Abstract
Studies of human infectious diseases have been limited by the paucity of functional models that mimic normal human physiology and pathophysiology. Recent advances in the development of multicellular, physiologically active organotypic cultures produced from embryonic and pluripotent stem cells, as well as from stem cells isolated from biopsies and surgical specimens are allowing unprecedented new studies and discoveries about host-microbe interactions. Here, we summarize recent developments in the use of organoids for studying human viral pathogens, including intestinal infections with human rotavirus, norovirus, enteroviruses and adenoviruses (intestinal organoids and enteroids), neuronal infections with Zika virus (cerebral organoids) and respiratory infections with respiratory syncytial virus in (lung bud organoids). Biologic discovery of host-specific genetic and epigenetic factors affecting infection, and responses to infection that lead to disease are possible with the use of organoid cultures. Continued development to increase the complexity of these cultures by including components of the normal host tissue microenvironment such as immune cells, blood vessels and microbiome, will facilitate studies on human viral pathogenesis, and advance the development of platforms for pre-clinical evaluation of vaccines, antivirals and therapeutics.
Keywords: Organoids, Enteroids, Viral tropism, Pathogenesis and Vaccines
Introduction
Studies of infectious agents have traditionally focused on in vitro culture systems using transformed cell lines and animal models. While these systems have enabled historic progress in the understanding of microbial pathogenesis and host-pathogen interactions, and facilitated the development of vaccines and therapeutics, the physiological relevance of these models for human pathogens and diseases can be limited. Routinely used cell lines are immortalized and cancer-derived and may not adequately reflect responses of normal human cells [1–3]. Many signaling pathways are altered in cancer cells and have profound effects on the core metabolism in cancer-derived lines [4]. Furthermore, cell lines are typically comprised of a single cell type and do not mimic the architecture, environmental complexity and functionality of tissues which are comprised of many different cell types. While some of these drawbacks are overcome in animal models, these have other restrictions for studying human infectious diseases. Many human pathogens display unique human specificity that precludes their study in animals while in other cases, animal models fail to reproduce human pathophysiology. Further, findings and effects seen in animal models do not always translate to humans, as has been observed in many different fields [5–7].
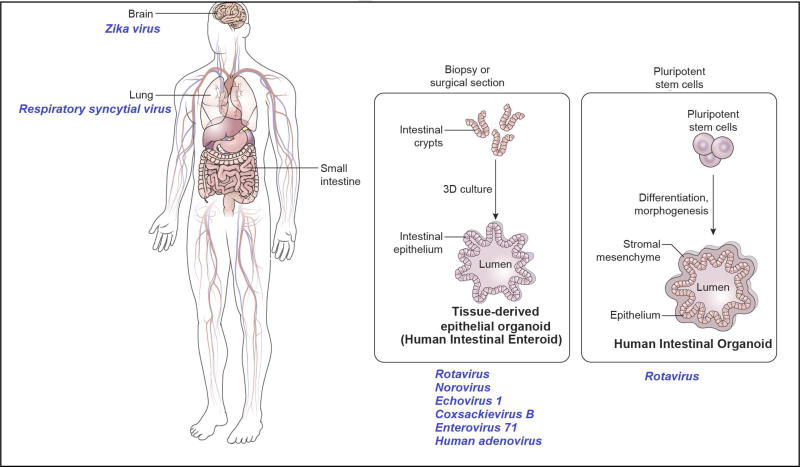
The development of in vitro organoid cultures offer remarkable new model systems to study infectious agents and disease pathogenesis [8]. While organotypic cultures comprised of three dimensional (3D) cell aggregates have been in existence for decades, the term organoids now largely refers to self-organizing, propagatable 3D cultures derived from stem cells that recapitulate the organization, functionality, and genetic signature of the specific tissue or organ and host from which they are derived. Organoids can be derived from embryonic and pluripotent stem cells (ESCs and PSCs, respectively) as well as stem cells isolated from specific human tissues. Through new understanding of defined developmental cues and growth factors, stem cells can be directed to grow ex vivo into organ-like structures. Unlike transformed cells lines and animal models, organoid cultures are multicellular, reflect the cellular heterogeneity of specific organs and are physiologically active; thus, they are increasingly being validated as relevant models for studies of infectious disease, particularly of human-specific pathogens. Organoids have been established for multiple organs including the intestine, stomach, esophagus, liver, kidneys, lungs, brain, prostate, pancreas, retina and ovary [8]. Although use of this new technology is in its infancy, paradigm shifting results and unexpected discoveries have been made with several human viruses (Figure 1) and select data are summarized below. Other recent articles address aspects of organoids for diseases studies that are not covered in this review [8–13].
Figure 1.
Human organoids currently used for studies of viral pathogens.
