Abstract
The use of vancomycin for treatment of serious infections caused by MRSA strains has resulted in emergence of vancomycin-resistant Staphylococcus aureus (VRSA) in clinical settings. Following our previous report of phenotypic VRSA in Nigeria, the current study attempts to determine the genetic basis underlying this resistance. Over a period of 6 months, non-duplicate clinical S. aureus isolates from 73 consecutive patients with infective conditions at Ladoke Akintola University of Technology Teaching Hospital, Osogbo were tested against a panel of eight selected antibiotics by disk diffusion test. The Epsilom test strip was used to determine vancomycin minimum inhibitory concentration (MIC) and polymerase chain reaction (PCR) assay to amplify nuc, mecA, vanA, and vanB genes. Of 73 isolates, 61 (83.6%) had MIC of ≤2 μg/ml, 11 (15.1%) had 4–8 μg/ml and 1 (1.4%) had 16 μg/ml. The mecA gene was detected in 5 (6.8%) isolates but none contained vanA or vanB genes. Both vancomycin-susceptible and intermediate isolates were resistant to multiple antibiotics, while the only vancomycin resistant isolate was resistant to all eight antibiotics. The result confirms the occurrence of phenotypic vancomycin intermediate-resistant S. aureus (VISA) and VRSA infections in Nigeria, but the molecular basis will require further investigation.
Keywords: VISA, VRSA, MRSA, phenotypic, molecular, E-test
Introduction
Staphylococcus aureus comprises Gram-positive aerobic or facultatively anaerobic cocci in clusters, occurring as normal flora of the skin, axilla, and anterior nares of man and animals. However, S. aureus is implicated in diseases of the skin and soft tissues, pneumonia, bloodstream infections, osteomyelitis, and endocarditis, as well as toxin-mediated syndromes such as toxic shock and food poisoning [1, 2]. The organism has developed resistance to a wide range of antimicrobial drugs, which complicates the treatment of these infections. S. aureus is also a common cause of hospital and community acquired infections. Mortality associated with severe S. aureus infections in the developing world far exceeds that in developed countries [3]. Recent studies have identified S. aureus as the main etiological agent of many infections in sub-Saharan Africa [4, 5], and a number of investigators have reported that S. aureus is among the most frequently encountered bacterial species in microbiology laboratories in Nigeria [6, 7]. Molecular data on this pathogen in Nigeria is however very limited.
As early as 1950, vancomycin, the first glycopeptide antibiotic to be discovered [8], was the drug of choice for therapy of infections caused by methicillin (meticillin)-resistant S. aureus (MRSA), but increase in vancomycin use has led to the emergence of two types of glycopeptide-resistant S. aureus. The first one, designated vancomycin intermediate-resistant S. aureus (VISA), is associated with a thickened and poorly cross-linked cell wall, resulting in accumulation of acyl-D-alanyl-D-alanine (X-D-Ala-D-Ala) targets at the periphery of the cell that sequester glycopeptides [9]. The second type, vancomycin-resistant S. aureus (VRSA), is due to acquisition of vanA operon from Enterococcus spp. [10, 11], carried by transposon Tn1546, resulting in high-level resistance [12].
Since the discovery of vancomycin-resistant Enterococcus (VRE) in the late 1980s [13], concern regarding the emergence of vancomycin resistance in isolates of MRSA by transfer of plasmids was always present. The first reports of reduced susceptibility of S. aureus to vancomycin were in 1997 [14, 15]. Since then, the threat of vancomycin resistance in S. aureus has been the topic of intense research and discussion. Although vancomycin resistance in S. aureus remains extremely rare, there is widespread concern that VRSA poses by far the greatest risk to patients due to the organism's virulent nature. Also, this virulent nature of S. aureus coupled with the limited options of MRSA infections treatment makes the emergence of VRSA a significant public health threat. Furthermore, there is the equally alarming threat of the risk of transmission of these organisms between patients.
In 2002, a fully vancomycin-resistant strain of MRSA emerged and was first reported from the United States of America [12, 16]. As expected, the vancomycin resistance gene cluster vanA has been acquired by these strains from vancomycin-resistant enterococci. Eleven cases of VRSA infection in the US have since been reported, with the majority from Michigan area (where seven cases were recovered), Pennsylvania, New York, and Delaware [12, 17] and from outside the US in India [18, 19] and Iran [20]. The responsible mechanism for vancomycin resistance in these strains has been found to be the acquisition of resistance plasmids carrying vanA or vanB operon. Except for the few [18–20] out of several reports, the genetic mechanisms of VRSA isolates from Iran and India [21–24] are not yet known. The first VRSA isolate in Europe was reported from Portugal in 2013 [25], and this isolate carried mecA and vanA genes probably acquired from VRE that coinfected the patient from whom the isolate was recovered.
