SUMMARY
Amyloids are fibrous protein assemblies that are often described as irreversible and intrinsically pathogenic. However, yeast cells employ amyloid-like assemblies of the RNA-binding protein Rim4 to control translation during meiosis. Here, we show that multi-site phosphorylation of Rim4 is critical for its regulated disassembly and degradation and that failure to clear Rim4 assemblies interferes with meiotic progression. Furthermore, we identify the protein kinase Ime2 to bring about Rim4 clearance via phosphorylation of Rim4’s intrinsically disordered region. Rim4 phosphorylation leads to reversal of its amyloid-like properties and degradation by the proteasome. Our data support a model in which a threshold amount of phosphorylation, rather than modification of critical residues, is required for Rim4 clearance. Our results further demonstrate that at least some amyloid-like assemblies are not as irreversible as previously thought. We propose that the natural pathways by which cells process these structures could be deployed to act on disease-related amyloids.
eTOC BLURB
Amyloids, fibrous protein assemblies associated with numerous diseases, are often referred to as being irreversible structures. Carpenter et al. demonstrate that, in coordination with meiotic development, budding yeast are able to disassemble and clear the amyloid-like translational repressor Rim4 by multi-site phosphorylation of residues within disordered regions of the protein.
INTRODUCTION
Amyloids are fibrous protein aggregates that are associated with numerous human diseases (Knowles et al., 2014). Protein assemblies that exhibit a subset of the biochemical properties associated with disease-related amyloids such as fiber formation, stable cross-β sheets, and/or resistance to ionic detergents are termed ‘amyloid-like’. The conformational switch into an amyloid-like state is often mediated by multivalent interactions among proteins containing low-complexity regions which are intrinsically disordered and can include nucleic acids (Han et al., 2012; Kato et al., 2012; Lin et al., 2015). Although the amyloid state often represents a pathological and irreversible form of protein aggregation, amyloid and amyloid-like assemblies can have specific functions in diverse biological processes (Fowler et al., 2007). These functions include melanin biosynthesis (Fowler et al., 2005) necrotic cell death signaling (Li et al., 2012), regulation of mRNA translation (Berchowitz et al., 2015; Khan et al., 2015), and formation of bacterial biofilms (Taglialegna et al., 2016). To regulate the processes governed by amyloid-like assemblies, cells have developed pathways and mechanisms that regulate their clearance and/or accumulation. Elucidating these mechanisms is important to advance our understanding of protein homeostasis as well as the diseases associated with amyloids. In this study, we investigated the mechanism by which yeast cells coordinate clearance of intracellular amyloid-like translational repressors with a precise developmental transition during meiosis.
Sexually reproducing organisms rely on the production of haploid cells called gametes in order to pass genetic material though successive generations. Gametes are produced via a specialized cell division called meiosis in which cells segregate their chromosomes twice without an intervening DNA synthesis phase. Homologous chromosomes are segregated in the first meiotic division, whereas sister chromatids are segregated during the second meiotic division. In comparison to mitotic cell proliferation, cells undergoing meiosis rely heavily on translational control to govern gene expression (i.e. the activation and repression of previously transcribed mRNA) (Kronja and Orr-Weaver, 2011; Nakamura and Seydoux, 2008). This is best exemplified by the fact that in most organisms in which gamete development is well characterized, mRNA transcription is shut down during the meiotic program.
Budding yeast cells employ amyloid-like structures to control translation during meiosis (Berchowitz et al., 2015). At the core of these structures is the RNA-binding protein Rim4. Rim4 is essential for meiosis and contains three N-terminal RNA-recognition motifs (RRMs) and two intrinsically disordered regions (IDRs) (Soushko and Mitchell, 2000). Rim4 also harbors a prion-like domain within its asparagine and proline-rich C-terminal IDR which is algorithmically-defined based on disorder and sequence similarity to yeast prions (Alberti et al., 2009; Si et al., 2003). During pre-meiotic G1, Rim4 aggregates into heterogeneously-sized multimeric structures that are resistant to the harsh detergent SDS, a hallmark property of amyloids. Based on this property we have defined Rim4 assemblies as amyloid-like. In vitro, Rim4 forms fibrils that bind thioflavin-T and contain β-sheets. Formation and clearance of amyloid-like Rim4 is a developmentally regulated process. When Rim4 is ectopically expressed in nutrient-rich conditions, it does not form SDS-resistant multimers and is non-functional. Starvation induces conversion of Rim4 monomers into its SDS-resistant form (termed assembly). After completion of the first meiotic division, Rim4 is rapidly degraded by an unknown mechanism (termed clearance). The amyloid-like form of Rim4 is the translationally repressive form of the protein. rim4 truncation mutants that cannot form amyloid-like assemblies are defective for translational repression but retain the ability to bind RNA (Berchowitz et al., 2015). Beyond yeast, amyloid-like assemblies are also critical features of mouse and frog meiosis highlighting the conserved use of these structures in sexual reproduction (Berchowitz et al., 2015; Boke et al., 2016).
Rim4 inhibits translation of mRNAs encoding protein products that can be toxic when expressed too early during meiosis, yet at the same time are necessary for late meiotic events (Berchowitz et al., 2013; Miller et al., 2012). One important Rim4 target is the mRNA encoding the B-type cyclin CLB3 which is normally translated only during meiosis II (Carlile and Amon, 2008). When CLB3 is inappropriately translated prior to meiosis I, cells undergo a catastrophic mitosis-like division rather than undergoing meiosis I (Carlile and Amon, 2008) illustrating the importance of Rim4-mediated translational control. In addition to CLB3, Rim4 binds to and represses translation of numerous mRNAs that encode proteins critical for late meiotic events. While many of these targets (such as CLB3) are translated precisely at meiosis II onset, several others are translated later (Berchowitz et al., 2013; Brar et al., 2012). To facilitate fine-tuning of translational activation, Rim4 interacts with RNA-binding proteins that act as accessory factors (Jin et al., 2017).
Inactivation of this translational control pathway depends on the precise clearance of Rim4 during the meiosis I-meiosis II transition which takes place in a 15 to 30-minute timeframe. We previously determined that Rim4 abundance is regulated by direct phosphorylation by the protein kinase Ime2 (Berchowitz et al., 2013). However, the manner by which phosphorylation contributes to Rim4 degradation and possibly reversibility was undetermined. The mechanisms by which cells employ phosphorylation to regulate membrane-less structures are likely conserved. In multicellular organisms, the dynamics of RNP granule assembly and clearance are regulated by hyperphosphorylation of granule constituents (Paix et al., 2014; Wippich et al., 2013). Furthermore, phosphomimetic mutations in the low-complexity region of the prototypical assembly-forming RNA binding protein FUS prevent its aggregation (Monahan et al., 2017). Here, we investigated how meiotic yeast cells disassemble and clear the SDS-resistant assemblies formed by Rim4. We demonstrate that cells clear these assemblies via multi-site phosphorylation. Furthermore, we find that failure to clear Rim4 assemblies imposes a severe meiosis II progression defect. Lastly, we show that the amyloid-like properties of Rim4 are reversed prior to proteasomal degradation. Taken together our observations illustrate that phosphorylation plays a critical role in the rapid reversibility and clearance of physiological amyloid-like assemblies.
RESULTS
Phosphorylation of Rim4 drives its clearance
Amyloids and amyloid-like assemblies are usually stable and long-lived. However, meiotic cells have a remarkable- and thus far unknown- mechanism for the abrupt clearance of amyloid-like Rim4 assemblies at the meiosis I-meiosis II transition. Several lines of evidence led us to hypothesize that phosphorylation is critical for Rim4 clearance. Rim4 is a direct target of the protein kinase Ime2, and Ime2 functions in late meiosis to antagonize Rim4-mediated translational repression (Berchowitz et al., 2013). Furthermore, Ime2 activity correlates with Rim4 degradation.
To begin to understand the role of phosphorylation in Rim4 clearance we sought to determine when and where Rim4 is phosphorylated. To this end, we purified Rim4 at hourly time points from cells undergoing synchronized meiosis using the NDT80 block-release system (Figure 1A). In this system, expression of the transcription factor NDT80 is driven from the GAL1-10 promoter (pGAL-NDT80) that in turn is controlled by an estradiol-regulatable Gal4-ER fusion (GAL4.ER). pGAL-NDT80 cells will arrest in meiotic G2 in sporulation medium lacking estradiol but will progress synchronously through the meiotic divisions upon estradiol addition (Benjamin et al., 2003; Carlile and Amon, 2008). We analyzed immunopurified Rim4 peptides by mass spectrometry (Figures 1B, 1S). Using this method, we were able to analyze ~6,000 Rim4 peptides (56% coverage) including ~1,000 Rim4 phosphopeptides in which we could determine the site of phosphorylation. We identified 39 unique Rim4 phosphorylation sites- 64% of the serine and threonine residues for which we had coverage. Many of these phosphorylation sites were specific to particular time points during meiosis.
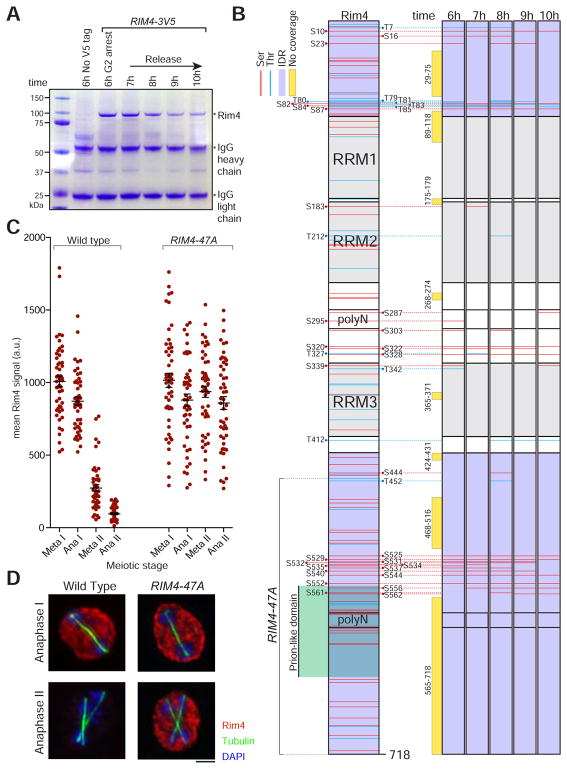
Figure 1. Rim4 is hyperphosphorylated during meiosis.
