Abstract
Case reports suggest an association between second‐generation antipsychotics (SGAs) and serotonin syndrome (SS). The US Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) was analyzed to generate hypotheses about how SGAs may interact with pharmacological targets associated with SS. FAERS was integrated with additional sources to link information about adverse events with drugs and targets. Using Proportional Reporting Ratios, we identified signals that were further investigated with the literature to evaluate mechanistic hypotheses formed from the integrated FAERS data. Analysis revealed common pharmacological targets perturbed in both SGA and SS cases, indicating that SGAs may induce SS. The literature also supported 5‐HT2A antagonism and 5‐HT1A agonism as common mechanisms that may explain the SGA‐SS association. Additionally, integrated FAERS data mining and case studies suggest that interactions between SGAs and other serotonergic agents may increase the risk for SS. Computational analysis can provide additional insights into the mechanisms underlying the relationship between SGAs and SS.
Study Highlights
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
✓ Case reports exist that associate second‐generation antipsychotics with serotonin syndrome. A systematic analysis to identify common targets associated with both serotonin syndrome and second‐generation antipsychotics has not been performed.
WHAT QUESTION DID THIS STUDY ADDRESS?
✓ Molecular mechanisms of serotonin syndrome and second‐generation antipsychotics were examined to determine their relationships. The molecular characterization relied on analysis of adverse events using FAERS reports and drug target data.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
✓ This study provides additional insight into the molecular mechanisms of serotonin syndrome and their possible association with second‐generation antipsychotic activity. 5‐HT1A agonism and 5‐HT2A antagonism were identified as potential mechanisms of second‐generation antipsychotic‐associated serotonin syndrome.
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE
✓ Bioinformatics tools can be used to detect drugs, drug pairs, and targets associated with adverse events.
Serotonin syndrome is a classification of potentially life‐threatening symptoms resulting from increased serotonin neurotransmission. This increase of serotonin can be the result of an overdose, drug interactions, increased therapeutic dose, or recreational drug use of a chemical that increases serotonin concentrations. Symptoms may include agitation, hallucinations, hyperthermia, tachycardia, and muscle twitching. First‐line treatment of serotonin syndrome includes removal of the involved drug(s) and supportive care, which can include benzodiazepines and cyproheptadine, a nonspecific serotonin antagonist, to counteract the increased serotonin synaptic levels.1 Serotonin syndrome can result from agonism of any of the seven families of serotonin receptors, although stimulation of 5‐HT1A and 5‐HT2A receptors have been primarily implicated.1
Second‐generation antipsychotic drugs were developed as an alternative to the first‐generation antipsychotics to treat a wide variety of conditions, including schizophrenia, bipolar disorder, and as an adjunctive treatment in major depressive disorder. Second‐generation antipsychotic drugs were intended to cause fewer side effects (e.g., extrapyramidal symptoms) than first‐generation antipsychotics. These drugs are known to antagonize the dopamine (D2) and serotonin (5‐HT2A) receptors, but many also act as partial agonists at the 5‐HT1A and/or 5‐HT1B receptors.2
In this study we performed a mechanistic analysis using computational methods to understand the association between serotonin syndrome and second‐generation antipsychotics. This mechanistic analysis examined several variables using informatics, including potential pharmacokinetic and pharmacodynamic interactions, single targets, and the possibility of confounding to create multiple hypotheses for the association between serotonin syndrome and second‐generation antipsychotics. A large number of serotonin syndrome cases have been reported to the US Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) across the second‐generation antipsychotics. Based on the initial profiles presented above, partial agonism of 5‐HT1A may be one mechanism. However, second‐generation antipsychotics are serotonin antagonists, and therefore mechanistically one might expect the class to not be associated with serotonin syndrome. Therefore, we performed a mechanistic analysis via computational and case analyses to determine if the signal was truly associated with second‐generation antipsychotics, what activity was responsible, and to identify other potential mechanisms through data mining. Additionally, this method can identify artificial inflations due to concomitant medications and other confounding factors. Data mining using a variety of informatics tools can quickly identify potential mechanisms, including those involving secondary targets, to further explore using a variety of in vivo and in vitro data. Analysis was performed with a bioinformatics tool, EFFECTTM, using the Proportional Reporting Ratio (PRR) to first hypothesize about potential mechanistic associations using FAERS data integrated with target and mechanistic data. Concomitant medications were additionally analyzed in FAERS to investigate the possibility of a drug–drug interaction. These hypotheses were further investigated with literature and case analyses to provide additional evidence for the mechanisms hypothesized by using integrated FAERS data.
MATERIALS AND METHODS
Molecular Health's EFFECT3 platform was used to perform adverse event analysis. The platform summarizes FAERS data integration results and their Proportional Reporting Ratio (PRR) characterization, as described below.
Adverse event data set
The analysis was performed using the public data set of adverse events collected by the FDA's FAERS. FAERS information used in this analysis included patients’ treatments (medications), indications (disease or condition), as well as reported adverse reactions and observed outcomes (e.g., “death” or “hospitalization”). Data included in this analysis were the publicly available FAERS data set from 2004 to 2016Q2. This data set contained 8.2 million reports for 6.8 million cases.
