Abstract
Systems that automate extraction of nucleic acid from cells or viruses in complex clinical matrices have tremendous value even in the absence of an integrated downstream detector. We describe our bench-top automated workstation that integrates our previously-reported extraction method – TruTip – with our newly-developed mechanical lysis method. This is the first report of this method for homogenizing viscous and heterogeneous samples and lysing difficult-to-disrupt cells using “MagVor”: a rotating magnet that rotates a miniature stir disk amidst glass beads confined inside of a disposable tube. Using this system, we demonstrate automated nucleic acid extraction from methicillin-resistant Staphylococcus aureus (MRSA) in nasopharyngeal aspirate (NPA), influenza A in nasopharyngeal swabs (NPS), human genomic DNA from whole blood, and Mycobacterium tuberculosis in NPA. The automated workstation yields nucleic acid with comparable extraction efficiency to manual protocols, which include commercially-available Qiagen spin column kits, across each of these sample types. This work expands the scope of applications beyond previous reports of TruTip to include difficult-to-disrupt cell types and automates the process, including a method for removal of organics, inside a compact bench-top workstation.
Keywords: Nucleic acid isolation, extraction, sample preparation
1. INTRODUCTION
The increased demand for timely diagnostic information to guide patient treatment has led to the development of numerous molecular sample-to-answer approaches. Despite that the quality of the purification process greatly impacts analysis, there has been less emphasis on stand-alone automated nucleic acid sample preparation systems (Jayamohan et al., 2017). Segregating automated sample preparation from analysis allows users the flexibility to tailor workflows to specific specimens of interest and to perform multiple types of downstream analysis on a single sample (e.g., sequencing, real-time PCR, and microarrays). Segregating sample preparation from analysis also frees analysis instruments for other laboratory needs (May, 2016). Additionally, automated sample preparation reduces – if not eliminates – lab-to-lab and technician-to-technician variability and makes molecular biology testing procedures more accessible to a wider range of users. Methods of automating sample processing can be addressed in many different ways depending upon the user skill level, the processing throughput, and the intended use environment, which can range from low resource clinics to high complexity laboratories.
Nucleic acid sample preparation devices are usually designed for lab-on-a-chip, point of care diagnostic devices (Cui et al., 2015; Govindarajan et al., 2012; Van Heirstraeten et al., 2014; Casavant et al., 2014; Strohmeier et al., 2015; Buser et al., 2015; Heiniger et al., 2016; Byrnes et al., 2015) or as a moderate to high-throughput laboratory and clinical tools (e.g., Qiagen QIAcube, Promega Maxwell®, ThermoFisher KingFisher™ Presto, Roche MagnaPure, and bioMérieux NucliSENS® easyMAG®). Point-of-care, integrated sample preparation methods and technology are necessarily developed around a specific intended use, sample type, or chemistry set (Clark et al., 2015; Nacham et al., 2016) and are therefore difficult to adapt or modify for large (mL) sample volumes (Ritzi-Lehnert, 2012), difficult-to-disrupt microorganisms (e.g. gram positive and acid fast bacilli), or complex clinical sample types (e.g., blood, nasopharyngeal aspirate [NPA], and nasopharyngeal swabs [NPS]) (Sin et al., 2014; Dineva et al., 2007). Commercially-available, automated workstations provide users the procedural flexibility to develop and validate extraction protocols for a wide range of sample types, but may still require off-line (manual) sample pretreatment (Brenov et al., 2009). Perhaps more limiting is that the automated systems themselves require significant capital investment and laboratory space, either of which may not be available at all institutions interested in performing molecular diagnostics. Therefore, there is still a clinical need for a flexible, compact, low-cost, moderate-throughput sample preparation workstation that can isolate nucleic acid from clinical sample types for analysis using most any molecular detection technology.
Claremont BioSolutions is a unique example of a company that has commercialized such a sample preparation system. They offer an eight-sample, flow-through processing system that includes a disposable electric motor for physical lysing (with beads) and subsequent nucleic acid purification (Doebler et al., 2009). As an alternative to the flow-through approach, we sought to develop a batch-type processing system that integrates conventional laboratory consumables (e.g., pipette tips and deep-well plates) to allow for procedural flexibility to more easily adapt to different applications.
