Abstract
Background:
Estrogen has played an important role in the development of breast cancer. ER-α PvuII gene polymorphism is in close association with the occurrence risk of breast cancer, but no consensus has been achieved currently.
Methods:
PubMed, Embase, China National Knowledge Infrastructure (CNKI) database, Wanfang database, and VIP database were retrieved to collect the case–control studies on association between ERα gene Pvu II polymorphism and breast cancer risk published before September 1, 2017. Newcastle–Ottawa Scale (NOS) was used to assess the quality of the literatures, Stata 14.0 software was applied for meta-analysis, and the pooled odds ratio (OR) and 95% confidence interval (95% CI) were calculated. The subgroup analysis was performed to assess the confounding factors, followed by assessment of publication bias and sensitivity analysis.
Results:
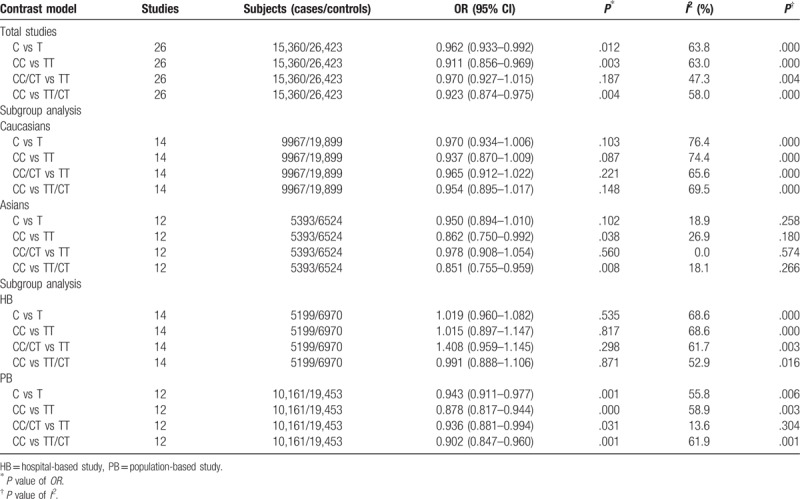
A total of 26 studies were enrolled in the analysis based on inclusion criteria, which included 15,360 patients and 26,423 controls. The results demonstrated that ERα gene Pvu II polymorphism was in significant association with the decrease of breast cancer risk in 3 genetic models (C vs T, OR = 0.962, 95% CI = 0.933–0.992, P = .012; CC vs TT, OR = 0.911, 95% CI = 0.856–0.969, P = .003; CC vs TT/CT, OR = 0.923, 95% CI = 0.874–0.975, P = .004). Subgroup analysis was conducted on the basis of ethnicity and source of controls, whose results illustrated that ERα gene Pvu II polymorphism was in significant association with the decrease of breast cancer risk in Asians rather than in Caucasians (CC vs TT, OR = 0.862, 95% CI = 0.750–0.922, P = .038; CC vs TT/CT, OR = 0.851, 95% CI = 0.755–0.959, P = .008). In population-based subgroup rather than in hospital-based subgroup, ERα gene Pvu II polymorphism was in significant association with the decrease of breast cancer risk in the allele model, homozygous model, dominant model, and recessive model (C vs T, OR = 0.943, 95% CI = 0.911–0.977, P = .001; CC vs TT, OR = 0.878, 95% CI = 0.817–0.944, P = .000; CC/CT vs TT, OR = 0.936, 95% CI = 0.881–0.994, P = .031; CC vs TT/CT, OR = 0.902, 95% CI = 0.847–0.960, P = .001).
Conclusion:
ERα gene Pvu II polymorphism exerts an important function in the progression of breast cancer.