FAERS data integration
FAERS data regarding treatments, indications, and adverse reactions were consolidated in the following way:
FAERS medication synonyms (coded in free‐text names) were mapped to drugs and compounds from DrugBank3 and PubChem.4
Drugs were further categorized according to the Anatomical Therapeutic Chemical (ATC) classification system.5
FAERS indications and reactions (coded in Medical Dictionary for Regulatory Activities (MedDRA) dictionary terms) were further contextualized by using the full hierarchical structure of MedDRA.6
Integration of molecular data
Using a drug‐centric integration process, FAERS was enhanced with chemical and biological data. Based on the FAERS medication‐to‐drug mapping, the link to biomolecules and molecular mechanisms involved in pharmacodynamics and pharmacokinetics was established via DrugBank.3 Additional information for the integrated molecular players (such as drug targets and metabolizing enzymes) was imported from UniProt,7 Reactome,8, 9 and BioCarta.10
Statistical characterization of the adverse event data
For the statistical characterization of the observed associations in FAERS between reported drugs, indications, reactions, outcomes, and molecular players, the established pharmacovigilance metric PRR was used. PRR quantifies the extent of a particular binary association with respect to the occurrence of each of its parts. The PRR metric was calculated using the approach described by van Puijenbroek et al.11 and shown in the below example.
Experiments: Definition of cohorts
Several approaches were used to analyze potential signals and the association between serotonin syndrome and second‐generation antipsychotics (see Figure 1). Using FAERS data, patient cohorts were defined as collections of adverse event cases that contain a drug or reaction of interest, or multiple drugs combined and examined as one group. The identified cohorts were then examined with respect to features such as reported drugs, indications, reactions, or drug classes, underlying targets and mechanisms, and their associations with serotonin syndrome. Cohort results are described in terms of observed case counts (i.e., number of cases a certain occurrence was observed in) and the PRR disproportionality score. Specifically:
The serotonin syndrome cohort: Cases were identified that reported serotonin syndrome as a reaction. This set of 6,935 reports was used to profile what is reported in FAERS about serotonin syndrome, to confirm the quality of the data integration process, and to examine how many patients experience serotonin syndrome had taken second‐generation antipsychotics. Finally, this set was used to identify those drug targets that were most frequently linked to serotonin syndrome cases.
The second‐generation antipsychotics cohort: Cases were identified that reported a second‐generation antipsychotic. The following drugs were considered: amisulpride, aripiprazole, asenapine, blonanserin, clozapine, iloperidone, lurasidone, melperone, olanzapine, paliperidone, perospirone, quetiapine, risperidone, sertindole, ziprasidone, zotepine, and cariprazine. This cohort was used to identify drug targets that linked to most second‐generation antipsychotic‐reporting cases and contrast them with those identified for serotonin syndrome.
Receptor agonism/antagonism cohorts: In order to identify their role with respect to the mechanisms underlying serotonin syndrome, the molecular targets perturbed in both the serotonin syndrome and second‐generation antipsychotic cohorts were examined further. For each target, the occurrence of serotonin syndrome was characterized in two cohorts—one when the target is inhibited (antagonism) and one when the target is induced (agonism). These cohorts were defined by looking at those cases that had the drugs associated with the activity of interest reported. For a drug‐target association to be considered, DrugBank was required to have both the “actions” (such as “inhibitor” or “antagonist”) and the “pharmacological action” annotations explicitly determined.
The selective serotonin reuptake inhibitors cohort: Selective serotonin reuptake inhibitors have a known association with serotonin syndrome. To compare selective serotonin reuptake inhibitor activity and serotonin syndrome statistical association with second‐generation antipsychotics, cases were identified where a selective serotonin reuptake inhibitor was reported. The following drugs were considered: duloxetine, nefazodone, desvenlafaxine, milnacipran, venlafaxine, levomilnacipran, dapoxetine, indalpine, fluvoxamine, citalopram, fluoxetine, paroxetine, sertraline, escitalopram, zimelidine, and etoperidone.
Figure 1.
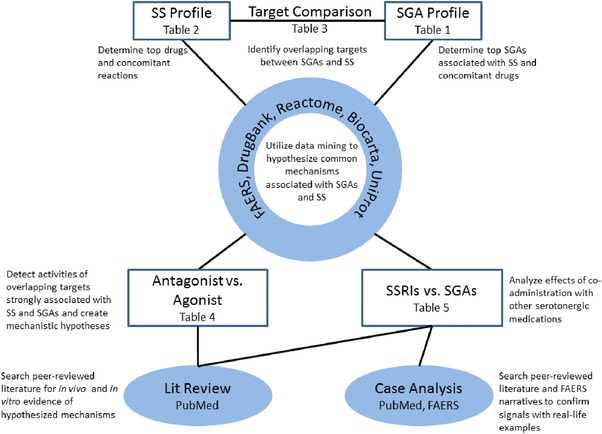
Methods of analyzing the relationship between serotonin syndrome and second‐generation antipsychotics. A variety of computational and expert analyses were used to explore the mechanistic relationship between serotonin syndrome and second‐generation antipsychotics. SS: Serotonin syndrome; SGA: Second‐generation antipsychotic; SSRI: Selective serotonin reuptake inhibitor.
RESULTS
FAERS analysis of serotonin syndrome cases
First, a FAERS analysis was performed to create a mechanistic hypothesis for the association between serotonin syndrome and second‐generation antipsychotics by using a bioinformatics tool that makes connections between adverse events and drugs, targets, and molecular pathways. A total of 6,935 cases of serotonin syndrome were reported to FAERS, and these cases were coreported with 892 total drugs, 816 disease indications, and 2,256 additional reactions.