Our approach to nucleic acid purification is the TruTip (Belgrader, 2010; Cooney and Belgrader, 2013), which is a pipette tip with an embedded porous sintered glass matrix that binds DNA using the chaotropic method first reported by Boom (Boom et al., 1990). The first TruTip publication describes the use of an electronic pipettor to sequentially aspirate and dispense chaotropic, wash, and elution buffers back and forth across the porous matrix (Chandler et al., 2012). This publication reports 98% accuracy of RNA extraction from influenza virions in NPS as compared to extraction using the easyMAG system. A later report describes the automation of the TruTip protocol using the epMotion liquid handling system for the same application (i.e., influenza RNA isolation from NPS) (Griesemer et al., 2013). Again, the TruTip extraction efficiency is comparable (R2 > 0.99 with respect to real-time PCR crossing points) to commercial methods (easyMAG and QIAcube instruments).
Here, we introduce an automated workstation that is designed for TruTip and additionally includes: a physical homogenization and lysis approach we call “MagVor” (Belgrader and Hindson, 2013), a pressurized air drying system for enhancing eluate purity, and a consumable design that consists of conventional laboratory disposables. Our objectives of this report are to demonstrate: (1) the feasibility of combining a mechanical lysis and homogenization approach with the TruTip extraction method; and (2) the integration of these two approaches in the format of an automated, bench-top workstation that can isolate nucleic acid from previously-reported sample types (e.g. influenza RNA in NPS) (Chandler et al., 2012; Griesemer et al., 2013; Holmberg et al., 2013b) as well as more challenging sample types (methicillin-resistant Staphylococcus aureus [MRSA] and M. tuberculosis [MTB] in NPA).
2. MATERIALS and METHODS
2.1. System Description
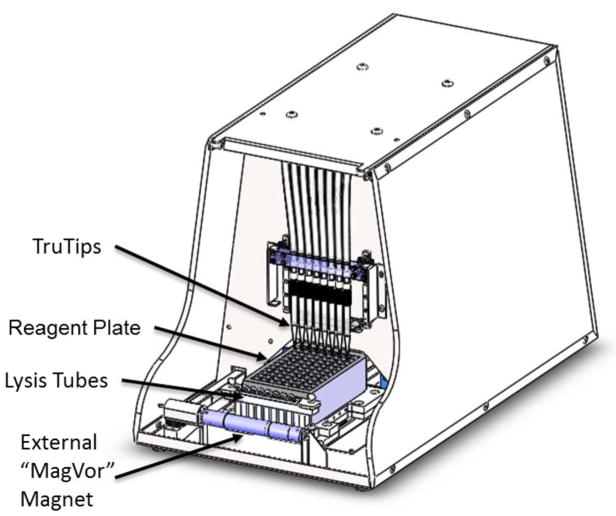
The design of the TruTip Automated Workstation (Akonni Biosystems, Frederick, MD) shown in Figure 1 includes: the integration of a homogenization and lysis method, an 8-channel TruTip pipetting sub-system with one degree of freedom in the vertical direction to raise and lower the TruTips in and out of the reagent wells, a one degree-of-freedom moving carriage in the horizontal direction to move the lysis tubes and reagent plate relative to the MagVor and TruTips, and a pressurized air drying sub-system to facilitate drying of the TruTips.
Figure 1.
Akonni’s TruTip Automated Workstation includes: an external magnet used for sample homogenization and cell lysis, a pipetting manifold, and an air pump (not shown). The consumables consist of lysis tubes that include a stir disk in a tube of glass beads, a reagent plate containing all necessary liquid reagents for extraction, and TruTips, which include a porous glass binding matrix inside of a standard pipette tip.
The consumables consist of lysis tubes (includes glass beads and a magnetic stir disk; Akonni Cat#402-00100; Akonni Biosystems, Frederick, MD), TruTips (1.2 mL SPT, Akonni Cat# 302-80021), and a deep-well reagent plate (USA Scientific Cat# 1896-2110; Ocala, FL) containing pre-metered Akonni catalog reagents. Consumables are placed on the carriage, which moves horizontally under the TruTips.