(A) Rim4 immunoprecipitation (IP) from meiotic cells. RIM4-3V5, pGAL-NDT80, GAL4.ER (B48) cells were induced to sporulate at 30°C. At 6 hours when cells had arrested in G2 due to the lack of Ndt80, cells were released from the G2 block by the addition of 1 μM β-estradiol. Lysates were prepared from G2-arrested cells (6 hour) and at 7, 8, 9 and 10 hours (post-release). G2-arrested pGAL-NDT80, GAL4.ER (A15055) was included as a no-tag control. IP was conducted under denaturing conditions using anti-V5 agarose beads. Bound factors were released by boiling in SDS-loading buffer and resolved by SDS-PAGE. Imperial blue-stained gels of the IPs are shown. Excised Rim4 bands were analyzed by mass spectrometry.
(B) Diagram of Rim4 protein and time course analysis of phosphorylation sites in meiosis by mass spectrometry. The left diagram shows the domain structure of Rim4 with all serine and threonine residues marked in red and blue, IDRs shaded in purple, and the prion-like domain shaded in green. Phosphorylated residues are denoted in black. The right diagrams show time course mapping of each phosphorylation site (phosphoserine in red and phosphothreonine in blue). Regions of no mass spectrometry coverage are shown as yellow bars adjacent to the diagrams.
(C, D) Phosphorylation of the Rim4 C-terminal IDR is required for its clearance. RIM4-3V5 (wild type, B48) and RIM4-47A-3V5 (A38075) strains harboring pGAL-NDT80, GAL4.ER, and CLB3-3HA were induced to sporulate 30°C. After 6 hours, cells were released from the G2 arrest. (C) Single-cell Rim4 levels in metaphase I, anaphase I, metaphase II, and anaphase II were determined by immunofluorescence (IF) (n = 50 cells for each meiotic stage). (D) The images show Rim4 protein (red), tubulin (green) and DNA (DAPI) in representative anaphase I and anaphase II cells.
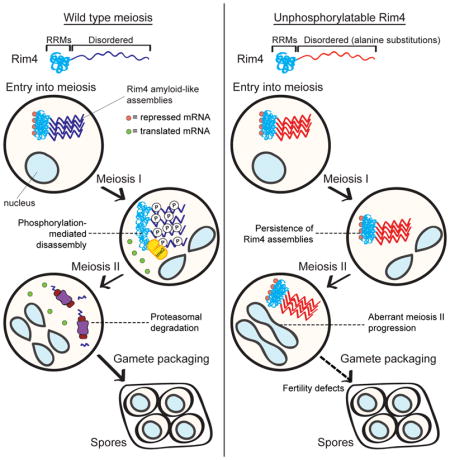
Because Rim4 is a heavily phosphorylated protein, we suspected that multi-site phosphorylation could lead to Rim4 clearance. We focused on phosphorylation in the C-terminal IDR due to its high degree of phosphorylation during meiosis and because it has previously been implicated in Rim4 clearance (Berchowitz et al., 2015). To test whether phosphorylation of the C-terminal IDR of Rim4 is important for Rim4 clearance, we mutated 47 C-terminal serine and threonine residues to alanine (henceforth RIM4-47A) and examined the phenotypic consequences. Wild type Rim4 was degraded efficiently after exit from meiosis I and prior to metaphase II. In contrast, Rim4-47A persisted well into meiosis II (Figures 1C, D). This result supports the hypothesis that phosphorylation within the Rim4 C-terminal IDR is important for its degradation. To examine whether Rim4-47A exhibits visually distinct aggregation properties compared to wild-type Rim4, we compared the abundance and distribution of Rim4-EGFP and Rim4-47A-EGFP in single cells (Figures S1A–C). We observed that Rim4-EGFP and Rim4-47A-EGFP exhibited no significant differences in either abundance or distribution in starvation-synchronized meiotic cells. These results strongly suggest that the effects of the alanine substitutions in Rim4-47A are restricted to clearance and do not affect its aggregation properties.
Aberrant Rim4 persistence results in hyper-repression of translation
We next examined the physiological consequences of aberrant Rim4 persistence on translational control and meiotic progression. As mentioned above, a critical Rim4 target is the B-type cyclin CLB3. We compared CLB3 translational control in wild type and RIM4-47A cells by measuring CLB3 mRNA and Clb3 protein in synchronized meiotic time courses. As shown previously, in wild type cells, CLB3 mRNA accumulates directly after NDT80 activation but Clb3 protein expression is restricted to meiosis II (Figures 2A, B). In contrast, RIM4-47A cells exhibit a significant delay in CLB3 translation activation as well as a reduction in maximum Clb3 protein levels (Figures 2A, B). Furthermore, RIM4-47A cells exhibit elevated CLB3 mRNA accumulation reflecting either increased CLB3 transcription and/or decreased mRNA decay. We also compared the clearance kinetics of Rim4 amyloid-like assemblies between wild-type and RIM4-47A by semi-denaturing detergent agarose gel electrophoresis (SDD-AGE). SDD-AGE allows for the resolution of amyloid-like structures based on their size and resistance to the harsh detergent SDS (Halfmann and Lindquist, 2008). Wild-type cells efficiently cleared amyloid-like Rim4 at the meiosis I-meiosis II transition. In contrast, RIM4-47A cells exhibited aberrant persistence of SDS-resistant structures well into meiosis II (Figure 2C).
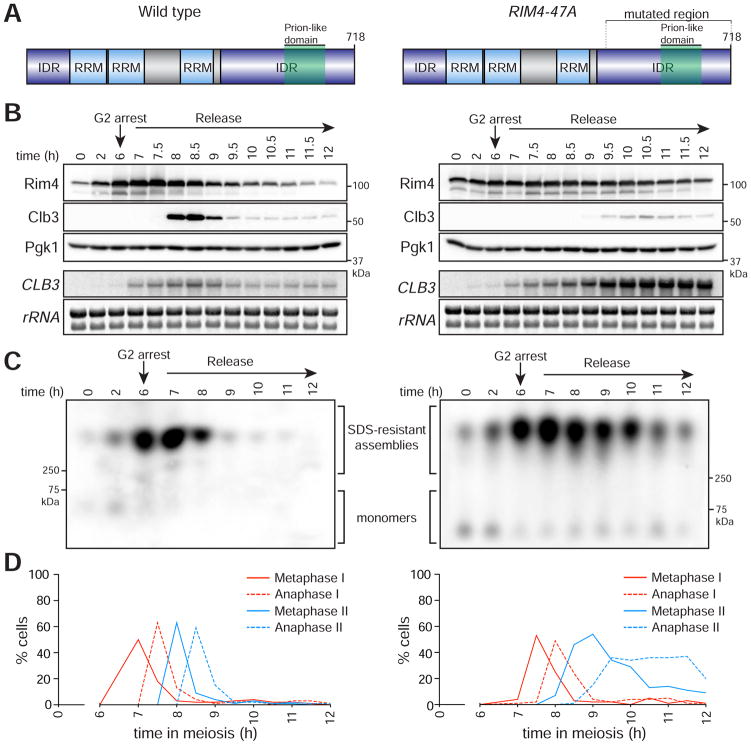
Figure 2. Phosphorylation-driven clearance of Rim4 assemblies is required for timely meiotic progression.
Left panel: wild type, right panel: RIM4-47A
(A) Diagrams of Rim4. The region mutated in the RIM4-47A allele is bracketed. This region harbors 47 serine and threonine residues mutated to alanine.
(B – D) RIM4-47A cells exhibit defective CLB3 translation, failure to clear SDS-resistant assemblies, and delayed meiotic progression. Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and either RIM4-3V5 (wild type, B48) or RIM4-47A-3V5 (A38075) were induced to sporulate 30°C. After 6 ho urs, cells were released from the G2 arrest. (B) Rim4-3V5, Clb3-3HA, and Pgk1 (loading control) protein and CLB3 mRNA and rRNA (loading control) levels were determined at the indicated times following β-estradiol addition. (C) Rim4’s ability to form SDS-resistant amyloid-like assemblies was analyzed by SDD-AGE as in (Halfmann and Lindquist, 2008) with minor modifications (see experimental procedures). (D) The percentage (n = 100 cells for each time point) of metaphase I, anaphase I, metaphase II, and anaphase II cells was determined by tubulin IF and DAPI staining.
Failure to properly disassemble and clear Rim4 amyloid-like assemblies had severe consequences on meiotic progression. RIM4-47A cells entered and exited meiosis I with kinetics similar to wild type. However, these cells exhibited significant progression delays in both metaphase II and anaphase II (Figure 2D). To quantify the meiotic progression defects in RIM4-47A cells, we compared meiotic progression in wild-type and RIM4-47A in live cells. We found that metaphase II and anaphase II were both significantly slowed (1.5 and 2.2-fold longer, respectively) in RIM4-47A cells compared to wild type (Figure S2A, B). These results strongly suggest that phosphorylation of the Rim4 C-terminal IDR is critical for clearance of Rim4 assemblies and relieving its translational inhibition of essential meiosis II targets. These results also inform whether or not Rim4 clearance and/or disassembly is required for activation of its mRNA targets. If mRNAs were released from Rim4 by a clearance-independent mechanism, CLB3 would be translated normally in the RIM4-47A background, which we do not observe. Thus, our results imply that Rim4 target mRNAs are not released into the translating pool prior to Rim4 clearance.
