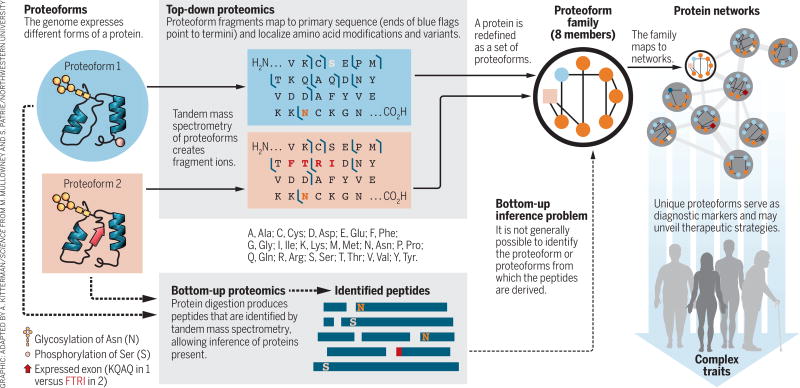
Proteoforms—the different forms of proteins produced from the genome with a variety of sequence variations, splice isoforms, and myriad posttranslational modifications (1)—are critical elements in all biological systems (see the figure, left). Yang et al. (2) recently showed that the functions of proteins produced from splice variants from a given gene—different proteoforms—can be as different as those for proteins encoded by entirely different genes. Li et al. (3) showed that splice variants play a central role in modulating complex traits. However, the standard paradigm of proteomic analysis, the “bottom-up” strategy pioneered by Eng and Yates some 20 years ago (4), does not directly identify proteoforms. We argue that proteomic analysis needs to provide the identities and abundances of the proteoforms themselves, rather than just their peptide surrogates. Developing new proteome-wide strategies to accomplish this goal presents a formidable but not insurmountable technological challenge that will benefit the biomedical community.
Figure. Identifying proteoforms within their families and protein networks.
Proteoforms underlie complex traits and molecular mechanisms in biology. Top-down (whole protein) and bottom-up (peptide) proteomics methods are compared.
The function of proteins can be strongly modulated by posttranslational modifications (PTMs) such as phosphorylation (consider kinase cascades), acetylation, methylation (consider histones), and many more of the >400 known PTMs in biology. These sources of variation combine to create a complex and largely uncharted world of natural proteins. Knowledge of the identities and quantities of these proteoforms present in dynamic biological systems is indispensable to development of a complete picture of functional regulation at the protein level.
Conventional proteomics digests protein mixtures into peptides, some of which are identified by tandem mass spectrometry (MS). Each identified peptide acts as a surrogate for the presence of the protein molecule from which it is derived. This strategy provides invaluable information on protein expression in complex systems. However, as many different gene products, isoforms, and proteoforms can contain the same peptide, direct information about the proteoforms present is lost (see the figure, bottom). This issue is the proteomic analog of the problem of “phasing” in genomics (5)—determining whether multiple alleles are present on the same segment of DNA. The step of digestion into peptides is essential to the success and robustness of the bottom-up strategy, as well-behaved peptides are more amenable to liquid chromatographic separation and MS analysis than are intact proteins. However, only inferences can be made as to the actual proteoform or proteoforms from which the identified peptide was derived (6).
An alternative approach is “top-down” proteomics, in which whole proteins are analyzed directly using tandem MS methods (see the figure, top left). Although great strides have recently been made in the topdown analysis of high-mass proteins (7) and complex proteomic samples (8), limitations remain to be addressed in the degree of sequence coverage and the ability to analyze low-abundance species. A complementary approach reported the proteome-wide identification of proteoforms in yeast, based primarily upon a high-accuracy determination of their intact mass, aided by a corollary measurement of the number of lysine residues in the molecule (9). Comparison of the measured masses and lysine counts with a theoretical database of possible yeast proteoforms yielded proteoform identifications. Further comparisons of all experimental masses with one another revealed related proteoforms differing by common PTMs, yielding more identifications. These pairwise relations (experimental:theoretical and experimental:experimental) were assembled into “families” of related proteoforms (see the figure, top center).