The most commonly reported adverse reactions along with serotonin syndrome included drug interaction (N: 2,097, PRR: 33.82), tremor (N: 869, PRR: 12.06), and hyperhidrosis (N: 747, PRR: 13.58). Tremor and hyperhidrosis are commonly associated with serotonin syndrome and are in accordance with symptoms on the Hunter Toxicity Criteria Decision Rules and the Sternbach Criteria.12 Additionally, drug interactions are frequently reported in serotonin syndrome cases; this finding agrees with a previous study performed with the French pharmacovigilance database that examined serotonin syndrome cases and found drug–drug interactions primarily responsible for a large proportion of cases.13
Finally, cases of serotonin syndrome reported with second‐generation antipsychotics were examined (Table 1). The strongest statistical signals were seen for lurasidone (N: 21, PRR: 8.71) and ziprasidone (N: 95, PRR: 6.27), while quetiapine had the most reports (N: 417, PRR: 5.23). An analysis of concomitant medications in cases of serotonin syndrome associated with second‐generation antipsychotics revealed several potential confounders, including many serotonergic agents, such as selective serotonin reuptake inhibitors, and potential serotonin syndrome treatments, such as benzodiazepines (Table 2). Serotonin syndrome occurrence was also found high within the overall second‐generation antipsychotic cohort (N: 1,168, PRR: 4.79).
Table 1.
Second‐generation antipsychotics and serotonin syndrome reports in FAERS
| Drug | N | PRR | PRR CI |
|---|---|---|---|
| Quetiapine | 417 | 5.23 | 4.74–5.77 |
| Risperidone | 269 | 4.49 | 3.97–5.07 |
| Olanzapine | 251 | 5.48 | 4.84–6.22 |
| Aripiprazole | 183 | 3.86 | 3.33–4.47 |
| Ziprasidone | 95 | 6.27 | 5.12–7.67 |
| Clozapine | 70 | 1.73 | 1.37–2.19 |
| Lurasidone | 21 | 8.71 | 5.69–13.34 |
| Asenapine | 16 | 2.53 | 1.55–4.13 |
| Paliperidone | 7 | 0.30 | 0.14–0.63 |
This list represents the second‐generation antipsychotics investigated in this study and the correlating case count, PRR score, and 95% confidence interval for the PRR score (PRR CI) for each drug with serotonin syndrome.
Table 2.
Top 20 concomitant drugs with second‐generation antipsychotics and serotonin syndrome
| Concomitant drug | N |
|---|---|
| Citalopram | 159 |
| Sertraline | 150 |
| Mirtazapine | 148 |
| Venlafaxine | 140 |
| Clonazepam | 139 |
| Lorazepam | 121 |
| Lithium | 118 |
| Fluoxetine | 112 |
| Paroxetine | 108 |
| Duloxetine | 104 |
| Escitalopram | 101 |
| Tramadol | 96 |
| Lamotrigine | 88 |
| Trazodone | 86 |
| Acetaminophen | 85 |
| Alprazolam | 75 |
| Ondansetron | 74 |
| Bupropion | 73 |
| Diazepam | 71 |
| Fentanyl | 63 |
This list represents the top reported drugs given when a case of serotonin syndrome is associated with a second‐generation antipsychotic. From this list, we identified that second‐generation antipsychotics and serotonin syndrome are often associated with another serotonergic drug. Total case count (N) is 1,140 reports.
FAERS analysis of molecular targets
Targets potentially involved in serotonin syndrome mechanisms were identified by looking at the drug‐target associations linked to the adverse events of the serotonin syndrome cohort. Similarly, targets involved in second‐generation antipsychotic adverse events were also examined; a second‐generation antipsychotic cohort was defined (consisting of AEs where at least one second‐generation antipsychotic was reported) and the targets perturbed by the drugs in each case were analyzed.
Table 3 juxtaposes the 20 most frequent targets linked to serotonin syndrome cases with the 20 most frequent targets linked to second‐generation antipsychotic cases. Top targets for second‐generation antipsychotics and serotonin syndrome included 5‐HT1A, 5‐HT2A, D2, and 5‐HT2C. Of these 20 individual targets, 11 targets overlapped between serotonin syndrome and second‐generation antipsychotics. We hypothesized that one or more of these 11 targets may be involved in the mechanisms associated with serotonin syndrome and second‐generation antipsychotics, and thus, these targets were selected for further computational analysis.
Table 3.