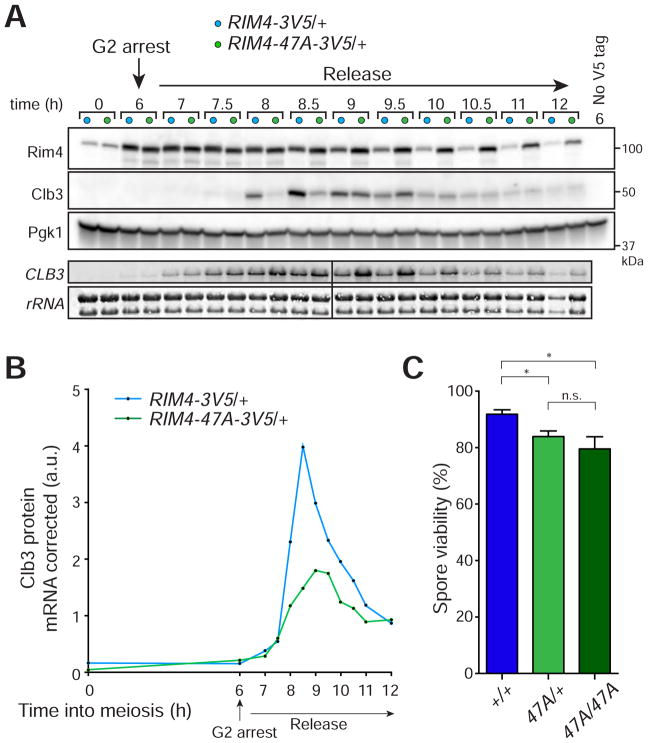
Our hypothesis that Rim4 clearance is mediated by phosphorylation of its C-terminal IDR predicts that RIM4-47A should be dominant. To test this, we compared translational control, assembly, and meiotic kinetics between wild type and RIM4-47A/+ cells as above. As with RIM4-47A/RIM4-47A cells, we found that RIM4-47A/+ cells exhibit delayed Clb3 protein synthesis, increased CLB3 mRNA accumulation and aberrant persistence of Rim4 assemblies (Figures 3A, 3B, S3). To assess if failure to clear Rim4 affects gamete viability we compared spore viability between RIM4-47A/RIM4-47A, RIM4-47A/+, and wild type strains. We found that presence of a single RIM4-47A allele was sufficient to cause a spore viability defect (Figure 3C). Interestingly, the meiotic progression phenotypes observed in RIM4-47A/+ were milder than those observed in the RIM4-47A/RIM4-47A background. This argues that the presence of Rim4-47A protein does not prevent wild type Rim4 from being cleared. Taken together these results indicate that non-clearable Rim4 has a dominant effect on protein synthesis and meiotic progression.
Figure 3. RIM4-47A is a dominant gain-of-function allele.
(A, B) A single copy of RIM4-47A is sufficient to cause CLB3 translational control defects. Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and either RIM4-3V5/+ (light blue, B119) or RIM4-47A-3V5/+ (light green, B120) were induced to sporulate at 30°C. After 6 hours, cells were released from the G2 arrest. (A) Rim4-3V5, Clb3-3HA, and Pgk1 (loading control) protein and CLB3 mRNA and rRNA (loading control) levels were determined at the indicated times following release from G2 arrest. (B) Clb3 protein levels corrected for CLB3 mRNA levels are plotted on the y-axis and time in meiosis is plotted on the x-axis. Analysis of Rim4 SDS-resistant assemblies and meiotic progression is shown in Figure S3.
(C) RIM4-47A cells exhibit reduced spore viability. Strains harboring RIM4-3V5/RIM4-3V5 (blue, B123) RIM4-47A-3V5/RIM4-3V5 (light green, B149), and RIM4-47A/RIM4-47A (green, B126) were sporulated on plates. Tetrads (n = 7 plates of 20 tetrads for each genotype) were dissected on YPD plates and spores were grown for 2 days. Shown is the percent viable spores. A Mann-Whitney test was used to statistically compare the means. * = P < 0.05
Rim4 clearance is regulated by multi-site phosphorylation
Although mutation of 47 Rim4 residues enhanced its ability to repress translation, we sought to identify the minimal essential residues in Rim4 that need to be modified to induce clearance. To this end we generated RIM4 phospho-mutant alleles in which we mutated 99, 52, 36, 27, 25, 21, 16 and 10 serine and threonine residues to alanine (rim4-99A, RIM4-52A, etc.) (Figure 4A). Importantly RIM4-21A contains no overlapping mutations with RIM4-25A and only a single overlapping mutation with RIM4-27A. To assay translational control in each mutant background, we analyzed CLB3 mRNA and Clb3 protein in synchronized meiotic time courses (Figure 4B, S4A–C). To capture the defective protein synthesis and mRNA hyper-accumulation phenotypes in a single measurement, we normalized Clb3 protein levels to CLB3 mRNA and compared the ratio to wild type at each time point.
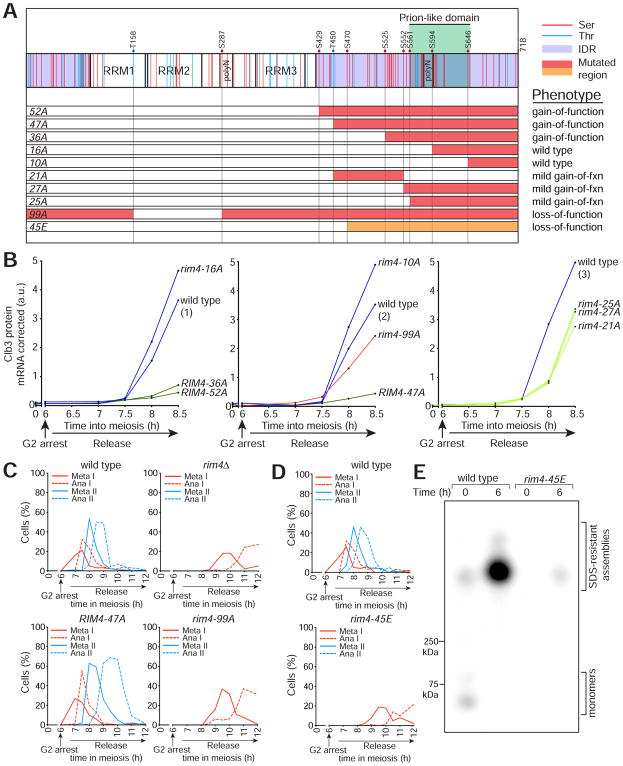
Figure 4. Rim4 clearance is regulated by multi-site phosphorylation.
(A) Diagram of Rim4 with serine and threonine residues denoted in red and blue, respectively. The IDRs are shaded in purple. Mutations contained in each RIM4 phospho-mutant allele are shaded in red or orange (S/T → A and S/T → E respectively). Translational control phenotypes (determined from (B)) are indicated on the right.
(B) CLB3 translational control analysis of RIM4 phospho-mutant alleles. Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and one of the following: RIM4-3V5 (wild type, blue, B48), rim4-99A-3V5 (red, A30860), RIM4-52A-3V5 (green, B67), RIM4-47A-3V5 (green, A38075), RIM4-36A-3V5 (green, B70), RIM4-27A-3V5 (light green, B152), RIM4-25A-3V5 (light green, B155), RIM4-21A-3V5 (light green, B129), rim4-16A-3V5 (blue, B73), or rim4-10A-3V5 (blue, A38072) were induced to sporulate 30°C. After 6 hours, cells were released from the G 2 arrest. Four strains were run per experiment, each containing a RIM4-3V5 (wild-type) control and three mutant strains. Clb3 protein levels corrected for CLB3 mRNA level are plotted on the y-axis and time in meiosis is plotted on the x-axis. Western and Northern blot source data is shown in Figure S4. (C–E) rim4-99A and rim4-45E are loss of function alleles. (C, D) Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and one of the following: RIM4-3V5 (wild type, B48), rim4Δ (B343), RIM4-47A-3V5 (A38075), rim4-99A-3V5 (A30860), and rim4-45E-3V5 (B346) were induced to sporulate at 30°C. After 6 hours, cells were released from the G2 arrest. Meiotic progression was analyzed by tubulin IF (n = 100 cells for each time point). (E) Rim4 SDS-resistant assemblies were analyzed by SDD-AGE in RIM4-3V5 (wild type) and rim4-45E-3V5 cells.
Strains harboring 52 and 36 mutated residues exhibited hyper-repressed CLB3 translation (decreased Clb3 protein and increased CLB3 mRNA) similar to a RIM4-47A mutant. Mutation of 16 and 10 residues had no detectable effect on CLB3 translation or meiotic progression. Intriguingly, strains harboring mutations in 27, 25, and 21 residues resulted in a mild gain-of-function phenotype. Because strains harboring non-overlapping stretches of serine/threonine mutations displayed similar intermediate phenotypes, we conclude that there exist no critical sites on Rim4 that must be modified to induce clearance. Rather, our results support a model of Rim4 regulation in which clearance is mediated by combinatorial accumulation of phosphorylation events. Once a threshold level of phosphorylation is reached, Rim4 clearance commences. Unsurprisingly, mutation of 99 putative phosphorylation sites in RIM4 resulted in a loss-of-function phenotype reminiscent of rim4Δ and distinctive from RIM4-47A as assessed by drastic meiotic progression failure (Figure 4C).
Our model that multi-site phosphorylation is important for Rim4 clearance predicts that phospho-mimetic mutations (alanine substituted with glutamic acid or aspartic acid) within the Rim4 IDR would cause premature clearance and thus loss of Rim4 function. This prediction was confirmed- a rim4 mutant allele harboring the 45 C-terminal serine and threonine residues to glutamic acid (rim4-45E) exhibited a severe meiotic progression defect (Figure 4D) and almost complete loss of SDS-resistant assemblies (Figure 4E).