The principle of MagVor, which stands for magnetically-induced vortexing is to decouple the consumable from the energy transducing mechanism to simplify the integration of a physical lysis device into an automated instrument. The energy transducing mechanism is a rotating magnet that is external to the reaction vessel. At high rotational speeds (2500 – 5000 rpm), the changing magnetic field causes rotation of a magnetic stir disk inside of a homogenization or lysis tube. When non-magnetic beads are included in the lysis vessel, the beads undergo chaotic mixing, similar to bead-beating. The result is sample homogenization and cell lysis. The rotating magnet is oriented horizontally – but perpendicular to the axis of carriage motion – and induces rotation of the stir disks inside 8 lysis tubes when in close proximity (~4 mm).
The TruTip pipetting sub-system includes eight plungers that travel in eight separate barrels segregated on a single block to create a positive air displacement when moved by a single stepper motor, which simultaneously controls all eight plungers. Tubing connects the pipettor block to the eight nozzles designed to hold the TruTips. Thus, plunger movement controls aspirating and dispensing of liquids in and out of the TruTips. The pressurized air-drying sub-system consists of a linear selection valve that opens a port to an air pump to provide controlled airflow through a single tip at a time to thoroughly dry the binding matrix of any residual wash buffers. Independent control of pressurized airflow across a single TruTip ensures that the differential pressure across each matrix is the same.
Operating software and a user interface were written in LabView (National Instruments, Austin, TX) to allow the user to control: 1) carriage motion, 2) pipettor aspiration/dispense volume and speed, 3) the air port selection valve, 4) airflow to dry the TruTips, and 5) MagVor rotational velocity.
2.2. Clinical Specimens and Organisms
De-identified NPS in universal transport medium and quantified influenza virus (in gene copies mL−1) were obtained from Wadsworth Center (New York State Department of Health). De-identified NPA were from Little Company of Mary Hospital (Evergreen Park, IL). Whole blood with K2EDTA preservative was obtained from Bioreclamation IVT (Baltimore, MD).
M. tuberculosis H37Ra and MRSA were obtained from the American Type Culture Collection (Manassas, VA; Cat# 25177 and 700699, respectively). Bacteria were grown on solid media and diluted in sterile saline to 1 McFarland (~108 cells mL−1). All primary clinical specimens, viruses, and cultivated microorganisms were stored at −80 °C until use. Cells or virus (50 μL) were spiked into 450 μL of the background respiratory specimen at a 10-fold dilution series to arrive at the titers in these studies.
2.3. MagVor Studies
A stand-alone MagVor testbed was constructed to evaluate the efficacy of sample processing both with MagVor at 5000 rpm for 10 minutes and without MagVor. MRSA and M. tuberculosis cells were added to 500 μL aliquots from pooled NPA at concentrations described in the Results section. Duplicates of each concentration were processed for each condition. DNA purification was performed with a manual TruTip extraction protocol using an 8-Channel Rainin Electronic Pipettor (Rainin, Cat# EA8-1200; Mettler-Toledo, Columbus, OH) with 1.2 mL SPT TruTips (Akonni, Cat# 302-80021), similar to previous reports (Chandler et al., 2012). Briefly, the MRSA protocol consisted of the following sequential aspirate-dispense pipetting cycles (denoted as X): 20X for 250 μL of sample and 500 μL of Lysis Buffer A (Akonni, Cat #301-20000), 10X for 500 μL of Wash Buffer A (Akonni, Cat #301-20020), 20X for air drying in an empty tube, and 10X for 50μL of Elution Buffer B (Akonni, Cat # #301-20050). The M. tuberculosis protocol consisted of the following sequential aspirate-dispense cycles: 20X for 250 μL of sample and 850 μL of Lysis Buffer A/95% Ethanol, 10X for 500 μL of Wash Buffer J (Akonni, Cat# 301-20026), 10X for 500 μL of Wash Buffer A, 20X for air drying in an empty tube, and 10X for 100 μL of Elution Buffer A2 (Akonni, Cat# 301-20042). The eluates were analyzed using the protocol described in the Real-Time PCR Detection section.
2.4. Pipetting Precision Studies
Liquid volume pipetting studies were designed to compare the accuracy and precision of the custom pipetting manifold across three systems. The instruments were programmed to aspirate 20 μL, 75 μL, 125 μL, or 500 μL of water from a deep-well reagent plate into 1.2 mL Rainin pipette tips (without aerosol filters or TruTip binding matrix). Aspirated liquid was automatically dispensed into pre-weighed 0.5 mL tubes (n ≥ 7 for each of the 12 conditions), and the quantity of dispensed water was measured with a AT261 Mettler-Toledo analytical balance (Columbus, OH).