Human Intestinal Organoids and Enteroids
The human gastrointestinal tract is a complex organ with a polarized epithelial layer that contains different cell types including enterocytes, enteroendocrine cells, tuft cells, goblet cells, Paneth cells and stem cells. Distinct regions of the intestine (duodenum, jejunum and ileum, proximal and distal colon) perform unique functions and demonstrate segment-specificity in terms of transport, protein expression and interactions with pathogens [14,15]. The breakthrough in intestinal organoid cultures began with the successful culture of murine epithelial organoids from Lgr5+ intestinal stem cells in 2009, followed by the development of organoids from human PSCs and biopsy samples [16–18]. Two types of intestinal organoids have been used for virus studies; organoids derived from PSCs, that are epithelial cultures associated with mesenchyme, and human tissue-derived organoids that are epithelial only cultures derived from stem cells isolated from biopsies or surgical tissues (also called mini-guts). To distinguish epithelial/mesenchymal cultures from epithelial only cultures, the nomenclature organoids and enteroids was proposed by the intestinal stem cell consortium in 2012 [19]. The terminology human intestinal organoids (HIOs) and human intestinal enteroids (HIEs) are used henceforth to describe these cultures. Apart from 3D cultures, HIEs can also be grown as monolayers that allow for easy access to apical and basolateral compartments as well as measurement of epithelial barrier function [20]. HIOs and HIEs have been documented to functionally recapitulate normal human gastrointestinal pathophysiology [21], and both 3D and monolayer cultures are being used to study enteric viruses including human rotavirus, norovirus, enteroviruses, and adenoviruses (Figure 1).
Rotaviruses are a leading cause of severe, dehydrating gastroenteritis in young children worldwide, resulting in about 215,000 death annually [22]. Live, attenuated rotavirus vaccines were introduced in 2006 and have significantly reduced the burden of rotavirus disease. However, these vaccines are less effective in developing countries with the greatest disease burden [23]. Both HIOs and HIEs have been used for studying rotavirus infections. Proof-of-principle studies showing utility of organoid cultures for human rotavirus infection was first shown with HIOs [24]. The replication of 12/13 clinical rotavirus isolates directly from stool samples also was seen in these cultures, with the majority of strains showing ~10 times greater viral replication when compared to a conventional monkey kidney epithelial cell line. Growth of clinical rotavirus isolates from stool is typically challenging and these data support the use of HIOs as a new tool for human rotavirus studies. An unanticipated finding on cell tropism from this study was the detection of virus replication not only in enterocytes but also in mesenchymal cells in the HIOs.
HIEs were subsequently demonstrated as a new model to study human rotavirus host restriction, pathophysiology, and innate epithelial responses to infection [25–27]. Contrary to commonly used cell lines, HIEs recapitulate in vivo observations of rotavirus host range restriction, with human strains showing greater replication efficiency than animal rotaviruses [25]. In vivo, rotavirus preferentially infects differentiated enterocytes at the tips of intestinal villi. While infections in enteroendocrine cells have been reported in mice and cancer cell lines, it was unknown whether human rotavirus strains infect enteroendocrine cells in vitro or in vivo. Studies in HIEs demonstrated that rotavirus infection not only occurs in differentiated enterocytes but also in enteroendocrine cells, confirming the tropism for this cell type. Differentiation of HIEs is key for virus replication and promotes the production of viroplasms and lipid droplets that are classic features of rotavirus replication. Importantly, HIEs are a unique physiologically responsive in vitro model of rotavirus infection; luminal expansion and fluid secretion are seen following treatment with rotavirus or with the viral enterotoxin NSP4, mimicking the pathophysiological observations of diarrhea. HIEs also provide new insight into innate immune responses of non-transformed human epithelial cells to rotavirus. Colon carcinoma cells respond to rotavirus infection with a strong type I interferon (IFN) response at both transcriptional and translational levels [28]. Unexpectedly, a paradox of transcriptional and functional epithelial IFN responses to human rotavirus infections is observed in HIEs [26]. While the dominant transcriptional pathway is a type III IFN response, there is minimal detection of protein and the endogenous response does not restrict virus replication, suggesting posttranscriptional antagonism of IFN response by the virus. By contrast, exogenous addition of IFNs restricts virus replication, with type I IFN having a much more potent effect than type III IFN. These data suggest that extra-epithelial sources of type I IFN may be critical for limiting enteric virus replication in vivo.
