Abstract
Background
Mannose receptor (MRC1/CD206) has been suggested to mediate allergic sensitization and asthma to multiple glycoallergens, including cockroach allergens.
Objective
We sought to determine the existence of a protective mechanism through which MRC1 limits allergic inflammation through its intronic miR-511-3p.
Methods
We examined MRC1-mediated cockroach allergen uptake by lung macrophages and lung inflammation using C57BL/6 wild-type (WT) and Mrc1−/− mice. The role of miR-511-3p in macrophage polarization and cockroach allergen–induced lung inflammation in mice transfected with adeno-associated virus (AAV)–miR-511-3p (AAV– cytomegalovirus–miR-511-3p–enhanced green fluorescent protein) was analyzed. Gene profiling of macrophages with or without miR-511-3p overexpression was also performed.
Results
Mrc1−/− lung macrophages showed a significant reduction in cockroach allergen uptake compared with WT mice, and Mrc1−/− mice had an exacerbated lung inflammation with increased levels of cockroach allergen–specific IgE and TH2/TH17 cytokines in a cockroach allergen–induced mouse model compared with WT mice. Macrophages from Mrc1−/− mice showed significantly reduced levels of miR-511-3 and an M1 phenotype, whereas overexpression of miR-511-3p rendered macrophages to exhibit a M2 phenotype. Furthermore, mice transfected with AAV–miR-511-3p showed a significant reduction in cockroach allergen–induced inflammation. Profiling of macrophages with or without miR-511-3p overexpression identified 729 differentially expressed genes, wherein expression of prostaglandin D2 synthase (Ptgds) and its product PGD2 were significantly downregulated by miR-511-3p. Ptgds showed a robust binding to miR-511-3p, which might contribute to the protective effect of miR-511-3p. Plasma levels of miR-511-3p were significantly lower in human asthmatic patients compared with nonasthmatic subjects.
Conclusion
These studies support a critical but previously unrecognized role of MRC1 and miR-511-3p in protection against allergen-induced lung inflammation.
Keywords: Mannose receptor, miR-511-3p, macrophage, asthma
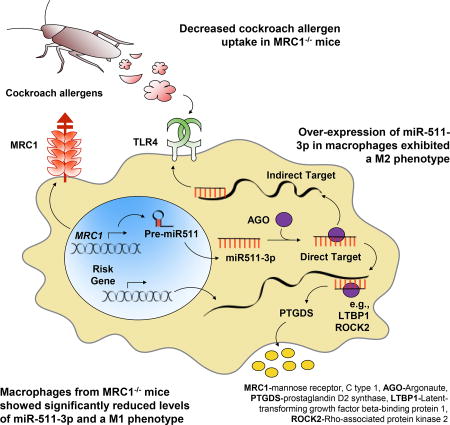
GRAPHICAL ABSTRACT
Mannose receptor (MRC1/CD206) is a member of the innate pattern recognition receptor family and a key C-type lectin receptor (CLR) expressed by macrophages.1,2 MRC1 contains 8 C-type lectin–like domains and acts as a phagocytic receptor for bacteria, fungi, parasites, viruses, and allergens.2–4 MRC1 has been suggested to mediate allergic sensitization and asthma to multiple glycoallergens,4,5 including cockroach allergens. In fact, sensitization to cockroach allergens has been identified as one of the strongest risk factors for the development of asthma.6 Indeed, the majority of school-aged children and adults with asthma have concomitant allergic sensitization; cockroach allergens are detectable in 85% of inner-city US homes, and 60% to 80% of inner-city children with asthma are sensitized to cockroach.7,8 Our recent study identified a functional interaction between MRC1 and cockroach allergens in fibrocytes.9 However, the role of MRC1 in cockroach allergen–induced inflammation and its signaling transduction is poorly understood.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that regulate gene expression programs by influencing the translation and stability of target mRNA, thus regulating gene expression and cell function.10–12 miRNAs have emerged as regulators of phagocyte activation,13,14 TH2 polarization,11,12 and pathogenic airway inflammation.12,15–17 Recent studies suggested that miRNAs might have the ability to shape the balance of M1 and M2 macrophage polarization (eg, miR-155 and miR-146) and skew the immune response.18–20
miR-511-3p, the active strand of miR-511, is an intronic miRNA encoded by both mouse and human Mrc1/MRC1 genes. miR-511-3p has been shown to be transcriptionally coregulated with Mrc1 in macrophages and to regulate macrophage activation.21,22 More recently, miR-511-3p has been found to control macrophage-mediated microbial responses and enhance intestinal inflammation.23
In this study we provided evidence supporting a critical role of MRC1 in allergen clearance, as a natural defense mechanism, and in limiting the progression and severity of cockroach allergen–induced allergic inflammation in a mouse model of asthma, in which its intronic miR-511-3p might play a crucial role.
METHODS
Mice
C57BL/6J and Mrc1 knockout (Mrc1−/−) mice were purchased from the Jackson Laboratory (Bar Harbor, Me) and maintained under specific pathogen-free conditions in the animal facility of the Johns Hopkins University School of Medicine. All mice were used at 6 to 9 weeks of age, and all experiments used age- and sex-matched control mice. The experimental protocols were reviewed and approved by the Animal Care and Use Committee at Johns Hopkins University School of Medicine.
Isolation of lung macrophages
Lung tissues harvested from naive and unsensitized wild-type (WT) and Mrc1−/− mice were minced and incubated for 45 minutes at 37°C in Dulbecco modified Eagle medium supplemented with 10 µg/mL DNAse I and 1 mg/mL collagenase D (Sigma-Aldrich, St Louis, Mo) in a shaking water bath. Digested lung tissues were then passed through a 70-µm nylon strainer (BD Biosciences, San Jose, Calif) to obtain a single-cell suspension. Cell suspensions were incubated with ammonium-chloride-potassium lysis buffer to lyse contaminating erythrocytes. Percentages of dendritic cells (DCs) and macrophages among those cells were analyzed by means of flow cytometry by gating on CD11C+CD45+ cells, encompassing all DC and macrophage populations and then focusing on the 2 subpopulations: DCs with F4/80−MHC-IIhi autofluorescence (AF)–low and alveolar macrophages with F4/80+MHC-IIloAFhi.24–26
Generation of bone marrow–derived macrophages
Mouse bone marrow (BM) cells were isolated from the marrow of mouse femurs and tibias of 6- to 8-week-old mice. BM cells were then cultured at a starting density of 5 × 105 cells/mL at 37°C in a humidified atmosphere containing 5% CO2 and differentiated into macrophages in Dulbecco modified Eagle medium containing 10% FBS, 1% penicillin/streptomycin (Life Technologies, Grand Island, NY), and 20 ng/mL recombinant murine macrophage colony-stimulating factor (M-CSF; R&D Systems, Minneapolis, Minn) for 7 days. The macrophage phenotype was confirmed by means of flow cytometry with antibodies specific for F4/80+ (BM8; eBioscience, San Diego, Calif) and CD11b+ (M1/70; eBioscience).
MRC1-mediated cockroach allergen uptake by macrophages
In vivo antigen uptake
Both WT and Mrc1−/− mice were injected intratracheally with 10 µg of fluorescence isothiocyanate (FITC)–labeled cockroach extract (CRE; 10 µg) for 24 hours. The FITC-labeled CRE in lung tissues was analyzed by means of immunofluorescent staining.
In vitro antigen uptake by macrophages
For cockroach allergen uptake by macrophages, alveolar macrophages sorted with a FACSAria (BD Biosciences) gated on the CD11C+CD45+F4/80+MHC-IIlowAFhi population were incubated with FITC-labeled Bla g 2 or CRE for 1 hour at 37°C and then washed in ice-cold PBS and fixed with 1% formaldehyde for flow cytometry.
Antigen uptake assay using a continuous time-lapse live cell confocal microscopy
For live cell microscopy imaging, alveolar macrophages or BMDMs were seeded on glass-bottom culture dishes (MatTek, Ashland, Mass) for 2 days. Before live cell microscopy, lysosomes and nuclei were stained in living cells with 50 nmol/L LysoTracker Red DND-99 and 250 ng/mL Hoechst 33342 (Invitrogen, Carlsbad, Calif), respectively, for 30 minutes in culture medium. Stained cells were then washed with 37°C cell-culture medium 3 times and then incubated in fresh medium for 5 minutes before subjecting the cells to live cell imaging. To perform live cell imaging, the cell-culture dish was then mounted onto the adapter in the stage of an inverted microscope, the Cell Observer System (ZEISS, Jena, Germany), with an environmental control chamber to provide 37°C and 5% CO2 (Carl Zeiss, Oberkochen, Germany). The antigen Bla g 2 labeled with FITC (10 ng/mL) was added to cultured macrophages on a cultured dish on the microscope stage once live cell imaging was begun. Images were continuously captured with the Zeiss LSM 780 confocal system through the inverted microscope with an NA1.4 Plan-Apochromat objective. Fluorescent signals were analyzed with Zen 2013 or AxioVision 4.2 software (Carl Zeiss).
Cockroach allergen–induced asthma mouse model
A cockroach allergen–induced asthma mouse model was established, as described previously, with some modifications.27,28 Briefly, mice were sensitized by means of intratracheal inhalation of 20 µg of CRE (B46; GREER Laboratories, Lenoir, NC) per mouse in 50 µL of PBS after achievement of light anesthesia with isoflurane on days 1, 2, 3, and 14 and challenged on days 20 and 22 with the same amount of CRE. Control mice received PBS during the sensitization and challenge phases. On day 23, mice were killed, and lung tissues were dissected and analyzed for inflammation. Bronchoalveolar lavage (BAL) fluid was harvested by means of 2 consecutive flushes of the lung with 1.0 mL of ice-cold PBS. Blood was taken to screen for serum antibodies against cockroach allergen.