Vancomycin resistance has been demonstrated to be inducible in vitro for many of the VRSA strains [26]. Also, in a majority of the cases, VRE strains have been isolated along with the VRSA strains from the same patients. This goes in favor of the popular theory that the Tn1546 plasmid which carries the vanA gene cluster found in such strains is acquired from VRE [18, 27]. In Nigeria, previous studies on vancomycin testing of clinical S. aureus isolates have reported full susceptibility with the disk diffusion, broth dilution, or automated Vitek-2 susceptibility test system [28–31]. Few studies [32–34], however, have reported clinical VISA isolates in some parts of the country, but only two studies [35, 36] have reported clinical VRSA strains using vancomycin agar screen, broth, or agar dilution methods.
Because S. aureus is a common cause of human infection, the emergence of VRSA is of great concern for the treatment of MRSA infections. There is therefore the need for continuous research on phenotypic identification and genetic detection of VRSA. This study was conducted to detect the biochemical and genetic characteristics of vancomycin resistance in S. aureus as a follow-up to a previous study [35] which reported phenotypic isolation of high level vancomycin-resistant S. aureus in Osogbo, Nigeria.
Materials and Methods
Study Design. The research is descriptive cross sectional, conducted over a 6-month period (July 2015 to January 2016) on patients who were either hospitalized or on outpatient treatment for infections at the Ladoke Akintola University of Technology (LAUTECH) Teaching Hospital, Osogbo, Nigeria. A total of 73 consecutive patients who gave informed consent were enrolled over the study period. Appropriate microbiological samples were collected from the patients and processed at the Bacteriology Laboratory of the hospital, while molecular characterization was done at the Molecular Microbiology Laboratory of the College of Health Sciences, LAUTECH, Osogbo. Relevant clinical data of each patient were collected into a designed proforma.
Isolation and Identification of S. aureus. S. aureus was identified by characteristic growth on Mannitol Salt Agar (MSA), positive catalase, tube coagulase, and deoxyribonuclease (DNAse) tests [37]. The isolates were confirmed by amplification of SA nuc gene on PCR.
Antibiotic Susceptibility Test by Disk Diffusion. The susceptibility of each S. aureus isolate and control strain (S. aureus ATCC 29213) to single antibiotic disk; amoxicillin, erythromycin, tetracycline, gentamicin, cotrimoxazole, chloramphenicol, fusidic, and novobiocin (Oxoid, UK) was performed in triplicate using the method of Bauer et al. [38]. The diameters of zone of inhibition that determine S. aureus susceptibility to each antibiotic were as follows: amoxicillin, ≥20 mm; erythromycin, ≥23 mm; tetracycline, ≥19 mm; gentamicin, ≥15 mm; cotrimoxazole, ≥16 mm; chloramphenicol, ≥18 mm; fusidic, ≥24 mm; and novobiocin, ≥16 mm [39].
Vancomycin E-test Method. The Minimum inhibitory concentration (MIC) of each S. aureus isolate and control strains (Enterococcus faecalis ATCC 29212 and S. aureus ATCC 29213) to vancomycin was determined in triplicate using vancomycin E-test (Epsilom test) strip (Solna, Sweden). The MIC for each isolate was compared with the interpretive standard of the Clinical and Laboratory Standards Institute (CLSI) to determine sensitivity or resistance. Isolate with vancomycin MIC of ≤2 μg/ml was categorized as vancomycin-susceptible S. aureus (VSSA), MIC 4–8 μg/ml as VISA and MIC of ≥16 μg/ml as VRSA [39]. The vancomycin MIC of S. aureus ATCC 29213 (negative control) was <2 μg/ml while that of E. faecalis ATCC 29212 (positive control strain) was 4 μg/ml.
Detection of van, nuc, and mecA genes by PCR. S. aureus vancomycin resistance genes (vanA and vanB), nuclease gene (nuc), and methicillin resistance gene (mecA) were detected by conventional singleplex polymerase chain reaction (PCR) assay.
1. DNA Extraction. DNA was extracted by the modification of simple crude extraction method previously described [31]. Briefly, 4 to 5 colonies of bacteria cells were scooped into Eppendorf tubes containing 500 µL of Tris Borate Ethylene diamine tetra acetic acid (TBE) buffer. This was boiled at 100 °C for 10 min after which it was cooled rapidly at –20 °C for 30 min. Three microliters of proteinase K was added to the lysed cells, and the mixture was incubated for 15–20 min at 55–60 °C. The enzyme was denatured at 100 °C for 10 min. The mixture was then centrifuged at 15,000 × g for 30 s. The supernatant containing the DNA was stored at –20 °C until use.
2. Amplification by PCR. S. aureus nuc gene was amplified with SANuc F primer (-gcgattgatggtgatacggtt-) and SANuc R primer (-agccaagccttgacgaactaaagc-). The vanA and vanB genes were amplified with primers VanA F (-gggaaaacgacaattgc-) and VanA R (-gtacaatgccgtta-) and primers VanB F (-tctgtttgaattgtctggtat-) and VanB R (-gacctcgtttagaacgatg-). The mecA gene was amplified with primer MecA F (-gtagaaatgactgaacgtccgataa-) and MecA R (-ccaattccacattgtttcggtctaa-).