Keywords: breast cancer, ERα gene, meta-analysis, polymorphism, Pvu II
1. Introduction
According to the global data statistics on cancer in 2012, breast cancer had become the most common malignancy and the leading cause for cancer-related death in females, and about 1,700,000 had been diagnosed with breast cancer, and 521,900 died of breast cancer annually.[1] In China, new cases with breast cancer account for 15% in females, with morbidity and mortality increasing annually.[2] At present, multiple studies have shown that genetic mutation, menopause status, family history, fertility, alcoholic consumption, smoking, and exposure to estrogen are risk factors for women with breast cancer, and play important roles in the pathogenesis and progression of breast cancer.[3–5]
Estrogens exert great functions in the development and progression of breast cancer, whose effects are mainly mediated by intracellular estrogen receptors (ERs). ER-α and ER-β, 2 types of ER, are important regulators for the actions of estrogens, in which ER-α gene can encode a transcription factor with an estrogen response element (ERE) DNA-binding domain and an estrogen-binding domain.[6]ERα gene, as a steroid hormone receptor gene, is located in chromosome 6 at 6p25.1, and ERα gene mutation can induce cell proliferation, regulate cell apoptosis by affecting protein expression, so as to participate in the development and progression of breast cancer.[7] Multiple single nucleotide polymorphisms (SNPs) in ER-α, as located in intron 1 of ER-α, have been studied in numerous clinical studies and have become one of the hot topics on tumor susceptibility, including rs2234693, also termed as ERα gene PvuII.[8] ER-α PvuII SNPs have found to be associated with numerous carcinoma included prostate cancer,[9] systemic lupus erythematosus,[10] Alzheimer disease,[11] etc. And ER-α PvuII polymorphisms have been verified to be in close connection with tumorigenesis. It has also been found that ER-α PvuII and Xbal polymorphisms are closely related with breast cancer,[12] in which ER-α PvuII polymorphism is proved to play an important role in breast cancer risk in pre-menopausal females .[13]
Although the association between ERα gene Pvu II polymorphism and breast cancer susceptibility has been extensively reported currently, there are great differences among study conclusions. Atoum et al[14] did not found any correlation between ERα gene Pvu II polymorphism and breast cancer risk in Jordaneses, but some investigators discovered that there was a significant correlation between ERα gene Pvu II polymorphism and breast cancer risk in Brazilians and Northern Indians and patients with p allele had lower morbidity of breast cancer.[15,16] Although meta-analysis have been conducted in pre-studies,[13,17–19] there are still new literatures on the association between ERα gene Pvu II polymorphism and breast cancer risk published. Considering the timeliness of meta-analysis and to provide a more accurate estimation of the association between ERα gene Pvu II polymorphism and breast cancer susceptibility, an updated meta-analysis on related case–control studies published in Open Access journals was conducted to further determine the association between ERα gene Pvu II polymorphism and breast cancer susceptibility by involving as many data as possible from published studies.
2. Materials and methods
2.1. Literature retrieval
Case–control studies on the association between ERα gene Pvu II polymorphism and breast cancer susceptibility included in medical databases such as PubMed, Embase, China Biomedicine (CBM) database, Wanfang database, VIP database, and China National Knowledge Infrastructure (CNKI) database were retrieved using terms such as “estrogen receptor α,” “ ERα,” “Pvu II,” “rs2234693,” “polymorphism,” “single nucleotide polymorphism,” “variation,” “breast carcinoma,” “breast cancer,” and “BC.” Simultaneously, references in corresponding literatures included in above databases were retrieved artificially based on the title of the literatures in order to screen more applicable literatures.
2.2. Inclusion and exclusion criteria
Inclusion criteria included studies published in English or Chinese language before September 1, 2017; case–control studies; studies that enrolled healthy populations in the control group; studies with full texts; studies on the distribution frequency of ERα gene Pvu II or studies that provided corresponding odds ratio (OR) value with clearly described data; studies focused on the association between breast cancer risk and ERα gene Pvu II polymorphism. Exclusion criteria included studies with incomplete data; studies in which patients with fibroadenoma of breast or nonbreast cancer were selected as study subjects; studies that focused on exploring the association between gene polymorphism and other factors such as disease progression, disease severity, phenotype modification, sensitivity to clinical treatment, or survival, etc; studies on family association; and studies in which systematic reviews or meta-analysis were also excluded. In overlapping studies, those with large sample size were selected.
2.3. Data extraction
In this study, the ERα gene Pvu II polymorphism in enrolled population was set as the primary index. Two researchers were assigned to collect literatures independently based on above inclusion criteria, exclusion criteria, and data extraction criteria, with following variables extracted from each study: the year of publication, name of the first author, source of the controls, ethnicity of enrolled population, gene detection method, and phenotype distribution data, etc. The disagreement in data extraction should be resolved by discussion or a third-party researcher when necessary. The data involving different ethnicities should be extracted independently based on ethnicity. The ethnicities included in this study contained Caucasians, Asians, and others.
2.4. Quality assessment
The quality of the case–control studies enrolled in this study was assessed by 2 investigators using Newcastle–Ottawa Scale (NOS). Primary contents to be assessed include selection of study subjects (4 scores in total); inter-group comparability (2 scores in total); exposure factors or outcomes (3 scores in total). Low-quality studies: 0 to 4 points; high-quality studies: 5 to 9 points.