Analysis of lung inflammation
Detailed methods were described previously.27 Mouse lungs were perfused with 20 mL of ice-cold PBS injected into the right ventricle followed by excision and fixation in 10% neutral buffered formalin. Sections (5 µm) were prepared from paraffin-embedded lungs and stained with hematoxylin and eosin (H&E) and periodic acid–Schiff (PAS), respectively. Images were obtained with a Nikon Eclipse Ti-U microscope equipped with DS-Fi2 camera (Nikon, Melville, NY). For analysis of BAL fluid, fluids were centrifuged at 1500 rpm at 4°C for 10 minutes and washed with PBS containing 1% FBS. Red blood cells in the pellet were lysed with ammonium-chloride-potassium lysis buffer. The cells were stained for eosinophils, macrophages, neutrophils, and lymphocytes by means of flow cytometry on a FACSCalibur cytometer (BD Biosystems), and data collected were analyzed with FlowJo software (TreeStar, Ashland, Ore).27 Eosinophils were defined as side scatter (SSC)high Siglec-F+Mac-3− cells, alveolar macrophages were identified as SSChighSiglec-F+Mac-3+ cells, granulocytes were recognized as SSChighGr-1+ cells, and lymphocytes were identified as forward scatter–low/SSClow cells expressing CD3 or CD19.
ELISA
Collected BAL fluid was assessed for the presence of cytokines by using Ready-Set-Go! ELISA sets (eBioscience) and PGD2 (Cayman Chemical, Ann Arbor, Mich), according to the manufacturer’s instructions. Results were read with a Bio-Rad Bio-Plex instrument (Bio-Rad Laboratories, Hercules, Calif). Serum cockroach allergen–specific IgE and IgG1 levels were analyzed by means of ELISA, according to our previously published work.27
Immunofluorescence
For immunofluorescent staining, nonspecific binding was blocked with 10% blocking serum in PBS for 1 hour at room temperature, and tissue samples were then incubated with primary antibodies to green fluorescent protein (GFP; clone 4B10) and F4/80 (clone BM8) overnight at 4°C. Secondary antibodies conjugated with fluorescence were added, and slides were incubated at room temperature for 1 hour. Isotype-matched negative control antibodies (R&D Systems) were used under the same conditions. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (Invitrogen). The sections were mounted with the ProLong Gold Anti-fade Kit (Molecular Probes, Grand Island, NY) and observed with a Nikon Eclipse Ti-U microscope equipped with DS-Fi2 camera (Nikon).
Mice transfected with adeno-associated virus–miR-511-3p
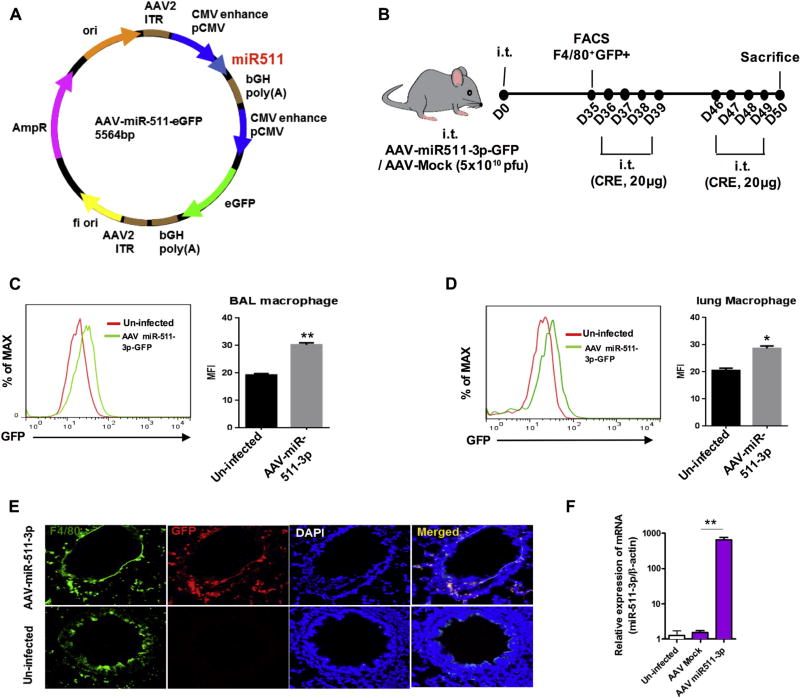
An adeno-associated virus (AAV) vector encoding the enhanced GFP reporter and expressing miR-511-3p (AAV–cytomegalovirus [CMV]–miR-511-3p–eGFP) driven by the CMV promoter was generated in SignaGen Laboratories, Rockville, Md. The AAV was generated by using standard triple-transfection methods and purification with CsCl gradient centrifugation. Titers were quantified by using silver stain after gel electrophoresis (SDS-PAGE) and quantitative PCR (qPCR). AAV at a dose of 5×1010 plaque-forming units in 20 µL of PBS per mouse was administered to mice through intratracheal injection. GFP expression in infected lung macrophages were detected on day 35 after AAV injection.
Lentivirus transduction
Bone marrow–derived macrophages (BMDMs) were plated at 1.0 × 106 cells/well in a 6-well plate 24 hours before lentivirus (LV) transduction in complete medium containing 20 ng/mL M-CSF. BMDMs were transduced with LV expressing miR-511-3p (LV miR-511-3p) or a mutated miR-511-3p (LV Mock), as described previously.21 After 24 hours, the medium was removed, and cells were washed with PBS and then continued to be cultured in complete medium containing 20 ng/mL of M-CSF for another 48 hours. Cells were harvested, and transduction was assessed by using FACS analysis.
RNA isolation and quantitative real-time PCR analysis
Total RNA was isolated with an RNeasy Plus Mini Kit (Qiagen, Hilden, Germany), and cDNA templates were synthesized with SuperScript II (Life Technologies). Quantitative RT-PCR was performed in triplicate by using SYBR Green Universal 2× qPCR Master Mix (Applied Biosystems, Foster City, Calif) on an ABI Prism 7300 detection system. Data were analyzed with the 2−ΔΔCT method, as described by Livak and Schmittgen.29 mRNA levels were normalized to the internal control gene (β-actin). Primer sequences are available on request.
Gene profiling of macrophages with or without miR-511-3p overexpression
Gene transcriptional profiling was performed in macrophages with or without miR-511-3p overexpression at the Sidney Kimmel Cancer Center Microarray Core facility at Johns Hopkins University. BMDMs were cultured and transfected with miR-511-3p mimic or a nontargeting control mimic (Life Technologies) by using Lipofectamine RNAiMax Transfection Reagent (Invitrogen) for 48 hours.
Isolation and quality assessment
Total RNA was prepared as described for the RNeasy Mini Kit (Qiagen) with on-column DNase I digestion. RNA quality was assessed by using a Nanodrop-1000 spectrometer for OD260/280 and OD260/230 ratio and the Bioanalyzer (Agilent Technologies, Santa Clara, Calif).
Microarray assay
MouseRef-8 v2 Expression BeadChip arrays (Illumina, San Diego, Calif) were used for microarray hybridization to examine global gene expression of the samples. The array targets 18,138 annotated mouse genes with 25,697 unique probes derived from the National Center for Biotechnology Information Reference Sequence (NCBI RefSeq) database (Build 36, Release 22), supplemented with probes derived from the Mouse Exonic Evidence Based Oligonucleotide (MEEBO) set, as well as exemplar protein-coding sequences described in the RIKEN FANTOM2 database.
Total RNA (500 ng) from each sample was amplified and labeled with the Illumina TotalPrep RNA Amplification Kit (AMIL1791; Ambion, Austin, Tex), as described in the instruction manual. For array assay, 750 ng of biotin-labeled cRNA was combined with hybridization buffer and hybridized to the array at 58°C for 16 to 20 hours. After hybridization, the hybridization cartridge was disassembled, and the array was washed with buffer at 55°C and blocked at room temperature. Bound biotinylated cRNA was stained with streptavidin-Cy3 and then washed. Dried arrays were stored in a dark box until scanned with the iScan System. Data were extracted with the Gene Expression Module in GenomeStudio Software.
Microarray data analysis
Raw microarray florescent signal data were subjected to filtering by using detection P value and z score normalization to obtain normalized probe signals; the sample quality was first analyzed by means of scatter plots, principal component analysis, and gene sample hierarchy clustering to exclude possible outliners. Gene expression change was calculated for each studies comparison by using the z ratio, which indicates the fold difference between experimental groups, and corrected for multiple comparison error level based on the false discovery rate (FDR). For individual genes with a 2-tailed z test, a P value of .05 or less, a z ratio of 1.5 or greater, and an FDR of 0.30 or less were considered significant changes.30 Furthermore, we used MedScan Reader 6 to search PubMed for all of the literature within 10 years for the interaction target of miR-511-3p on Mrc1 networks and build up their connections by using the pathway studio 10 and rnen11 database.
Analysis of miRNA-mRNA interaction
A synthetic miR-511-3p containing a biotin at the 3′ end (or a negative control cel-miR-39 biotinylated miRNA; synthesized by Integrated DNA Technologies, Coralville, Iowa) was annealed with a complementary strand, and 20 ng of the duple was incubated with cell lysates of RAW 264.7 (mouse leukaemic monocyte macrophage cell line) (10 million cells) stimulated with IL-4 (10 ng/mL) for 24 hours, and miRNA-mRNA complexes were isolated by using streptavidin beads. qPCR was performed to identify bound mRNA, and enrichment was analyzed with miR-511-3p versus negative control miRNA.
Plasma miR-511-3p in asthmatic and nonasthmatic patients
Study population
Detailed descriptions of the study subjects have been describes previously.31,32 For these subjects, asthma was defined by history of asthma and a documented history of physician-diagnosed asthma, and allergy status was determined based on a clinical history of aeroallergen sensitivity and a multiallergen IgE test (ImmunoCAP or skin prick test). The study of human subjects was approved by the Institutional Review Boards of Penn State College of Medicine Institute.