HIEs are also promising new tools for studies of rotavirus vaccines and antivirals. The infectivity of a rotavirus vaccine (RV1, also called Rotarix®) is attenuated in HIEs compared to a laboratory-adapted rotavirus strain belonging to the same genotype [25]. While the attenuation of rotavirus vaccines in vivo is well established, this is the first demonstration of attenuated infectivity with vaccine viruses in vitro and supports the use of HIEs as a biological model to address fundamental questions on rotavirus vaccines. In particular, HIEs are beginning to be used to understand how genetic polymorphisms in expression of intestinal glycans affects susceptibility to rotavirus strains and vaccines since these cultures retain the genetic makeup of individual donors. This is significant because several recent studies demonstrate a role of histoblood group antigens (HBGAs) in susceptibility to specific human, but not animal, rotavirus strains. HBGA biosynthesis is determined by genetically-encoded glycosyltransferases such as fucosyltransferase-2 (FUT2, secretor gene). Individuals with a functional FUT2 are called secretors while those lacking a functional FUT2 are referred to as non-secretors. Several recent epidemiology studies indicate that non-secretors are less susceptible to diarrhea with specific human rotaviruses, raising new questions on the role of secretor status in susceptibility to infections and to live, attenuated rotavirus vaccines [29]. There are no significant differences in virus replication between HIEs from secretors and non-secretors, suggesting that differences in HBGA expression may be more critical for disease presentation than for infection susceptibility. However, heterogeneity in replication is seen in HIE lines from different donors, mirroring differences in susceptibility between individuals. RV1 vaccine replication is seen in both secretor and non-secretor HIE lines suggesting that secretor status may not be a critical factor in vaccine take. Indeed, there were no differences in the incidence of severe rotavirus diarrhea between secretor and non-secretor RV1 vaccine recipients in a clinical trial in Bangladesh, although natural resistance to disease among unvaccinated non-secretors impacted efficacy estimates. In this setting, vaccine efficacy is low, highlighting other still to be determined factors affecting vaccine response in developing countries [30]. Mimicking heterogeneity in people, one of six HIE lines showed poor infectivity with RV1, opening up the possibility to use HIEs to identify additional host factors that influence vaccine susceptibility.
Finally, HIEs can be used to evaluate virus neutralization and antivirals with responses to IFN alpha, ribavirin and mycophenolic acid showing significant reductions in rotavirus infectivity in HIEs [31,32]. Neutralization was more efficient in HIEs compared to responses in Caco-2 (immortalized human colonic adenocarcinoma) cells using one monoclonal antibody suggesting HIEs might uncover new mechanisms of virus neutralization and differential responses to IFN alpha and ribavirin were detected when both detection of a response and sensitivity of response were evaluated using patient-derived human rotavirus strains (Yin et al., 2015). These results suggest HIEs may be useful for personalized evaluation of the efficacy of antiviral therapy.
The successful cultivation of human noroviruses in HIE monolayers highlights the potential for these cultures to be used for previously non-cultivable human pathogens [33]. Human noroviruses are the leading cause of non-bacterial food borne gastroenteritis worldwide and have replaced rotavirus as a leading cause of diarrheal illness in children in many countries [34–36]. For over 4 decades since their discovery, attempts to cultivate human noroviruses in transformed cell lines and in small animal models remained unsuccessful. This major barrier limited the understanding of norovirus pathogenesis and development of effective interventions. In 2016, multiple human norovirus strains were successfully cultivated in HIEs. Significant replication is seen in differentiated HIEs from the duodenum, jejunum and ileum. Importantly, the susceptibility of HIEs from different individuals to support replication of norovirus genotypes confirmed known epidemiological differences based on HBGA expression; these data emphasize the biological relevance and potential of using HIEs to cultivate human pathogens and understand genetically-determined host-virus interactions. In another example of unexpected discovery of host factors modulating virus infections, strain-specific requirements for replication with bile, a critical component of the intestinal milieu, was identified as being essential for the cultivation of some norovirus strains and simply enhancing other strains. The discovery of HIEs as an intestinal cultivation model for human noroviruses and demonstration of virus neutralization using immune sera as well as virus inactivation provides proof-of-principle for the use of HIEs as pre-clinical tools for testing antivirals, evaluating vaccine correlates of protections, as well as a new tool to model disease pathogenesis.
New insights into enteroviral infections in the gastrointestinal tract and induction of immune responses have been obtained using enteroid models. Enteroviruses are a significant cause of human infections worldwide and are primarily transmitted by the fecal-oral route. Previous studies of enteroviruses in murine models were limited by the requirement for intraperitoneal injections and ablation of host immune cells. Infections in colon carcinoma cell lines are not associated with the induction of strong antiviral responses, suggesting attenuation of host innate immunity [37]. Infection of fetal HIEs with diverse enteroviruses including coxsackievirus B, echovirus 11 and enterovirus showed varying degrees of susceptibility between the different viruses, and the induction of antiviral and inflammatory signaling pathways in a virus-specific manner [38]. While echovirus 11 infection induced the differential expression of 350 transcripts, coxsackievirus B induced changes in only 13 transcripts. Similar to rotavirus and norovirus, a new discovery on cell type specificity of infection was observed in fetal HIEs for echovirus 11 with infections noted in enterocytes and enteroendocrine cells but goblet cells. A point of note is that these findings were obtained using enteroids derived from premature fetal small intestine and it remains to be determined whether other outcomes will be obtained if HIEs from older children and adults are tested [13].