The reaction was set up in a PCR vial, after adding the master mix, the forward and reverse primers, and the extracted DNA. A 25 µL of master mix contained 4 µL of 10× buffer, 0.5 µL MgCl2, 3 µL dNTPs, and 0.2 µL Taq polymerase. The PCR vial was then placed in the PCR machine (Prime Therma cycler, UK). For amplification of nuc gene, initial denaturation occurred at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s and elongation at 72 °C for 30 s, and a final extension at 72 °C for 7 min [40]. For amplification of mecA gene, initial denaturation was at 94 °C for 4 min, followed by 30 cycles of denaturation at 94 °C for 45 s, annealing at 50 °C for 30 sec and elongation at 72 °C for 1 min, and a final extension at 72 °C for 3 min [41]. For amplification of vanA gene, initial denaturation occurred at 94 °C for 2 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 54 °C for 1 min, elongation at 72 °C for 1 min, and a final extension at 72 °C for 10 min [42]. For the amplification of vanB gene, initial denaturation occurred at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 25 s, annealing at 52 °C for 40 s, elongation at 72 °C for 1 min, and final extension at 72 °C for 6 min [43].
3. Gel Electrophoresis. At the completion of the amplification of each gene, the PCR amplicons were resolved on agarose gel electrophoresis. Agarose gel (1%), into which 0.5 µL ethidium bromide had been added, was first prepared. The gel plates were then placed inside electrophoresis tank (Weal Tek Corp., Taiwan) which contained 1× TBE solution. A 5 µL of amplicon was mixed with 5 µL Orange G (loading buffer) and loaded to the well of the agarose gel. The power supply was adjusted to 100 volts for 25 min. For each run, a 100 base-pair molecular weight DNA standard (New England Biolabs “NEB”) was used to determine the size of each PCR amplicon. The DNA bands were then visualized with a short wave ultraviolet trans-illuminator and photographed using gene gel bio-imaging system (UVP Imaging System, Upland, CA, USA) and products analyzed.
4. Results and Interpretation. Expected band for S. aureus nuc gene is approximately 276 bp, vanA gene 732 bp, vanB gene 807 bp, and mecA gene 310 bp.
Data Entry and Statistical Analysis. All data (clinical and laboratory) were entered into Window 7 laptop computer with GraphPad statistical software. Frequency tables were generated, and relationship between variables tested with χ2 with significant value set atp < 0.05.
Ethics Statement. Informed consent was obtained from each patient-participant. Approval of the hospital Ethics and Research Committee (ERC) was obtained prior to conduct of the study.
Results
Non-duplicate S. aureus was isolated from 73 consecutive patients with different infective conditions as shown in Table 1. Non-surgical wound infections accounted for the largest percentage (30.1%); followed by otitis media (12.3%); pelvic inflammatory disease (10.9%); bacterial conjunctivitis (8.2%); surgical site infection, osteomyelitis, urinary tract infection, and abscess (5.5%); septic arthritis (4.2%); and others (9.6%), while necrotizing fasciitis and blood stream infection constituted the lowest percentage (1.4%).
Table 1.
Distribution of Staphylococcus aureus isolates by clinical infection types
| Clinical infections | Staphylococcus aureus isolates | |||||
|---|---|---|---|---|---|---|
| Methicillin susceptibility | Vancomycin susceptibility | Total (%) | ||||
| MSSA | MRSA | VSSA | VISA | VRSA | ||
| Non-surgical wound infections | 20 | 2 | 18 | 4 | 0 | 22 (30.1) |
| Surgical site infection | 3 | 1 | 3 | 0 | 1 | 4 (5.5) |
| Septic arthritis | 3 | 0 | 3 | 0 | 0 | 3 (4.2) |
| Osteomyelitis | 4 | 0 | 4 | 0 | 0 | 4 (5.5) |
| Otitis media | 8 | 1 | 7 | 2 | 0 | 9 (12.3) |
| Urinary tract infection | 3 | 1 | 1 | 3 | 0 | 4 (5.5) |
| Bacteria conjunctivitis | 6 | 0 | 6 | 0 | 0 | 6 (8.2) |
| Necrotizing fasciitis | 1 | 0 | 1 | 0 | 0 | 1 (1.4) |
| Blood stream infection | 1 | 0 | 1 | 0 | 0 | 1 (1.4) |
| Abscess | 4 | 0 | 4 | 0 | 0 | 4 (5.5) |
| Pelvic inflammatory disease | 8 | 0 | 6 | 2 | 0 | 8 (10.9) |
| Others (non-specified) | 7 | 0 | 7 | 0 | 0 | 7 (9.6) |
| Total (%) | 68 (93.2) | 5 (6.8) | 61 (83.6) | 11 (15.1) | 1 (1.4) | 73(100) |
MSSA, methicillin-sensitive Staphylococcus aureus; MRSA, methicillin-resistant Staphylococcus aureus; VSSA, vancomycin-sensitive Staphylococcus aureus; VISA, vancomycin intermediate-resistant Staphylococcus aureus; VRSA, vancomycin-resistant Staphylococcus aureus.