Measurement of circulating miR-511-3p
Blood was isolated by means of venipuncture in a purple-top tube containing EDTA, left at room temperature for 15 to 30 minutes, and then centrifuged at 2000g in a clinical centrifuge to collect plasma.32 RNA was then extracted from 500 µL of banked plasma with addition of synthetic cel-miR-39 as a spiked-in small RNA for normalization control.33 Isolated RNA was reverse transcribed to cDNA with the qScript microRNA cDNA Synthesis Kit (Quanta BioSciences, Gaithersburg, Md), which added a universal adapter sequence to the 3′ end. Quantification of miRNAs was performed by using quantitative RT-PCR on the CFX384 Real-Time System (Bio-Rad Laboratories) with primers specific to the miR-511-3p sequence and the 3′ adapter. miRNA levels were expressed as the copy number per microliter determined by a standard curve generated from multiple dilution of known concentrations of synthesized miR-39.
Statistical analysis
All values are expressed as means ± SEMs for each group. Statistical significance for normally distributed samples was assessed by using an independent 2-tailed Student t test with Prism software (GraphPad Software, La Jolla, Calif). One-way ANOVA with the Tukey correction for multiple comparisons was used to compare miR-511-3p expression across the 3 human subject groups. GraphPad Prism software was used for all statistical calculations. Differences with P values of less than .05 were considered statistically significant.
RESULTS
MRC1 mediates cockroach allergen uptake by macrophages
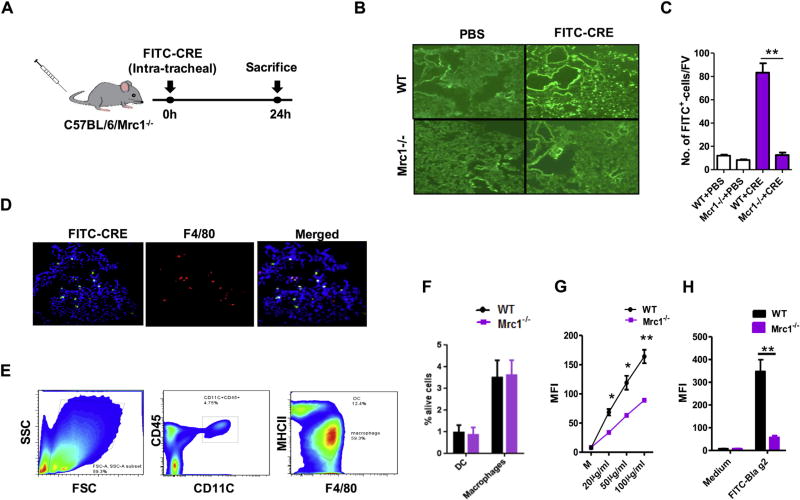
To examine whether MRC1 on macrophages mediated cockroach allergen uptake, WT or Mrc1−/− mice were injected with FITC-labeled CRE (10 µg) intratracheally, and the FITC-labeled CRE (FITC-CRE) in lung tissues was analyzed 24 hours after administration (Fig 1, A). A significant reduction in CRE uptake was observed in Mrc1−/− compared with WT mice (Fig 1, B and C).
FIG 1.
MRC1 mediates cockroach allergen uptake by macrophages. A, Experimental setup for cockroach allergen uptake in mice. B, Representative of FITC-CRE in lung tissues from WT and Mrc1−/− mice (n = 3 per group). Images are depicted at ×10 magnification. C, Amount of FITC-CRE in lung tissues of WT and Mrc1−/− mice. D, FITC-CRE uptake by macrophages (F4/80+) in lung tissues (merged). Images are depicted at ×20 magnification. E, Flow cytometric gating strategy for analysis of macrophages and DCs in dispersed lung cell suspensions. FSC, Forward scatter. F, Percentage of DCs and macrophages in lung tissues of WT and Mrc1−/− mice. G, Sorted lung macrophages from WT and Mrc1−/− mice were cocultured with various doses of FITC-CRE for 30 minutes, and the CRE-uptake was analyzed by using flow cytometry. H, A similar experiment as in Fig 1, G, was repeated by using FITC–Bla g 2. I, Uptake of FITC–Bla g 2 by sorted lung macrophages from WT and Mrc1−/− mice was observed by using a continuous time-lapse live cell confocal microscopy. Data are expressed as means ± SEMs. DIC, Differential interference contrast; MM, minute; SS, second. *P <.05 and **P < .01.
We next examined the change in CRE uptake by macrophages, the most abundant inflammatory cell type in asthmatic airways.34,35 A large number of cells showed colocalization of FITC-CRE with F4/80+ macrophages (Fig 1, D), suggesting that macrophages might be the major cell type contributing to the difference in antigen-uptake by WT and Mrc1−/− mice.
We then assessed whether cockroach allergen uptake was different in macrophages from WT and Mrc1−/− mice. Alveolar macrophages were collected from naive unsensitized WT and Mrc1−/− mice by means of FACS (Fig 1, E). CD11c+CD45+ cells were gated to encompass all DC and macrophage populations, in which 2 subpopulations were further gated: F4/80−MHC-IIhiAFlow (DCs) and F4/80+MHC-IIloAFhi (alveolar macrophages).24–26 Greater numbers of alveolar macrophages compared with DCs (approximately 3-fold) were detected in both WT and Mrc1−/− mice. Percentages of DCs and alveolar macrophages were relatively unchanged in Mrc1−/− compared with WT mice (Fig 1, F). Significantly less FITC-CRE was detected in the macrophages sorted from Mrc1−/− mice compared with those from WT mice after the alveolar macrophages were cultured with various dosages of FITC-CRE for 30 minutes (Fig 1, G). Reduced antigen uptake was also observed in alveolar macrophages from Mrc1−/− mice when the cells were cultured with the FITC-labeled cockroach allergen Bla g 2 (FITC–Bla g 2; Fig 1, H).
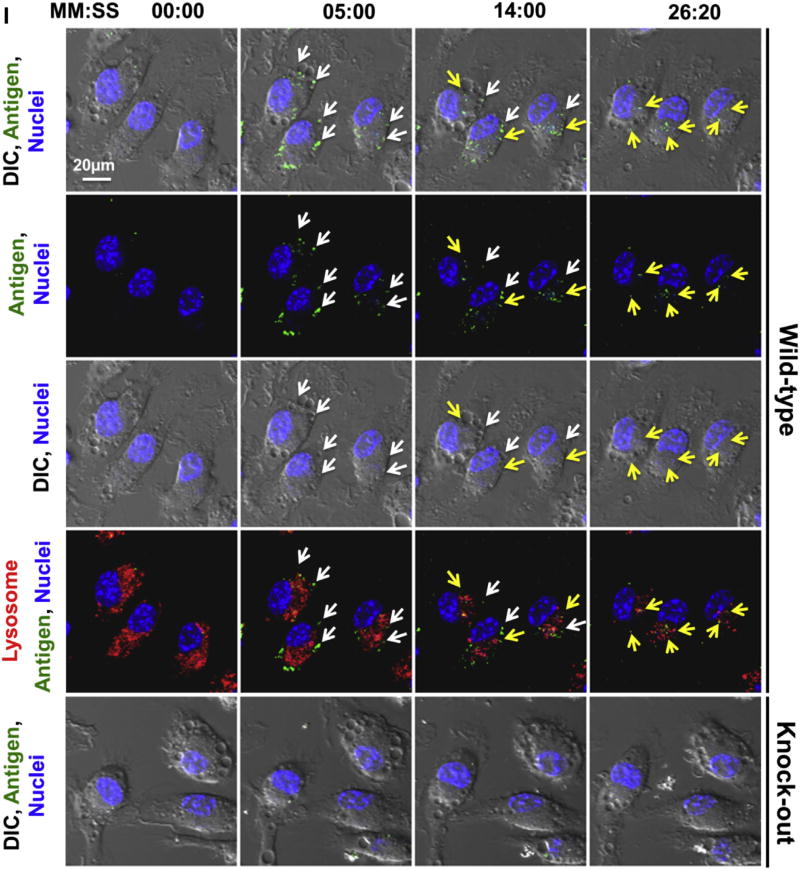
Furthermore, the role of MRC1 in mediating cockroach allergen uptake was also confirmed by using a continuous time-lapse live cell confocal microscopy (Fig 1, I). By continuously monitoring the macrophages after incubating with FITC–Bla g 2, we observed a sequential process: accumulation of the cockroach allergen in the cell, association with the cell vesicles, and movement of the allergen-containing vesicles from the cell boundary toward the inner part of the cell (see Video E1 in this article’s Online Repository at www.jacionline.org). There was no specific association of FICT–Bla g 2 and lysosome signal at all time points. Importantly, we observed a significant reduction in antigen uptake by macrophages from Mrc1−/− mice (see Video E2 in this article’s Online Repository at www.jacionline.org).
Together, these studies suggest that MRC1 can mediate the cockroach allergen uptake and clearance by lung macrophages.