2.5. Statistical data analysis
STATA 14.0 software was used to analyze the extracted data. Hardy–Weinberg equilibrium (HWE) analysis was performed using χ2 test. Allele model, homozygous model, dominant model, and recessive model were used, and pooled OR values and 95% confidence intervals (95% CIs) were used to assess the association between ERα gene Pvu II polymorphism and breast cancer susceptibility. The heterogeneity was assessed by Q and I2 statistics. There was no heterogeneity if P > .1 or I2 < 50%, for which fixed-effect model (Mantel–Haenszel method) was used. There was heterogeneity if P < .1 and I2 > 50%, for which random-effect (DerSimonian Laird) model was used for analysis. Subgroup analysis was performed on the basis of ethnicity and source of controls. Funnel plot was applied to detect whether there was publication bias in the enrolled literatures. There was publication bias if the funnel plot was asymmetric. Egger test was adopted to test the severity of publication bias. The stability of results was analyzed and assessed by sensitivity analysis. Two-tailed test was applied, with a P < .05 level of significance.
3. Results
3.1. General information of enrolled literatures
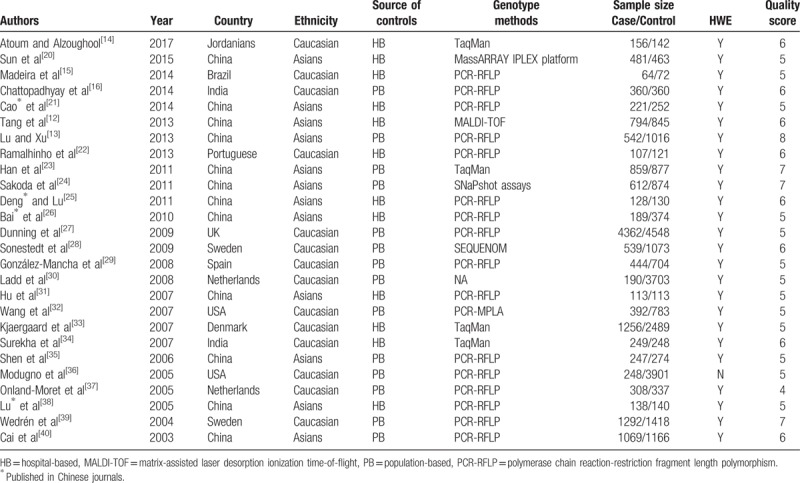
A total of 409 articles were retrieved using the keywords, as shown in Fig. 1. A total of 367 articles were excluded according to the titles and abstracts, among which 211 were significantly irrelevant, 146 were overlapped, 5 were reviews, and 5 were animal or cell experiments; 7 were about meta-analysis and 9 were not related with Pvu II polymorphisms. Finally, 26 full-text articles with detailed study information enrolling 15,360 cases and 26,423 controls were obtained. The primary information of studies enrolled in this meta-analysis is summarized in Table 1.[20–40]
Figure 1.
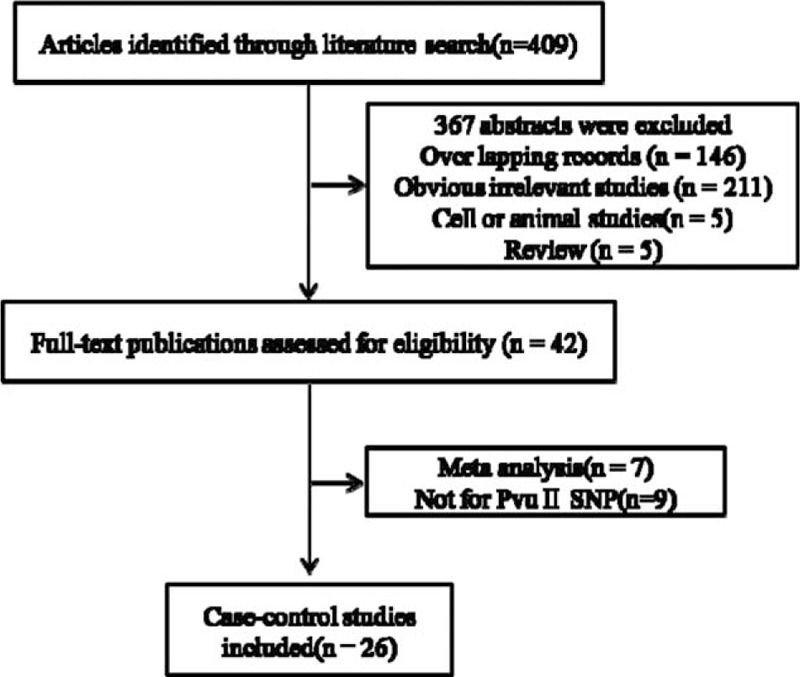
Retrieval flow chart of eligible case–control studies enrolled in the meta-analysis.
Table 1.
Characteristics of literatures enrolled in the meta-analysis.
