Abstract
Metal homeostasis poses a major challenge to microbes, which must acquire scarce elements for core metabolic processes. Methanobactin, an extensively modified copper-chelating peptide, was one of the earliest natural products shown to enable microbial acquisition of a metal other than iron. We describe the core biosynthetic machinery responsible for the characteristic posttranslational modifications that grant methanobactin its specificity and affinity for copper. A heterodimer comprising MbnB, a DUF692 family iron enzyme, and MbnC, a protein from a previously unknown family, performs a dioxygen-dependent four-electron oxidation of the precursor peptide (MbnA) to install an oxazolone and an adjacent thioamide, the characteristic methanobactin bidentate copper ligands. MbnB and MbnC homologs are encoded together and separately in many bacterial genomes, suggesting functions beyond their roles in methanobactin biosynthesis.
Metals are necessary for many biological processes, and strategies to facilitate their acquisition have evolved across the domains of life. Metal scarcity presents a particular challenge, whether due to low environmental levels of soluble metals or metal-limiting conditions imposed by other organisms (1, 2). Iron acquisition, particularly by pathogenic bacteria in environments where iron is sequestered by the host, is the canonical example of high-affinity metal uptake in low-metal environments (3). Instead of relying on passive import of scarce Fe(II), bacteria secrete natural products known as siderophores to bind Fe(III) sequestered in insoluble iron oxides or in host proteins; the Fe(III)-siderophore complex is then imported, and the iron is made available to the organism. In addition, some of these compounds play additional roles in metal regulation and protection from toxicity of metals other than iron (4). There is increasing evidence for the existence of similar processes for other metals. Staphylopine, recently discovered in Staphylococcus aureus, is involved in the import of a range of divalent metals (5), and nickelophores [ranging from simple amino acids (6) to more complex organic molecules (7)] are thought to be present in both S. aureus and Helicobacter pylori. Yersiniabactin has been shown to bind copper as well as iron (8). Similarly, despite iron-responsive regulation, delftibactin binds gold (9), and further reexamination of other siderophores suggests that many have roles as non-iron metallophores (10). Some of the first identified non-iron metallophores belong to the family of copper-binding compounds known as methanobactins (Mbns) (11).
Mbns were initially discovered in obligate methanotrophs (12), which oxidize their sole carbon source, methane, using the copper-dependent particulate methane monooxygenase or, when starved for copper, the iron-dependent soluble methane monooxygenase. Under copper-limiting conditions, methanotrophs also produce and secrete Mbns, peptidic compounds that chelate Cu(I) directly or Cu(II) via a reductive process with an unknown mechanism, resulting in Cu(I)Mbn. The high affinity and specificity of Mbns for Cu(I) is conferred by a pair of bidentate ligands, each comprising a nitrogen heterocycle (most commonly an oxazolone ring) and an adjacent thioamide or enethiol, which chelate the metal in a distorted tetrahedral geometry (Fig. 1A and fig. S1) (11).
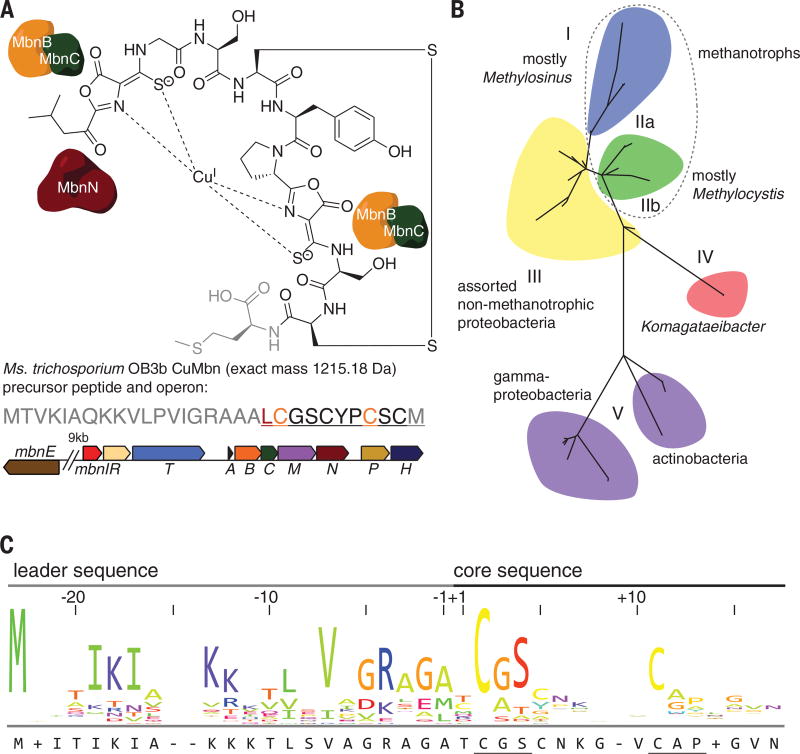
Fig. 1. Mbn structure, operon organization, phylogeny, and precursor peptide sequence.