New discoveries in human adenovirus biology have also recently been uncovered using HIEs [39]. These DNA viruses cause a number of diseases including gastroenteritis, respiratory infections and conjunctivitis [40]. While many human adenovirus species replicate in the gastrointestinal tract, and are shed in the feces, only a subset of human adenovirus species F (serotypes Ad 40 and 41) are tropic for the intestinal tract and are known to cause gastroenteritis in humans. Prototype strains and clinical isolates of both enteric and non-enteric adenoviruses have now been shown to replicate in ileal HIEs. Unlike for rotavirus and norovirus, differentiation of HIEs is not required for successful replication of adenoviruses. Comparison of viral replication in monolayer cultures of HIEs and in the classically used immortalized A549 lung cells show that sensitivity to type I and type III IFNs for both respiratory and enteric human adenoviruses is seen only in HIEs, and not in A549 cells, despite comparable upregulation of interferon inducible genes in both culture types. These results indicate that HIEs adopt an IFN-induced antiviral state capable of limiting human adenovirus infection more similar to primary airway cells than immortalized cells [41]. Finally, striking serotype-specific differences in sensitivity to antiviral peptides and a new cell tropism for adenovirus were discovered in HIEs. The respiratory human adenovirus 5 was found to preferentially infect goblet cells and was sensitive to the antiviral effects of human defensin 5 while the enteric adenovirus was resistant. Addressing pending questions on enteric adenoviruses including cell type tropism of clinical isolates and development of a cell culture system for isolation of patient samples are now feasible with the use of HIEs.
Cerebral Organoids
Given the complex organization of the developing cortex with different progenitor and neuronal layers, the development of 3D brain organoids from human PSCs that mimic human brain development is a significant breakthrough in neuroscience [42]. Brain organoids have been used extensively in recent times for studies with Zika virus (ZIKV), a mosquito-borne Flavivirus. In 2016, the WHO declared a global health emergency due to the link between the ZIKV outbreak and the increase of incidence of microcephaly observed in Brazil [43]. The pathogenesis of ZIKV and how the virus affects brain neurons have now been assessed in a number of studies using organoids derived from ESCs and PSCs. A causal role for ZIKV in fetal brain malformations was shown when the growth and viability of brain organoids was found to be reduced following ZIKV infection [44–47]. Reduction in the overall size of ESC-derived cerebral organoids correlated with ZIKV copy number. The expression of Toll-like-Receptor 3 (TLR3) and specific neuronal genes were upregulated following ZIKV infection and inhibition of TLR3 was demonstrated to reduce the effects of ZIKV infection [46]. This was a striking finding because TLR3 has previously been implicated in many neuroinflammatory and neurodegenerative disorders [48]. Forebrain-specific organoids were generated using miniaturized spinning bioreactors and exposure to ZIKV at different developmental stages showed tropism towards neural progenitor cells including radial glial cells over intermediate neural progenitors or immature neurons [47]. Providing insight into viral factors affecting clinical presentation, the NS2A protein of ZIKV reduces radial glial cell proliferation and causes defects in adherens junctions in human forebrain organoids [49].
Intrinsic differences in the pathogenicity and virulence of ZIKV strains from different lineages are thought to contribute in part to differences in clinical presentations [50]. Cerebral organoids have now been used to analyze differences in the pathogenesis of different ZIKV strains and host restriction [45]. A ZIKV strain from Brazil was found to have a more profound effect on neuronal layer thickness in human cerebral organoids compared to an African strain and did not affect cerebral organoids derived from non-human primates. The pattern and outcome of infection in PSC-derived brain organoids were also shown to vary between more recent American and Asian clinical ZIKV isolates when compared to an extensively passaged ZIKV strain [51]. Another area where organoids have been used in ZIKV studies is in the examination of factors with long-term implications. The effects of ZIKV infection on the epigenome have been tested using cerebral organoids and changes in DNA methylation were observed suggesting potential long-term implications of ZIKV infection [52]. Interestingly, preferential infection of stem cells by ZIKV was also indicated in prostate organoids where replication was mostly observed in stromal cells [53]. Finally, brain organoids have been used for the evaluation of therapeutics and interventions against ZIKV [54]. A drug repurposing screen of about 6,000 compounds including approved drugs, drug candidates in clinical trials and other pharmacologically active compounds was first performed in neuronal cells for cytotoxicity and inhibition of ZIKV-induced increases in caspase-3 activity. A pan-caspase inhibitor, Emricasan was found to protect human cortical neural progenitors in both monolayer and 3D forebrain organoid cultures. The number of studies using organoids for ZIKV in a short interval of time validate their use as a promising tool for evaluation of viral pathogenesis, clinical presentation and drug discovery.
Lung Organoids
Lung bud organoids containing the mesoderm and pulmonary endoderm were generated from human PSCs and develop into branching airway and early alveolar structures after in vivo transplantation (xenotransplantation) under the mouse kidney capsule as well as in Matrigel 3D cultures [55]. Resembling lung buds at the second trimester of gestation, these organoids can be infected with respiratory syncytial virus (RSV), the most common cause of bronchiolitis and pneumonia in children younger than 1 year of age in the United States. Shedding of swollen, infected cells into the lumen of organoids was seen at day 2 post infection. These findings were consistent with previous studies in human airway epithelial cell lines where RSV-infected cells were demonstrated to swell and detach from the epithelium and was consistent with observations in pathology specimens. There is currently no model that recapitulates the morphological characteristics of RSV disease and lung bud organoids may provide a new developmental model for infectious lung diseases.