Table 2 shows the distribution of S. aureus isolates by methicillin and vancomycin susceptibility; 61 (83.6%) of the isolates were VSSA, 11 (15.1%) were VISA, and 1 (1.4%) was VRSA. A total of 68 (93.2%) isolates were methicillin-susceptible S. aureus (MSSA; mecA negative), while 5 (6.8%) were MRSA (mecA positive). The prevalence of MRSA in the study was 6.8%.
Table 2.
Distribution of Staphylococcus aureus isolates by methicillin and vancomycin susceptibility
| Staphylococcus aureus isolates | Vancomycin susceptibility (E-test) MIC | Total (%) | ||
|---|---|---|---|---|
| ≤2 μg/ml (VSSA) | 4–8 μg/ml (VISA) | ≥16 μg/ml (VRSA) | ||
| MSSA | 61 | 7 | 0 | 68 (93.2) |
| (mecA gene negative) | ||||
| MRSA | 0 | 4 | 1 | 5 (6.8) |
| (mecA gene positive) | ||||
| Total (%) | 61 (83.6) | 11 (15.1) | 1 (1.4) | 73(100) |
MSSA, methicillin-sensitive Staphylococcus aureus; MRSA, methicillin-resistant Staphylococcus aureus; VSSA, vancomycin-sensitive
Staphylococcus aureus; VISA, vancomycin intermediate-resistant
Staphylococcus aureus; VRSA, vancomycin-resistant Staphylococcus aureus.
Table 3 shows the antibiotic susceptibility of VSSA isolates assessed by the disk diffusion method to selected antibiotics. Most of the VSSA isolates were resistant to commonly used antibiotics in the environment but were mostly sensitive to novobiocin (78.7%) and gentamicin (73.8%).
Table 3.
Antibiotic susceptibility of VSSA isolates by disk diffusion method to selected antibiotics
| Antibiotic disk | Susceptibility profile of VSSA isolates (n = 61) | |
|---|---|---|
| Sensitive (%) | Resistant (%) | |
| Amoxicillin | 8 (13.1) | 53 (86.9) |
| Erythromycin | 4 (6.6) | 57 (93.4) |
| Tetracycline | 20 (32.8) | 41 (67.2) |
| Gentamicin | 45 (73.8) | 16 (26.2) |
| Cotrimoxazole | 18 (29.5) | 43 (70.5) |
| Chloramphenicol | 28 (45.9) | 33 (54.1) |
| Fusidic acid | 26 (42.6) | 35 (57.4) |
| Novobiocin | 48 (78.7) | 13 (21.3) |
Table 4 displays antibiotic susceptibility of VISA isolates applying the disk diffusion method to selected antibiotics. Almost all the VISA isolates were resistant to the antibiotics tested. Only few, however, were sensitive to gentamicin (36.4%), tetracycline (9.1%), and cotrimoxazole (9.1%). The only VRSA isolate (with vancomycin MIC = 16 μg/ml) was resistant to all antibiotic disks tested in the study.
Table 4.
Antibiotic susceptibility of VISA isolates by disk diffusion method to selected antibiotics
| Antibiotic disk | Susceptibility profile of VISA isolates (n = 11) | |
|---|---|---|
| Sensitive (%) | Resistant (%) | |
| Amoxicillin | 0 | 11(100) |
| Erythromycin | 0 | 11(100) |
| Tetracycline | 1 (9.1) | 10 (90.9) |
| Gentamicin | 4 (36.4) | 7 (63.6) |
| Cotrimoxazole | 1 (9.1) | 10 (90.9) |
| Chloramphenicol | 0 | 11(100) |
| Fusidic acid | 0 | 11(100) |
| Novobiocin | 0 | 11(100) |
Table 5 shows the comparison of resistant profile of VSSA and VISA isolates to selected antibiotics. Both VSSA and VISA isolates showed high in vitro resistance to multiple antibiotics tested with the VISA isolates being more significantly resistant to gentamicin (p = 0.0302), chloramphenicol (p = 0.0049), fusidic acid (p = 0.0056), and novobiocin (p < 0.0001). The only vancomycin-resistant isolate was resistant to all antibiotics tested.
Table 5.