(A) Structure of CuMbn from Ms. trichosporium OB3b (top); sequence of the MbnA precursor peptide (middle) and Mbn operon (bottom) from that species. The operon includes genes related to import (the periplasmic binding protein MbnE and the TonB-dependent transporter MbnT), export (the MATE multidrug exporter MbnM), regulation (the sigma/anti-sigma factor pair MbnIR), biosynthesis (the precursor peptide MbnA, the hypothetical proteins MbnB and MbnC, and the aminotransferase MbnN), and two genes of unknown function (MbnP and MbnH). Color coding of the genes in the operon indicates previously proposed roles: MbnB (orange) and MbnC (green) are responsible for modifying the two orange cysteines in the peptide to form oxazolone-thioamide pairs in the final natural product, whereas MbnN (dark red) is responsible for a transamination reaction at the leucine (dark red) in the peptide and the final natural product. The final methionine, depicted in gray, is sometimes missing from the compound. (B) Phylogenetic tree for Mbn operons. All Mbn operons contain mbnA, mbnB, and mbnC genes, and phylogenetic analyses of all three genes support the subgroups illustrated here. (C) MbnA sequence logo, illustrating the leader and core peptide sequences and the conserved cysteine modification motifs. Amino acid abbreviations: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; Y, Tyr.
Operons encoding the biosynthetic machinery for Mbns, first identified and shown to be copper-regulated in the methanotroph Methylosinus (Ms.) trichosporium OB3b (13, 14), are present in a range of methanotrophic and nonmethanotrophic bacteria (Fig. 1, A and B) (11, 15). The gene encoding the precursor peptide (MbnA) was recognized because the predicted C-terminal amino acid sequence corresponds to the peptidic Mbn backbone (13). Disruption of this gene abolishes Mbn production (16), providing evidence that Mbns are ribosomally produced, posttranslationally modified natural products (RiPPs), a class that also includes microcins, lasso peptides, lanthipeptides, thiopeptides, and pyrroloquinoline quinone (15, 17). Like the precursor peptides in most other RiPP families, MbnAs consist of an N-terminal leader peptide, which is cleaved in the final natural product, and a C-terminal core peptide, to which posttranslational modifications are directed (Fig. 1C).
The locations of oxazolone rings and their neighboring thioamide groups in the peptidic backbones of mature Mbns indicate that these groups are installed at cysteine residues (15). Bioinformatic analysis of all MbnA sequences reveals that the modified cysteines are part of conserved motifs, wherein the cysteine is followed by a small, typically hydrophobic residue (usually glycine or alanine, but occasionally serine) and then by a larger, more frequently hydrophilic residue (primarily serine and threonine) (Fig. 1C). In Ms. trichosporium OB3b MbnA, there are two modification sites, at Cys21 and Cys27 (residue numbering from the full-length peptide). Notably, Mbn operons lack genes associated with oxazolone or thioamide biosynthesis, indicating that the pathway(s) for their generation in Mbns must be biochemically distinct from nonenzymatic oxazolone biosynthesis in jadomycins (18) and the unrelated microbial enzymatic thioamide formation pathways for closthioamide, thioviridamide, and methyl coenzyme M reductase (19–21).
Mbn operons, which vary in the composition and arrangement of their genes, have been divided into five major groups on the basis of their content as well as sequence-based phylogenetic analyses of three core components present in all operons (Fig. 1B and fig. S2). Also present are genes encoding proteins with predicted or, in some cases, experimentally established roles in regulation, transport, and secondary posttranslational modifications (Fig. 1B and fig. S2) (15). Of particular interest are two genes encoding uncharacterized proteins designated MbnB and MbnC. These proteins are encoded within every Mbn operon identified to date, along with the precursor peptide MbnA, and we previously proposed (15) that they are responsible for the posttranslational modifications of MbnA that form the copper-binding ligands of Mbns. For the structurally characterized Mbns (and most predicted Mbns), there are two heterocyclethioamide moieties in the final compound (fig. S1) (13, 22–24). The second heterocycle is uniformly an oxazolone; the identity of the first varies, but it is predicted that all originate from oxazolone precursors (15). Formation of each oxazolonethioamide moiety requires a net four-electron oxidation. The responsible enzymes would thus be expected to contain a redox cofactor.