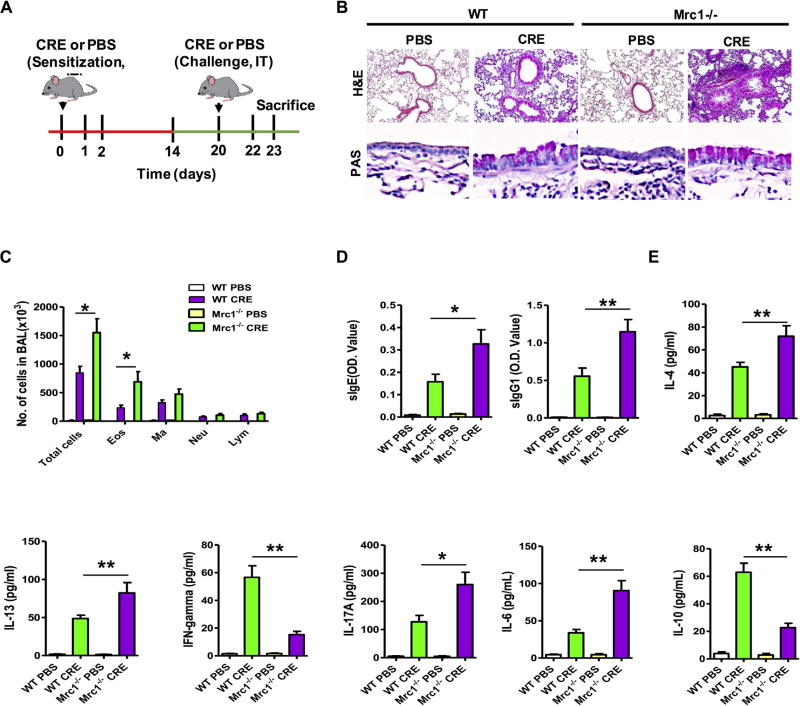
Mrc1-deficient mice had exacerbated lung inflammation
We next examined whether MRC1 plays a role in mediating cockroach allergen–induced allergic inflammation. To this aim, we develop a mouse model of asthma with WT and Mrc1−/− mice using a protocol of cockroach allergen–induced lung inflammation (Fig 2, A). Compared with PBS-challenged mice, asthma pathologies were developed in CRE-challenged WT and Mrc1−/− mice, as indicated by a significant recruitment of inflammatory cells to the lung with dense peribronchial infiltrates and goblet cell hyperplasia in the histologic examination (Fig 2, B, H&E and PAS). Compared with WT mice, CRE-treated Mrc1−/− mice displayed exacerbation of peribronchial inflammation and goblet cell hyperplasia. Furthermore, CRE-treated Mrc1−/− mice displayed significantly increased numbers of total inflammatory cells, especially with eosinophils, among all analyzed cell types in the BAL fluid (Fig 2, C). These CRE-sensitized and challenged Mrc1−/− mice also produced higher serum titers of cockroach allergen–specific IgE and IgG1 (Fig 2, D). Furthermore, BAL fluids from CRE-sensitized and challenged Mrc1−/− mice had increased levels of IL-4, IL-13, IL-17, and IL-6 but attenuated levels of IFN-γ and IL-10 (Fig 2, E). These data suggest that MRC1 might play a protective role in cockroach allergen–triggered airway inflammation.
FIG 2.
Mrc1 mediates cockroach allergen–induced lung inflammation. A, Protocol for cockroach allergen– induced mouse model of asthma. Green line, Sensitization phase; red line, challenge phase. B, Paraffin-embedded tissue sections of lungs were stained with H&E (upper panel) and PAS (lower panel). Original magnification ×20. C, BAL fluid total and differential cell counts of PBS- and CRE-challenged WT and Mrc1−/− mice. D, Serum levels of cockroach allergen–specific IgE and IgG1. E, Cytokine levels in BAL fluid. For Fig 2, C–E, there were 8 to 16 mice pooled from 3 independent experiments. Data are expressed as means ± SEMs. *P < .05 and **P < .01.
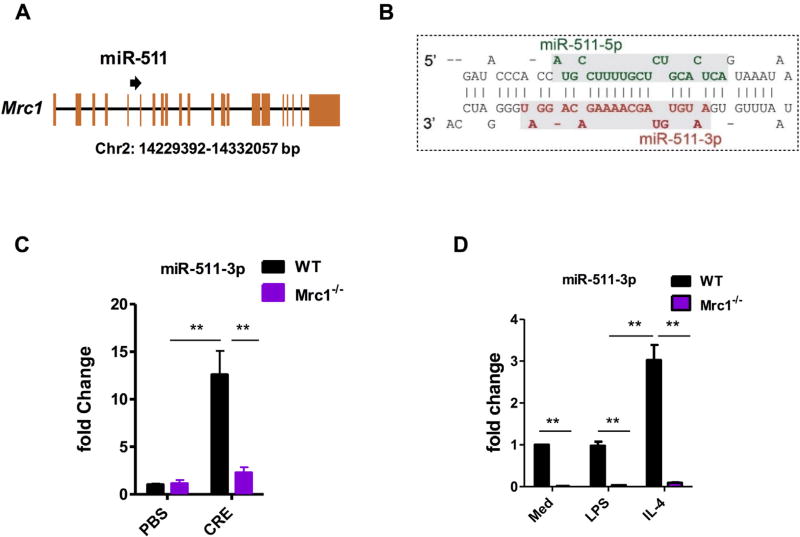
miR-511-3p was transcriptionally correlated with Mrc1
miR-511-3p, an intronic miRNA encoded in intron 5 of the Mrc1 gene (Fig 3, A), is the active strand of miR-511 (Fig 3, B). miR-511-3p and Mrc1 are transcriptionally coregulated in macrophages.2,21,36 To confirm the coregulation between miR-511-3p and Mrc1 in macrophages reported by Squadrito et al,21 we examined the expression of miR-511-3p in lung macrophages from our cockroach allergen–induced mouse model and macrophages differentiated from WT and Mrc1−/− BMDMs. miR-511-3p expression was increased in lung macrophages from CRE-treated WT mice, but the increased expression was not observed in those from Mrc1−/− mice after CRE challenge (Fig 3, C). A similar pattern was also seen for miR-511-3p in BMDMs, and miR-511-3p was virtually undetectable in BMDMs from Mrc1−/− mice (Fig 3, D). These results further strengthen the significant correlation between the levels of miR-511-3p and magnitude of Mrc1 expression, as well as suggest that miR-511-3p might be responsible for mediating MRC1’s regulatory effects.
FIG 3.
miR-511-3p is transcriptionally correlated with Mrc1. A, miR-511 is located in intron 5 of the Mrc1 gene on mouse chromosome 2. B, miR-511-3p is the active strand (red line) of mouse pre-miR-511. C, Expression of miR-511-3p in lung macrophages of PBS- or CRE-treated WT and Mrc1−/− mice. D, Relative expression levels of miR-511-3p in BMDMs from WT and Mrc1−/− mice polarized in M1 and M2 by LPS or IL-4, respectively (n = 3 samples per condition). Data were expressed as means ± SEMs. **P < .01.
MRC1 plays a role in macrophage polarization through miR-511-3p
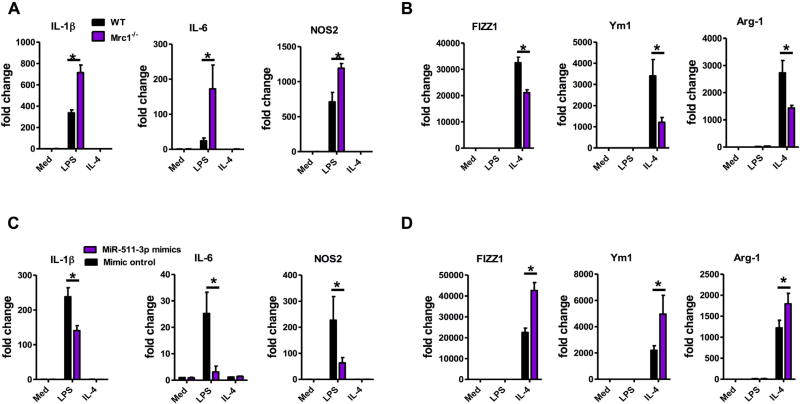
miRNAs have the ability to shape the balance of M1 and M2 macrophage polarization,22,37,38 raising the possibility that MRC1 controls macrophage polarization through miR-511-3p. To test this possibility, BMDMs were treated with either IL-4 to promote M2 polarization or LPS for M1 polarization. The expression of M1 (IL-1β, IL-6, and NOS2) and M2 (Fizz1, Ym1, and Arg1) genes was determined by using quantitative RT-PCR. Compared with BMDMs from WT mice, Mrc1−/− BMDMs displayed an increased extent of IL-1β, IL6, and NOS2 induction and a reduced extent of Fizz1, Ym1, and Arg1 induction on LPS and IL-4 stimulation, respectively (Fig 4, A and B). In agreement with these observations, knockdown of Mrc1 increased the expression of M1-associated genes in LPS-treated and reduced the expression of M2-associated genes in IL-4–treated BMDMs (see Fig E1 in this article’s Online Repository at www.jacionline.org). Accordingly, miR-511-3p overexpression with miR-511-3p mimic reduced the expression of IL-1β, IL6, and NOS2 and enhanced that of Fizz1, Ym1, and Arg1 in LPS- and IL-4–treated BMDMs, respectively (Fig 4, C and D) when compared with BMDMs transfected with control mimic. These results imply that both Mrc1 and miR-511-3p have an effect on macrophage polarization and activation, which can control allergen-induced inflammatory responses.
FIG 4.
Mrc1 regulates macrophage inflammatory polarization through miR-511-3p. A and B, Real-time PCR analysis of M1 (Fig 4, A) and M2 (Fig 4, B) markers in BMDMs isolated from WT and Mrc1−/− mice and treated with LPS (M1) and IL-4 (M2). C and D, Real-time PCR analysis of M1 (Fig 4, C) and M2 (Fig 4, D) markers in WT BMDMs transfected with control mimic or miR-511-3p mimic. Data are representative of 3 independent experiments (n = 3 samples per condition). Data represent means ± SEMs. *P < .05.
miR-511-3p protected against cockroach allergen–induced lung inflammation
Next, we tested whether miR-511-3p mediates protection from allergen-induced inflammation. We generated an AAV vector expressing miR-511-3p (AAV–CMV–miR-511-3p–eGFP; Fig 5, A) and infected mice according to the experimental approach in Fig 5, B. Expression of miR-511-3p in AAV-infected mice was confirmed on day 35 after infection by detecting GFP expression with flow cytometry in BAL fluid (Fig 5, C) and lung (Fig 5, D) macrophages. AAV-infected macrophages were also detected in lung tissues by costaining with F4/80 and GFP (Fig 5, E). Overexpression of miR-511-3p was confirmed in lung tissues of AAV–miR-511-3p–GFP–infected mice compared with those infected with AAV-Mock or uninfected (Fig 5, F). Those AAV-infected mice were then used for generating cockroach allergen–induced mouse model of asthma, according to the protocol illustrated in Fig 5, B. As expected, no significant changes were observed between CRE-challenged uninfected and AAV-Mock–infected mice. However, CRE-triggered peribronchial inflammation (H&E) and goblet cell hyperplasia (PAS) were substantially suppressed in the AAV–miR-511-3p–infected mice compared with those AAV-Mock–infected control mice (Fig 6, A).