Comparison of resistant profile of VSSA and VISA isolates to selected antibiotics
| Antibiotic disk | Resistance of VSSA (%) (n = 61) | Resistance of VISA (%) (n = 11) | p Value |
|---|---|---|---|
| Amoxicillin | 53 (86.9) | 11(100) | 0.3439 |
| Erythromycin | 57 (93.4) | 11(100) | 1.000 |
| Tetracycline | 41 (67.2) | 10 (90.9) | 0.1579 |
| Gentamicin | 16 (26.2) | 7 (63.6) | 0.0302* |
| Cotrimoxazole | 43 (70.5) | 10 (90.9) | 0.2674 |
| Chloramphenicol | 33 (54.1) | 11(100) | 0.0049* |
| Fusidic acid | 35 (57.4) | 11(100) | 0.0056* |
| Novobiocin | 13 (21.3) | 11(100) | < 0.0001* |
*Significant difference.
All S. aureus isolates, including the 11 VISA and 1 VRSA strains, were positive for nuc gene (Figure 1). Five isolates were mecA gene positive which confirmed them to be MRSA (Figure 2) and were either VISA or VRSA. All the VISA and VRSA isolates were vanA and vanB gene negative.
Figure 1.
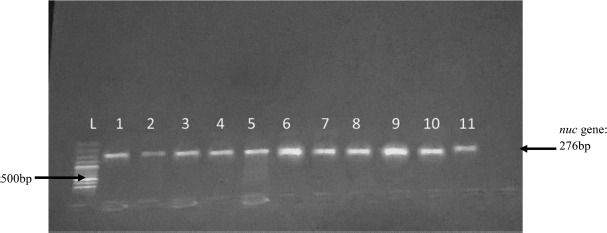
Gel electrophoresis of PCR amplification of nuc gene. L = 100 bp ladder 1, 2, 3, 4, 5, 7, 8, 9, 10, 11 = nuc positive isolates; 6 = nuc positive control; not labeled = nuc negative control (PCR water)
Figure 2.
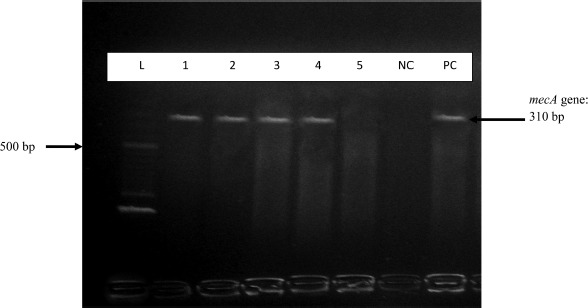
Gel electrophoresis of PCR amplification of mecA gene. L = 100 bp ladder; 1, 2, 3, 4 = mecA positive isolates; 5 = mecA negative isolate; NC = mecA negative control; PC = mecA negative control (PCR water)
Discussion
Since VISA was first reported in the 1997 [14, 15] and VRSA in 2002 [16], the threat of vancomycin resistance in S. aurleus has been of great concern and a threat to public health. This has made the study of VISA and VRSA a topic of intensive research, although there have been limited reports of clinical infections caused by VRSA worldwide [12, 19, 21, 23], while infections caused by VISA continued to be reported from different parts of the world [44–47]. As there have also been previous reports of phenotypic resistance of MRSA to vancomycin in our environment [32–36], the present study was designed to investigate the molecular basis of this resistance.
This study detected VRSA in 1 (1.4%) out of 73 S. aureus isolates, which was isolated from a surgical site infection (SSI) and resistant to all antibiotics tested in the study. The strain displayed a vancomycin MIC value of 16 μg/ml and carried a mecA gene but did not contain vanA or vanB gene. In our previous study [35], we isolated VRSA strains (12.2%, 6 of 49 S. aureus isolates) with vancomycin MIC of >256 μg/ml from patients with wound infection and chronic osteomyelitis, who had been on prolonged hospitalization. Similarly, there was a 5.3% VRSA prevalence rate reported among clinical patients in a hospital in Eastern part of Nigeria [36] and 5.4% positivity rate in fresh and fermented milk obtained from animals in Northern Nigeria [48]. The importance of all these findings is that VRSA isolates are increasingly emerging in several parts of the world including Nigeria. There remains a public health threat from these emerging S. aureus strains that can no longer be ignored.
The molecular basis of all VRSA isolates is yet to be clearly elucidated, but it has been shown in some that phenotypic resistance is conferred by induction of the vanA operon found on transposon Tn1546 and carried by Inc18-like plasmid from coinfecting vancomycin-resistant enterococci [12]. The transfer of this plasmid requires the presence of a pSK41-like plasmid in recipient S. aureus [49]. In our study, however, the only VRSA isolate neither contains vanA nor vanB gene that has been described to be present in all confirmed VRSA isolates reported so far [12]. It is possible that other van operons (vanC, vanD, vanE, vanG, or vanL) found in enterococci may also be transferrable to S. aureus subsequently conferring vancomycin resistance to the S. aureus recipient strain. Although there was no coinfection of enterococci in the patient from whom our VRSA isolate was recovered from, we believe that the range of van operons routinely tested in S. aureus exhibiting resistance phenotype to vancomycin should be expanded to other van operons aside vanA and vanB.