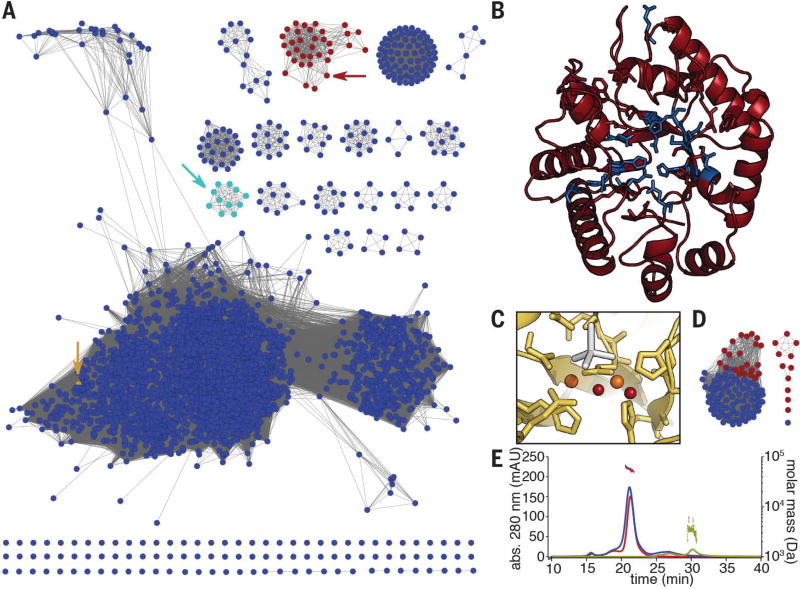
MbnBs are members of the uncharacterized DUF692 subfamily (Fig. 2A). Although these proteins are distantly related to characterized members of the TIM barrel family 15, including divalent metal-dependent enzymes such as xylose isomerase and endonuclease IV (25), no DUF692 family member has been functionally characterized, despite their presence in a wide range of Gram-positive and Gram-negative bacteria. An unpublished crystal structure (PDB: 3BWW) of one family member from Haemophilus (Hs.) somnus 129Pt revealed a diiron cluster at the center of the TIM barrel fold, with ligands corresponding to two of the three metal-binding sites observed in endonuclease IV enzymes. Two of the three ligands required for the third endonuclease IV metal binding site are present in the sequence of the Hs. somnus 129Pt protein but were disordered in the structure. The metal-binding residues observed in the Hs. somnus 129Pt structure are strictly conserved throughout the DUF692 family, including MbnBs, and a plausible structure for Ms. trichosporium OB3b MbnB was generated by modeling its sequence onto the experimentally determined structure of the Hs. somnus 129Pt protein, even though MbnBs constitute a distinct subfamily within the DUF692 family (Fig. 2, B and C). Less is known about MbnC. Four years after the initial bioinformatics study, this predicted protein still has no identifiable domains, and members of this family are found only in Mbn operons or separate from mbnB in Pseudomonas species (Fig. 2D). Neither MbnB nor MbnC contains a RiPP precursor protein recognition element, which is present in leader peptide–dependent modifying enzymes from many RiPP families (26).
Fig. 2. Bioinformatic and biochemical analyses of the MbnBC heterodimer.
(A) Sequence similarity network for the DUF692 family. MbnBs are depicted in red, MbnXs (found only in the fifth Mbn subgroup) are in teal, and the structurally characterized Hs. somnus 129Pt protein sequence (PDB: 3BWW) is in yellow. Arrows indicate the location of the relevant nodes. (B) The predicted structure of Ms. trichosporium OB3b MbnB (red), as modeled against 3BWW using iTasser (RMSD, 4.2 ± 2.8 Å). Residues in blue are strictly conserved in MbnBs. (C) Diiron site as observed in the 3BWW structure (yellow). All amino acids coordinating the two irons (orange) are strictly conserved in MbnBs. Two water molecules (red) are also present in the coordination sphere in the crystal structure, along with a cacodylate molecule (gray) from the crystallization buffer. (D) Sequence similarity network for the broader MbnC family. Red circles, MbnCs; blue circles, related non-MbnC genes found exclusively in Pseudomonas species. (E) SEC-MALS analysis of MbnA (yellow; 3 kDa predicted, 3 kDa observed), MbnBC (red; 58 kDa predicted, 55 kDa observed), and MbnABC (blue; 61 kDa predicted, 58 kDa observed) indicates that MbnB and MbnC form a heterodimeric complex that can bind MbnA.
Separate heterologous overexpression of either MbnB or MbnC from several species failed to yield soluble protein. However, simultaneous coexpression of MbnB and MbnC from a single vector (27) yielded soluble, stable heterodimeric complexes (MbnBC; 58.2 kDa for the Ms. trichosporium OB3b proteins with intact affinity tags) that could be purified via affinity and size exclusion chromatographies (Fig. 2E and fig. S3). Consistent with the proposed role of MbnBC in Mbn biosynthesis, addition of MbnA resulted in the formation of a heterotrimeric MbnA-MbnBC complex, detectable by size exclusion chromatography with multiangle light scattering (SEC-MALS) (Fig. 2E) as well as denaturing and native polyacrylamide gel electrophoresis (fig. S3). Moreover, purified MbnBC complexes from six different species (table S1 and fig. S4) contained iron, potentially satisfying the expected requirement for a redox cofactor.