Ime2 kinase triggers Rim4 clearance via phosphorylation of the Rim4 C-terminal IDR
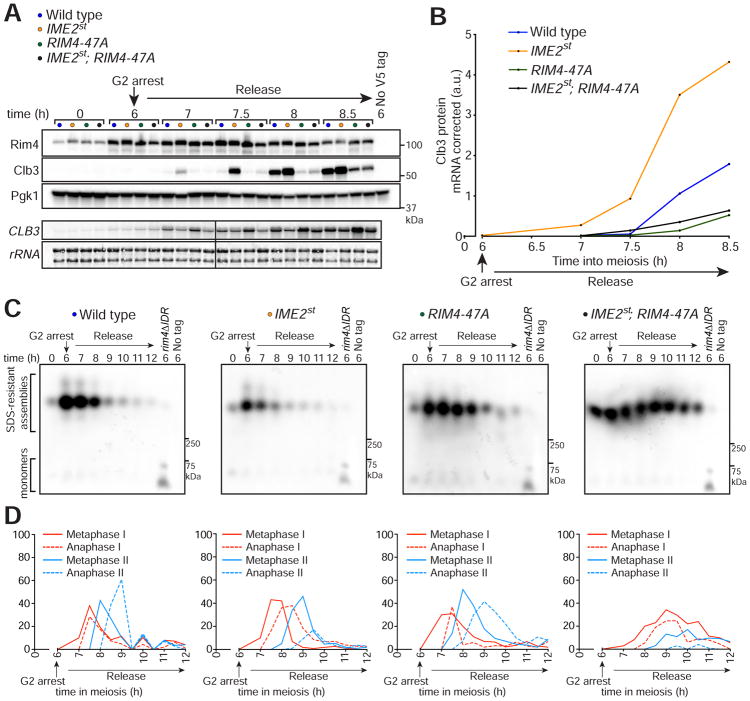
Ime2 kinase activity plays a central role in the developmental regulation of Rim4. We previously showed that expression of a hyperactive IME2 allele, IME2st (Sia and Mitchell, 1995), results in premature Rim4 clearance and de-repression of CLB3 translation during meiosis I (Berchowitz et al., 2013). To test the hypothesis that Ime2 is the kinase that triggers Rim4 clearance via phosphorylation of the Rim4 C-terminal IDR, we analyzed CLB3 translational control and Rim4 assembly in synchronized meiotic cultures of wild type, IME2st, RIM4-47A, and IME2st; RIM4-47A double mutant cells. Confirming earlier results, we found that cells harboring IME2st exhibited increased CLB3 translational efficiency and cells harboring RIM4-47A exhibited decreased translation compared to wild type (Figures 5A–D, S5). CLB3 translational efficiency in IME2st; RIM4-47A double mutant cells was decreased (compared to wild type) in a manner phenotypically indistinguishable from RIM4-47A single mutant cells (Figures 5A, 5B). This result shows that Rim4 clearance (and possibly assembly and reversibility of its amyloid-like properties) is mediated almost entirely through phosphorylation of its C-terminal IDR.
Figure 5. Ime2 regulates Rim4 clearance via phosphorylation of the Rim4 C-terminal IDR.
(A, B) Epistatic analysis of IME2st and RIM4-47A translational control phenotypes. Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and either RIM4-3V5 (wild type, blue, B48), IME2st (yellow, A33024), RIM4-47A-3V5 (green, A38075), or IME2st; RIM4-47A-3V5 (black, B5) were induced to sporulate at 30°C. After 6 hours, cells were releas ed from the G2 arrest. (A) Rim4-3V5, Clb3-3HA, and Pgk1 (loading control) protein and CLB3 mRNA and rRNA (loading control) levels were determined at the indicated times. (B) Clb3 protein levels corrected for CLB3 mRNA levels are plotted on the y-axis and time in meiosis is plotted on the x-axis. Western and Northern blot data for later time points is shown in Figure S5.
(C) Rim4 SDS-resistant assemblies were analyzed by SDD-AGE.
(D) Meiotic progression was analyzed by tubulin IF (n = 100 cells for each time point).
We next assayed clearance of Rim4 amyloid-like assemblies in the various mutants by SDD-AGE. Mirroring the above results we found that Rim4 assemblies were prematurely degraded in the IME2st background and aberrantly persisted in the RIM4-47A background compared to wild type (Figure 5C). In agreement with our observations on CLB3 translation, we found that Rim4 assemblies persisted in the IME2st; RIM4-47A double mutant (Figure 5C). Combined with the observation that Rim4 is a direct target of Ime2 (Berchowitz et al., 2013), these results reveal that Ime2 phosphorylates the C-terminal IDR of Rim4 in order to induce clearance of Rim4 amyloid-like assemblies. We also noted that IME2st; RIM4-47A double mutant cells exhibited a more severe meiotic progression defect and increased persistence of Rim4 assemblies compared to RIM4-47A single mutant cells. The severe meiotic progression defect we observed in IME2st; RIM4-47A cells could be due to the combined deleterious effects of two dominant and pleiotropic alleles. The increased persistence of Rim4 assemblies in IME2st; RIM4-47A cells is likely due to delayed meiotic progression combined with variability in the SDD-AGE assay.
Rim4 assemblies are degraded by the proteasome and are disassembled prior to clearance
We next sought to determine how Rim4 assemblies are degraded precisely at the meiosis I-meiosis II transition. Because Rim4-EGFP fusion proteins do not accumulate in the vacuole after meiosis I (Berchowitz et al., 2015), we reasoned that autophagy was unlikely to play a major role in Rim4 clearance. We focused on the proteasome as a mechanism for Rim4 clearance due to the rapidity by which Rim4 is degraded. However, such a clearance mechanism likely requires disassembly of amyloid-like Rim4 to facilitate its entry into the proteasome. Furthermore, Rim4 disassembly would also be necessary to release bound mRNAs into the translating pool.
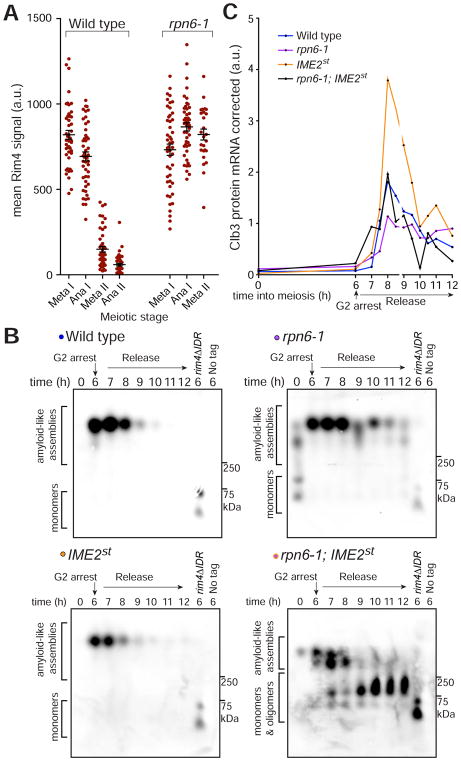
To determine whether impaired proteasome function affects Rim4 clearance, we analyzed Rim4 protein and CLB3 translational control in cells harboring a temperature-sensitive allele of RPN6 (rpn6-1). RPN6 encodes a lid component of the 26S proteasome and is essential for its activity (Isono et al., 2005; Santamaría et al., 2003). We found that Rim4 degradation was impaired in the rpn6-1 mutants, regardless of whether or not the cultures were transferred to the restrictive temperature (Figures S6A, S6B). To eliminate bias from the meiotic progression defects we observed in the rpn6-1 cells, we analyzed Rim4 protein levels in single cells. In support of proteasome-mediated degradation of Rim4, we found that, in rpn6-1 cells, Rim4 persisted in the rare cells that entered meiosis II (Figure 6A).
Figure 6. Rim4 assemblies are disassembled prior to proteasomal degradation.
(A) The proteasome is critical for Rim4 clearance. Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and RIM4-3V5 and either RPN6 (wild type) or rpn6-1 were induced to sporulate at 30°C. After 6 hours, cells were released from the G2 arrest. Single-cell Rim4 levels in metaphase I, anaphase I, metaphase II, and anaphase II were determined by immunofluorescence (IF) (n = 48 for each meiotic stage except rpn6-1 metaphase II in which n = 26 and anaphase II in which n = 0). CLB3 translational control and meiotic progression analysis is shown in Figures S6A, S6B.
(B, C) Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, RIM4-3V5 and either RPN6 (wild type, blue, B48), rpn6-1 (purple, B207), IME2st (yellow, A33024), or rpn6-1; IME2st (black, B251) were grown as in (A). (B) Rim4 SDS-resistant assemblies were analyzed by SDD-AGE. (C) CLB3 translational control analysis. Clb3 protein levels corrected for CLB3 mRNA levels are plotted on the y-axis and time in meiosis is plotted on the x-axis. Quantification of mRNA-corrected Clb3 protein levels is shown. Source data and meiotic progression analysis is shown in Figures S6C, S6D.
We next determined whether phosphorylation causes the disassembly of Rim4 prior to its degradation. We reasoned that if we combined a mutant that causes premature clearance and hyper-phosphorylation of Rim4 (IME2st) with a proteasome mutant that blocks Rim4 degradation (rpn6-1), we may be able to observe accumulation of monomeric and oligomeric Rim4 intermediates. To this end, we assessed whether Rim4 intermediates accumulate in rpn6-1; IME2st double mutant cells during meiosis by SDD-AGE. As expected, we observed persistent Rim4 assemblies in rpn6-1 cells and premature clearance in IME2st single mutant cells. In contrast, rpn6-1; IME2st double mutant cells accumulated Rim4 monomers and oligomers demonstrating that Rim4 assemblies are indeed disassembled as a consequence of phosphorylation (Figure 6B). When we analyzed translational control in these mutants, we found that although CLB3 translation was inhibited in rpn6-1 cells (likely due to delayed progression into meiosis II), CLB3 was translated prematurely in rpn6-1; IME2st double mutant cells (Figure 6C, S6C, S6D). This result is clarified by our SDD-AGE results which show persistent full-sized Rim4 assemblies in rpn6-1 cells but not in rpn6-1; IME2st double mutant cells. The absence of repression-competent Rim4 assemblies explains the CLB3 translation we observe in the double mutant. This result also provides further support of our conclusion that it is the SDS-resistant form of Rim4 that is the repressive form of the protein. Taken together, these results reveal that Rim4 assemblies are disassembled post-phosphorylation and degraded by the proteasome.