The top 20 targets associated with serotonin syndrome or second‐generation antipsychotics (excluding CYPs) in integrated FAERS data
| 20 Top targets associated with serotonin syndrome | 20 Top targets associated with second‐generation antipsychotics | ||||
|---|---|---|---|---|---|
| PRR | PRR | ||||
| Target | N | PRR CI | Target | N | PRR CI |
| Sodium‐dependent serotonin transporter | 6,135 | 8.47 | 5‐HT1a receptor | 274,178 | 27.28 |
| 8.39–8.54 | 27.17–27.39 | ||||
| Multidrug resistance protein 1 | 5,719 | 1.68 | 5‐HT2a receptor | 274,178 | 23.34 |
| 1.66–1.69 | 23.26–23.43 | ||||
| Sodium‐dependent noradrenaline transporter | 5,023 | 8.07 | Alpha‐2C adrenergic receptor | 274,178 | 26.41 |
| 7.95–8.19 | 26.31–26.51 | ||||
| Sodium‐dependent dopamine transporter | 4,492 | 8.67 | D2 dopamine receptor | 274,178 | 26.01 |
| 8.52–8.82 | 25.91–26.11 | ||||
| Muscarinic acetylcholine receptor M1 | 3,487 | 4.98 | Alpha‐2a adrenergic receptor | 273,537 | 19.52 |
| 4.87–5.10 | 19.45–19.58 | ||||
| Alpha‐1a adrenergic receptor | 3,397 | 4.43 | Histamine H1 receptor | 272,980 | 14.59 |
| 4.32–4.53 | 14.55–14.64 | ||||
| Histamine H1 receptor | 3,362 | 4.60 | Alpha‐1a adrenergic receptor | 272,967 | 13.48 |
| 4.49–4.72 | 13.44–13.52 | ||||
| 5‐HT2a receptor | 3,055 | 5.43 | D1a dopamine receptor | 272,725 | 64.48 |
| 5.29–5.58 | 64.08–64.88 | ||||
| 5‐HT2c receptor | 3,014 | 5.97 | D3 dopamine receptor | 272,721 | 62.79 |
| 5.81–6.14 | 62.41–63.17 | ||||
| Muscarinic acetylcholine receptor M3 | 2,983 | 4.88 | D4 dopamine receptor | 272,662 | 112.80 |
| 4.75–5.02 | 111.88–113.72 | ||||
| Serum albumin | 2,871 | 1.41 | 5‐HT2c receptor | 272,177 | 28.82 |
| 1.37–1.45 | 28.70–28.94 | ||||
| Kappa‐type opioid receptor | 2,760 | 5.63 | Alpha‐2b adrenergic receptor | 272,127 | 27.90 |
| 5.46–5.79 | 27.79–28.01 | ||||
| Mu‐type opioid receptor | 2,634 | 4.50 | Alpha‐1b adrenergic receptor | 267,103 | 17.74 |
| 4.37–4.64 | 17.68–17.80 | ||||
| Delta‐type opioid receptor | 2,495 | 4.51 | 5‐HT1d receptor | 266,733 | 59.03 |
| 4.37–4.65 | 58.67–59.38 | ||||
| 5‐HT1a receptor | 2,487 | 4.77 | Multidrug resistance protein 1 | 230,210 | 1.76 |
| 4.62–4.92 | 1.75–1.76 | ||||
| Alpha‐2a adrenergic receptor | 2,485 | 4.02 | 5‐HT7 receptor | 217,891 | 58.44 |
| 3.90–4.15 | 58.05–58.84 | ||||
| Alpha‐1‐acid glycoprotein 1 | 2,465 | 2.99 | Muscarinic acetylcholine receptor M1 | 216,532 | 10.91 |
| 2.89–3.08 | 10.88–10.95 | ||||
| Muscarinic acetylcholine receptor M2 | 2,450 | 3.98 | 5‐HT6 receptor | 215,175 | 83.58 |
| 3.85–4.11 | 82.90–84.26 | ||||
| D2 dopamine receptor | 2,368 | 4.43 | 5‐HT1b receptor | 214,824 | 28.64 |
| 4.29–4.58 | 28.49–28.78 | ||||
| Alpha‐1b adrenergic receptor | 2,260 | 3.55 | Muscarinic acetylcholine receptor M3 | 212,787 | 13.05 |
| 3.43–3.67 | 13.0–13.10 | ||||
This list depicts the top targets associated with second‐generation antipsychotics or serotonin syndrome in integrated FAERS data. Targets that are shared between the two cohorts that were used for mechanistic hypothesis generation are bolded. The association of some of these targets resulted from the multiple target activities of the drugs analyzed. PRR CI: 95% confidence interval of the PRR score.
The 11 overlapping targets in Table 3 were individually analyzed to determine if agonism or antagonism of each target is primarily responsible for the serotonin syndrome signal. Individual cohorts of each target's agonists and antagonists were analyzed for serotonin syndrome occurrence (Table 4). The strongest signals were seen for 5‐HT1A agonism, 5‐HT1A antagonism, 5‐HT1B agonism, 5‐HT2A antagonism, 5‐HT2C agonism, and alpha‐2A antagonism. Based on this analysis, we hypothesized that second‐generation antipsychotics could be associated with serotonin syndrome through at least one of these mechanisms. These six target mechanisms were individually investigated for their associations with second‐generation antipsychotics and serotonin syndrome in peer‐reviewed literature to determine if one or more hypothesized mechanisms linked the two entities. This literature analysis provided further support for two proposed mechanisms: 5‐HT1A agonism and 5‐HT2A antagonism (Figure 2).2, 14
Table 4.