Challenges and Future Directions
While organoids have several advantages over transformed cell lines and animal models, key challenges remain to be addressed in order to improve their utility for studying viruses (Box 1). One limitation is that current organoid models are devoid of components of the normal host microenvironment such as immune cells and blood vessels. Co-culture models incorporating other cell types including endothelial cells and immune cells is an area of ongoing development [56–58]. In the case of intestinal organoid cultures, the production of scaffolds and platforms that will allow proper organization of the villus-crypt axis is another ongoing area of research [59,60]. Recapitulating the organ microenvironment and complexity will significantly advance the biological relevance of organoids for pathogen studies. Practical considerations including variability in reagents used for culturing organoids, complexity of techniques and costs are factors that limit the wide use of organoids. While HIEs and HIOs are relatively easy to maintain, the growth factors required to grow these cultures are expensive and lot-to-lot variability in components such as Matrigel are considerations for reproducibility of findings [9]. Similarly, culturing cerebral organoids requires specialized expertise and resources [61]. Current organoid culture methods are relatively low-throughput culture systems that limits their use in high-throughput applications such as drug screens. Maturation status of organoids is another important factor for consideration in viral studies. After 6 months of culture in Matrigel, lung organoids were found to display structures and expression signatures that correlate with second trimester of human gestation. RNAseq analysis of HIOs revealed a distinctly fetal transcriptional signature although in vivo transplantation of these cultures results in the expression of more adult tissue-like markers [62]. It remains to be seen whether maturation differences contribute significantly to differences in susceptibility and response to viruses and for the potential applications for organoids as pre-clinical models for evaluation of interventions.
Box 1. Strategies for enhancing the relevance of organoids for viral studies.
Inclusion of immune cells required to evaluate host response to infections
Development of co-culture models to incorporate the microbiome
Addition of vasculature to current organoid models
In-depth characterization of “age” of the organoid (fetal, adult-like) and evaluation of relevance for disease studies
Development of tools to improve accessibility to organoid cultures (such as centralized, high throughput organoid banks) for pre-clinical studies
Conclusions
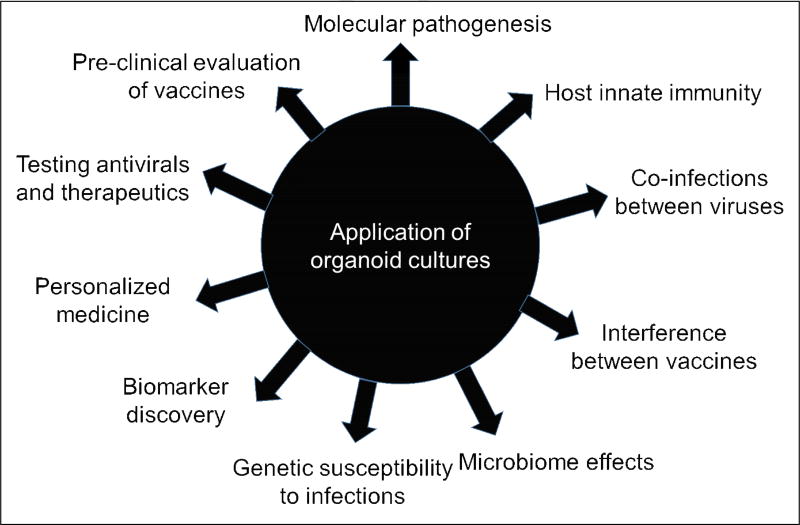
The development of organoids representing different sites of infection is an exciting advance that can facilitate studies and new discoveries about host-virus interactions including molecular pathogenesis of viral infections, novel cell tropisms, new insights into host innate immune responses and characterizing interactions between pathogens (Figure 2). Importantly, human organoids/enteroids provide a functional and physiologically relevant model for pre-clinical evaluation of vaccines, antivirals and other therapeutics to mitigate disease symptoms. Use of organoids derived from human tissues also provides a new pathway for evaluating personalized therapies. Organoid cultures are transformative tools for studies of viruses, including previously non-cultivatable viruses, and exhibit enormous potential to model disease and develop interventions. While this article focuses on applications for human viruses, we predict future breakthroughs in understanding animal viruses using similar organoid technology. Significant efforts are being made not only for the organoids described here but also for other systems in order to improve the breadth of pathogens that can be studied [10] and contribute to long-term goals of developing broadly applicable, physiologically relevant platforms for infectious disease studies.
Figure 2.