Supplementation with 200 to 500 µM ferrous ammonium sulfate during expression increased the iron loading, and in our model construct of MbnBC from Ms. trichosporium OB3b, purified protein samples contained 1.3 ± 0.2 iron atoms per heterodimer (table S1). 57Fe-Mössbauer spectra of MbnBC are complex, but for aerobically purified protein from three species, a mixture of exchange-coupled di- and trinuclear clusters was observed in addition to smaller amount(s) of unidentified species (fig. S5 and supplementary materials). Native top-down mass spectrometry (nTDMS) of MbnBC from six species showed that, as anticipated, all the iron was associated with the MbnB subunit (figs. S6 and S7). A notable subset of the population appeared to have as many as three iron ions per MbnB subunit, indicating that a fraction of the MbnB protein remained in the metal-free (apo) state, which was also observed by native mass spectrometry (figs. S6 and S7). The heterogeneity implied by the noninteger iron stoichiometry and observed by both Mössbauer spectroscopy and nTDMS implied that only a fraction of the purified MbnBC complexes were in the active state. Reduced Fe(II) is commonly required to initiate the oxidation of aliphatic carbon centers (28, 29), and less than 5% of the total iron in aerobically purified MbnBC was in this form. Purification of MbnBC under O2-free conditions resulted in a variable increase in Fe(II) levels at the expense of diminished iron loading.
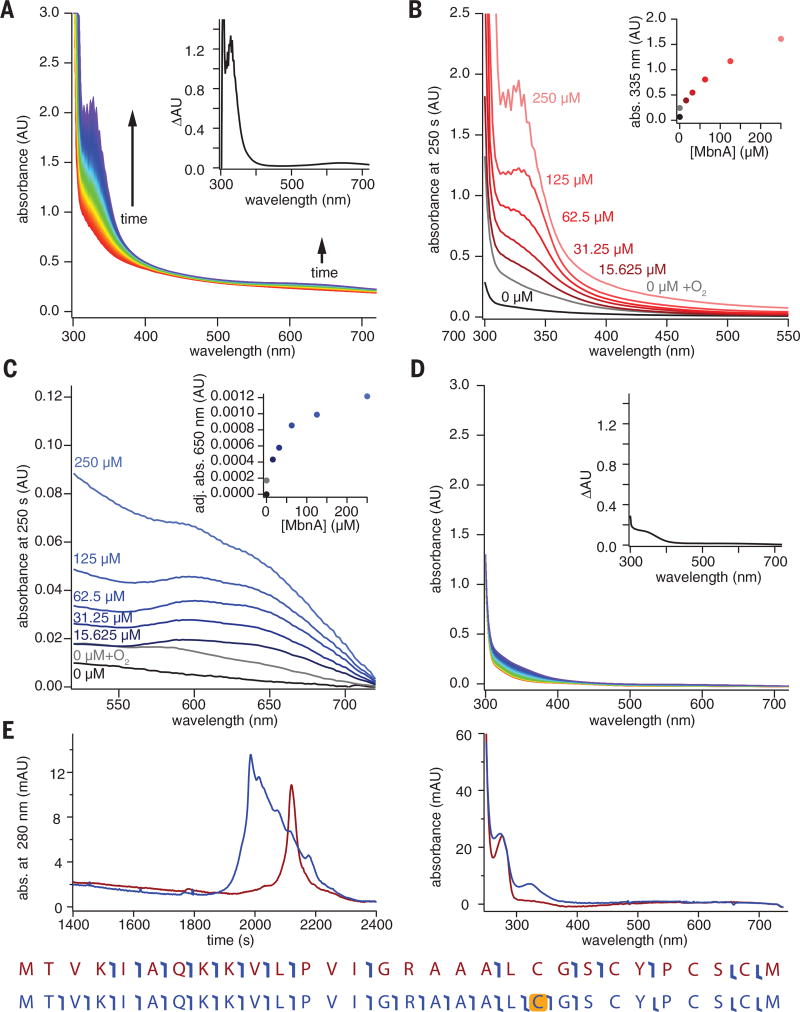
Both aerobically and anaerobically produced MbnBC react with MbnA in the presence of O2, without the addition of external reductant, to yield a product characterized by a sharp absorption feature at 335 nm and a broad feature at 650 nm (Fig. 3A and fig. S8). Despite its decreased iron content, the anaerobically produced MbnBC was more active, consistent with the involvement of an Fe(II) species in the active form of the cofactor. In the presence of excess O2, the 335-nm feature increased in intensity with increasing quantities of substrate (Fig. 3B and fig. S8), whereas the intensity of the 650-nm feature was limited by MbnBC concentration (Fig. 3C and fig. S8). This latter feature likely reflects a charge-transfer transition associated with the complex of MbnA with the MbnBC iron cofactor. When MbnA was omitted from the reaction, a broader, weaker feature with a wavelength of maximum absorption (λmax) of 340 nm developed more slowly (Fig. 3D and fig. S9). This feature resembles those arising from oxo-to-ferric charge-transfer transitions in nonheme dinuclear Fe enzymes (30) and likely derives from a species generated by unproductive oxidation of the Fe(II) in the MbnBC cofactor in the absence of substrate. Similarly, incubation of MbnA with MbnBC in the absence of O2 did not yield the intense 335-nm feature, consistent with a role for O2 in production of the chromophore. Minor spectral changes observed under these conditions likely reflect low levels of residual O2 in the deoxygenated reaction buffers.
Fig. 3. Modification of Ms. trichosporium OB3b MbnA by MbnBC.