3.2. Meta-analysis results
The meta-analysis results indicated that a significant association between ERα gene Pvu II polymorphism and the decrease of breast cancer risk was found in allele model, homozygous model, and recessive model (C vs T, OR = 0.962, 95% CI = 0.933–0.992, P = .012; CC vs TT, OR = 0.911, 95% CI = 0.856–0.969, P = .003; CC vs TT/CT, OR = 0.923, 95% CI = 0.874–0.975, P = .004) rather than in dominant model (CC/CT vs TT, OR = 0.970, 95% CI = 0.927–1.015, P = .187), as shown in Fig. 2.
Figure 2.
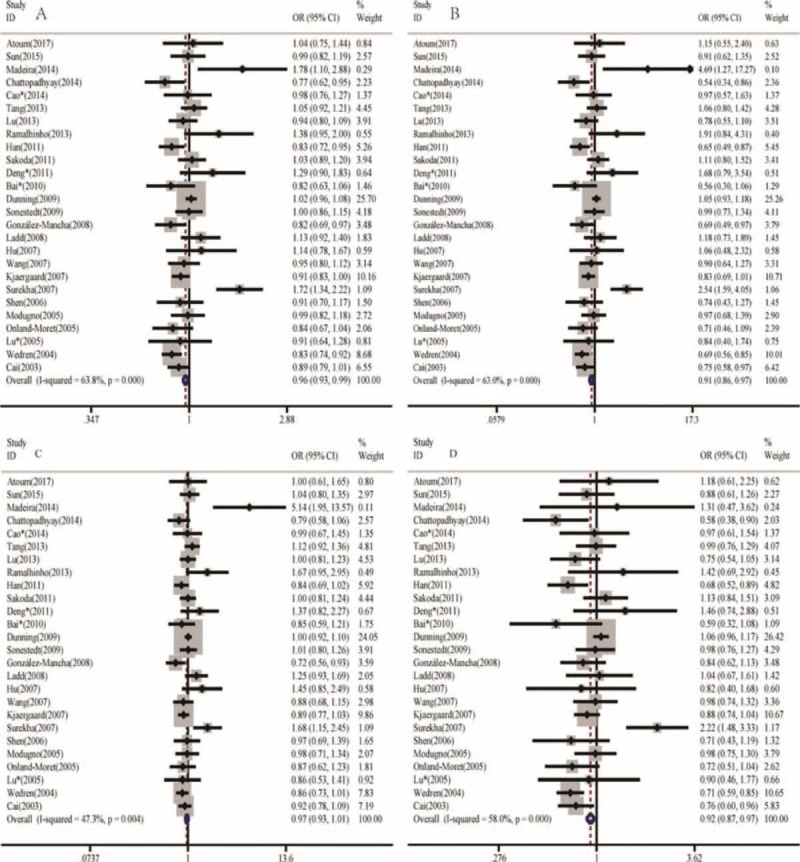
Significant association between ERα gene Pvu II polymorphism and breast cancer risk. (A) Allele model (C vs T, P = .012), (B) Homozygote model (CC vs TT, P = .003), (C) Dominant model (CC/CT vs TT, P = .187), and (D) Recessive model (CC vs TT/CT, P = .004).
Subgroup analysis was conducted based on ethnicity, and the results suggested that in Asians, a significant association between ERα gene Pvu II polymorphism and the decrease of breast cancer risk was shown in homozygous model and recessive model, whereas in Caucasians, no association was found between ERα gene Pvu II polymorphism and the decrease of breast cancer risk. In population-based subgroup rather than in hospital-based subgroup, the results of analysis based on the source of healthy controls demonstrated that a significant association between ERα gene Pvu II polymorphism and the decrease of breast cancer risk was found in allele model, homozygous model, dominant model, and recessive model, as summarized in Table 2.
Table 2.
Meta-analysis results of association between ERα gene Pvu II polymorphism and breast cancer risk.
3.3. Analysis of publication bias and sensitivity
Funnel plot and Egger test were used to analyze the publication bias in above 4 models, but no significant publication bias was found, as shown in Fig. 3A and B. The Egger results were C vs T: t = 0.71, P = .487; CC vs TT: t = 0.56, P = .582; CC/CT vs TT: t = 0.74, P = .465; CC vs TT/CT: t = 0.53, P = .601. The sensitivity analysis also indicated stable results, as shown in Fig. 3C.
Figure 3.