FIG 5.
Cockroach allergen–induced mouse model with AAV–miR-511-3p–infected mice. A, Full sequence map for AAV–CMV–miR-511-3p–eGFP. B, Experimental setup for mice infected with AAV–CMV–miR-511-3p–eGFP or AAV-CMV-eGFP (AAV-Mock) had cockroach allergen–induced asthma. i.t., Intratracheal. C and D, Flow cytometric analysis for expression of AAV–miR-511-3p–eGFP in BAL fluid (Fig 5, C) and lung (Fig 5, D) macrophages of uninfected and AAV–miR-511-3p–eGFP–infected mice on day 35. E, Detection of AAV–miR-511-3p–eGFP (red) in lung macrophages (F4/80, green) of uninfected and infected mice by means of immunofluorescence staining. Nuclei were stained with, 6′-diamidino-2-phenylindole (DAPI; blue). Original magnification ×20. F, Expression of miR-511-3p in lung tissues of AAV–miR-511-3p, AAV-Mock, and uninfected mice detected by using RT-PCR. Data represent means ± SEMs. *P < .05 and **P < .01.
FIG 6.
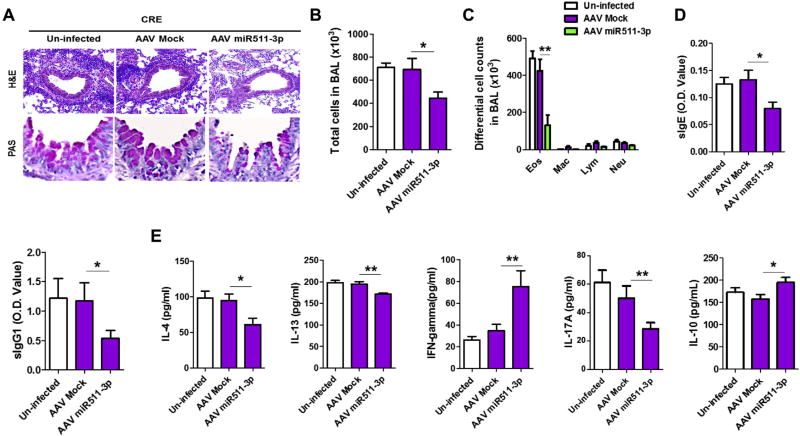
miR-511-3p protects against cockroach allergen–induced lung inflammation. A, Paraffin-embedded tissue sections of lungs from PBS- and CRE-challenged uninfected mice and CRE-challenged AAV–miR-511-3p– and AAV-Mock–infected mice were stained with H&E (upper panel) and PAS (lower panel). Original magnification ×20. B and C, BAL total (Fig 6, B) and differential cell (Fig 6, C) counts of CRE-challenged mice. D, Serum levels of cockroach allergen–specific IgE and IgG1 in CRE-challenged mice. E, Levels of cytokines in BAL fluid of CRE-challenged mice. For Fig 6, B–E, there were 9 to 13 mice pooled from 3 independent experiments. Data represent means ± SEMs. *P < .05 and **P < .01.
Furthermore, BAL fluids from AAV–miR-511-3p–infected mice displayed a reduced number of total inflammatory cells (Fig 6, B), with a prominent reduction in eosinophil numbers (Fig 6, C). Sera from those mice had lower levels of CRE-specific IgE and IgG1 (Fig 6, D). Similarly, BAL fluids from CRE-treated AAV–miR-511-3p–infected mice had lower levels of IL-4, IL-13, and IL-17A but higher levels of IFN-γ and IL-10 (Fig 6, E). Collectively, the results suggest that miR-511-3p can protect against allergen-induced lung allergic inflammation.
Gene profiling of macrophages with or without miR-511-3p overexpression
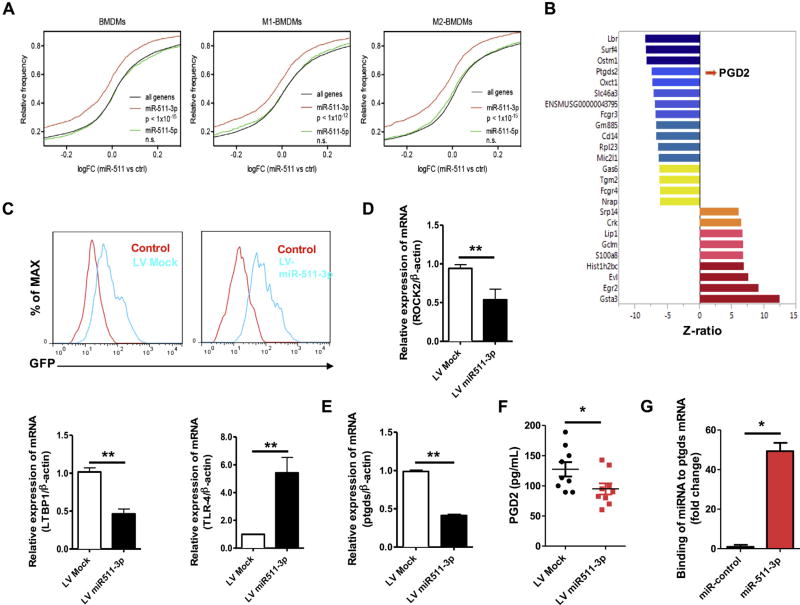
Overexpression of miR-511-3p can regulate gene expression and subsequent macrophage functions. Thus we performed transcriptional profiling of BMDMs transfected with or without miR-511-3p mimic using the MouseRef-8 v2.0 BeadChip. Remarkably, the predicted target genes of miR-511-3p were globally downregulated by miR-511-3p overexpression in BMDMs, but predicted target genes of miR-511-5p were not affected (Fig 7, A). Results showed that a total of 729 genes were identified to be differentially expressed (z ratio ≥ 1.5, P ≤ .05, FDR < 0.3), including 400 downregulated and 329 upregulated genes. The heat map depicts expression levels and patterns of the top-ranking genes (n = 25) identified as significantly different between BMDMs transfected with miR-511-3p or control mimic (n = 3 per group; Fig 7, B), which include those that are most downregulated (eg, Lbr, Surf4, Ostm1, prostaglandin D2 synthase [Ptgds], and CD14) and upregulated (eg, Gsta3, Egr2, and S100a8) by miR-511-3p. It was noted particularly that expression of Ptgds (encoding hematopoietic prostaglandin D synthase), which catalyzes the conversion of PGH2 to PGD2,39 was downregulated significantly in BMDMs overexpressing miR-511-3p. Furthermore, we searched PubMed for all of the literature within 10 years for miR-511-3p targets using MedScan Reader 6 and generated a miR-511-3p interactive network using the pathway Studio 10 and RNEN11 database (see Fig E2 in this article’s Online Repository at www.jacionline.org). Among these, both Rock2 and Ltbp1 were significantly downregulated, but Tlr4 and Cebpa were upregulated by miR-511-3p (see Table E1 in this article’s Online Repository at www.jacionline.org).
FIG 7.
Gene profiling of BMDMs with or without miR-511-3p mimic. A, Cumulative distribution of fold changes in miR-511 versus control BMDMs (n = 3) for all genes and miR-511-3p and miR-511-5p target genes. B, Heat map depicting expression levels and patterns of the top-ranking genes (n = 25) identified as significantly different between BMDMs by miR-511-3p. The z ratio is given on the x-axis. C, Conformation of LV-miR-511-3p–infected BMDMs by using flow cytometry. D, Expression of ROCK2, LTBP1, and TLR4 in LV–miR-511-3p–transfected BMDMs. E, Expression of Ptgds in LV–miR-511-3p–transfected BMDMs. F, PGD2 levels in supernatants of BMDMs with LV-miR-511-3p. G, Binding of miR-511-3p to the Ptgds mRNA by using an affinity-based assay. For Fig 7, D, E, and G, there were 3 samples per condition. Data represent means ± SEMs. *P < .05 and **P < .01.