Such “proteoform families” offer a new and more detailed way of viewing the proteome (see the figure, top right). To extend the strategy to mammalian genomes, RNA sequencing can be used to construct sample-specific proteoform databases that capture the genetic variation and extent of splicing patterns in the sample (10, 11). Integrating such proteogenomic data with synergistic information obtained from bottom-up (for PTM identification and localization), top-down (for protein identification and PTM localization), and intact mass measurements (for proteoform identification) can provide the comprehensive analysis needed to broadly identify and quantify proteoforms in complex samples.
The question of how many proteoforms exist in nature quickly arises in this discussion (12). This question may prove impossible to answer fully, as errors in transcription and translation can produce numerous low-abundance proteoforms, perhaps as few as only a single molecule per cell, or even a single molecule in a large population of cells. We currently can only detect proteoforms present at concentrations above the instrumental detection limits of existing mass spectrometers, although the advent of single-molecule nanopore or other strategies for proteoform identification may change that landscape in the future.
However, the number and variety of proteoforms expressed in biological systems appear to be far below the calculated combinatorial possibilities (12). Garcia and coworkers have pioneered MS methods for histone proteoform analysis, finding much smaller numbers of histone proteoform variants than the maximal number of combinatorial possibilities would suggest (13). Similarly, in a deep study of histone H4 proteoforms by Coon and co-workers, only 74 were identified (14). This stands in striking contrast to the ~3 million possibilities that are theoretically possible from the combinatorial explosion of known site-specific modifications (14). This difference may simply indicate that many or most proteoforms are not detectable with current technology, and that we are only able to see at present the few of those that are most abundant. Alternatively, nature may only make and use a small subset of the proteoforms that are theoretically possible, as deduced from the combinatorial possibilities offered by considering all of the various possible PTM combinations. Understanding which of these explanations is correct, or perhaps a blend of both, will require improved technologies that can reveal proteoforms at ever lower abundance.
Proteoform analyses will become increasingly straightforward as information is accrued and archived on the proteoforms that actually exist in nature and can be observed. Establishing a comprehensive atlas of identified proteoforms for human and other species has begun, and over time this atlas will begin to yield transformative insights into the levels and roles of proteoform complexity present in biological systems. As proteoforms are tightly linked to the functioning of cells and tissues that underlie complex phenotypes, their identification and quantification will provide critical insights into the fundamental workings of biological systems (see the figure, top right). Proteoforms should also help identify key diagnostic markers and therapeutic targets and thereby provide greater statistical power for deciphering human disease phenotypes.
Acknowledgments
We thank J. Loo, J. Chamot-Rooke, L. Pasa-Tolic, Y. Ge, and Y. Tsybin for their comments and suggestions and D. Walt for pointing out the potential effects of errors in transcription and translation. The Proteoform Atlas is supported by a grant from the Paul G. Allen Family Foundation (http://repository.topdown-proteomics.org; Award 11715). We thank the National Institute of General Medical Sciences for their support under grants 1R01GM114292 (L.M.S.) and P41 GM108569 (N.L.K.). The authors are members of the Consortium for Top Down Proteomics.
REFERENCES AND NOTES
- 1.Smith LM, et al. Nat. Methods. 2013;10:186. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X, et al. Cell. 2016;164:805. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li YI, et al. Science. 2016;352:600. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eng JK, McCormack AL, Yates JR. J. Am. Soc. Mass Spectrom. 1994;5:976. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 5.Browning SR, Browning BL. Nat. Rev. Genet. 2011;12:703. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nesvizhskii AI, Aebersold R. Mol. Cell. Proteomics. 2005;4:1419. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Han X, Jin M, Breuker K, McLafferty FW. Science. 2006;314:109. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 8.Tran JC, et al. Nature. 2011;480:254. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shortreed MR, et al. J. Proteome Res. 2016;15:1213. doi: 10.1021/acs.jproteome.5b01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, et al. J. Proteome Res. 2012;11:1009. doi: 10.1021/pr200766z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans VC, et al. Nat. Methods. 2012;9:1207. doi: 10.1038/nmeth.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aebersold R, et al. Nat. Chem. Biol. 2018;14:206. doi: 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Z-F, Arnaudo AM, Garcia BA. Annu. Rev. Analyt. Chem. 2014;7:113. doi: 10.1146/annurev-anchem-071213-015959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phanstiel D, et al. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4093. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]