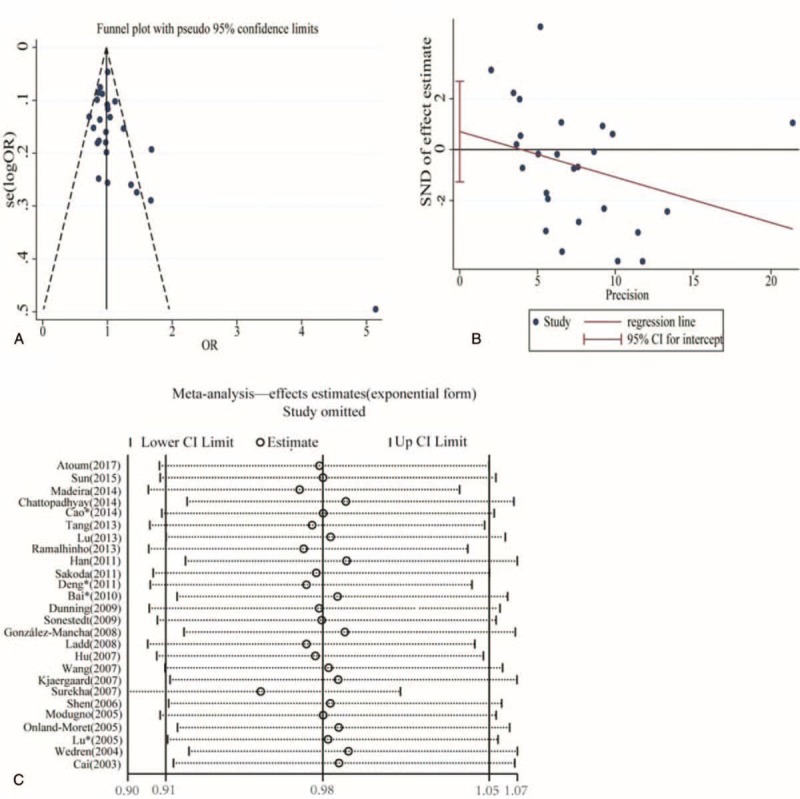
Publication bias and sensitivity analysis of studies included in the meta-analysis of dominant model. (A) Funnel plot, (B) Egger test (P = .465), (C) Sensitivity analysis.
4. Discussion
Breast cancer is the most common malignancy in females, which affect females’ physiological and psychological health seriously. At present, genetic polymorphism has become one of the hot topics in the studies on risk factors of breast cancer. Although the association between ERα gene PvuII polymorphism and breast cancer risk has been reported widely, the conclusions vary in different studies due to the difference of study design, study methods, regions and ethnicities, etc. Multiple studies have reported that patients carried with T allele exhibit relatively low morbidity of breast cancer.[15,16,22] However, Cai et al[40] pointed out that TT could increase the risk of breast cancer. Some researchers believed that there was no association between ERα gene PvuII polymorphism and breast cancer risk.[14,20] Therefore, this study mainly analyzed 26 studies on the association between ERα gene Pvu II polymorphism and breast cancer susceptibility through meta-analysis, and obtained more accurate and objective conclusion by reducing randomization error and increasing the power of test.
This meta-analysis mainly enrolled 15,360 cases and 26,423 controls, and the results revealed that the breast cancer susceptibility decreased significantly in people carried with ERα gene Pvu II CC genotype or C allele, which was similar to the conclusions of Lu et al,[13] Zhang et al,[17] and Li et al.[18] However, how ERα gene intron Pvu II affect the receptor function to participate in the pathogenesis of breast cancer is still unclear. A study has shown that Pvu IIare near to ERα gene promotor, which may impact the stability of other genes or mRNA transcription so as to affect the expressions of other genes.[41] In addition, some introns may regulate the expression levels of proteins by regulating the gene transcription.[42] Another study has reported that compared with T allele, C allele could produce the function-binding site of transcription factor B-myb, so as to influence the transcription level of ERα gene.[43] Liu et al[44] reported that ERα played an important role in the active regulation of p53, as the combination of ERα and p53 could inhibit the function of wild p53 so that the wild p53could not suppress the tumor growth and metastasis of ERα-positive breast cancer. However, whether CC phenotype participates in the development and progression of breast cancer via these pathways needs to be further studied and verified.
Factors such as ethnicity, region, living environment, and age exert certain function in the development and progression of breast cancer. We found in the subgroup analysis based on ethnicity that there was significant association between ERα gene Pvu II polymorphism and the decrease of breast cancer risk in Asians rather than in Caucasians. Lu et al,[13] Zhang et al,[17] and Li et al[18] also reported the significant association between ERα gene Pvu II polymorphism and the decrease of breast cancer risk in Asians. This difference might be caused by the genetic heterogeneity among different ethnicities, or was connected with interaction between genes, the linkage disequilibrium between SNPs sites, regions, living environment, and lifestyles. There are significant differences in the distribution of Pvu II polymorphism in European, Asian, and African populations. Most genetic phenomenon, such as natural selection, mutation, random shift, genetic hitchhiking, or gene flow, can cause large amount of linkage disequilibrium, which may also occur in different ethnicities or populations. In addition, intergenetic interaction and gene–environment interaction may also trigger distinct contributions of gene to tumorigenesis among different ethnicities.[45]
The difference in selection of controls may lead to between-study heterogeneity. Therefore, this study conducted a subgroup analysis based on the source of controls, and the results illustrated that in population-based subgroup rather than hospital-based subgroup, there was a significant association between ERα gene Pvu II polymorphism and the decrease of breast cancer risk. The morbidity of breast cancer may increase greatly in hospital-based controls in the future, which may trigger significant difference between different subgroups. However, Li et al[13] did not show that the association between Pvu II polymorphism and breast cancer risk was in any association with the source of controls.