DISCUSSION
Numerous proteins harbor regions, such as prion-like domains, that can confer increased propensity to form amyloids and amyloid-like structures. In this study, we illuminate a mechanism by which cells disassemble and degrade amyloid-like structures in the context of a developmental process: meiosis. We demonstrate that disassembly and clearance of Rim4 amyloid-like translational repressors is governed by a thresholding effect of phosphorylation events within the IDR of the protein. Furthermore, timely clearance of Rim4 amyloid-like assemblies is critical for progression through meiosis II. Finally, we demonstrate that the amyloid-like properties of Rim4 are reversed prior to degradation by the proteasome.
Cells regulate the aggregation-prone properties of many RNA-binding proteins
In humans and yeast, RRM-containing proteins are significantly over-represented (greater than 10-fold enrichment) within proteins harboring algorithmically-defined prion-like domains (King et al., 2012). Thus, the functions of many RNA-binding proteins could be profoundly influenced by their increased propensity to form amyloid-like structures. Regulating the balance of functional assemblies versus pathological aggregation is an important problem for cells to overcome. Uncontrolled aggregation of many of these proteins such as FUS and TDP-43 is thought to be a major driver of neurodegenerative disease. On the other hand, cells can clearly take advantage of these properties. The formation of RNA stress granules in mammalian cells requires the prion-like domain of an RRM-containing protein (TIA-1) (Gilks et al., 2004). Additionally, upon exposure to heat stress, many RNA-binding proteins assemble into protein aggregates which can be rapidly reversed upon return to non-stressful temperatures (Wallace et al., 2015). This type of regulation could be of elevated importance during gamete development due to the reliance on translational control (which requires RNA-binding proteins) during this process. For example, hyperphosphorylation of CPEB4 during Xenopus meiosis inhibits the protein from forming phase-separated droplets and is important for meiotic progression (Guillén-Boixet et al., 2016).
Clearance of RNP assemblies and aggregated RNA-binding proteins can be achieved by autophagy (Buchan et al., 2013) and/or the proteasome (Osaka et al., 2016). Both pathways have distinct advantages and disadvantages depending on the cellular context and possibly depending on whether bound RNA must be released prior to protein degradation. Proteasome-mediated degradation likely requires disassembly whereas autophagy does not. However, the disassembly process inherent to proteasome-based mechanisms could facilitate the release of bound mRNA. Envisioning a mechanism by which autophagic degradation pathways would allow mRNA release is more difficult. Because Rim4 target mRNAs are likely released to be translated precisely when Rim4 assemblies are cleared, a proteasome-mediated clearance mechanism is advantageous. We speculate that other RNP assemblies in which RNA must be liberated prior to clearance will also employ the proteasome.
How does phosphorylation lead to disassembly of amyloid-like Rim4?
While we do not know whether a fraction of Rim4 is in a droplet form, the protein forms SDS-insoluble (i.e. amyloid-like) assemblies in vivo which distinguishes it biochemically from what has been described for the phase-separated droplet and hydrogel-forming protein FUS (Kato et al., 2012). The detergent insolubility of Rim4 is comparable to yeast prions such as those formed by Sup35 and Ure2 (Alberti et al., 2009; Kryndushkin et al., 2003). This is notable because in contrast to what has been reported for droplets and hydrogels, amyloid-like structures are often referred to as irreversible. Yeast cells’ ability to rapidly clear these structures presents a remarkable feat of protein quality control. How does Rim4 phosphorylation ultimately lead to its degradation via the proteasome? We know from our proteasome inactivation experiments that Rim4 must be disassembled prior to degradation which probably requires chaperones. Chaperone-mediated processing is well documented for amyloid-forming yeast prions (Moriyama et al., 2000; Shorter and Lindquist, 2004). The Hsp104 chaperone, which can process and fragment Sup35 fibrils (Shorter and Lindquist, 2004), is however unlikely to play a major role in Rim4 clearance; hsp104Δ strains do not exhibit meiotic dysfunction. Intriguingly, the Hsp70-family members SSA3 and SSA4 both drastically increase their expression levels as meiosis progresses (Primig et al., 2000) leading us to hypothesize that these chaperones could play an important role in Rim4 clearance.
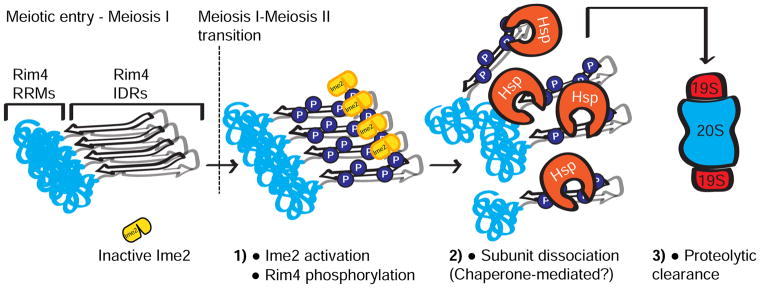
The proteasome, which has been shown to recruit chaperones and/or have chaperone-like activity (Braun et al., 1999; Hjerpe et al., 2016), could also play a direct role in Rim4 disassembly. Consistent with this idea is our observation that SDS-resistant Rim4 assemblies persist in rpn6-1 cells. Interestingly, this disassembly defect is suppressed by overexpressing Ime2. We interpret this observation to mean that Rim4 hyperphosphorylation alleviates the disassembly defect of rpn6-1 mutants. It is however also possible that hyper-phosphorylation by Ime2st per se leads to Rim4 disassembly. Irrespective of whether chaperones or the proteasome facilitate Rim4 aggregate disassembly, our results lead to a model where cumulative phosphorylation events within the C-terminal IDRs of Rim4 assemblies facilitate access by chaperones and/or proteasomes (Figure 7). Chaperones and/or proteasomes then unfold and remove Rim4 subunits which are then degraded via the proteasome.
Figure 7. Model of Rim4 clearance.
Another non-mutually exclusive interpretation of our results is that the proteasome is required for Ime2 activation- but not Ime2st. Ime2 kinase activity is developmentally regulated; the protein persists throughout meiosis but kinase activity is restricted to pre-meiotic S-phase and after the meiosis I- meiosis II transition (Berchowitz et al., 2013). The manner by which Ime2 is activated is not known and could require proteasome-mediated degradation of a negative regulator. In rpn6-1 cells, Ime2 is possibly never activated and thus Rim4 assemblies persist. IME2st cells exhibit constitutively active Ime2 which would bypass this requirement. A putative negative regulator could bind the C-terminal autoinhibitory region of Ime2- the region that is deleted to generate Ime2st. Our findings raise the question of how Ime2 kinase accesses the Rim4 IDR. How does a protein kinase phosphorylate residues that are buried within an amyloid-like structure? One speculative possibility is that Ime2 could phosphorylate solvent-exposed residues which are rapidly disassembled and degraded thus exposing other residues.
Could phosphatases be involved in regulation of Rim4 assembly? When Rim4 is expressed in cells growing in rich medium, the protein is unassembled as indicated by the lack of SDS-resistance and lack of activity as a translational repressor. However, when Rim4 is expressed in nutrient-starved cells, it efficiently assembles into its amyloid-like form. How starvation regulates Rim4 assembly is unknown. In light of our results, we hypothesize that starvation-induced phosphatases could function as pro-assembly factors, which would act early in meiosis. Such a mechanism could be conserved- deletion of PP2A phosphatases causes instability of P-granules in developing C. elegans embryos (Paix et al., 2014). Likewise, mitosis-specific kinases could play antagonistic roles in Rim4 assembly. An alternative and non-mutually exclusive route to starvation-mediated Rim4 assembly could be via cytoplasmic acidification. In yeast cells, acidification of the cytoplasm in response to nutrient deprivation has been shown to cause mobility decreases and assembly of numerous proteins (Munder et al., 2016). While sporulating yeast cells are known to increase local extracellular pH by secreting bicarbonate (Ohkuni et al., 1998), it will be interesting to determine if sporulation is also accompanied with decreased intracellular pH.
What causes the phenotypes in meiotic cells unable to clear Rim4 assemblies?
Meiotic cells that express unclearable Rim4 variants display several intriguing meiotic phenotypes. While decreased Clb3 is easily explained by the failure to degrade a translational repressor, the causes of increased CLB3 mRNA are less clear. Increased mRNA synthesis or decreased turnover could simply be the result of being stuck in a stage of the cell cycle (the MI-MII transition) at a time when CLB3 transcription is high and decay is minimal. Alternatively, failure to translate critical factors (such as Clb3) might be sensed by the cell. This could activate a positive feedback loop to hyperactivate transcription to overcome this problem.
Rim4 alanine substitution mutants show no detectable progression phenotypes until meiosis II onset after which point they exhibit lengthy delays and decreased spore viability. What causes the meiotic progression defects observed in these mutants? One possible explanation is that these phenotypes could be due to a failure to translate gene products that are essential for meiotic progression. The anaphase II delay phenotype could also be due to hyperactivation of cyclin-dependent kinase (CDK) activity (obviously not including Clb3-CDK), pathological persistence of amyloid-like Rim4, and/or failure to clear critical factors such as Shugoshin (Sgo1) which would cause a metaphase II delay. Further investigation of these phenomena offers the opportunity to understand the factors required for proper timing and progression of the later events of gametogenesis including meiosis II and spore packaging.
Because the IDRs of several RNA-binding proteins have been shown to function in processes such as RNA-binding and scaffolding for ubiquitination factors (Calabretta and Richard, 2015), alanine substitutions within the Rim4 C-terminal IDR could affect Rim4 function outside of the ablation of phosphorylation sites. Thus we cannot exclude the possibility that the phenotypes we observe in unclearable RIM4 mutants are, to some degree, caused by the mutations per se rather than their inability to be phosphorylated. However, the loss-of-function and Rim4 instability phenotypes of the rim4-45E mutant provide additional support for our conclusion that Rim4 phosphorylation is critical for clearance.