(A) Reaction of 100 µM anaerobic MbnBC with 125 µM MbnA and 900 µM O2 over a period of 250 s; two prominent features are observed at 335 and 650 nm. Inset shows a difference spectrum between the final (250 s) and initial (0 s) time points. (B) UV-vis absorption spectra of the feature at 335 nm with different concentrations of MbnA. Inset shows the absorbance at 335 nm at different concentrations of MbnA. (C) UV-vis absorption spectra of the feature at 650 nm at different concentrations of MbnA; inset shows absorbance at 650 nm adjusted via a dropline correction using values at 640 and 660 nm. (D) Reaction of 100 µM anaerobically prepared MbnBC containing 140 µM Fe with 900 µMO2 over a period of 250 s reveals the formation of a broad spectral feature between 300 and 400 nm. Inset shows a difference spectrum between the final (250 s) and initial (0 s) time points. (E) LC-UV-MS-MS shows that the retention time (left chromatogram) of the modified peptide (blue) changes relative to the unmodified peptide (red), that the absorption spectrum of the modified peptide maintains the 335-nm feature (right spectrum), and that the presence of the 335-nm feature is associated with a mass shift of −4 Da that can be assigned to the first of the two modifiable cysteines (yellow highlight, bottom). Angled blue “flags” in the peptide sequence maps indicate the location of fragment ions observed during tandem MS.
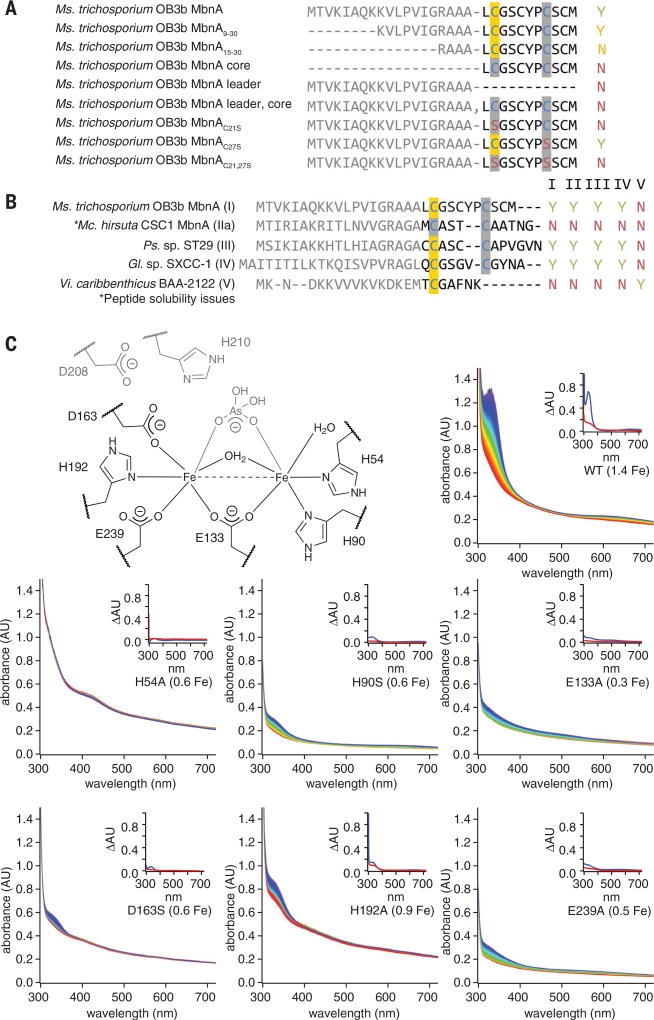
Analysis of MbnA before and after exposure to MbnBC and O2 by coupled liquid chromatography, ultraviolet-visible (UV-vis) absorption spectroscopy, and tandem mass spectrometry (LC-UV-MS-MS) showed that the 335-nmfeature is associated with MbnA and correlates with a mass shift of −4.031 Da localized to Cys21, which is the position of the N-terminal oxazolone ring (OxaA) in the final natural product (Fig. 3E and fig. S10). This mass shift matches that predicted for the installation of a single oxazolone-thioamide group. In support of this assignment, addition of 100 to 500 mM HCl resulted in the decay of the 335-nm feature in modified MbnA, as expected if the modification is indeed an oxazolone-thioamide group (fig. S11) (13, 24). Although species with mass shifts of −2 and −6 Da could also be detected in many reactions, fragmentation analyses indicated that these species corresponded to intramolecular disulfide formation between unmodified cysteines and modification together with intramolecular disulfide bond formation, respectively. Consistent with modification of Cys21, a Cys21 → Ser substitution eradicated peptide modification as observed via UV-vis spectroscopy and mass spectrometry, whereas a Cys27 → Ser substitution had no effect (Fig. 4A and figs. S12 and S13). MbnBC additionally did not modify the core peptide alone, nor did it react with the leader and core peptides added in trans, and even the truncation of the leader peptide resulted in diminished activity (Fig. 4A and figs. S12 and S13).
Fig. 4. Correlation of MbnBC activity with proposed diiron active site ligands and MbnA sequence.