2.5. TruTip Drying Studies
Drying studies were performed to evaluate the presence of residual ethanol in the final eluate. TruTips were subjected to the following mixtures containing ethanol: 95% ethanol/Lysis Buffer F (Akonni, Cat# 301-20006), Akonni Wash Buffer J, and Wash Buffer K (Akonni, Cat# 301-20027). After binding and wash cycles, the TruTips were dried for either 0, 15, 30, 45, or 60 seconds using the pressurized air pump on the workstation (n=3 per condition). Alternatively, the TruTips were dried by either 30 or 60 aspiration and dispense cycles in an empty well (n=3 per condition). Elution Buffer A2 was then passed through the matrix and collected. The eluate was analyzed with the NanoDrop 1000 (ThermoFisher, Waltham, MA) for ethanol concentration using a colorimetric assay (NAD-ADH Reagent, Sigma, St. Louis, MO, Cat# N7160-10X16ML). Samples with varied drying conditions were compared to standards prepared with known quantities of ethanol in Elution Buffer A2 (50% – 0.01%) to determine the ethanol content in the final eluate.
2.6. Cross-Contamination Studies
Cross-contamination experiments were designed to evaluate the potential for contamination across parallel lanes and between successive runs. Two extractions were performed in a single reagent plate with positive samples (105 cells mL−1 M. tuberculosis in NPA, 100 μL whole blood, or 105 virions mL−1 influenza A in NPS) in rows A, C, E, and G; and negatives (water) in rows B, D, F, and H for columns 1 through 6. The pattern was transposed for the second half of the plate (columns 7 through 12). Extraction protocols followed the procedures described in the Automated Extraction Protocols section for the respective sample types. The eluates were analyzed using the protocol described in the Real-Time PCR Detection section.
2.7. Automated Extraction Protocols
Protocols using the automated MagVor and TruTip system were developed to isolate four different nucleic acid targets from three different clinical sample matrices. All steps for the following four protocols were automated on the workstation after adding the sample to the lysis tube, placing all consumables on the carriage, and initiating the protocol through the software interface.
1. MRSA in NPA
NPA specimens were pooled and subsequently segregated into 500 μL aliquots. MRSA cells were added to the 500 μL aliquots at concentrations and replicates described in the Results section. Each aliquot was then transferred to a lysis tube, and placed in the receptacle on the carriage. After executing the protocol (script) from the software interface, all of the following steps were automated. First, MagVor processed the sample at 5000 rpm for 10 minutes. Two-hundred and fifty microliters (250 μL) of the processed sample was then combined with 500 μL of Lysis Buffer A. This sample mixture was aspirated and dispensed through the matrix of a 1.2 mL SPT TruTip for 20 aspiration and dispense pipetting cycles. After this DNA binding step, 500 μL of Wash Buffer A was aspirated and dispensed through the TruTip matrix for 10 pipetting cycles. Each TruTip matrix was then dried in an empty well using pressurized air for 45 seconds. Finally, 100 μL of Elution Buffer A2 was aspirated and dispensed through the TruTip matrix for 10 pipetting cycles.
2. Influenza A in NPS
Influenza A virions were added to 500 μL aliquots from pooled NPS at concentrations and replicates described in the Results section. One-hundred twenty-five microliters (125 μL) of NPS aliquots were then combined with 375 μL of Lysis Buffer D (Akonni, Cat# 301-20004). This mixture was added to a lysis tube, which included a stir disk but no beads. After executing the protocol (script) from the software interface, all of the following steps were automated. First, the sample was actively mixed with MagVor for one minute at 5000 rpm. The sample was then incubated for 10 minutes at room temperature. The mixture was then combined with 375 μL of 95% ethanol. This entire mixture was then aspirated and dispensed through the matrix of a 1.2 mL SPT TruTip for 7 pipetting cycles. After this RNA binding step, 500 μL of Wash Buffer D (Akonni, Cat# 301-20021) was aspirated and dispensed through the TruTip matrix for five pipetting cycles, followed by five pipetting cycles of 500 μL of Wash Buffer E (Akonni, Cat# 301-20022). Each TruTip matrix was then dried in an empty well using forced air for 45 seconds. Finally, 50 μL of Elution Buffer A2 was aspirated and dispensed through the TruTip matrix for five pipetting cycles.