To validate these findings, we transduced BMDMs with LVs expressing either miR-511-3p (LV-miR-511-3p) or a mutated form (LV-Mock).21 The LV transfection was confirmed by means of flow cytometry (Fig 7, C). Several selected candidate genes were validated in BMDMs with LV–miR-511-3, including the most upregulated (eg, Gata3 and Egr2) or downregulated (eg, Lbr and Surf4) genes from gene array analysis (see Fig E3 in this article’s Online Repository at www.jacionline.org) and target genes within the interactive network (Rock2, Ltbp1, and Tlr4; Fig 7, D). Downregulation of Ptgds by miR-511-3p was also confirmed (Fig 7, E). Consistently, the level of PGD2, the catalytic product of Ptgds, was significantly lower in the supernatants of BMDMs with miR-511-3p overexpression compared with control values (Fig 7, F). Additionally, to determine that Ptgds was a direct target of miR-511-3p, we used an affinity-based assay to show binding of miR-511-3p to the Ptgds mRNA (see Fig E4 in this article’s Online Repository at www.jacionline.org).31 We found a significant enrichment in Ptgds mRNA in miR-511-3p but not in the negative control miRNA pulldown (Fig 7, G), indicating that there was a significant interaction between miR-511-3p and Ptgds.
miR-511-3p expression is lower in asthmatic patients
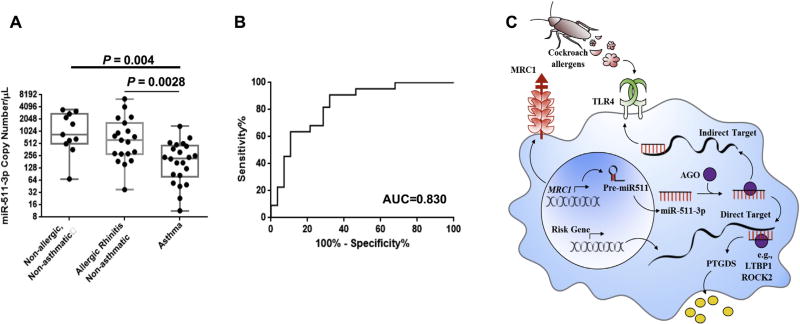
To determine the potential importance of miR-511-3p in human allergic asthma, we measured the expression of miR-511-3p in plasma of human asthmatic patients and nonasthmatic control subjects. To exclude allergic sensitization as a confounder, we compared miR-511-3p expression between allergic asthmatic patients, allergic subjects without asthma, and nonallergic subjects without asthma. Detailed characteristics of study subjects are presented in Table E2 in this article’s Online Repository at www.jacionline.org. Levels of plasma miR-511-3p were significantly lower in allergic asthmatic patients compared with those in allergic nonasthmatic subjects (P = .0028) and nonallergic nonasthmatic subjects (P = .004; Fig 8, A).
FIG 8.
Plasma levels of miR-511-3p. A, Plasma levels of miR-511-3p were detected in asthmatic (n = 22), allergic, nonasthmatic (n = 17), and nonallergic and nonasthmatic (n = 12) samples by using real-time PCR analysis. B, A receiver operating curve analysis to determine the sensitivities and specificities of miR-511-3p in the study population. Data represent means ±SEMs. AUC, Area under the curve. C, Proposed model of the role of Mrc1 in mediating CRE-induced lung inflammation through miR-511-3p.
The ability of miR-511-3p to differentiate asthmatic from nonasthmatic subjects was assessed by using a receiver operating curve analysis, which yielded an area under the curve of 0.83 (Fig 8, B). The receiver operating curves helped determine the sensitivity and specificity of the miRNA at various cutoff values. By using the optimal cutoff points, sensitivity and specificity were 81.8% and 71.4%, respectively (data not shown).
DISCUSSION
Complex allergens contain multiple innate immune-activating components that often trigger the activation of mucosal innate immune cells and subsequently promote TH2-polarized adaptive immune responses.40 In this study, for the first time, we showed that MRC1 on macrophages exerted a protective effect in mediating not only cockroach allergen uptake and clearance but also controls the genesis and limits the progression of allergic inflammation. MRC1, a member of the CLRs, on fibrocytes has been reported to mediate recognition of cockroach allergen, one of the most common allergens containing mannose-terminating glycans, which might contribute to innate immune responses.9 Also, previous work has suggested that MRC1 on DCs mediates the uptake of diverse native allergens and determination of allergen-induced T-cell polarization.4 In particular, MRC1-mediated Bla g 2 uptake on DCs is through the C-type lectin-like carbohydrate recognition domains 4 to 7.5 In this study we observed a significant reduction in cockroach allergen uptake in Mrc1−/− compared with WT mice, with a large number of macrophages that were colocalized with the inhaled cockroach allergen in the lungs of WT mice. Thus it is possible that reduction of inhaled cockroach allergen uptake in Mrc1−/− mice might be due to MRC1 deficiency in macrophages.
Alveolar macrophages (CD11c+CD45+F4/80+MHC-IIlo AFhi)24–26 were obtained from WT and Mrc1−/− mice to further confirm the significance of MRC1 in macrophages in cockroach allergen uptake. We first determined whether other CLRs (eg, SIGN-R1, Dectin-1, Dectin-2, and CD205) are differentially expressed in those macrophages from WT and Mrc1−/− mice, contributing to cockroach allergen uptake in these mice (see Fig E5 in this article’s Online Repository at www.jacionline.org). Except for MRC1, we found no differences for tested CLRs.
We next confirmed the MRC1-mediated allergen uptake by macrophages. As expected, Mrc1−/− macrophages showed significant reduction in allergen uptake by means of in vitro analysis with either CRE or the purified natural cockroach allergen Bla g 2. Importantly, this finding was further supported by continuous time-lapse live cell confocal microscopy. Of interest, these allergens, which are taken up, are accumulated and associated with cell vesicles but not the lysosomal compartment and then gradually moved from the cell boundary toward the inner parts of the cell. Although the ultimate fate of cockroach allergens is presently unknown, these studies suggest that macrophages in the lungs might play a critical and seemingly fundamental role in “scavenging” and internalizing the inhaled cockroach allergens for clearance.
Considering the fact that MRC1 lacks any known signaling motif, the signaling cascades of MRC1 remain unknown. MRC1 has been suggested to play no role in disease severity of mice infected with various pathogens, including fungi and parasites.41,42 Also, it has been shown to promote glomerulonephritis43 in part through augmented Fc-mediated function and to enhance experimental colitis23 through enhanced miR-511-3p–controlled activation of LPS–Toll-like receptor 4 (TLR4) signaling. Furthermore, Emara et al4 suggested the importance of MRC1 in recognition of a major cat allergen, Fel d 1, and its subsequent influence on specific antibody response. However, our findings in this study suggest that Mrc1 is important in triggering active immunity and host defense mechanisms, as in the case of microbial infection, whereas in the context of allergen recognition and its subsequent allergic response, MRC1 can be protective. This is consistent with previous findings by Juncadella et al44 demonstrating that deletion of the small GTPase Rac1 in airway epithelial cells in a mouse model led to defective engulfment of allergens but exacerbated inflammation with reduced IL-10 levels. Interestingly, our CRE-challenged Mrc1−/− mice also showed impaired production of IL-10 and PTGDS, which might confer protection against allergic responses. Although the discrepancies among those reported studies remain unclear, it is likely that different allergens, adjuvants, and experimental protocols in different models, microbial versus allergic responses, and the location and microenvironment of macrophages might contribute to the discrepancies. In this study we provided evidence supporting a critical role of MRC1 in allergen clearance as a natural defense mechanism and in limiting the progression and severity of cockroach allergen–induced allergic inflammation in a mouse model of asthma.
Importantly, we made the novel finding that miR-511-3p, which is encoded by MRC1, plays a critical role in MRC1-mediated downstream allergic immune responses. Especially, the Mrc1-mediated allergic inflammation could be related to the involvement of miR-511-3p in controlling macrophage polarization and activation. It has been evident that miRNAs control the balance of M1 and M2 macrophage polarization and modulate downstream immune responses.18–20,22,37,38 It is possible that MRC1 can modulate allergen-induced inflammation through controlling the balance of inflammation and anti-inflammation by modulating macrophage polarization after allergen challenge. Indeed, BMDMs from Mrc1−/− mice or BMDMs with Mrc1 knockdown mice displayed altered responses to M1 and M2 stimuli, suggesting that Mrc1 and miR-511-3p control macrophage polarization and activation, which might contribute to the protection of allergen-induced inflammatory responses.45 However, although BMDMs are broadly used surrogates for analysis of macrophage polarization, it will be essential to validate these findings in cultured alveolar macrophages (in vitro–differentiated alveolar macrophages)46 and human lung macrophages. Intriguingly, we found that plasma levels of miR-511-3p were significantly lower in asthmatic patients compared with those in nonasthmatic subjects, regardless of allergic status, indicating that miR-511-3p can protect against asthma. Although the sensitivity and specificity are expected to vary depending on the primer sequences, the results from the analysis of miR-511-3p were informative in obtaining the relative concentrations of miR-511-3p in samples for comparison. The relative quantity of plasma miR-511-3p levels in human subjects would provide potential relevance to our understanding of its role in allergic inflammation.
Additionally, we analyzed whether there were any associations between miR-511-3p and clinical features of asthma (eg, lung function, controller medication use, asthma control, atopy, and peripheral eosinophilia). No associations were observed, which might be limited by the small sample size and relatively well-controlled asthmatic population. Thus a larger sample size is needed for the determination of the association between miR-511-3p and specific asthma phenotypes.
One of the most significant observations we made in this study is that miR-511-3p protects against cockroach allergen–induced lung inflammation. In particular, we investigated the effect of miR-511-3p on cockroach allergen-induced lung inflammation by delivering the AAV–miR-511-3p vector to mouse lungs. In this study we tested several different AAVs and selected the AAV9 vector, which was encoded with the enhanced GFP reporter and driven by the CMV promoter because of its high transfection efficiency. In contrast to Mrc1−/− mice with increased lung inflammation, AAV–miR-511-3p–infected mice showed significant reduction in lung inflammation compared with AAV-control–infected mice. These findings suggest that miR-511-3p might play a role in protecting the lungs from cockroach allergen–induced inflammation.
Finally, we explored the mechanisms underlying miR-511-3p– mediated macrophage polarization and allergic inflammation and performed transcriptional profiling of BMDMs transfected with or without miR-511-3p. A total of 729 miR-511-3p–regulated genes were identified, including those most downregulated (eg, Lbr, Surf4, Ostm1, Ptgds, and Cd14) and upregulated (eg, Gsta3, Egr2, and S100a8) by miR-511-3p. These genes might be involved in Mrc1-regulated homeostatic state through antigen clearance and intronic miR-511-3p. Among them, S100A8, as an alarmin or danger-associated molecular pattern molecule,47 has antimicrobial activity in stimulating innate immune cells through binding to pattern recognition receptors, such as TLR4.48 PTGDS is involved in the generation of PGD2, a key lipid mediator of allergic inflammation.49–51 We not only confirmed the downregulation of PTGDS and its product PGD2 in BMDMs by miR-511-3p but also demonstrated a direct interaction between miR-511-3p and them RNA of Ptgds by using a well-established affinity-based isolation method, which is more specific than luciferase reporter assays,31 implicating that Ptgds might be one of the major targets for miR-511-3p. Furthermore, it was noted that Squadrito et al21 have previously identified Rho-associated coiled-coil containing protein kinase 2 (ROCK2) and latent TGF-β binding protein 1 (LTBP1) as direct targets of miR-511-3p, and both genes were also significantly downregulated by miR-511-3p overexpression in macrophages in our study.