Eleven (15.1%) of the S. aureus isolates in this study were VISA (vancomycin MIC of 4–8 μg/ml) involved in different types of infection. The mechanism of resistance in VISA is yet to be fully explained although sequestering of glycopeptide (vancomycin and teicoplanin) in the periphery of the cell due to thickening of the wall from accumulation of peptidoglycan precursor, acyl-D-alanyl-D-alanine dipeptide, has been reported to reduce penetration of the drug to its target site [9]. The genetic basis of this remains largely unexplained as no gene or operon has been linked with VISA, although frequent exposure of patients to high dosages of different antibiotics including vancomycin, which may cause chromosomal mutations, has been implicated. Our 11 VISA isolates neither contain vanA nor vanB gene, although four of them carried the mecA gene. This result is in line with other studies from Nigeria [33, 35] and Iran [21, 47].
We observed in this study that the prevalence of MRSA was 6.8%, all of which were VISA and VRSA strains and resistant to most of the antibiotics used in the study. The only VRSA strain was resistant to all the antibiotics tested including amoxicillin, erythromycin, tetracycline, gentamicin, cotrimoxazole, chloramphenicol, fusidic acid, and novobiocin. This finding is supported by a previous report [50] in which VRSA and heterogeneous vancomycin-resistant S. aureus (hVRSA) isolates were resistant to 21 different antibiotics including methicillin, tetracycline, gentamicin, and erythromycin. The efficacy of vancomycin in treatment of severe MRSA infections is gradually being eroded by the emergence of VISA and VRSA strains. Moreover, these strains have been found to be resistant to multiple antibiotics thereby making antibiotic selection more difficult in this era of decreasing pipeline of new antibiotic discovery. Although the occurrence of VRSA strain is thought to be geographically restricted due to several biological constraints that limit the spread of the van plasmids between strains [12], the potential for spread should not be underestimated especially with discovery that S. aureus harboring the pSK41-like plasmid readily accept van plasmid [49].
Conclusions
In conclusion, this study confirms the occurrence of multiple antibiotic resistant VISA and emergence of VRSA isolates involved in clinical infections in Nigeria, whereas the genetic basis remains to be elucidated, however. There is further need to curtail further emergence and possible spread of these strains by reducing the glycopeptide pressure through appropriate prescribing and initiating infection control measures.
Acknowledgments
The authors wish to acknowledge the technical assistance of Mr. O.A. Adefioye, Assistant Chief Medical Laboratory Scientist, Department of Medical Microbiology and Parasitology, College of Health Sciences, Ladoke Akintola University of Technology, Osogbo.
Footnotes
Funding Sources
The authors received no funding for this project.
Authors' Contribution
BTB was involved in study design, sample and data collection, laboratory analysis and production of initial draft of the manuscript. OOA was involved in co-supervision of the study. TSS designed and supervised the study, and was involved in correction of initial manuscript draft. All authors agreed to the final draft.
Conflict of Interest
The authors have no competing interests.
References
- 1.Projan SJ, Novick RP. The molecular basis of pathogenicity. Crossley KB, Archer GL, editors. The Staphylococci in Human Diseases. New York: Churchill Livingstone; 1997. p. 55–81. [Google Scholar]
- 2.Perez-Vazquez M, Vindel A, Marcos C, Oteo J, Cuevas O, Trincado P, et al. Spread of invasive Spanish Staphylococcus aureus spa-type t067 associated with a high prevalence of aminoglycoside modifying enzyme ant (49)-Ia and the efflux pump genes mrsA/mrsB. J Antimicrob Chemother. 2009;63:21–31. [DOI] [PubMed] [Google Scholar]
- 3.Nickerson EK, Wuthiekanun V, Wongsuvan G, Limmathurosakul D, Srisamang P, Mahavanakul W. Factors predicting and reducing mortality in patients with invasive Staphylococcus aureus disease in a developing country. PLoS ONE. 2009;4:e6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulu A, Moges F, Tessema B, Kassu A. Pattern and multiple drug resistance of bacterial pathogens isolated from wound infection at University of Gondar Teaching Hospital, North West Ethiopia. Ethiop Med J. 2006;44:125–31. [PubMed] [Google Scholar]
- 5.Nantanda R, Hildenwall H, Peterson S, Kaddu-Mulindwa D, Kalyesubula I, Tumwine JK. Bacterial aetiology and outcome in children with severe pneumonia in Uganda. Ann Trop Paediatr. 2008;28:253–60. [DOI] [PubMed] [Google Scholar]