The pathways that meiotic cells use to clear amyloid-like assemblies could also clear disease-associated aggregates
Determining how Rim4 amyloid-like assemblies are cleared by meiotic cells has implications for our understanding of amyloid-related diseases. Hyperphosphorylated forms of disease-associated amyloids such as those formed by tau and aggregates such as those formed by TDP-43 are associated with neurodegenerative disease progression (Billingsley and Kincaid, 1997; Neumann et al., 2006) suggesting that this is a typical cellular response to the presence of amyloids and protein aggregates. However, unlike Rim4, tau and TDP-43 aggregate despite their phosphorylation. Perhaps cells that accumulate hyper-phosphorylated tau and TDP-43 lack a critical chaperone or exhibit proteasomal dysfunction. The pathways and mechanisms by which cells process and clear amyloid-like structures represent potential drug targets. Further study of yeast’s remarkable ability to clear amyloid-like assemblies has the potential to uncover previously unknown factors that have the ability to disassemble or prevent formation of disease-related amyloids.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Luke Berchowitz (leb2210@cumc.columbia.edu).
Experimental Model and Subject Details
Yeast strain construction
All experiments performed in this study were performed using diploid Saccharomyces cerevisiae strains derived from the SK1 background and are described in the Key Resources Table. RIM4-3V5, IME2st, IME2-3V5, and IME2st-3V5 is described in (Berchowitz et al., 2013; Carlile and Amon, 2008); pGAL-NDT80 and GAL4.ER are described in (Benjamin et al., 2003); CLB3-3HA is described in (Carlile and Amon, 2008); and rim4Δ271C-3V5 and SPC42-mCherry are described in (Berchowitz et al., 2015). pTUB1-GFP-TUB1 is described in (Straight et al., 1997) and was integrated into the URA3 locus. rim4-99A, RIM4-52A, RIM4-47A, RIM4-47A, RIM4-36A, RIM4-27A, RIM4-25A, RIM4-21A, rim4-16A, rim4-10A, and rim4-45E were all constructed by a PCR-based method outlined in (Rock et al., 2013). Briefly, plasmids containing the RIM4 CDS with all serines and threonines mutated to alanine (pLB90) or glutamic acid (pLB119) (synthesized by Genscript) and the HIS3 selectable marker were used as a template to generate PCR products which were transformed into the RIM4 locus. rim4Δ was created by the PCR-based mutagenesis method outlined in (Longtine et al., 1998). rpn6-1 was originally described in (Isono et al., 2005) and was generated in SK1 by PCR-based mutagenesis. All strains used in this study were verified by sequencing.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α-HA.11 Clone:16B12 | BioLegend | Cat# 901514; RRID:AB_2565336 |
| α-Pgk1 (Phosphoglycerate Kinase monocolonal) | Novex (Life Technologies) | Part# 459250; RRID:AB_2532235 |
| α-v5 monoclonal | Invitrogen | Cat# 46-0705; RRID:AB_2556564 |
| α-mouse HRP-conjugated secondary | GE Healthcare | Cat# NA931-1ML |
| α-V5-coupled agarose | Sigma Aldrich | Cat# A7345-1ml |
| Rat α-tubulin alpha | Bio-Rad | Cat# MCA77G |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Acid-washed glass beads, 425–600μm | Sigma Aldrich | G8772-500G |
| Imperial Blue Protein Stain | Thermo Fisher | Cat# 24615 |
| Halt Protease Inhibitor | Thermo Fisher | Cat# 1861279 |
| 0.5mm dia Zirconia/silica beads | BioSpec Products | Cat# 11079105z |
| Hybond H+ membrane | GE Healthcare | Prod# RPN203B |
| Illustra Probequant Columns | GC Healthcare | Prod#28903408 |
| ProlongGold w/DAPI | LifeTechnologies | Cat#P36935 |
| Critical Commercial Assays | ||
| Amersham MegaPrime DNA Labeling Kit | GE Healthcare | Prod#RPN1604 |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae: Strain background: SK1 (ho::LYS2, lys2, ura3, leu2::hisG, his3::hisG, trp1::hisG) | ATCC (Kane and Roth, 1974) | ATCC: 204722 |
|
A15055: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX |
Berchowitz/Amon labs | N/A |
|
A33024: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-3V5::HIS3MX6/RIM4-3V5::HIS3MX6 IME2st/IME2st |
Berchowitz/Amon labs | N/A |
|
A37489: MATa/α RIM4-EGFP::HIS3/RIM4-EGFP::HIS3 HTB1-mCherry-HISMX6/+ |
Berchowitz/Amon labs | N/A |
|
A38060: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-99A-3V5::HIS3MX6/RIM4-99A-3V5::HIS3MX6 |
This paper | N/A |
|
A38072: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-10A-3V5::HIS3MX6/RIM4-10A-3V5::HIS3MX6 |
This paper | N/A |
|
A38075: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-47A-3V5::HIS3MX6/RIM4-47A-3V5::HIS3MX6 |
This paper | N/A |
|
A38932: MATa/α rim4-47A-EGFP::HIS3/rim4-47A-EGFP::HIS3 HTB1-mCherry-HISMX6/HTB1-mCherry-HISMX6 |
This paper | N/A |
|
B5: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-47A-3V5::HIS3MX6/RIM4-47A-3V5::HIS3MX6 IME2st/IME2st |
This paper | N/A |
|
B48: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-3V5::HIS3MX6/RIM4-3V5::HIS3MX6 |
Berchowitz/Amon labs | N/A |
|
B67: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-52A-3V5::HIS3MX6/RIM4-52A-3V5::HIS3MX6 |
This paper | N/A |
|
B70: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-36A-3V5::HIS3MX6/RIM4-36A-3V5::HIS3MX6 |
This paper | N/A |
|
B73: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-16A-3V5::HIS3MX6/RIM4-16A-3V5::HIS3MX6 |
This paper | N/A |
|
B119: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-3V5::HIS3MX6/+ |
This paper | N/A |
|
B120: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-47A-3V5::HIS3MX6/+ |
This paper | N/A |
|
B123: MATa/α ura3::pTUB1-GFP-TUB1::URA3/ura3::pTUB1-GFP-TUB1::URA3 SPC42-mCherry::NATMX6/SPC42-mCherry::NATMX6 ZIP1-GFP(700)/ZIP1-GFP(700) RIM4-3V5::HIS3MX6/RIM4-3V5::HIS3MX6 |
This paper | N/A |
|
B126: MATa/α ura3::pTUB1-GFP-TUB1::URA3/ura3::pTUB1-GFP-TUB1::URA3 SPC42-mCherry::NATMX6/SPC42-mCherry::NATMX6 ZIP1-GFP(700)/ZIP1-GFP(700) RIM4-47A-3V5::HIS3MX6/RIM4-47A-3V5::HIS3MX6 |
This paper | N/A |
|
B129: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-21A-3V5::HIS3MX6/RIM4-21A-3V5::HIS3MX6 |
This paper | N/A |
|
B149: MATa/α ura3::pTUB1-GFP-TUB1::URA3/ura3::pTUB1-GFP-TUB1::URA3 SPC42-mCherry::NATMX6/SPC42-mCherry::NATMX6 ZIP1-GFP(700)/ZIP1-GFP(700) RIM4-47A-3V5::HIS3MX6/RIM4-3V5::HIS3MX6 |
This paper | N/A |
|
B152: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-27A-3V5::HIS3MX6/RIM4-27A-3V5::HIS3MX6 |
This paper | N/A |
|
B155: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-25A-3V5::HIS3MX6/RIM4-25A-3V5::HIS3MX6 |
This paper | N/A |
|
B197: MATa/α ura3::pTUB1-GFP-TUB1::URA3/ura3::pTUB1-GFP-TUB1::URA3 SPC42-mCherry::NATMX6/SPC42-mCherry::NATMX6 RIM4-3V5::HIS3MX6/RIM4-3V5::HIS3MX6 |
This paper | N/A |
|
B198: MATa/α ura3::pTUB1-GFP-TUB1::URA3/ura3::pTUB1-GFP-TUB1::URA3 SPC42-mCherry::NATMX6/SPC42-mCherry::NATMX6 RIM4-47A-3V5::HIS3MX6/RIM4-47A-3V5::HIS3MX6 |
This paper | N/A |
|
B207: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-3V5::HIS3MX6/RIM4-3V5::HIS3MX6 RPN6::rpn6-1::HIS3MX6/RPN6::rpn6-1::HIS3MX6 |
This paper | N/A |
|
B251: MATa/α ura3::pGPD1-GAL4(848).ER::URA3/ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1/pGAL-NDT80::TRP1 CLB3-3HA::KANMX6/CLB3-3HA::KANMX RIM4-3V5::HIS3MX6/RIM4-3V5::HIS3MX6 RPN6::rpn6-1::HIS3MX6/RPN6::rpn6-1::HIS3MX6 IME2st/IME2st |
This paper | N/A |
|
B343: MATa/α ura3::pGPD1-GAL4(848).ER::URA3 ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1, pGAL-NDT80::TRP1 CLB3-3HA:KANR CLB3-3HA:KANR rim4Δ::HIS3MX6 rim4Δ::HIS3MX6 |
This paper | N/A |
|
B346: MATa/α ura3::pGPD1-GAL4(848).ER::URA3 ura3::pGPD1-GAL4(848).ER::URA3 pGAL-NDT80::TRP1, pGAL-NDT80::TRP1 CLB3-3HA:KANR CLB3-3HA:KANR rim4-45E-3V5::HIS3MX6 rim4-45E-3V5::HIS3MX6 |
This paper | N/A |
| Oligonucleotides | ||
| Primer: CLB3 Northern blot probe forward: AGC ATA AGT AAG CCA AAA GTC GC | (Carlile and Amon, 2008) | N/A |
| Primer: CLB3 Northern blot probe reverse: C TAA TGC TAT CCA CTT CGC TAC G | (Carlile and Amon, 2008) | N/A |
| Recombinant DNA | ||
| RIM4-allA-HIS3 plasmid (pLB90) | This paper | N/A |
| RIM4-allE-HIS3 plasmid (pLB119) | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism | GraphPad software | https://www.graphpad.com/scientific-software/prism/ |
| Proteome Discoverer | Thermo Fisher | OPTON-30795 |
| Mascot version 2.4.1 | Matrix Science | http://www.matrixscience.com/ |
| Other | ||
| CellAsic ONIX microfluidic system | Millipore Sigma | Model EV-261 |
| CellAsic ONIX microfluidic diploid yeast plates | Millipore Sigma | Cat #: Y04D-02-5PK |
| DeltaVision Elite Microcope | GE Healthcare | Prod #: 29065728 |
Yeast media details and culture conditions
To generate meiotic cultures, strains were inoculated in YEPD (1% yeast extract, 2% peptone, 2% dextrose) and grown overnight with shaking at 30°C. The following morning, the cells were diluted in BYTA (1% yeast extract, 2% tryptone, 1% potassium acetate, 50 mM potassium pthalate) to OD600 = 0.3 and grown overnight with shaking at 30°C. The following morning, cells were washed once with water and resuspended in sporulation (SPO) medium (0.3% potassium acetate [pH 7.0], 0.02% raffinose) at OD600 = 1.8 and grown with shaking at 30°C. pGAL-NDT80, GAL4.ER strains were released from G2 arrest by the addition of 1 μM β-estradiol at 6 hr.