(A) Summary of activity observed via UV-vis absorption spectroscopy and LC-MS for 125 µM Ms. trichosporium OB3b MbnA variants with 100 µM Ms. trichosporium OB3b MbnBC and excess O2. Yes (Y) indicates that modification is observed; No (N) indicates no modification. Modified cysteines localized via tandem MS are highlighted in yellow. (B) Summary of activity observed via UV-vis absorption spectroscopy and LC-MS for 125 µM MbnA substrate peptides and 100 µM MbnBC enzyme complexes from the different Mbn operon families in the presence of excess (~900 µM) O2. (C) Stopped-flow UV-vis absorption spectra over a period of 250 s for reactions between 125 µM MbnA, excess (~900 µM) O2, and 100 µM wild-type Ms. trichosporium OB3b MbnBC or one of six enzyme variants with alanine mutations in predicted active-site residues (the diiron site from the Hs. somnus 129Pt structure, labeled with Ms. trichosporium OB3b residue numbers, is provided for reference). Spectra are colored in rainbow order according to time, with the earliest spectra in red and the latest spectra in blue. Iron content per heterodimer is included in each label. Insets depict difference spectra for the enzyme in the presence of oxygen (red) or oxygen and the MbnA substrate peptide (blue).
Despite this marked sensitivity to the presence of valid leader peptides and modification sites, MbnBCs are otherwise promiscuous. MbnBCs from Mbn groups I to IV were all capable of modifying heterologous MbnA groups I to IV, despite the substantial differences in leader and core peptide sequences. MbnA from group V has the most divergent sequence (11), and the MbnBC from this group could interact only with the group V MbnA (Fig. 4B and fig. S14). All observed MbnA modifications were associated with the same mass shift of −4 Da observed in the Ms. trichosporium MbnABC system (fig. S15), indicating that a detailed analysis of this system can be generalized to the larger MbnBC family.
Although the mass shift and acid hydrolysis data are consistent with a single oxazolonethioamide moiety in modified MbnA, the 335-nm absorption feature differs from the 392-nm feature of OxaA in mature Mbns. This discrepancy is attributable to structural differences between the modified MbnA product and mature Mbn. In mature Mbn, the leader peptide is cleaved and the N-terminal primary amine is converted to a carbonyl group by the pyridoxal-5′-phosphate (PLP)–dependent aminotransferase MbnN (15). This modification extends the conjugation of the oxazolone ring, accounting for the bathochromic shift in the spectral feature; OxaB moieties in all Mbns lack such extended conjugation, and their absorption features fall within the 325- to 345-nm range (13, 23, 24, 31). Consistent with this explanation, disruption of mbnN in Ms. trichosporium OB3b resulted in accumulation of a previously unknown copper-binding compound that exhibited a single major spectral feature with λmax of 332 nm and a mass shift of +1 Da with respect to CuMbn, reflecting the presence of the N-terminal primary amine in place of the carbonyl group (fig. S16).
The formation of this oxazolone-thioamide moiety is dependent on the presence of the iron cofactor. Mutagenesis of the proposed iron ligands in Ms. trichosporium OB3b MbnB (fig. S17) diminished or eradicated both iron incorporation and modification of MbnA (Fig. 4C). Native mass spectrometry confirmed that diminished iron loading of MbnBC measured by inductively coupled plasma optical emission spectrometry (ICP-OES) correlated with reduced metal binding by MbnB (fig. S18 and table S1). Activity could also be eliminated in the wild-type enzyme by removal of the iron or complete oxidation with excess H2O2 before reaction with MbnA and O2 (fig. S19). The clear requirements for reduced iron and O2 (and no other reductants or cofactors) suggest that MbnBC is an oxidase that activates O2 for cleavage of the three aliphatic C–H bonds on Cα and Cβ of the first modifiable cysteine of MbnA to promote the cyclization/migration reaction necessary to complete the biosynthesis of the oxazolone-thioamide group.
The observed conversion of Cys21 to an oxazolonethioamide appears to be an O2-dependent four-electron oxidation involving multiple aliphatic carbons; such reactions are mediated by several mononuclear (32, 33) and dinuclear (34, 35) iron enzymes. A hallmark of all these enzymes is the ability to extract all necessary electrons from the substrate, allowing them to mediate multiple turnovers in the absence of a cosubstrate. By contrast, related iron enzymes that promote two-electron oxidations uniformly require reducing cosubstrates (29, 36). Because MbnBC performed multiple turnovers in reactions lacking an obvious reductant (figs. S8, S9, and S20), it may mediate a single four-electron oxidation rather than two sequential two-electron oxidations. As the MbnA or O2 concentration increased, the transition from the single-turnover regime (characterized by a single-exponential kinetic phase in development of the 335-nm feature) to the multiple-turnover regime (characterized by multiphasic kinetics) was observed at an MbnA concentration markedly lower than the total iron present in the anaerobically prepared proteins (figs. S8 and S9); this finding is potentially consistent with an active cofactor that contains more than one iron ion. The presence of a multinuclear iron (Fe2 or Fe3) cluster is consistent with the nTDMS analysis and the observation by Mössbauer spectroscopy of multinuclear iron species in aerobically purified MbnBC. Moreover, examination of the metal centers in xylose isomerase, endonuclease IV, and the Hs. somnus 129Pt DUF692 protein—the closest structurally characterized relatives of MbnB—suggests that a dinuclear or perhaps even trinuclear cluster is distinctly possible (fig. S21). By analogy to other nonheme iron enzymes that mediate four-electron oxidations, MbnA processing could be initiated by abstraction of a hydrogen atom from the Cβ of the first modifiable cysteine by a superoxo(di/tri)iron(III) intermediate (fig. S22).