Although researchers in pre-study have reported the association between ERα gene Pvu II polymorphism and breast cancer risk,[13,18,24] this study was more in the number of articles enrolled and larger in sample size, which comparatively reduced the influence of contingency on the results of meta-analysis. Therefore, the conclusions of this study were more persuasive and accurate.
However, this study also had some limitations described as follows. First, the studies enrolled lacked of data of family history, smoking, alcoholic consumption, age, and other environmental exposure factors; therefore, its OR values were nonadjusted data. Second, stratified analysis was not conducted based on pathological patterns, as the studies enrolled lacked of complete data on pathological patterns. Third, this study did not analyze the influence of interaction of different genes and interaction of genes with environment on the association of gene polymorphism and breast cancer risk. Fourth, the selection bias of controls by researchers in studies could not be avoided. Therefore, a comprehensive analysis of larger sample size and sample information is needed in the future so as to obtain more accurate conclusion on the association between ERα gene Pvu II polymorphism and breast cancer susceptibility.
In conclusion, this meta-analysis has found that ERα gene Pvu II polymorphism has participated in the development and progression of breast cancer. However, larger sample size, more accurate sample information, and more strict and reasonable study design are needed in the future to comprehensively verify the association between ERα gene Pvu II polymorphism and breast cancer risk.
Author contributions
Conceptualization: Shun-e Yang.
Data curation: Yan Li.
Formal analysis: Shun-e Yang.
Investigation: Zhen-hui Zhao.
Methodology: Cui-zhen Zhang.
Resources: Zhen-hui Zhao, Shun-e Yang.
Software: Zhen-hui Zhao.
Supervision: Zhen-hui Zhao, Shun-e Yang.
Validation: Zhen-hui Zhao, Shun-e Yang.
Visualization: Shun-e Yang.
Writing – original draft: Zhen-Lian Zhang.
Footnotes
Abbreviations: CBM = China Biomedicine, CI = confidence interval, CNKI = China National Knowledge Infrastructure, ER = estrogen receptors, ERα = estrogen receptor α, ERE = estrogen response element, HB = hospital-based, HWE = Hardy–Weinberg equilibrium, MALDI-TOF = matrix-assisted laser desorption ionization time-of-flight, NOS = Newcastle–Ottawa Scale, OR = odds ratio, PB = population-based study, PB = population-based, PCR-RFLP = polymerase chain reaction-restriction fragment length polymorphism, SNPs = single nucleotide polymorphisms.
Z-LZ and C-ZZ contributed equally to this article.
Funding/support: This study was approved by Science & Technology Department of Xinjiang Uygur Autonomous Region Natural Science Foundation Surface Project (No.: 2013211A068).
The authors have no conflicts of interest to disclose.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [2].Chen WQ, Zheng RS, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [3].Goss PE, Strasser-Weippl K, Lee-Bychkovsky BL, et al. Challenges to effective cancer control in China, India, and Russia. Lancet Oncol 2014;15:489–538. [DOI] [PubMed] [Google Scholar]
- [4].Li L, Ji J, Wang JB, et al. Attributable causes of breast cancer and ovarian cancer in China: reproductive factors, oral contraceptives and hormone replacement therapy. Chin J Cancer Res 2012;24:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].McCormack VA, Boffetta P. Today's lifestyles, tomorrow's cancers: trends in lifestyle risk factors for cancer in low- and middle-income countries. Ann Oncol 2011;22:2349–57. [DOI] [PubMed] [Google Scholar]
- [6].Mallepell S, Krust A, Chambon P, et al. Paracrine signaling through the epithelial estrogen receptor alpha is required for proliferation and morphogenesis in the mammary gland. Proc Natl Acad Sci U S A 2006;103:2196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhang W, Yu YY. Polymorphisms of short tandem repeat of genes and breast cancer susceptibility. Eur J Surg Oncol 2007;33:529–34. [DOI] [PubMed] [Google Scholar]