- 6.Ambe JP, Gasi IS, Mava Y. Review of neonatal infections in University of Maiduguri Teaching Hospital: common bacterial pathogens seen. Nig J Clin Pract. 2007;10:290–3. [PubMed] [Google Scholar]
- 7.Ubani UA. Common bacterial isolates from infected eyes. J Nig Optometr Ass. 2009;15:40–7. [Google Scholar]
- 8.Murray BE, Nannini EC. Glycopeptides (vancomycin and teicoplanin), streptogramins (quinupristin-dalfopristin) and lipopeptides (daptomycin). Mandell GL, Bennett JE, Dolin R, editors. Principles and Practice of Infectious Diseases. Churchill Livingstone: Philadelphia; 2010. p. 449–68. [Google Scholar]
- 9.Cui L, Ma X, Sato K, Okuma K, Tenover FC, Maimizuka EM, et al. Cell wall thickening is a common feature of vancomycin-resistant in Staphylococcus aureus. J Clin Microbiol. 2003;41:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium. J Bacteriol. 1992;174:2582–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arthur M, Reynolds PE, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–7. [DOI] [PubMed] [Google Scholar]
- 12.Périchon B, Courvalin P. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009;53:4580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. New Engl J Med. 1988;319:157–61. [DOI] [PubMed] [Google Scholar]
- 14.Hiramatsu K, Hanaki H, Ito T, Yabuta K, Oguri T, Tenover FC. Methicillin resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–6. [DOI] [PubMed] [Google Scholar]
- 15.Center for Disease Control and Prevention. Staphylococcus aureus with reduced susceptibility to vancomycin, United States. Morb. Mortal. Wkly Rep.. 1997; 46 p. 765–6. [PubMed] [Google Scholar]
- 16.Sievert DM, Boulton ML, Stolman G, Johnson D, Stobierski MG, Downes FP, et al. Staphylococcus aureus resistant to vancomycin. Morb. Mortal. Wkly Rep. 2002; 51 p. 565–7. [PubMed] [Google Scholar]
- 17.Walters M, Lonsway D, Rasheed K, Albrecht V, McAllister S, Limbago B, et al. Investigation and control of vancomycin resistant Staphylococcus aureus: a guide for health departments and infection control personnel. Atlanta, GA; 2015. Available from: http://www.cdc.gov/hai/pdfs/VRSA-Investigation-Guide-05_12_2015.pdf. [Google Scholar]
- 18.Chakraborty SP, KarMahapatra S, Bal M, Roy S. Isolation and identification of vancomycin resistant Staphylococcus aureus from post operative pus sample. Al Ameen J Med Sci. 2011;4:152–68. [Google Scholar]
- 19.Thati V, Shivannavar CT, Gaddad SM. Vancomycin resistance among methicillin resistant Staphylococcus aureus isolates from intensive care units of tertiary care hospitals in Hyderabad. Indian J Med Res. 2011;134:704–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azimian A, Havaei SA, Fazeli H, Naderi M, Ghazvini K, Samiee SM, et al. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a university hospital in northeastern Iran. J Clin Microbiol. 2012;50:3581–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saderi H, Owlia P, Shahrbanooie R. Vancomycin resistance among clinical isolates of Staphylococcus aureus. Arch Iranian Med. 2005;8:100–3. [Google Scholar]
- 22.Aligholi M, Emaneini M, Jabalameli F, Shahsavan S, Dabiri H, Sedaght H. Emergence of high-level vancomycin-resistant Staphylococcus aureus in the Imam Khomeini Hospital in Tehran. Med Prin Pract. 2008;17:432–4. [DOI] [PubMed] [Google Scholar]
- 23.Tiwari HK, Sen MR. Emergence of vancomycin resistant Staphylococcus aureus from tertiary care hospital from northern part of India. BMC Infect Dis. 2006;6:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha B, Singh AK, Ghosh A, Bal M. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J Med Microbiol. 2008;57:72–9. [DOI] [PubMed] [Google Scholar]
- 25.Melo-Cristino J, Resina C, Manuel V, Lito L, Ramirez M. First case of infection with vancomycin-resistant Staphylococcus aureus in Europe. The Lancet. 2013;382:205. [DOI] [PubMed] [Google Scholar]
- 26.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flanagan SE, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–71. [DOI] [PubMed] [Google Scholar]
- 27.Tenover FC, Moellering RCJ. The rationale for revising the clinical and laboratory standards institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin Infect Dis. 2007;44:1208–15. [DOI] [PubMed] [Google Scholar]
- 28.Okesola AO, Oni AA, Bakare RA. Prevalence and antibiotic sensitivity pattern of methicillin-resistant Staphylococcus aureus in Ibadan, Nigeria. J Hosp Infect. 1999;41:74–5. [DOI] [PubMed] [Google Scholar]