How MbnBC proceeds to the second modification site remains unclear, as does the mechanism of leader peptide loss (fig. S23). The enzyme complex may require leader peptide cleavage to install the C-terminal oxazolone-thioamide group, although neither modification site in the core peptide can be modified without the leader (Fig. 4A and figs. S12 and S13). The only genes found in all Mbn operons are mbnA, mbnB, and mbnC. Mbn operons encode no proteases, and MbnA leader peptides lack known protease recognition motifs at the cleavage location. It may be that a protease not encoded in the operon is required. Alternatively, the missing protease could be MbnC, for which neither a role nor a cofactor requirement has been established. However, addition of a wide range of potential cofactors as well as lysate from Mbn-producing Ms. trichosporium OB3b cells to MbnABC reactions in vitro failed to enable leader peptide cleavage (figs. S24 and S25). In addition, coexpression of MbnBC from Vibrio caribbenthicus BAA-2122 with C-terminally His6-tagged MbnA from the same species resulted in the same mass shift and UV-visible spectral features observed in vitro (fig. S26). This may indicate that the protease, a cofactor, or a cosubstrate for the protease is absent or incorrectly assembled during heterologous expression. Additional work will be necessary to fully reconstruct the biosynthetic pathway in heterologous expression systems or in vitro.
The involvement of a metalloenzyme in oxazolone and thioamide biosynthesis is unprecedented, and both components of the MbnBC complex belong to previously uncharacterized protein families. Similar to RiPP systems such as the cyanobactins and lantibiotics (37, 38), the promiscuity of the Mbn biosynthetic system suggests that it might be amenable to engineering efforts, which could facilitate the development of natural and non-natural Mbns as drugs for Wilson disease and other disorders involving copper accumulation (39–41). Furthermore, the new understanding of these enzymes sets the stage for elucidating the biosynthesis and function of the Mbn-like natural products from nonmethanotrophic bacteria.
Supplementary Material
Acknowledgments
Supported by NIH grants GM118035 (A.C.R.), R01AT009143 (N.L.K.), U54-GM094662, U54 GM093342, P01 GM118303 (S.C.A.), R00GM111978 (M.-E.P.), and F32GM110934 (L.M.K.D.), as well as NSF grants MCB0842366 (A.C.R.) and MCB1330784 (J.M.B. and C.K.) and the Price Foundation (S.C.A.). G.E.K. was supported by American Heart Association grant 14PRE20460104. L.M.K.D. was additionally funded by a Postdoctoral Enrichment Program grant from the Burroughs Wellcome Fund. L.F.S. was supported by NIH grant T32GM105538 and a Howard Hughes Medical Institute Gilliam Fellowship. O.S.S. was supported by NSF GRFP grant 2014171659 and R.J.M. under NSF grant DGE1255832. At Northwestern University, the Quantitative Bio-element Imaging Center is supported by NASA Ames Research Center grant NNA06CB93G, the Proteomics Center for Excellence is supported by National Resource for Translational and Developmental Proteomics under NIH grant P41 GM108569 and National Cancer Institute (NCI) grant CCSG P30 CA060553, the Integrated Molecular Structure Education and Research Center is supported by Northwestern University and the State of Illinois, the International Institute for Nanotechnology, and the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF grant NNCI-1542205), and the Keck Biophysics Facility is supported in part by NCI Cancer Center Support Grant CA060553. Some work was performed at the Albert Einstein Anaerobic Structural and Functional Genomics Resource. All data are presented in the supplementary materials.
Footnotes
All authors declare no conflicts of interest.