Targets of interest and serotonin syndrome in integrated FAERS data
| Agonist | Antagonist | |||||
|---|---|---|---|---|---|---|
| Target | Case count with serotonin syndrome | PRR | PRR CI | Case count with serotonin syndrome | PRR | PRR CI |
| Targets associated with serotonin syndrome and second‐generation antipsychotics from FAERS | ||||||
| 5‐HT1A | 639 | 8.36 | 7.71–9.07 | 110 | 7.39 | 6.13–8.92 |
| 5‐HT1B | 261 | 7.06 | 6.24–7.99 | N/A | N/A | N/A |
| 5‐HT2A | 4 | 2.26 | 0.85–6.03 | 2,326 | 7.74 | 7.37–8.14 |
| 5‐HT2C | 389 | 9.39 | 8.48–10.39 | 225 | 5.87 | 5.14–6.70 |
| M1 | 259 | 4.76 | 4.21–5.39 | 354 | 3.07 | 2.76–3.42 |
| M3 | 2 | 0.61 | 0.15–2.45 | 238 | 1.24 | 1.09–1.41 |
| Alpha‐1A | 104 | 1.36 | 1.12–1.65 | 169 | 1.11 | 0.95–1.29 |
| Alpha‐2A | 163 | 1.47 | 1.26–1.71 | 526 | 16.87 | 15.44–18.42 |
| D2 | 164 | 2.69 | 2.30–3.14 | 1,369 | 5.05 | 4.76–5.36 |
| H1 | N/A | N/A | N/A | 508 | 1.79 | 1.62–1.95 |
| Targets Known to be Associated with Serotonin Syndrome | ||||||
|---|---|---|---|---|---|---|
| SERT | N/A | N/A | N/A | 2,044 | 15.20 | 14.43–16.0 |
| Mu‐type opioid receptor | 2,270 | 6.11 | 5.81–6.42 | 42 | 1.64 | 1.21–2.23 |
| 5‐HT3A | 3 | 2.51 | 0.81–7.78 | 896 | 9.82 | 9.16–10.54 |
This list represents the targets of interest investigated in this study and the correlating case count, PRR score, and 95% confidence interval for the PRR score (PRR CI) for each target with serotonin syndrome.
Figure 2.
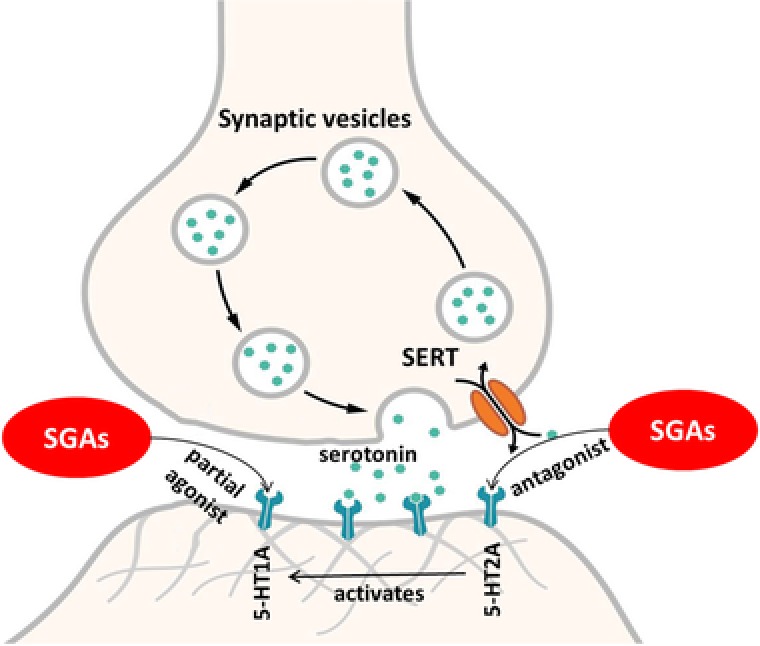
Mechanisms of serotonin syndrome. Depiction of the actions of the second‐generation antipsychotics on the serotonergic system. SGA: Second‐generation antipsychotic; SERT: Serotonin transporter. © User: Splette/Wikimedia Commons/CC‐BY‐SA‐4.0 (https://commons.wikimedia.org/wiki/File:SynapseSchematic_unlabeled.svg).
FAERS analysis of concomitant medications
The possibility of a drug–drug interaction or confounding was also investigated by examining concomitant medications. Confounding may occur if the second‐generation antipsychotics are often seen in cases of serotonin syndrome by chance, but it is truly another drug that is associated with serotonin syndrome mechanistically. A drug–drug interaction may occur if two drugs may independently be associated with serotonin syndrome, but the risk of serotonin syndrome increases when both drugs are taken together. Concomitant medications were analyzed as described previously (Table 2), and this analysis noted several serotonergic agents, particularly selective serotonin reuptake inhibitors were often taken concomitantly with second‐generation antipsychotics in serotonin syndrome cases. Therefore, the significant statistical signal noted in FAERS for second‐generation antipsychotics and serotonin syndrome may be attributable to confounding or a drug–drug interaction. To determine the relationship between second‐generation antipsychotics, serotonergic drugs, such as selective serotonin reuptake inhibitors, and serotonin syndrome, four additional cohorts were created: FAERS cases that include selective serotonin reuptake inhibitors but not second‐generation antipsychotics, FAERS cases that include second‐generation antipsychotics but not selective serotonin reuptake inhibitors, FAERS cases that include both selective serotonin reuptake inhibitors and second‐generation antipsychotics, and a cohort that includes all FAERS cases with selective serotonin reuptake inhibitors, similar to the original second‐generation antipsychotic cohort. All cohorts were then analyzed for their relationship with serotonin syndrome (Table 5).
Table 5.