Method Details
Denaturing protein sample preparation for immunoblot and immunoprecipitation
Samples were prepared by resuspending the pellet of 4 ml SPO culture in 5% TCA, incubating overnight at 4° C, washing with acetone, and breaking cells using 50 μl acid-washed glass beads (Sigma), 100 μl lysis Buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8], 2.75 mM DTT, Halt protease inhibitors (Thermo Fisher)), and a 45 sec process in a FastPrep-24 (MP Biomedicals) at max speed. 50 μL Loading Buffer (9% SDS, 0.75 mM Bromophenol blue, 187.5 mM Tris-HCl [pH 6.8], 30% glycerol, and 810 mM β-mercaptoethanol) was added, samples were heated at 100°C for 5 mins, and centrifuged 5 mins at 20,000 g.
Immunoblots
Polyacrylamide gels were run on a midi gel system (BioRad) with SDS Running Buffer (190 mM glycine, 25 mM Trizma base, 3.5 mM, 1% SDS), using 10% gels loaded with 4 μl sample per well. They were transferred using a semi-dry transfer apparatus (Biorad) to a nitrocellulose membrane. α-HA.11 (BioLegend) was used at 1: 1,000, α-Pgk1 (Novex) was used at 1: 20,000, α-v5 (Invitrogen) was used at 1: 2,000. α-mouse HRP-conjugated secondary antibody (GE Healthcare) was used at 1: 10,000, 1: 20,000, and 1: 40,000 for respective primaries.
Immunoprecipitation
1,350 μl denaturing IP buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 2mM MgCl2, 1% NP-40, 10% glycerol, 2% BSA) was added to 150 μl denatured protein sample (see above) and then clarified by centrifugation at 15,000 rcf for 15 mins at 4°C. 20 μL of equilibrated α-V5-coupled agarose (Invitrogen) was added and the samples were incubated with rotation for 2 hours at 4°C. Beads were collected by centrifugation and washed 2x in high salt wash buffer (50 mM Tris-HCl [pH 7.4]; 1 M NaCl; 1 mM EDTA; 1% NP-40; 0.1% SDS; 0.5% sodium deoxycholate) and 2x in low-salt wash buffer (20 mM Tris-HCl [pH 7.4]; 10 mM MgCl2; 0.2% Tween-20). Bound factors were released by boiling in 20 μl SDS sample buffer for 5 min, separated on polyacrylamide gels, and stained with Imperial Blue (Thermo Fisher).
Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE)
SDD-AGE was adapted from previous protocols (Halfmann and Lindquist, 2008). 2 ml samples of sporulating culture were harvested by centrifugation and snap frozen in liquid nitrogen. Cell pellets were resuspended in 200 μl lysis buffer (100 mM Tris/HCl [pH 8.0], 20 mM NaCl, 2 mM MgCl2, 50 mM β-mercaptoethanol, 1% Triton-X, 2x Halt protease inhibitors (Thermo)) and broken using a FastPrep-24 and 0.5 mm zirconia/silica beads (BioSpec). The lysates were clarified twice by centrifugation at 2,500 rcf for 5 min at 4°C. 4x Sample loading buffer (fin al concentration 0.5x TAE, 5% glycerol, 2% SDS, bromophenol blue) was added to lysates which were incubated for 10 min at room temperature. Following incubation, 20 μl of each sample was separated on a 1.7% agarose 0.5x TAE + 0.1% SDS gel for 16 hours at 25 V at 4°C using 0.5x TAE supp lemented with 0.1% SDS as the running buffer. A peristaltic pump was used to recirculate the running buffer at 3 ml/min. The gel was blotted to nitrocellulose (GE Healthcare) by capillary transfer in 1X TBS and processed by the immunoblot procedure outlined above. α-v5 (Invitrogen) was used at 1: 2,000 with α-mouse HRP-conjugated secondary antibody (GE Healthcare) at 1: 5,000.
Light Microscopy
Acquisition of images was conducted using a DeltaVision microscope at 100x magnification (Applied Biosystems). Immunofluorescence was performed as described in (Visintin et al., 1999). We used a 1: 200 dilution of a mouse α-V5 antibody to detect Rim4 and a 1: 100 dilution of a rat α-tubulin antibody to visualize spindles. Immunofluorescence samples were mounted in ProlongGold that included DAPI (Life Technologies). Mean intensity of Rim4 signal was quantified in a 15 pixel diameter circle region of interest (nucleus and vacuole excluded) using ImageJ software.
Live-imaging of cells harboring Spc42-mCherry and GFP-Tub1 was done using a CellAsic microfluidics chip and manifold pump (Millipore). Strains were induced to sporulate at 30°C and were incubated with shaking for 3 hours. Then, 200 μl of culture was pipetted into the microfluidics device, which was then affixed to the microscope stage. SPO medium was pumped through the device throughout the course of the experiment. Seven 0.3 μm z-stacks were taken every 10 minutes. Exposure times were 0.01 ms for FITC (10% T) and 0.05 ms for mCherry (10% T). Max intensity projections were generated with Worx software that is built in to the Deltavision software.
Northern Blot Analysis
Samples were harvested as 2 ml of SPO culture, centrifuged, and flash-frozen in liquid nitrogen. Pellets were resuspended in 400 μl TES Buffer (10 mM Tris-HCl [pH 7.5], 10 mM EDTA, and 0.5% SDS), 400 μl acid phenol: chloroform 5: 1 (Ambion), and 50 μl 0.5 mm zirconia/silica beads (BioSpec). Cells were lysed by shaking at 1400 rpm for 30 mins at 65°C in a Thermomixer (Eppendorf) followed by centrifugation for 5 mins at 13,000 g, extraction to 1 ml 100% ethanol and 40 μl sodium acetate [pH 5.5], and precipitation at −20°C overnight.
Samples were centrifuged at 13,000 g for 20 mins, washed with 1 ml 80% ethanol, centrifuged at 13,000 g for 5 mins, and dried. RNA pellets were resuspended in 25 μl DEPC water at 37°C with shaking at 1000 rpm for 15 mins and concentrations were determined using a NanoDrop (Thermo Fisher). 22 μl denaturing mix (15 μl formamide, 5.5 μl formaldehyde, and 1.5 μl 10x MOPS) was added to 8 μg total RNA in 8 μl and heated at 55°C for 15 mins. 20 μL of sample (~5 μg) was resolved on a denaturing agarose gel (1.9% agarose, 3.7% formaldehyde, 1x MOPS buffer) for 2.5 hours at 80 V. The gel was blotted to a Hybond membrane (GE Healthcare) by capillary transfer in 10x SSC (1.5 M NaCl, 0.15 M trisodium citrate dihydrate, [pH 7]). The membrane was incubated in hybridization Buffer (0.25 M Na-Phosphate [pH 7.2], 0.25 M NaCl, 1 mM EDTA, 7% SDS, and 5% dextran sulfate) at 65°C probed with 32P-labeled CLB3 DNA probes prepared via Amersham Megaprime DNA labeling kit (GE Healthcare) and Illustra ProbeQuant columns (GE Healthcare), transferred to a phosphor screen, and imaged on a Typhoon imager (GE Healthcare).
Identification of Rim4 phosphorylation sites by mass spectrometry
Proteins were run on an SDS-PAGE gel and stained with Imperial Blue (Thermo Fisher) according to manufacturer’s instructions. Gel bands were excised and subjected to mass spectrometry. After destaining with 40% ethanol/10% acetic acid, proteins were reduced with 20 mM dithiothreitol (Sigma) for 1 h at 56°C and then alkylated with 60 mM iodoa cetamide (Sigma) for 1 h at 25°C in the dark. Proteins were then digested with 12.5 ng/μl modified trypsin (Promega) in 50 μl 100 mM ammonium bicarbonate [pH 8.9] at 25°C overnight. Peptides we re extracted by incubating the gel pieces with 50% acetonitrile/5% formic acid then 100 mM ammonium bicarbonate, repeated twice followed by incubating the gel pieces with 100% acetonitrile then 100 mM ammonium bicarbonate, repeated twice. Each fraction was collected, combined, and reduced to near dryness in a vacuum centrifuge.
Phosphorylated peptides were enriched as described in (Ficarro et al., 2009). In brief, nickel was removed from the Ni-NTA agarose (Qiagen) with 100 mM EDTA. The NTA agarose was then incubated with 100 mM FeCl3. The peptides were acidified and incubated with the Fe-NTA agarose for 1 h at room temperature. Phosophopeptides were eluted with 250 mM sodium phosphate.