References
- 1.Butler A. Science. 1998;281:207–210. doi: 10.1126/science.281.5374.207. [DOI] [PubMed] [Google Scholar]
- 2.Palmer LD, Skaar EP. Annu. Rev. Genet. 2016;50:67–91. doi: 10.1146/annurev-genet-120215-035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nairz M, Schroll A, Sonnweber T, Weiss G. Cell. Microbiol. 2010;12:1691–1702. doi: 10.1111/j.1462-5822.2010.01529.x. [DOI] [PubMed] [Google Scholar]
- 4.Miethke M, Marahiel MA. Microbiol. Mol. Biol. Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghssein G, et al. Science. 2016;352:1105–1109. doi: 10.1126/science.aaf1018. [DOI] [PubMed] [Google Scholar]
- 6.Chivers PT, Benanti EL, Heil-Chapdelaine V, Iwig JS, Rowe JL. Metallomics. 2012;4:1043–1050. doi: 10.1039/c2mt20139a. [DOI] [PubMed] [Google Scholar]
- 7.Cherrier MV, Cavazza C, Bochot C, Lemaire D, Fontecilla-Camps JC. Biochemistry. 2008;47:9937–9943. doi: 10.1021/bi801051y. [DOI] [PubMed] [Google Scholar]
- 8.Chaturvedi KS, Hung CS, Crowley JR, Stapleton AE, Henderson JP. Nat. Chem. Biol. 2012;8:731–736. doi: 10.1038/nchembio.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston CW, et al. Nat. Chem. Biol. 2013;9:241–243. doi: 10.1038/nchembio.1179. [DOI] [PubMed] [Google Scholar]
- 10.Johnstone TC, Nolan EM. Dalton Trans. 2015;44:6320–6339. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dassama LMK, Kenney GE, Rosenzweig AC. Metallomics. 2017;9:7–20. doi: 10.1039/c6mt00208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HJ, et al. Science. 2004;305:1612–1615. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 13.Krentz BD, et al. Biochemistry. 2010;49:10117–10130. doi: 10.1021/bi1014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenney GE, Sadek M, Rosenzweig AC. Metallomics. 2016;8:931–940. doi: 10.1039/c5mt00289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenney GE, Rosenzweig AC. BMC Biol. 2013;11:17. doi: 10.1186/1741-7007-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semrau JD, et al. Environ. Microbiol. 2013;15:3077–3086. doi: 10.1111/1462-2920.12150. [DOI] [PubMed] [Google Scholar]
- 17.Arnison PG, et al. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rix U, et al. J. Am. Chem. Soc. 2004;126:4496–4497. doi: 10.1021/ja031724o. [DOI] [PubMed] [Google Scholar]
- 19.Behnken S, Hertweck C. Appl. Microbiol. Biotechnol. 2012;96:61–67. doi: 10.1007/s00253-012-4285-8. [DOI] [PubMed] [Google Scholar]
- 20.Izawa M, Kawasaki T, Hayakawa Y. Appl. Environ. Microbiol. 2013;79:7110–7113. doi: 10.1128/AEM.01978-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nayak DD, Mahanta N, Mitchell DA, Metcalf WW. eLife. 2017;6:e29218. doi: 10.7554/eLife.29218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behling LA, et al. J. Am. Chem. Soc. 2008;130:12604–12605. doi: 10.1021/ja804747d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Ghazouani A, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:8400–8404. doi: 10.1073/pnas.1112921109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenney GE, et al. J. Am. Chem. Soc. 2016;138:11124–11127. doi: 10.1021/jacs.6b06821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagano N, Orengo CA, Thornton JM. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 26.Burkhart BJ, Hudson GA, Dunbar KL, Mitchell DA. Nat. Chem. Biol. 2015;11:564–570. doi: 10.1038/nchembio.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolia NH, Joshua-Tor L. Nat. Methods. 2006;3:55–64. doi: 10.1038/nmeth0106-55. [DOI] [PubMed] [Google Scholar]
- 28.Wallar BJ, Lipscomb JD. Chem. Rev. 1996;96:2625–2658. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- 29.Costas M, Mehn MP, Jensen MP, Que L., Jr Chem. Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 30.Brown CA, Remar GJ, Musselman RL, Solomon EI. Inorg. Chem. 1995;34:688–717. [Google Scholar]
- 31.Kim HJ, Galeva N, Larive CK, Alterman M, Graham DW. Biochemistry. 2005;44:5140–5148. doi: 10.1021/bi047367r. [DOI] [PubMed] [Google Scholar]
- 32.Tamanaha E, et al. J. Am. Chem. Soc. 2016;138:8862–8874. doi: 10.1021/jacs.6b04065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peck SC, et al. J. Am. Chem. Soc. 2017;139:2045–2052. doi: 10.1021/jacs.6b12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing G, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6130–6135. doi: 10.1073/pnas.0508473103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wörsdörfer B, et al. Proc. Natl. Acad. Sci. U.S.A. 2013;110:18874–18879. doi: 10.1073/pnas.1315927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trehoux A, Mahy J-P, Avenier F. Coord. Chem. Rev. 2016;322:142–158. [Google Scholar]
- 37.Sardar D, Lin Z, Schmidt EW. Chem. Biol. 2015;22:907–916. doi: 10.1016/j.chembiol.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Heel AJ, Mu D, Montalbán-López M, Hendriks D, Kuipers OP. ACS Synth. Biol. 2013;2:397–404. doi: 10.1021/sb3001084. [DOI] [PubMed] [Google Scholar]
- 39.Ala A, Walker AP, Ashkan K, Dooley JS, Schilsky ML. Lancet. 2007;369:397–408. doi: 10.1016/S0140-6736(07)60196-2. [DOI] [PubMed] [Google Scholar]
- 40.Summer KH, et al. J. Trace Elem. Med. Biol. 2011;25:36–41. doi: 10.1016/j.jtemb.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Lichtmannegger J, et al. J. Clin. Invest. 2016;126:2721–2735. doi: 10.1172/JCI85226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.