3. Human genomic DNA (gDNA) in whole blood
Aliquots of 100 μL blood were combined with 380 μL Lysis Buffer F, and 40 μL of Proteinase K (Qiagen, Germany, Cat# 19131). This reagent was added to a lysis tube, which included a stir disk but no beads. After executing the protocol (script) from the software interface, all of the following steps were automated. First, the sample was actively mixed with MagVor for one minute at 5000 rpm. The sample was then incubated at room temperature for 10 minutes and combined with 500 μL of 95% ethanol. The mixture was then aspirated and dispensed through the matrix of a 1.2 mL SPT TruTip for 10 pipetting cycles. After this DNA binding step, 500 μL of Wash Buffer J was aspirated and dispensed through the TruTip matrix for 10 pipetting cycles, followed by two repetitions of five pipetting cycles of 500μL of Wash Buffer K (repeated twice). Each TruTip matrix was then dried in an empty well using forced air for 45 seconds. Finally, 100μL of Elution Buffer A2 was aspirated and dispensed through the TruTip matrix for five pipetting cycles.
4. M. tuberculosis in NPA
NPA specimens were pooled and subsequently segregated into 500 μL aliquots. M. tuberculosis cells were added to the 500 μL aliquots at concentrations and replicates described in the Results section. Each aliquot was then transferred to a lysis tube, and placed in the receptacle on the carriage. M. tuberculosis cells were added to 500 μL aliquots from pooled NPA at concentrations and replicates described in the Results section. After executing the protocol (script) from the software interface, all of the following steps were automated. First, MagVor processed the sample at 5000 rpm for 10 minutes. Two-hundred and fifty microliters (250 μL) of the processed sample was transferred into a mixture of 350 μL of Lysis Buffer B (Akonni, Cat# 301-20002) and 500 μL of ethanol. The mixture was then aspirated and dispensed through the matrix of a 1.2 mL SPT TruTip for 20 pipetting cycles. After this DNA binding step, 500 μL of Wash Buffer J was aspirated and dispensed through the TruTip matrix for 10 pipetting cycles, followed by 10 pipetting cycles of 500 μL of Wash Buffer A. Each TruTip matrix was then dried in an empty well using forced air for 45 seconds. Finally, 100 μL of Elution Buffer A2 was aspirated and dispensed through the TruTip matrix for five pipetting cycles.
2.8. Reference Extraction Protocol
To compare recovery of the automated workstation to a standard method, extractions were performed using the manual Qiagen spin column protocols (n=8 for each specimen type at each input titer). Specifically, manufacturer’s instructions were followed for the Qiagen QIAamp RNA Viral Mini Kit (Cat# 52904) to process Influenza in NPS, the Qiagen QIAamp DNA Blood Mini Kit (Cat# 51104) for whole blood, and the Qiagen QIAamp DNA Mini Kit (Cat# 51304) for MRSA samples. There is no commercial kit for MTB DNA extraction from NPA. Thus, for MTB spiked in NPA, we used the BD (Franklin Lakes, NJ) GeneOhm lysis tubes (Cat# 441243), which was vortexed for 5 minutes, then heated at 95°C for 2 minutes. Two hundred microliters (200 μL) of crude lysate was processed using the Qiagen QIAamp DNA Blood Mini Kit (manufacturer recommended kit for MTB extractions). The eluates were analyzed using the protocol described in the Real-Time PCR Detection section.
2.9. Real-Time PCR Detection
Real-time PCR assays were performed on a Roche 480 LightCycler (Basel, Switzerland).