Taken together, we hypothesized that miR-511-3p is critical in the cockroach allergen–MRC1 axis in allergen-induced allergic inflammation. miR-511-3p might control macrophage polarization and shape the immune response by inhibiting its direct targets (eg, PTGDS, ROCK2, and LTBP1) or regulating the expression of indirect targets (eg, TLR4 and C/EBPα; Fig 8, C). Previous studies have suggested that ROCK2 is highly expressed in MRC1+ tumor-associated macrophages,21 and inhibition of ROCK2 in M2 macrophages inhibited M2 (eg, Fizz 1 and Ym1) but promoted M1 phenotypes (NOS2).21,52 However, these studies did not provide direct evidence for the role of ROCK2 in macrophage polarization for those undifferentiated or at an early differentiation phase. Furthermore, these different findings can result from different experimental models of disease and parameters analyzed. Thus detailed studies are clearly needed to clarify whether any of these candidate genes, including ROCK2 and LTBP1, can serve as targets in mediating miR-511-3p effects on macrophage polarization and subsequently allergic inflammation in patients with allergic diseases.
In summary, our findings provided evidence that Mrc1 plays a critical role in cockroach antigen uptake. Importantly, for the first time, we demonstrated that MRC1 influences cockroach allergen–induced allergic inflammation in mouse model of asthma, which might function through miR-511-3p. Although we cannot rule out possible involvement of other regulatory pathways independent of miR-511-3p, our results from a combined analysis of cellular transfection and gene knockout models support the functional importance of miR-511-3p in mediating MRC1’s effect. Future studies will apparently focus on the mechanistic investigation into the role of miR-511-3p in the signaling cascades of MRC1 and address the question of whether miR-511-3p is also sufficient to confer MRC1’s regulatory function. Collectively, these studies provide a basis for exploring the mechanisms underlying the Mrc1/miR-511-3p modulation of macrophage activation, function, and macrophage-driven lung inflammation and ultimately lead to the discovery of novel therapeutic targets for allergic asthma.
METHODS
MRC1 mediates cockroach allergen uptake by macrophages
Uptake of FITC–Bla g 2 by sorted lung macrophages from WT and Mrc1−/− mice was observed by using a continuous time-lapse live cell confocal microscopy. In brief, FITC–Bla g 2 was added to cultured macrophages in an environmental control chamber to provide 37°C and 5% CO2 and then continuously observed for 30 minutes. Lysosomes and nuclei were stained in living cells with LysoTracker Red DND-99 and Hoechst 33342 (Invitrogen), respectively. Images were captured at 00:00, 05:00, 14:00, and 26:20 (minutes:seconds) by using a Zeiss LSM 780 confocal inverted microscope, respectively. Yellow and white arrows indicate the vesicles containing FITC–Bla g 2 with or without association of LysoTracker signal.
For gene profiling of macrophages with or without miR-511-3p overexpression, sample quality assessment and microarray analysis were conducted at the Sidney Kimmel Cancer Center Microarray Core Facility at Johns Hopkins University.
RNA isolation and quality assessment
Total RNA was prepared as described for the RNeasy Mini Kit (Qiagen) with on-column DNase I digestion. RNA quality was assessed by using the Nanodrop-2000 spectrometer for OD260/280 and OD260/230 ratio and the Bioanalyzer (Agilent Technologies).
Microarray assay
MouseRef-8 v2 Expression BeadChip arrays (Illumina) were used for microarray hybridizations to examine global gene expression of the samples. The array targets 18,138 annotated mouse genes with 25,697 unique probes derived from the National Center for Biotechnology Information Reference Sequence (NCBI RefSeq) database (Build 36, Release 22) supplemented with probes derived from the Mouse Exonic Evidence Based Oligonucleotide (MEEBO) set, as well as exemplar protein-coding sequences described in the RIKEN FANTOM2 database.
In brief, 500 ng of total RNA from each sample was amplified and labeled with the Illumina TotalPrep RNA Amplification Kit (AMIL1791; Ambion), as described in the instruction manual. For array assay, 750 ng of biotin-labeled cRNA was combined with hybridization buffer and hybridized to the array at 58°C for 16 to 20 hours. After hybridization, the hybridization cartridge was disassembled, and the array was washed with buffer at 55°C and blocked at room temperature. Bound biotinylated cRNA was stained with streptavidin-Cy3 and then washed. Dried arrays were stored in a dark box until scanned with the iScan System. Data were extracted with Gene Expression Module in GenomeStudio Software.
Generating BM chimeric mice with miR-511-3p overexpression
BMs were harvested by flushing the femurs and the tibias of 6-week-old female C57BL/6 mice. Lineage-negative cells (BM-Lin− cells) enriched in hematopoietic stem and progenitor cells (HSPCs) were isolated from BM by using a cell purification kit (STEMCELL Technologies, Vancouver, British Columbia, Canada). Approximately 1 × 106 HSPCs per milliliter were prestimulated for 6 hours in serum-free medium containing a cocktail of cytokines and then transduced with miR-511–overexpressing or mutant LVs with a dose equivalent to 1 × 108 transducing units/mL for 12 hours. After transduction, 1×106 cells were infused into the tail veins of lethally whole-body irradiated mice (5.5 Gy in 2 doses 3 hours apart) for the generation of chimeric mice. Five weeks after transplantation, the chimeric mice were used to investigate the pulmonary inflammatory response by using well-established protocols. Briefly, mice were sensitized with CRE (20 µg administered intratracheally) on days 36, 37, 39, and 46 and then challenged on days 52 and 54. The mice were killed 24 hours after the last challenge.
Supplementary Material
Key messages.
MRC1 is involved in cockroach allergen antigen binding, antigen recognition, and macrophage polarization.
miR-511-3p is coregulated with MRC1 and contributes to macrophage polarization.
miR-511-3p can serve as a marker for diagnosis or therapeutic target of asthma.
Acknowledgments
Supported by grants from the US National Institutes of Health (NIH; RO1ES021739, R21 AI109062, and R21 AI121768) and the National Science Foundation of China (NSFC; 81628001; to P.G.); the Intramural Research Program of the National Institutes of Health, National Institute on Aging; the China 1000 Young Talents Plan Program (to Y.Z.); and Fudan Children’s Hospital and Fudan University for Initial Funding, NSFC 81671561, and MOST 2016YFC1305102 (to Y.Z.); and by grants from the National Health Research Institutes (EOPP10-014 and EOSP07-014 to S.-K.H.) and the Ministry of Health (EODOH01; to S.-K.H.); Academia Sinica (AS-105-TP-B05 to SK Huang); and Kaohsiung Medical University “Aim for the Top Universities Grant” and “the Talent Plan” (KMU-TP-105A04 and KMU-SH000184 to SK Huang), Taiwan.
We acknowledge the support of Dr De Palma (Swiss Federal Institute of Technology, Lausanne) for providing LVs expressing miR-511-3p (LV miR-511-3p) or a miR-511-3p-mut (LV Mock).
