness but enhances virulence in some strains of pandemic E. coli lineages. Front Microbiol. 2016;7:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernebro J. Fighting bacterial infections-future treatment options. Drug Resist Updat. 2011;14:125–39. [DOI] [PubMed] [Google Scholar]
- 61.Gould IM, Bal AM. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence. 2013;4:185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kern WV. New antibacterial agents on the market and in the pipeline. Internist (Berl). 2015;56, 1255–63. [DOI] [PubMed] [Google Scholar]