- [8].Li T, Zhao J, Yang J, et al. A meta-analysis of the association between ESR1 genetic variants and the risk of breast cancer. PLoS One 2016;11:e0153314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gu Z, Wang G, Chen W. Estrogen receptor alpha gene polymorphisms and risk of prostate cancer: a meta-analysis involving 18 studies. Tumour Biol 2014;35:5921–30. [DOI] [PubMed] [Google Scholar]
- [10].Cai L, Zhang JW, Xue XX, et al. Meta-analysis of associations of IL1 receptor antagonist and estrogen receptor gene polymorphisms with systemic lupus erythematosus susceptibility. PLoS One 2014;9:e109712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang T. Meta-analysis of PvuII, XbaI variants in ESR1 gene and the risk of Alzheimer's disease: the regional European difference. Neurosci Lett 2014;574:41–6. [DOI] [PubMed] [Google Scholar]
- [12].Tang LY, Chen LJ, Qi ML, et al. Effects of passive smoking on breast cancer risk in pre/post-menopausal women as modifed by polymorphisms of PARP1 and ESR1. Gene 2013;524:84–9. [DOI] [PubMed] [Google Scholar]
- [13].Li LW, Xu L. Menopausal status modifes breast cancer risk associated with ESR1 PvuII and XbaI polymorphisms in Asian women: a HuGE review and meta-analysis. Asian Pac J Cancer Prev 2012;13:5105–11. [DOI] [PubMed] [Google Scholar]
- [14].Atoum MF, Alzoughool F. Reduction in breast cancer susceptibility due to XbaI gene polymorphism of alpha estrogen receptor gene in Jordanians. Breast Cancer (Dove Med Press) 2017;9:45–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Madeira KP, Daltoe RD, Sirtoli GM, et al. Estrogen receptor alpha (ERS1) SNPs c454-397T > C (PvuII) and c454-351A > G (XbaI) are risk biomarkers for breast cancer development. Mol Biol Rep 2014;41:5459–66. [DOI] [PubMed] [Google Scholar]
- [16].Chattopadhyay S, Siddiqui S, Akhtar MS, et al. Genetic polymorphisms of ESR1, ESR2, CYP17A1, and CYP19A1 and the risk of breast cancer: a case control study from North India. Tumor Biol 2014;35:4517–27. [DOI] [PubMed] [Google Scholar]
- [17].Lu H, Chen D, Hu LP, et al. Estrogen receptor alpha gene polymorphisms and breast cancer risk: a case-control study with meta-analysis combined. Asian Pac J Cancer Prev 2014;14:6743–9. [DOI] [PubMed] [Google Scholar]
- [18].Zhang Y, Zhang M, Yuan X, et al. Association between ESR1 PvuII, XbaI, and P325P polymorphisms and breast cancer susceptibility: a meta-analysis. Med Sci Monit 2015;21:2986–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xu H, Linfei J, Chenhui T, et al. Association of three single nucleotide polymorphisms of ESR1 with breast cancer susceptibility: a meta-analysis. J Biomed Res 2017;31:213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sun MY, Du HY, Zhu AN, et al. Genetic polymorphisms in estrogen-related genes and the risk of breast cancer among Han Chinese women. Int J Mol Sci 2015;16:4121–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cao LQ, Li H, Liu L, et al. Er(gene polymorphism and breast cancer risk among females in sichuan province: a case-control study. J Cancer Control Treat 2014;27:171–5. [Google Scholar]
- [22].Ramalhinho AC, Marques J, Fonseca-Moutinho J, et al. Genetic polymorphims of estrogen receptor alpha 2397 PvuII (T > C) and 2351 XbaI (A > G) in a portuguese population: prevalence and relation with breast cancer susceptibility. Mol Biol Rep 2013;40:5093–103. [DOI] [PubMed] [Google Scholar]
- [23].Han J, Jiang T, Bai H, et al. Genetic variants of 6q25 and breast cancer susceptibility: two-stage fine mapping study in a Chinese population. Breast Cancer Res Treat 2011;129:901–7. [DOI] [PubMed] [Google Scholar]
- [24].Sakoda LC, Blackston CR, Doherty JA, et al. Selected estrogen receptor 1 and androgen receptor gene polymorphisms in relation to risk of breast cancer and fibrocystic breast conditions among Chinese women. Cancer Epidemiol 2011;35:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Deng LL, Lu YF. Research on polymorphism of estrogen ( receptor sites xba i and pvu ii in relation to breast cancer. Chinese J Oncol Prev Treat 2011;3:19–22. [Google Scholar]
- [26].Bai YH, Lu H, Huang YZ, et al. Association between polymorphisms of estrogen receptor alpha and vitamin d receptor gene and breast cancer risk. Chinese J Pub Heal 2010;26:1525–7. [Google Scholar]