Interaction of second‐generation antipsychotics with selective serotonin reuptake inhibitors in serotonin syndrome cases
| Total number of cases | Number of cases of serotonin syndrome | PRR | PRR CI | |
|---|---|---|---|---|
| Second‐generation antipsychotics | 275,543 | 1,168 | 4.79 | 4.50–5.10 |
| Second‐generation antipsychotics w/o selective serotonin reuptake inhibitors | 208,643 | 326 | 1.56 | 1.39–1.74 |
| Second‐generation antipsychotics w/ selective serotonin reuptake inhibitors | 66,900 | 842 | 13.89 | 12.93–14.92 |
| Selective serotonin reuptake inhibitors w/o second‐generation antipsychotics | 403,641 | 4,390 | 27.30 | 26.0–28.66 |
| Selective serotonin reuptake inhibitors | 470,541 | 5,232 | 41.27 | 39.08–43.59 |
This table shows the cohorts created for selective serotonin reuptake inhibitors and second‐generation antipsychotics to analyze their relationships with serotonin syndrome. Five cohorts were created for this query: second‐generation antipsychotics, which includes all FAERS cases with second‐generation antipsychotics, second‐generation antipsychotics without selective serotonin reuptake inhibitors, which includes FAERS cases that include second‐generation antipsychotics but not selective serotonin reuptake inhibitors, second‐generation antipsychotics with selective serotonin reuptake inhibitors, which includes FAERS cases that include both selective serotonin reuptake inhibitors and second‐generation antipsychotics, selective serotonin reuptake inhibitors without second‐generation antipsychotics, which includes FAERS cases with selective serotonin reuptake inhibitors but not second‐generation antipsychotics, and selective serotonin reuptake inhibitors, which includes all FAERS cases with selective serotonin reuptake inhibitors. PRR CI: 95% confidence interval of the PRR score.
The cohort including all FAERS cases with selective serotonin reuptake inhibitors displayed a strong signal for serotonin syndrome (N: 5,232, PRR: 41.27) compared with the second‐generation antipsychotics (N: 1,168, PRR: 4.79). Notably, a cohort for second‐generation antipsychotics without selective serotonin reuptake inhibitors had a weakened signal (N: 326, PRR: 1.56). Additionally, by restricting FAERS data to only serotonin syndrome cases with both second‐generation antipsychotics and selective serotonin reuptake inhibitors, the signal increased significantly compared with second‐generation antipsychotics alone (N: 842, PRR: 13.89). These three cohorts indicate that cases involving both selective serotonin reuptake inhibitors and second‐generation antipsychotics are more disproportionally associated with serotonin syndrome compared with cases involving second‐generation antipsychotics alone. This suggests a potential drug–drug interaction, as there is a signal for serotonin syndrome when second‐generation antipsychotics are evaluated alone, but the disproportionality score increases when second‐generation antipsychotics and selective serotonin reuptake inhibitors are given together. Additionally, a cohort of selective serotonin reuptake inhibitors without second‐generation antipsychotics had a weaker signal for serotonin syndrome (N: 4,390, PRR: 27.30) compared with the general selective serotonin reuptake inhibitors, indicating that cases with second‐generation antipsychotics heavily contribute to the signal seen for selective serotonin reuptake inhibitors. As before, this suggests a drug–drug interaction, as the removal of second‐generation antipsychotics lowers the disproportionality score and suggests that the two drug classes increase each other's risk for serotonin syndrome. These fluctuations in signal strength in the second‐generation antipsychotic and selective serotonin reuptake inhibitor cohorts indicate that second‐generation antipsychotics and selective serotonin reuptake inhibitors may act synergistically to induce serotonin syndrome. Further analysis was performed with the literature, which supported that pharmacodynamic and pharmacokinetic interactions between second‐generation antipsychotics and other drugs, especially serotonergic agents, may increase the risk for serotonin syndrome.15, 16, 17, 18
DISCUSSION
This study identified drugs and the common receptors that they target that are associated with serotonin syndrome and examined potential mechanisms for the association between serotonin syndrome and second‐generation antipsychotics. This analysis integrated FAERS data with molecular data from other sources to produce hypotheses about the mechanisms that may explain the association between serotonin syndrome and second‐generation antipsychotics.
The FAERS analysis demonstrated that the function of second‐generation antipsychotic targets overlap with receptor functions that are commonly associated with serotonin syndrome. Eleven targets of interest were identified that overlapped between second‐generation antipsychotics and serotonin syndrome, which were further narrowed down to six targets and activities of interest (agonism of 5‐HT1A, 5‐HT1B, and 5‐HT2C and antagonism of 5‐HT1A, 5‐HT2A, and alpha‐2A) based on their association with serotonin syndrome. Additionally, analysis of concomitant medications with FAERS demonstrated a potentially synergistic effect between second‐generation antipsychotics and selective serotonin reuptake inhibitors, as the disproportionality score for serotonin syndrome was higher for second‐generation antipsychotics and selective serotonin reuptake inhibitors combined than for second‐generation antipsychotics individually. Put together, analysis of the integrated FAERS data alone generated two hypotheses: first, six targets and activities of interest were hypothesized to be associated with both second‐generation antipsychotics and serotonin syndrome, and second, concomitant administration of additional serotonergic medications may increase the risk of serotonin syndrome with second‐generation antipsychotics. Further literature investigation supported these hypotheses.