The peptides were separated by reverse phase HPLC using an EASY- nLC1000 (Thermo) over a 75 min gradient before nanoelectrospray using a QExactive mass spectrometer (Thermo). The mass spectrometer was operated in a data-dependent mode. The parameters for the full scan MS were: resolution of 70,000 across 350–2000 m/z, AGC 3e6, and maximum IT 50 ms. The full MS scan was followed by MS/MS for the top 10 precursor ions in each cycle with a NCE of 28 and dynamic exclusion of 30 s. Raw mass spectral data files (.raw) were searched using Proteome Discoverer (Thermo) and Mascot version 2.4.1 (Matrix Science). Mascot search parameters were: 10 ppm mass tolerance for precursor ions; 10 mmu for fragment ion mass tolerance; 2 missed cleavages of trypsin; fixed modification was carbamidomethylation of cysteine; variable modifications were oxidized methionine, serine phosphorylation, threonine phosphorylation, and tyrosine phosphorylation. Only peptides with a Mascot score greater than or equal to 25 and an isolation interference less than or equal to 30 were included in the data analysis.
Quantification and Statistical Analysis
Blot quantification
Immunoblot and Northern blot experiments were quantified using FIJI (ImageJ) software. Signal intensity was normalized to a loading control (Pgk1 for immunoblots and rRNA for Northerns).
Microscopy Quantification and Analysis
Spindle morphologies were classified as in (Lee and Amon, 2003). n = 100 cells for each time point and the data are plotted as % cells in each cell cycle phase. Metaphase I cells were defined as cells with a single DAPI mass spanned by a short, thick, bipolar, meiotic spindle (approximately 1–2 μm in length). Anaphase I cells were defined as cells with two distinct (though not always separated) DAPI masses, and a single long spindle that spans both DAPI masses. Metaphase II cells were defined as cells with two separate DAPI masses with each spanned by a bipolar, short, thick, meiotic spindle. Anaphase II cells were defined as cells with four distinct (though not always separated) DAPI masses with two long spindles.
To analyze Rim4 levels in single cells, mean intensity of Rim4 IF signal was quantified in a 15 pixel diameter circle region of interest (vacuole excluded, background corrected) using ImageJ software. To compare differences between independent paired experiments, we used a Mann-Whitney test. For Figure S1 n = 50 cells
Supplementary Material
HIGHLIGHTS.
Clearance of amyloid-like Rim4 is governed by multi-site phosphorylation
Ime2 drives Rim4 clearance by phosphorylation of Rim4 intrinsic disordered region
Amyloid-like Rim4 is disassembled prior to degradation by the proteasome
Failure to clear amyloid-like Rim4 interferes with progression through meiosis II
Acknowledgments
We thank Raphaelle Laureau, Rodney Rothstein, David Phizicky, and Michael Smith for critical reading of the manuscript. We thank Amanda Del Rosario for mass spectrometry analysis of Rim4. We thank Soni Lacefield for providing the pTUB1-GFP-TUB1 plasmid. This research is supported by the Alice Bohmfalk Charitable Trust, the Amyloidosis Foundation, and NIH grant R35 GM124633-01 to L.E.B. A.A. is an investigator of the Howard Hughes Medical Institute. The authors declare no conflicts of interest.
Footnotes
AUTHOR CONTRIBUTIONS
L.E.B. and A.A. conceived of the study. L.E.B. and K.C. designed the experiments. K.C. led the experimental procedures. K.C., R.B.B., J.Y., and L.E.B. performed and analyzed the experiments. L.E.B. wrote the paper with input from all authors.
DECLARATION OF INTERESTS
All authors declare no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes & Development. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Gajadhar AS, van Werven FJ, De Rosa AA, Samoylova ML, Brar GA, Xu Y, Futcher B, Weissman JS, White FM, et al. A developmentally regulated translational control pathway establishes the meiotic chromosome segregation pattern. Genes & Development. 2013;27:2147–2163. doi: 10.1101/gad.224253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, Amon A. Regulated Formation of an Amyloid-like Translational Repressor Governs Gametogenesis. Cell. 2015;163:406–418. doi: 10.1016/j.cell.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingsley ML, Kincaid RL. Regulated phosphorylation and dephosphorylation of tau protein: effects on microtubule interaction, intracellular trafficking and neurodegeneration. Biochemical Journal. 1997;323:577–591. doi: 10.1042/bj3230577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boke E, Ruer M, Wühr M, Coughlin M, Lemaitre R, Gygi SP, Alberti S, Drechsel D, Hyman AA, Mitchison TJ. Amyloid-like Self-Assembly of a Cellular Compartment. Cell. 2016;166:637–650. doi: 10.1016/j.cell.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-Resolution View of the Yeast Meiotic Program Revealed by Ribosome Profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel PM, Finley D, Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabretta S, Richard S. Emerging Roles of Disordered Sequences in RNA-Binding Proteins. Trends Biochem Sci. 2015;40:662–672. doi: 10.1016/j.tibs.2015.08.012. [DOI] [PubMed] [Google Scholar]
- Carlile TM, Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, Marto JA. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal Chem. 2009;81:4566–4575. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid–from bacteria to humans. Trends in Biochemical …. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional Amyloid Formation within Mammalian Tissue. PLoS Biol. 2005;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén-Boixet J, Buzon V, Salvatella X, Méndez R. CPEB4 is regulated during cell cycle by ERK2/Cdk1-mediated phosphorylation and its assembly into liquid-like droplets. Elife. 2016;5:e19298. doi: 10.7554/eLife.19298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Screening for amyloid aggregation by Semi-Denaturing Detergent-Agarose Gel Electrophoresis. J Vis Exp. 2008 doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, et al. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell. 2016;166:935–949. doi: 10.1016/j.cell.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, Saito N, Kamata N, Saeki Y, Toh-e A. Functional analysis of Rpn6p, a lid component of the 26 S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J Biol Chem. 2005;280:6537–6547. doi: 10.1074/jbc.M409364200. [DOI] [PubMed] [Google Scholar]
- Jin L, Zhang K, Sternglanz R, Neiman AM. Predicted RNA Binding Proteins Pes4 and Mip6 Regulate mRNA Levels, Translation, and Localization during Sporulation in Budding Yeast. Molecular and Cellular Biology. 2017;37:e00408–e00416. doi: 10.1128/MCB.00408-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MR, Li L, Pérez-Sánchez C, Saraf A, Florens L, Slaughter BD, Unruh JR, Si K. Amyloidogenic Oligomerization Transforms Drosophila Orb2 from a Translation Repressor to an Activator. Cell. 2015;163:1468–1483. doi: 10.1016/j.cell.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TPJ, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- Kronja I, Orr-Weaver TL. Translational regulation of the cell cycle: when, where, how and why? Philosophical Transactions of the Royal Society B: Biological Sciences. 2011;366:3638–3652. doi: 10.1098/rstb.2011.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Lee BH, Amon A. Role of Polo-like Kinase CDC5 in Programming Meiosis I Chromosome Segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsaio Y, Damko E, Moquin D, Walz T, McDermott A, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Molecular Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Miller MP, Unal E, Brar GA, Amon A. Meiosis I chromosome segregation is established through regulation of microtubule-kinetochore interactions. Elife. 2012;1:e00117. doi: 10.7554/eLife.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O’Meally R, Dignon GL, Conicella AE, Zheng W, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. Embo J. 2017:e201696394. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Molecular and Cellular Biology. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder MC, Midtvedt D, Franzmann T, Nüske E, Otto O, Herbig M, Ulbricht E, Müller P, Taubenberger A, Maharana S, et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife. 2016;5:59. doi: 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Seydoux G. Less is more: specification of the germline by transcriptional repression. Development. 2008;135:3817–3827. doi: 10.1242/dev.022434. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Ohkuni K, Hayashi M, Yamashita I. Bicarbonate-mediated social communication stimulates meiosis and sporulation of Saccharomyces cerevisiae. Yeast. 1998;14:623–631. doi: 10.1002/(SICI)1097-0061(199805)14:7<623::AID-YEA264>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Osaka M, Ito D, Suzuki N. Disturbance of proteasomal and autophagic protein degradation pathways by amyotrophic lateral sclerosis-linked mutations in ubiquilin 2. Biochem Biophys Res Commun. 2016;472:324–331. doi: 10.1016/j.bbrc.2016.02.107. [DOI] [PubMed] [Google Scholar]
- Paix A, Lambrus BG, Calidas D, Betzig E, Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically-disordered proteins in C. elegans. Elife. 2014 doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Rock JM, Lim D, Stach L, Ogrodowicz RW, Keck JM, Jones MH, Wong CCL, Yates JR, Winey M, Smerdon SJ, et al. Activation of the Yeast Hippo Pathway by Phosphorylation-Dependent Assembly of Signaling Complexes. Science. 2013;340:871–875. doi: 10.1126/science.1235822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría PG, Finley D, Ballesta JPG, Remacha M. Rpn6p, a Proteasome Subunit from Saccharomyces cerevisiae, Is Essential for the Assembly and Activity of the 26 S Proteasome. J Biol Chem. 2003;278:6687–6695. doi: 10.1074/jbc.M209420200. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004 doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A Neuronal Isoform of the Aplysia CPEB Has Prion-Like Properties. Cell. 2003 doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Sia RA, Mitchell AP. Stimulation of later functions of the yeast meiotic protein kinase Ime2p by the IDS2 gene product. Molecular and Cellular Biology. 1995;15:5279–5287. doi: 10.1128/mcb.15.10.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soushko M, Mitchell AP. An RNA-binding protein homologue that promotes sporulation-specific gene expression in Saccharomyces cerevisiae. Yeast. 2000;16:631–639. doi: 10.1002/(SICI)1097-0061(200005)16:7<631::AID-YEA559>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Taglialegna A, Lasa I, Valle J. Amyloid Structures as Biofilm Matrix Scaffolds. J Bacteriol. 2016;198:2579–2588. doi: 10.1128/JB.00122-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Wallace EWJ, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell. 2015;162:1286–1298. doi: 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual Specificity Kinase DYRK3 Couples Stress Granule Condensation/Dissolution to mTORC1 Signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.