Application of organoid cultures for virus studies
Acknowledgments
The authors would like to acknowledge support from grants U18-TR000552, UH3-TR00003, U19-AI116497, R01-AI080656, R01 AI-105101 P30-DK56338 from the National Institutes of Health and the Food Research Initiative Competitive grant 2011-68003-30395 from the US Department of Agriculture, National Institute of Food and Agriculture.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* Of special interest
** Of outstanding interest
- 1.Alge CS, Hauck SM, Priglinger SG, Kampik A, Ueffing M. Differential protein profiling of primary versus immortalized human RPE cells identifies expression patterns associated with cytoskeletal remodeling and cell survival. J Proteome Res. 2006;5:862–878. doi: 10.1021/pr050420t. [DOI] [PubMed] [Google Scholar]
- 2.Pan C, Kumar C, Bohl S, Klingmueller U, Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol Cell Proteomics. 2009;8:443–450. doi: 10.1074/mcp.M800258-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun D, Lennernas H, Welage LS, Barnett JL, Landowski CP, Foster D, Fleisher D, Lee KD, Amidon GL. Comparison of human duodenum and Caco-2 gene expression profiles for 12,000 gene sequences tags and correlation with permeability of 26 drugs. Pharm Res. 2002;19:1400–1416. doi: 10.1023/a:1020483911355. [DOI] [PubMed] [Google Scholar]
- 4.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 5.Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6:114–118. [PMC free article] [PubMed] [Google Scholar]
- 6.Martic-Kehl MI, Schibli R, Schubiger PA. Can animal data predict human outcome? Problems and pitfalls of translational animal research. Eur J Nucl Med Mol Imaging. 2012;39:1492–1496. doi: 10.1007/s00259-012-2175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren HS, Tompkins RG, Moldawer LL, Seok J, Xu W, Mindrinos MN, Maier RV, Xiao W, Davis RW. Mice are not men. Proc Natl Acad Sci U S A. 2015;112:E345. doi: 10.1073/pnas.1414857111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutta D, Heo I, Clevers H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Blutt SE, Crawford SE, Ramani S, Zou WY, Estes MK. Engineered Human Gastrointestinal Cultures to Study the Microbiome and Infectious Diseases. Cell Mol Gastroenterol Hepatol. 2018;5:241–251. doi: 10.1016/j.jcmgh.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutta D, Clevers H. Organoid culture systems to study host-pathogen interactions. Curr Opin Immunol. 2017;48:15–22. doi: 10.1016/j.coi.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 12.Hill DR, Spence JR. Gastrointestinal Organoids: Understanding the Molecular Basis of the Host-Microbe Interface. Cell Mol Gastroenterol Hepatol. 2017;3:138–149. doi: 10.1016/j.jcmgh.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanik WE, Mara MA, Mihi B, Coyne CB, Good M. Stem Cell-Derived Models of Viral Infections in the Gastrointestinal Tract. Viruses. 2018;10 doi: 10.3390/v10030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foulke-Abel J, In J, Yin J, Zachos NC, Kovbasnjuk O, Estes MK, de Jonge H, Donowitz M. Human Enteroids as a Model of Upper Small Intestinal Ion Transport Physiology and Pathophysiology. Gastroenterology. 2016;150:638–649. e638. doi: 10.1053/j.gastro.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajan A, Vela L, Zeng XL, Yu X, Shroyer N, Blutt SE, Poole NM, Carlin LG, Nataro JP, Estes MK, et al. Novel Segment- and Host-Specific Patterns of Enteroaggregative Escherichia coli Adherence to Human Intestinal Enteroids. MBio. 2018;9 doi: 10.1128/mBio.02419-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. Use of intestinal stem cells to generate epithelial only human intestinal enteroid cultures. [DOI] [PubMed] [Google Scholar]
- 17*.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. Use of pluripotent stem cells to generate human intestinal organoids with epithelial cells and mesenchyme. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 19.Stelzner M, Helmrath M, Dunn JC, Henning SJ, Houchen CW, Kuo C, Lynch J, Li L, Magness ST, Martin MG, et al. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1359–1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, Ciorba MA, Stappenbeck TS. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2015;64:911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zachos NC, Kovbasnjuk O, Foulke-Abel J, In J, Blutt SE, de Jonge HR, Estes MK, Donowitz M. Human Enteroids/Colonoids and Intestinal Organoids Functionally Recapitulate Normal Intestinal Physiology and Pathophysiology. J Biol Chem. 2016;291:3759–3766. doi: 10.1074/jbc.R114.635995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD World Health Organization-Coordinated Global Rotavirus Surveillance N. Global, regional, and national estimates of rotavirus mortality in children <5 Years of age, 2000–2013. Clin Infect Dis. 2016;62(Suppl 2):S96–S105. doi: 10.1093/cid/civ1013. [DOI] [PubMed] [Google Scholar]
- 23.Jonesteller CL, Burnett E, Yen C, Tate JE, Parashar UD. Effectiveness of Rotavirus Vaccination: A Systematic Review of the First Decade of Global Postlicensure Data, 2006–2016. Clin Infect Dis. 2017;65:840–850. doi: 10.1093/cid/cix369. [DOI] [PubMed] [Google Scholar]