- [27].Dunning AM, Healey CS, Baynes C, et al. Association of ESR1 gene tagging SNPs with breast cancer risk. Hum Mol Genet 2009;18:1131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sonestedt E, Ivarsson MIL, Harlid S, et al. The protective association of high plasma enterolactone with breast cancer is reasonably robust in women with polymorphisms in the estrogen receptor alpha and beta genes. J Nutr 2009;139:993–1001. [DOI] [PubMed] [Google Scholar]
- [29].González-Mancha R, Galán JJ, Crespo C, et al. Analysis of the ERalpha germline PvuII marker in breast cancer risk. Med Sci Monit 2008;14:CR136–43. [PubMed] [Google Scholar]
- [30].Ladd AMGZ, Vasquez AA, Rivadeneira F, et al. Estrogen receptor alpha polymorphisms and postmenopausal breast cancer risk. Breast Cancer Res Treat 2008;107:415–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hu Z, Song CG, Lu JS, et al. A multigenic study on breast cancer risk associated with genetic polymorphisms of ER Alpha, COMT and CYP19 gene in BRCA1/BRCA2 negative Shanghai women with early onset breast cancer or affected relatives. J Cancer Res Clin Oncol 2007;133:969–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang J, Higuchi R, Modugno F, et al. Estrogen receptor alpha haplotypes and breast cancer risk in older Caucasian women. Breast Cancer Res Treat 2007;106:273–80. [DOI] [PubMed] [Google Scholar]
- [33].Kjaergaard AD, Ellervik C, Tybjaerg-Hansen A, et al. Estrogen receptor alpha polymorphism and risk of cardiovascular disease, cancer, and hip fracture: cross-sectional, cohort, and case-control studies and a meta-analysis. Circulation 2007;115:861–71. [DOI] [PubMed] [Google Scholar]
- [34].Surekha D, Vishnupriya S, Rao DN, et al. PvuII polymorphism of estrogen receptor-( gene in breast cancer. Indian J Hum Genet 2007;13:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shen Y, Li DK, Wu J, et al. Joint effects of the CYP1A1 MspI, ERalpha PvuII, and ERalpha XbaI polymorphisms on the risk of breast cancer: results from a population-based case-control study in Shanghai, China. Cancer Epidemiol Biomarkers Prev 2006;15:342–7. [DOI] [PubMed] [Google Scholar]
- [36].Modugno F, Zmuda JM, Potter D, et al. Association of estrogen receptor a polymorphisms with breast cancer risk in older Caucasian women. Int J Cancer 2005;116:984–91. [DOI] [PubMed] [Google Scholar]
- [37].Onland-Moret NC, van Gils CH, Roest M, et al. The estrogen receptor alpha gene and breast cancer risk (The Netherlands). Cancer Causes Control 2005;16:1195–202. [DOI] [PubMed] [Google Scholar]
- [38].Lu X, Li B, Wei JM, et al. The Xba I and Pvu II gene polymorphisms of the estrogen receptor ( gene in Chinese women with breast cancer. Chin J Surg 2005;43:290–3. [PubMed] [Google Scholar]
- [39].Wedrén S, Lovmar L, Humphreys K, et al. Oestrogen receptor alpha gene haplotype and postmenopausal breast cancer risk: a case control study. Breast Cancer Res Treat 2004;6:R437–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cai Q, Shu XO, Jin F, et al. Genetic polymorphisms in the estrogen receptor alpha gene and risk of breast cancer: results from the Shanghai Breast Cancer Study. Cancer Epidemiol Biomarkers Prev 2003;12:853–9. [PubMed] [Google Scholar]
- [41].Goessl C, Plaschke J, Pistorius S, et al. An intronic germline transition in the HNPCC gene hMSH2 is associated with sporadic colorectal cancer. Eur J Cancer 1997;33:1869–74. [DOI] [PubMed] [Google Scholar]
- [42].Aronow B, Lattier D, Silbiger R, et al. Evidence for a complex regulatory array in the first intron of the human adenosine deaminase gene. Genes Dev 1989;3:1384–400. [DOI] [PubMed] [Google Scholar]
- [43].Herrington DM, Howard TD, Brosnihan KB, et al. Common estrogen receptor polymorphism augments effects of hormone replacement therapy on E-Selectin but not C-reactive protein. Circulation 2002;105:1879–82. [DOI] [PubMed] [Google Scholar]
- [44].Liu W, Konduri SD, Bansal S, et al. Estrogen receptor-alpha binds p53 tumor suppressor protein directly and represses its function. J Biol Chem 2006;281:9837–40. [DOI] [PubMed] [Google Scholar]
- [45].Goldstein DB, Weale ME. Population genomics: linkage disequilibrium holds the key. Curr Biol 2001;11:R576–9. [DOI] [PubMed] [Google Scholar]