Abbreviations used
- AAV
Adeno-associated virus
- AF
Autofluorescence
- BAL
Bronchoalveolar lavage
- BM
Bone marrow
- CLR
C-type lectin receptor
- CMV
Cytomegalovirus
- CRE
Cockroach extract
- DC
Dendritic cell
- FDR
False discovery rate
- FITC
Fluorescence isothiocyanate
- GFP
Green fluorescent protein
- H&E
Hematoxylin and eosin
- LTBP1
Latent TGF-β binding protein 1
- LV
Lentivirus
- M-CSF
Macrophage colony-stimulating factor
- miRNA
MicroRNA
- MRC1
Mannose receptor
- PAS
Periodic acid–Schiff
- PTGDS
Prostaglandin D2 synthase
- qPCR
Quantitative PCR
- ROCK2
Rho-associated coiled-coil containing protein kinase 2
- SSC
Side scatter
- TLR4
Toll-like receptor 4
- WT
Wild-type
Footnotes
Disclosure of potential conflict of interest: The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Pomares L. The mannose receptor. J Leukoc Biol. 2012;92:1177–86. doi: 10.1189/jlb.0512231. [DOI] [PubMed] [Google Scholar]
- 3.Boskovic J, Arnold JN, Stilion R, Gordon S, Sim RB, Rivera-Calzada A, et al. Structural model for the mannose receptor family uncovered by electron microscopy of Endo180 and the mannose receptor. J Biol Chem. 2006;281:8780–7. doi: 10.1074/jbc.M513277200. [DOI] [PubMed] [Google Scholar]
- 4.Emara M, Royer PJ, Abbas Z, Sewell HF, Mohamed GG, Singh S, et al. Recognition of the major cat allergen Fel d 1 through the cysteine-rich domain of the mannose receptor determines its allergenicity. J Biol Chem. 2011;286:13033–40. doi: 10.1074/jbc.M111.220657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Royer PJ, Emara M, Yang C, Al-Ghouleh A, Tighe P, Jones N, et al. The mannose receptor mediates the uptake of diverse native allergens by dendritic cells and determines allergen-induced T cell polarization through modulation of IDO activity. J Immunol. 2010;185:1522–31. doi: 10.4049/jimmunol.1000774. [DOI] [PubMed] [Google Scholar]
- 6.Togias A, Fenton MJ, Gergen PJ, Rotrosen D, Fauci AS. Asthma in the inner city: the perspective of the National Institute of Allergy and Infectious Diseases. J Allergy Clin Immunol. 2010;125:540–4. doi: 10.1016/j.jaci.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 7.Sohn MH, Kim KE. The cockroach and allergic diseases. Allergy Asthma Immunol Res. 2012;4:264–9. doi: 10.4168/aair.2012.4.5.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou J, Li WQ, Li Q, Li XQ, Zhang JT, Liu GQ, et al. Two functional microRNA-126s repress a novel target gene p21-activated kinase 1 to regulate vascular integrity in zebrafish. Circ Res. 2011;108:201–9. doi: 10.1161/CIRCRESAHA.110.225045. [DOI] [PubMed] [Google Scholar]
- 9.Tsai YM, Hsu SC, Zhang J, Zhou YF, Plunkett B, Huang SK, et al. Functional interaction of cockroach allergens and mannose receptor (CD206) in human circulating fibrocytes. PLoS One. 2013;8:e64105. doi: 10.1371/journal.pone.0064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol. 2011;187:3362–73. doi: 10.4049/jimmunol.1101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson LJ, Patel S, Bhakta NR, Choy DF, Brightbill HD, Ren X, et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol. 2014;15:1162–70. doi: 10.1038/ni.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106:5282–7. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das A, Ganesh K, Khanna S, Sen CK, Roy S. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J Immunol. 2014;192:1120–9. doi: 10.4049/jimmunol.1300613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comer BS, Camoretti-Mercado B, Kogut PC, Halayko AJ, Solway J, Gerthoffer WT. MicroRNA-146a and microRNA-146b expression and anti-inflammatory function in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2014;307:L727–34. doi: 10.1152/ajplung.00174.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci U S A. 2009;106:18704–9. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solberg OD, Ostrin EJ, Love MI, Peng JC, Bhakta NR, Hou L, et al. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med. 2012;186:965–74. doi: 10.1164/rccm.201201-0027OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem. 2012;287:21816–25. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1) J Biol Chem. 2011;286:1786–94. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Squadrito ML, Pucci F, Magri L, Moi D, Gilfillan GD, Ranghetti A, et al. miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell Rep. 2012;1:141–54. doi: 10.1016/j.celrep.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Squadrito ML, Etzrodt M, De Palma M, Pittet MJ. MicroRNA-mediated control of macrophages and its implications for cancer. Trends Immunol. 2013;34:350–9. doi: 10.1016/j.it.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinsbroek SE, Squadrito ML, Schilderink R, Hilbers FW, Verseijden C, Hofmann M, et al. miR-511-3p, embedded in the macrophage mannose receptor gene, contributes to intestinal inflammation. Mucosal Immunol. 2016;9:960–73. doi: 10.1038/mi.2015.113. [DOI] [PubMed] [Google Scholar]
- 24.Soroosh P, Doherty TA, Duan W, Mehta AK, Choi H, Adams YF, et al. Lung-resident tissue macrophages generate Foxp31 regulatory T cells and promote airway tolerance. J Exp Med. 2013;210:775–88. doi: 10.1084/jem.20121849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol. 2013;49:503–10. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaynagetdinov R, Sherrill TP, Kendall PL, Segal BH, Weller KP, Tighe RM, et al. Identification of myeloid cell subsets in murine lungs using flow cytometry. Am J Respir Cell Mol Biol. 2013;49:180–9. doi: 10.1165/rcmb.2012-0366MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao P, Zhou Y, Xian L, Li C, Xu T, Plunkett B, et al. Functional effects of TGF-beta1 on mesenchymal stem cell mobilization in cockroach allergen-induced asthma. J Immunol. 2014;192:4560–70. doi: 10.4049/jimmunol.1303461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu T, Zhou Y, Qiu L, Do DC, Zhao Y, Cui Z, et al. Aryl hydrocarbon receptor protects lungs from cockroach allergen-induced inflammation by modulating mesenchymal stem cells. J Immunol. 2015;195:5539–50. doi: 10.4049/jimmunol.1501198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Zhang P, Abdelmohsen K, Liu Y, Tominaga-Yamanaka K, Yoon JH, Ioannis G, et al. Novel RNA- and FMRP-binding protein TRF2-S regulates axonal mRNA transport and presynaptic plasticity. Nat Commun. 2015;6:8888. doi: 10.1038/ncomms9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panganiban RP, Pinkerton MH, Maru SY, Jefferson SJ, Roff AN, Ishmael FT. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol. 2012;1:154–65. [PMC free article] [PubMed] [Google Scholar]
- 32.Panganiban RP, Wang Y, Howrylak J, Chinchilli VM, Craig TJ, August A, et al. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol. 2016;137:1423–32. doi: 10.1016/j.jaci.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 33.Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fehervari Z. Alveolar macrophages in asthma. Nat Immunol. 2014;16:64. [Google Scholar]
- 35.Lee YG, Jeong JJ, Nyenhuis S, Berdyshev E, Chung S, Ranjan R, et al. Recruited alveolar macrophages, in response to airway epithelial-derived MCP-1/CCL2, regulate airway inflammation and remodeling in allergic asthma. Am J Respir Cell Mol Biol. 2015;52:772–84. doi: 10.1165/rcmb.2014-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karo-Atar D, Itan M, Pasmanik-Chor M, Munitz A. MicroRNA profiling reveals opposing expression patterns for miR-511 in alternatively and classically activated macrophages. J Asthma. 2015;52:545–53. doi: 10.3109/02770903.2014.988222. [DOI] [PubMed] [Google Scholar]
- 37.Saha B, Momen-Heravi F, Kodys K, Szabo G. MicroRNA cargo of extracellular vesicles from alcohol-exposed monocytes signals naive monocytes to differentiate into M2 macrophages. J Biol Chem. 2016;291:149–59. doi: 10.1074/jbc.M115.694133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baer C, Squadrito ML, Laoui D, Thompson D, Hansen SK, Kiialainen A, et al. Suppression of microRNA activity amplifies IFN-gamma-induced macrophage activation and promotes anti-tumour immunity. Nat Cell Biol. 2016;18:790–802. doi: 10.1038/ncb3371. [DOI] [PubMed] [Google Scholar]
- 39.Urade Y, Hayaishi O. Prostaglandin D2 and sleep/wake regulation. Sleep Med Rev. 2011;15:411–8. doi: 10.1016/j.smrv.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Wills-Karp M, Nathan A, Page K, Karp CL. New insights into innate immune mechanisms underlying allergenicity. Mucosal Immunol. 2010;3:104–10. doi: 10.1038/mi.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SJ, Zheng NY, Clavijo M, Nussenzweig MC. Normal host defense during systemic candidiasis in mannose receptor-deficient mice. Infect Immun. 2003;71:437–45. doi: 10.1128/IAI.71.1.437-445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swain SD, Lee SJ, Nussenzweig MC, Harmsen AG. Absence of the macrophage mannose receptor in mice does not increase susceptibility to Pneumocystis carinii infection in vivo. Infect Immun. 2003;71:6213–21. doi: 10.1128/IAI.71.11.6213-6221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chavele KM, Martinez-Pomares L, Domin J, Pemberton S, Haslam SM, Dell A, et al. Mannose receptor interacts with Fc receptors and is critical for the development of crescentic glomerulonephritis in mice. J Clin Invest. 2010;120:1469–78. doi: 10.1172/JCI41560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juncadella IJ, Kadl A, Sharma AK, Shim YM, Hochreiter-Hufford A, Borish L, et al. Apoptotic cell clearance by bronchial epithelial cells critically influences airway inflammation. Nature. 2013;493:547–51. doi: 10.1038/nature11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spence S, Fitzsimons A, Boyd CR, Kessler J, Fitzgerald D, Elliott J, et al. Suppressors of cytokine signaling 2 and 3 diametrically control macrophage polarization. Immunity. 2013;38:66–78. doi: 10.1016/j.immuni.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 46.Kuroda E, Ozasa K, Temizoz B, Ohata K, Koo CX, Kanuma T, et al. Inhaled fine particles induce alveolar macrophage death and interleukin-1alpha release to promote inducible bronchus-associated lymphoid tissue formation. Immunity. 2016;45:1299–310. doi: 10.1016/j.immuni.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 47.Gao P, Grigoryev DN, Rafaels NM, Mu D, Wright JM, Cheadle C, et al. CD14, a key candidate gene associated with a specific immune response to cockroach. Clin Exp Allergy. 2010;40:1353–64. doi: 10.1111/j.1365-2222.2010.03561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogl T, Eisenblatter M, Voller T, Zenker S, Hermann S, van Lent P, et al. Alarmin S100A8/S100A9 as a biomarker for molecular imaging of local inflammatory activity. Nat Commun. 2014;5:4593. doi: 10.1038/ncomms5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joo M, Sadikot RT. PGD synthase and PGD2 in immune response. Mediators Inflamm. 2012;2012:503128. doi: 10.1155/2012/503128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura T, Maeda S, Horiguchi K, Maehara T, Aritake K, Choi BI, et al. PGD2 deficiency exacerbates food antigen-induced mast cell hyperplasia. Nat Commun. 2015;6:7514. doi: 10.1038/ncomms8514. [DOI] [PubMed] [Google Scholar]
- 51.Jandl K, Stacher E, Bálint Z, Sturm EM, Maric J, Peinhaupt M, et al. Activated prostaglandin D2 receptors on macrophages enhance neutrophil recruitment into the lung. J Allergy Clin Immunol. 2016;137:833–43. doi: 10.1016/j.jaci.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zandi S, Nakao S, Chun KH, Fiorina P, Sun D, Arita R, et al. ROCK-isoform-specific polarization of macrophages associated with age-related macular degeneration. Cell Rep. 2015;10:1173–86. doi: 10.1016/j.celrep.2015.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.