- 24*.Finkbeiner SR, Zeng XL, Utama B, Atmar RL, Shroyer NF, Estes MK. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio. 2012;3:e00159–00112. doi: 10.1128/mBio.00159-12. First description of intestinal organoids for infection with human rotaviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Saxena K, Blutt SE, Ettayebi K, Zeng XL, Broughman JR, Crawford SE, Karandikar UC, Sastri NP, Conner ME, Opekun AR, et al. Human Intestinal Enteroids: a New Model To Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J Virol. 2016;90:43–56. doi: 10.1128/JVI.01930-15. Established HIEs as non-transformed cell culture models to understand human intestinal physiology and pathophysiology and the epithelial response to enteric viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Saxena K, Simon LM, Zeng XL, Blutt SE, Crawford SE, Sastri NP, Karandikar UC, Ajami NJ, Zachos NC, Kovbasnjuk O, et al. A paradox of transcriptional and functional innate interferon responses of human intestinal enteroids to enteric virus infection. Proc Natl Acad Sci U S A. 2017;114:E570–E579. doi: 10.1073/pnas.1615422114. Type III IFN pathway identified as the dominant transcriptional response of HIEs following rotavirus infection but with a limited role in restricting virus replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou WY, Blutt SE, Crawford SE, Ettayebi K, Zeng XL, Saxena K, Ramani S, Karandikar UC, Zachos NC, Estes MK. Human Intestinal Enteroids: New Models to Study Gastrointestinal Virus Infections. Methods Mol Biol. 2017 doi: 10.1007/7651_2017_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frias AH, Jones RM, Fifadara NH, Vijay-Kumar M, Gewirtz AT. Rotavirus-induced IFN-beta promotes anti-viral signaling and apoptosis that modulate viral replication in intestinal epithelial cells. Innate Immun. 2012;18:294–306. doi: 10.1177/1753425911401930. [DOI] [PubMed] [Google Scholar]
- 29.Kambhampati A, Payne DC, Costantini V, Lopman BA. Host genetic susceptibility to enteric viruses: a systematic review and metaanalysis. Clin Infect Dis. 2016;62:11–18. doi: 10.1093/cid/civ873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee B, Dickson DM, deCamp AC, Colgate ER, Diehl SA, Uddin MI, Sharmin S, Islam S, Bhuiyan TR, Alam M, et al. Histoblood Group Antigen Phenotype Determines Susceptibility to Genotype-specific Rotavirus Infections and Impacts Measures of Rotavirus Vaccine Efficacy. J Infect Dis. 2018 doi: 10.1093/infdis/jiy054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Y, Wang Y, Dang W, Xu L, Su J, Zhou X, Wang W, Felczak K, van der Laan LJ, Pankiewicz KW, et al. Mycophenolic acid potently inhibits rotavirus infection with a high barrier to resistance development. Antiviral Res. 2016;133:41–49. doi: 10.1016/j.antiviral.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 32*.Yin Y, Bijvelds M, Dang W, Xu L, van der Eijk AA, Knipping K, Tuysuz N, Dekkers JF, Wang Y, de Jonge J, et al. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antiviral Res. 2015;123:120–131. doi: 10.1016/j.antiviral.2015.09.010. Use of intestinal organoids to evaluate antivirals to rotavirus. [DOI] [PubMed] [Google Scholar]
- 33**.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. First in vitro intestinal culture system for the previously noncultivatable human noroviruses showing robust replication of multiple virus isolates and strain-specific requirements for bile. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14:725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koo HL, Neill FH, Estes MK, Munoz FM, Cameron A, Dupont HL, Atmar RL. Noroviruses: The Most Common Pediatric Viral Enteric Pathogen at a Large University Hospital After Introduction of Rotavirus Vaccination. J Pediatric Infect Dis Soc. 2013;2:57–60. doi: 10.1093/jpids/pis070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Payne DC, Vinje J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, Hall CB, Chappell J, Bernstein DI, Curns AT, et al. Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med. 2013;368:1121–1130. doi: 10.1056/NEJMsa1206589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris KG, Coyne CB. Enter at your own risk: how enteroviruses navigate the dangerous world of pattern recognition receptor signaling. Cytokine. 2013;63:230–236. doi: 10.1016/j.cyto.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Drummond CG, Bolock AM, Ma C, Luke CJ, Good M, Coyne CB. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc Natl Acad Sci U S A. 2017;114:1672–1677. doi: 10.1073/pnas.1617363114. Proc Natl Acad Sci U S A 2017, 114:1672–1677. Using fetal intestinal organoids for the culture of Echovirus 11, Coxsackie virus B and Enterovirus 71 showed differences in susceptibility, antiviral response and tropism for intestinal cell types between the enteroviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Holly MK, Smith JG. Adenovirus infection of human enteroids reveals interferon sensitivity and preferential infection of goblet cells. J Virol. 2018 doi: 10.1128/JVI.00250-18. HIEs were identified as a new platform to support robust replication of respiratory and enteric adenoviruses; respiratory serotype showed a surprising preference for goblet cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wold WSM, Ison MG. Adenoviruses. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 6. Wolters Kluwer Health/Lippincott Williams & Wilkins; 2013. [Google Scholar]