First, the six target mechanisms of interest that were identified with integrated FAERS data were further investigated using peer‐reviewed literature. This literature analysis provided further support for two proposed mechanisms: 5‐HT2A antagonism and 5‐HT1A agonism. The literature supports that second‐generation antipsychotics are 5‐HT2A antagonists.2 There is evidence that antagonism of 5‐HT2A may cause selective activation of 5‐HT1A, which may in turn increase the sensitivity of this receptor to serotonin.19, 20, 21 Additionally, data have shown that 5‐HT2A antagonists enhance the effects of a 5‐HT1A agonist.22, 23 Additionally, many second‐generation antipsychotics have also been shown to be 5‐HT1A agonists14 5‐HT1A agonism has been implicated in increased serotonin levels and an increased risk for serotonin syndrome.24 These two mechanisms, in combination, may increase serotonin levels, increase receptor sensitivity to serotonin, or increase the effects of other drugs (such as selective serotonin reuptake inhibitors), potentially leading to serotonin syndrome.
Second, based on our findings of potential interactions with concomitant medications, including serotonergic medications, we additionally investigated the associations between second‐generation antipsychotics and metabolic enzymes that may lead to drug–drug interactions using the literature. Many second‐generation antipsychotics are metabolized via CYP3A4, CYP2D6, or CYP1A2 and are substrates and inhibitors of P‐glycoprotein.15, 16, 17 Many other drugs also interact with these enzymes and transporters, including several serotonergic agents.18 These interactions could lead to increased concentrations of various drugs or serotonin levels, which could be associated with serotonin syndrome. Finally, FAERS and literature case analyses demonstrated that second‐generation antipsychotics alone can be associated with serotonin syndrome, but in most reported cases serotonin syndrome occurs when second‐generation antipsychotics are given with other serotonergic agents.20, 25, 26
This analysis combines FAERS data with several other data sources to connect drugs, adverse events, and targets, among many other entities. In this analysis, FAERS data were used for hypothesis generation. These data and the signals generated are used regularly at the FDA to evaluate safety concerns, and the large amount of data (over 8 million reports) contained in public FAERS from healthcare professionals, consumers, and manufacturers includes valuable information. FAERS and similar postmarket reporting databases have previously been used successfully for adverse event analysis.27, 28, 29 However, FAERS also has limitations, including confounded cases, reporting biases, and a lack of confirmation that the case even occurred. For example, serotonin syndrome could be miscoded in FAERS as neuroleptic malignant syndrome, or vice versa. These limitations can all affect the statistics calculated from FAERS. FAERS should not be used to infer causality. Thus, FAERS may be used for hypothesis generation, but identified signals should be further investigated for confirmation, as performed here.
Future work includes the integration of a systems biology approach to further examine targets, proteins, and genes that may be associated with a variety of adverse events and drugs. Additionally, off‐target prediction is under investigation for routine integration into this methodology to evaluate secondary or off‐target effects that may be responsible for adverse events. Similar methodologies have previously been successfully employed to analyze adverse events.30, 31
This study demonstrated that informatics tools and analyses can quickly identify drugs, targets, drug–drug interactions, and mechanisms that may be associated with adverse events. Computational analysis performed with FAERS and supporting pharmacokinetic data from the literature and case studies show that second‐generation antipsychotics are associated with serotonin syndrome. Further research should be performed to elucidate the mechanism of 5‐HT2A/5‐HT1A interaction and serotonin levels after administration of second‐generation antipsychotics to better clarify the risk of serotonin syndrome with second‐generation antipsychotics. Additionally, drug–drug interaction studies may explain the magnitude of increased risk for serotonin syndrome when second‐generation antipsychotics are taken concomitantly with other serotonergic drugs. Confirmation of these mechanisms may impact patient treatment and result in new combinations of drugs, lower dosages of drug regimens, or increased patient monitoring to reduce the risk of serotonin syndrome.
Conflict of Interest
Theodoros Soldatos and David Jackson are employees of Molecular Health, GmbH.
Acknowledgments and disclaimer
This work was performed as part of a Research Collaboration Agreement between the FDA and Molecular Health GmbH, Heidelberg, Germany. The findings and conclusions in this article reflect the views of the authors and should not be construed to represent the FDA's views or policies. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services.
Source of Funding
No funding was received for this work.
Author Contributions
R.R., T.S., and K.B. wrote the article; R.R., T.S., and K.B. designed the research; R.R. and T.S. performed the research; R.R., T.S., and K.B. analyzed the data; D.J. contributed new reagents/analytical tools.
References
- 1. Boyer, E.W. Serotonin syndrome (serotonin toxicity). In (ed. Traub S.J.), UpToDate. (UpToDate, Waltham, MA, 2016). [Google Scholar]
- 2. Mauri, M.C. , et al. Clinical pharmacology of a typical antipsychotics: an update. EXCLI J. 13, 1163–1191 (2014). [PMC free article] [PubMed] [Google Scholar]
- 3. Wishart, D.S. , et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim, S. , et al. PubChem substance and compound databases. Nucleic Acids Res. 44, D1202–D1213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. WHO Collaborating Centre for Drug Statistics Methodology . Anatomical Therapeutic Chemical (ATC) Classification System. (Oslo, Norway, 2015). [Google Scholar]
- 6. International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals of Human Use . Medical Dictionary for Regulatory Activities (MedDRA). McLean, VA. [Google Scholar]
- 7. UniProt, C. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fabregat, A. , et al. The reactome pathway knowledgebase. Nucleic Acids Res. 44, D481–D487 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Milacic, M. , et al. Annotating cancer variants and anti‐cancer therapeutics in reactome. Cancers (Basel). 4, 1180–1211 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. National Cancer Institute . BioCarta San Diego, CA. Available from: https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways.