- 29.Ghebremedhin B, Olugbosi MO, Raji AM, Layer F, Bakare RA, Konig B, et al. Emergence of community associated methicillin resistance Staphylococcus aureus strain with unique resistance profile in Southwest Nigeria. J Clin Microbiol. 2009;47:2975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shittu A, Oyedara O, Abegunrin F, Okon K, Raji A, Taiwo S, et al. Characterization of methicillin susceptible and -resistant staphylococci in the clinical setting: a multi-centre study in Nigeria. BMC Infect Dis. 2012;12:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olowe OA, Kukoyi OO, Taiwo SS, Ojurongbe O, Opaleye OO, Bolaji OS, et al. Phenotypic and molecular characteristics of methicillin-resistant Staphylococcus aureus isolates from Ekiti State, Nigeria. Infect Drug Resist. 2013;6:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olayinka BO, Olayinka AT, Onaolapo JA, Olurinola PF. Pattern of resistance to vancomycin and other antimicrobial agents in staphylococcal isolates in a University Teaching Hospital. Afr J Clin Exper Microbiol. 2005;6:21–7. [Google Scholar]
- 33.Onolitola OS, Olayinka BO, Salawu MJ, Yakubu SE. Nasal carriage of methicillin resistant Staphylococcus aureus with reduced vancomycin susceptibility (MRSA RVS) by healthy adults in Zaria. J Trop Microbiol Biotechnol. 2007;3:19–22. [Google Scholar]
- 34.Ogbolu DO, Terry Alli OA, Bello LA, Ibrahim AO. Emergence of vancomycin intermediate Staphylococcus aureus (VISA) in clinical isolates of methicillin resistant S. aureus from Southwestern region of Nigeria. Int J Trop Dis Hlth. 2015;10:1–5. [Google Scholar]
- 35.Taiwo SS, Bamigboye TB, Odaro S, Adefioye OA, Fadiora SO. Vancomycin intermediate and high level resistant Staphylococcus aureus clinical isolates. Microbiol Res. 2011;3:22–5. [Google Scholar]
- 36.Alo M, Ugah U, Okoro N. Epidemiology of vancomycin resistant Staphylococcus aureus among clinical isolates in a tertiary hospital in Abakaliki, Nigeria. Am J Epidemiol Infect Dis. 2013;1:24–6. [Google Scholar]
- 37.Barrow GI, Feltham RKA. Cowan and Steel Manual for the Identification of Medical Bacteria. Cambridge: Cambridge University Press; 1993. [Google Scholar]
- 38.Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6. [PubMed] [Google Scholar]
- 39.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 26th informational supplement M100-S26. Wayne, PA: CLSI; 2016. [Google Scholar]
- 40.Asad UK, Ayesha S, Anju T, Shazia Z, Mohd A, Sukhminderit K, et al. Amplification of mecA gene in multi-drug resistant Staphylococcus aureus strains from hospital personnel. J Infect Dev Countr. 2007;1:289–95. [PubMed] [Google Scholar]
- 41.Anand KB, Agrawal P, Kumar S, Kaplla K. Comparison of cefoxitin disc diffusion test, oxacillin screen agar, and PCR for mecA gene for detection of MRSA. Indian J Med Microbiol. 2009;27:27–9. [PubMed] [Google Scholar]
- 42.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptides resistance genotypes and identification on the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werner G. Intra-hospital dissemination of quinupristin/dalfopristin and vancomycin-resistant Enterococcus faecium in a pediatric ward of German hospital. J Antimicrob Chemother. 2003;52:113–5. [DOI] [PubMed] [Google Scholar]
- 44.Menezes GA, Harish BN, Sujatha S, Vinothini K, Parija SC. Emergence of vancomycin-intermediate Staphylococcus species in southern India. J Med Microbiol. 2008;57:911–2. [DOI] [PubMed] [Google Scholar]
- 45.Howden BP, Davies JK, Johnson PDR, Stinear TP, Grayson ML. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. 2010;23:99–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marques JB, Dalmolin TV, Bonez PC, Agertt VA, Anraku de Campos MM, Santos RCV. Detection of Staphylococcus aureus with an intermediate profile to vancomycin (VISA) isolate from Santa Maria, RS. Braz J Microbiol. 2013;44:277–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farhadian A, Nejad QB, Peerayeh SN, Rahbar M, Vaziri F. Determination of vancomycin and methicillin resistance in clinical isolates of Staphylococcus aureus in Iranian hospitals. Br Microbiol Res J. 2014;4:454–61. [Google Scholar]
- 48.Umaru GA, Kabir J, Umoh VJ, Bello M, Kwaga JKP. Occurrence of vancomycin resistant Staphylococcus aureus (VRSA) in fresh and fermented milk in Nigeria: a preliminary report. Int J Pub Hlth Epidemiol. 2013;3:54–8. [Google Scholar]
- 49.Zhu W, Clark N, Patel JB. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob Agents Chemother. 2013;57:212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wotton M, Howe RA, Walsh TR, Bennett PM, McGowan AP. In vitro activity of 21 antimicrobials against vancomycin-resistant Staphylococcus aureus (VRSA) and hetero-VRSA (hVRSA). J Antimicrob Agents Chemother. 2002;50:760–1. [DOI] [PubMed] [Google Scholar]