- 41.Zheng Y, Stamminger T, Hearing P. E2F/Rb Family Proteins Mediate Interferon Induced Repression of Adenovirus Immediate Early Transcription to Promote Persistent Viral Infection. PLoS Pathog. 2016;12:e1005415. doi: 10.1371/journal.ppat.1005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. Development of first brain organoids from pluripotent stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heymann DL, Hodgson A, Sall AA, Freedman DO, Staples JE, Althabe F, Baruah K, Mahmud G, Kandun N, Vasconcelos PF, et al. Zika virus and microcephaly: why is this situation a PHEIC? Lancet. 2016;387:719–721. doi: 10.1016/S0140-6736(16)00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–818. doi: 10.1126/science.aaf6116. ZIKV was shown to abrogate neurogenesis during human brain development. [DOI] [PubMed] [Google Scholar]
- 45**.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–271. doi: 10.1038/nature18296. ZIKV infection of human brain organoids resulted in a reduction of proliferative zones and disrupted cortical layers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46**.Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, Rana TM. Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell. 2016;19:258–265. doi: 10.1016/j.stem.2016.04.014. Demonstrated a link between TLR3 upregulation and reduction in cerebral organoid volume. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. Miniaturized spinning bioreactors used to develop forebrain specific enteroids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okun E, Griffioen KJ, Mattson MP. Toll-like receptor signaling in neural plasticity and disease. Trends Neurosci. 2011;34:269–281. doi: 10.1016/j.tins.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoon KJ, Song G, Qian X, Pan J, Xu D, Rho HS, Kim NS, Habela C, Zheng L, Jacob F, et al. Zika-Virus-Encoded NS2A Disrupts Mammalian Cortical Neurogenesis by Degrading Adherens Junction Proteins. Cell Stem Cell. 2017;21:349–358. e346. doi: 10.1016/j.stem.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simonin Y, van Riel D, Van de Perre P, Rockx B, Salinas S. Differential virulence between Asian and African lineages of Zika virus. PLoS Negl Trop Dis. 2017;11:e0005821. doi: 10.1371/journal.pntd.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, et al. Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell. 2017;20:397–406. e395. doi: 10.1016/j.stem.2016.12.005. Emphasizes the importance of choosing appropriate strains of ZIKV for cellular studies based on differences between clinical isolates and long-term passage viruses. [DOI] [PubMed] [Google Scholar]
- 52*.Janssens S, Schotsaert M, Karnik R, Balasubramaniam V, Dejosez M, Meissner A, Garcia-Sastre A, Zwaka TP. Zika Virus Alters DNA Methylation of Neural Genes in an Organoid Model of the Developing Human Brain. mSystems. 2018;3 doi: 10.1128/mSystems.00219-17. Potential for long term implications of ZIKV infection through epigenetic changes in brain organoids following infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53*.Spencer JL, Lahon A, Tran LL, Arya RP, Kneubehl AR, Vogt MB, Xavier D, Rowley DR, Kimata JT, Rico-Hesse RR. Replication of Zika Virus in Human Prostate Cells: A Potential Source of Sexually Transmitted Virus. J Infect Dis. 2018;217:538–547. doi: 10.1093/infdis/jix436. Replication of ZIKV in prostate organoids demonstrates a potential source for sexual transmission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. Brain organoids used to screen drugs and inhibitors of ZIKV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55**.Chen YW, Huang SX, de Carvalho A, Ho SH, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova JL, Bhattacharya J, et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol. 2017;19:542–549. doi: 10.1038/ncb3510. Lung bud organoids infected with RSV mimic swelling and detachement of infected cells similar to human lungs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noel G, Baetz NW, Staab JF, Donowitz M, Kovbasnjuk O, Pasetti MF, Zachos NC. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci Rep. 2017;7:45270. doi: 10.1038/srep45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noel G, Doucet M, Nataro JP, Kaper JB, Zachos NC, Pasetti MF. Enterotoxigenic Escherichia coli is phagocytosed by macrophages underlying villus-like intestinal epithelial cells: modeling ex vivo innate immune defenses of the human gut. Gut Microbes. 2017;0 doi: 10.1080/19490976.2017.1398871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci Rep. 2018;8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Gunasekara DB, Reed MI, DiSalvo M, Bultman SJ, Sims CE, Magness ST, Allbritton NL. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials. 2017;128:44–55. doi: 10.1016/j.biomaterials.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y, Zhou W, Roh T, Estes MK, Kaplan DL. In vitro enteroid-derived three-dimensional tissue model of human small intestinal epithelium with innate immune responses. PLoS One. 2017;12:e0187880. doi: 10.1371/journal.pone.0187880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qian X, Nguyen HN, Jacob F, Song H, Ming GL. Using brain organoids to understand Zika virus-induced microcephaly. Development. 2017;144:952–957. doi: 10.1242/dev.140707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finkbeiner SR, Hill DR, Altheim CH, Dedhia PH, Taylor MJ, Tsai YH, Chin AM, Mahe MM, Watson CL, Freeman JJ, et al. Transcriptome-wide Analysis Reveals Hallmarks of Human Intestine Development and Maturation In Vitro and In Vivo. Stem Cell Reports. 2015 doi: 10.1016/j.stemcr.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]