- 11. van Puijenbroek, E.P. , Bate, A. , Leufkens, H.G. , Lindquist, M. , Orre, R. , & Egberts, A.C. A comparison of measures of disproportionality for signal detection in spontaneous reporting systems for adverse drug reactions. Pharmacoepidemiol. Drug Saf. 11, 3–10 (2002). [DOI] [PubMed] [Google Scholar]
- 12. Dunkley, E.J. , Isbister, G.K. , Sibbritt, D. , Dawson, A.H. , & Whyte, I.M . The hunter serotonin toxicity criteria: simple and accurate diagnostic decision rules for serotonin toxicity. QJM. 96, 635–642 (2003). [DOI] [PubMed] [Google Scholar]
- 13. Abadie, D. , Rousseau, V. , Logerot, S. , Cottin, J. , Montastruc, J.L. , & Montastruc, F. Serotonin syndrome: Analysis of cases registered in the french pharmacovigilance database. J. Clin. Psychopharmacol. 35, 382–388 (2015). [DOI] [PubMed] [Google Scholar]
- 14. Schreiber, R. , & Newman‐Tancredi, A . Improving cognition in schizophrenia with antipsychotics that elicit neurogenesis through 5‐HT(1A) receptor activation. Neurobiol. Learn. Mem. 110, 72–80 (2014). [DOI] [PubMed] [Google Scholar]
- 15. Kennedy, W.K. , Jann, M.W. , & Kutscher, E.C. Clinically significant drug interactions with atypical antipsychotics. CNS Drugs. 27, 1021–1048 (2013). [DOI] [PubMed] [Google Scholar]
- 16. Moons, T. , de Roo, M. , Claes, S. , & Dom, G . Relationship between P‐glycoprotein and second‐generation antipsychotics. Pharmacogenomics. 12, 1193–1211 (2011). [DOI] [PubMed] [Google Scholar]
- 17. O'Brien, F.E. , Dinan, T.G. , Griffin, B.T. , & Cryan, J.F. Interactions between antidepressants and P‐glycoprotein at the blood‐brain barrier: clinical significance of in vitro and in vivo findings. Br. J. Pharmacol. 165, 289–312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schellander, R. , & Donnerer, J. Antidepressants: clinically relevant drug interactions to be considered. Pharmacology. 86, 203–215 (2010). [DOI] [PubMed] [Google Scholar]
- 19. Stahl, S.M. Basic psychopharmacology of antidepressants, part 1: Antidepressants have seven distinct mechanisms of action. J. Clin. Psychiatry. 59, 5–14 (1998). [PubMed] [Google Scholar]
- 20. Duggal, H.S. , & Fetchko, J . Serotonin syndrome and atypical antipsychotics. Am. J. Psychiatry. 159, 672–673 (2002). [DOI] [PubMed] [Google Scholar]
- 21. Dvir, Y. , & Smallwood, P . Serotonin syndrome: a complex but easily avoidable condition. Gen. Hosp. Psychiatry. 30, 284–287 (2008). [DOI] [PubMed] [Google Scholar]
- 22. Backus, L.I. , Sharp, T. , & Grahame‐Smith, D.G. Behavioural evidence for a functional interaction between central 5‐HT2 and 5‐HT1A receptors. Br. J. Pharmacol. 100, 793–799 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marek, G.J. , Carpenter, L.L. , McDougle, C.J. , & Price, L.H. Synergistic action of 5‐HT2A antagonists and selective serotonin reuptake inhibitors in neuropsychiatric disorders. Neuropsychopharmacology. 28, 402–412 (2003). [DOI] [PubMed] [Google Scholar]
- 24. Mills, K.C. Serotonin syndrome. A clinical update. Crit. Care Clin. 13, 763–783 (1997). [DOI] [PubMed] [Google Scholar]
- 25. Lin, P.Y. , Hong, C.J. , & Tsai, S.J. Serotonin syndrome caused by ziprasidone alone. Psychiatry Clin. Neurosci. 64, 338–339 (2010). [DOI] [PubMed] [Google Scholar]
- 26. Kohen, I. , Gordon, M.L. , Manu, P . Serotonin syndrome in elderly patients treated for psychotic depression with atypical antipsychotics and antidepressants: two case reports. CNS Spectr. 12, 596–598 (2007). [DOI] [PubMed] [Google Scholar]
- 27. Voss, E.A. , Boyce, R.D. , Ryan, P.B. , van der Lei, J. , Rijnbeek, P.R. , & Schuemie, M.J. Accuracy of an automated knowledge base for identifying drug adverse reactions. J. Biomed. Inform. 66, 72–81 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang, Z. , Clark, N.R. , & Ma'ayan A. Drug‐induced adverse events prediction with the LINCS L1000 data. Bioinformatics. 32, 2338–2345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gurulingappa, H. , Toldo, L. , Rajput, A.M. , Kors, J.A. , Taweel, A. , & Tayrouz, Y . Automatic detection of adverse events to predict drug label changes using text and data mining techniques. Pharmacoepidemiol. Drug Saf. 22, 1189–1194 (2013). [DOI] [PubMed] [Google Scholar]
- 30. Liu, T. , & Altman, R.B. Relating essential proteins to drug side‐effects using canonical component analysis: a structure‐based approach. J. Chem. Inf. Model. 55, 1483–1494 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mestres, J. , Seifert, S.A. , & Oprea, T.I. Linking pharmacology to clinical reports: cyclobenzaprine and its possible association with serotonin syndrome. Clin. Pharmacol. Ther. 90, 662–665 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]