For MRSA studies, real-time PCR primers and probes (adapted from Zimmerman et al., 2012 for real-time assay) were designed to amplify and detect the tufA gene. Two microliters (2 μL) of eluate was added to 23 μL of: 1× PCR mix from the QuantiTect Probe PCR kit (Qiagen, Cat# 204343), 0.6 mg/ml BSA (Sigma, Cat# B6917), 0.4 μM forward primer (5′-GCATTAAAAGCATTAGAAGGCG-3′), 0.4 μM reverse primer (5′-CGGCCTGTAGCAACAGTACC-3′), and 0.2 μM probe (5′ -6FAM-CATGATGCCAGTTGAGGACGTATTCT-BHQ1-3′). The 25 μL reaction mixture was heat denatured for 15 min at 95°C and then cycled for 40 cycles of [15 s at 95°C, 60 s at 60°C].
For M. tuberculosis studies, real-time PCR primers and probes were designed to amplify and detect the IS6110 insertion complex (primers and probes are reported in Savelkoul et al., 2006). Five microliters (5 μL) of eluate was added to 20 μL of: a LightCycler FastStart DNA Master HybProbe kit (Roche, Cat# 03003248001), 2.5 mM MgCl2, 0.45 μM forward primer (5′ GGG-TAG-CAG-ACC-TCA-CCT-ATG-3′), 1.35 μM reverse primer (5′ - AGC-GTA-GGC-GTC-GGT-GA - 3′), and 0.25 μM minor groove binding probe (5′ - 6FAM-TCG-CCT-ACG-TGG-CCT-TT-mgb). The 25 μL reaction mix was heat denatured for 10 min at 95°C and then cycled for 45 cycles of [15 s at 95°C, 60 s at 60°C].
For influenza A studies, real-time PCR primers and probes (reported by de-Paris et al., 2012) were used to amplify and detect a segment of the matrix protein in H3N2 influenza A. Five microliters (5 μL) of eluate was added to 20 μL of: 1× Superscript III RT PCR Kit w/Platinum Taq (Invitrogen, Carlsbad, CA, Cat# 12574-026), 0.1 mg/mL BSA, 0.8 μM forward primer (5′-GAC CRA TCC TGT CAC CTC TGA C-3′), 0.8 μM reverse primer (5′-AGG GCA TTY TGG ACA AAK CGT CTA-3′), and 0.025 μM probe (5′-6 FAM TGC AGT CCT CGC TCA CTG GGC ACG-BHQ1-3′). The 25 μL reaction mix was heated to 50°C for 30 minutes for reverse transcription, heated to 95°C for 2 minutes for denaturation, then cycled 45 times [15 s at 95°C, 30 s at 55°C].
For detection of human DNA in blood, 2 μL of eluate was added to 23 μL of the Applied Biosystems (Foster City, CA) Quantifiler Human DNA Quantification Kit (Cat # 4343895), which targets the human telomerase reverse transcriptase gene (hTERT).
Crossing Points (Cp) were determined using the second derivative analysis method in the LightCycler 480 software. Average Cp and standard deviations were calculated using Microsoft Excel (Seattle, WA). Statistical comparisons between the automated workstation and Qiagen Kits were evaluated using one-way analysis of variance (ANOVA) and a t-test using JMP software (SAS, Cary, NC).
2.10. Nanodrop
To assess the purity of the nucleic acid isolated from blood, purified eluates of human genomic DNA were analyzed using the Nanodrop 1000 UV-Vis Spectrophotometer (ThermoFisher). Absorbance ratios at 260/280 nm and 260/230 nm were averaged from two runs (n=16) using two different blood specimens.
3. RESULTS
A series of functional tests, both biological and non-biological, were performed to test the performance of each subsystem (MagVor, pipetting and drying), prior to testing the full extraction protocol. First, experiments were performed to highlight the impact of the MagVor to overall DNA recovery. Data in Tables 1 and 2 show an increase in DNA recovery from MRSA and M. tuberculosis in an NPA background with the addition of MagVor, with a more noticeable increase in DNA recovery at higher titers. Nucleic acid recovery was linearly correlated with bacterial titer (R2 = 0.98 and 0.99 for MRSA and M. tuberculosis with MagVor treatment, respectively). Assuming 100% amplification efficiency, a ΔCp of 3.3 corresponds to a 10-fold difference in measured DNA concentration. MagVor treatment therefore improved DNA recovery by at least 4-fold for MRSA and M. tuberculosis at ≥ 105 cells mL−1 NPA, and approximately 2-fold at titers ≤ 104 cells mL−1.
Table 1.
The effect on recovery when using MagVor to process NPA spiked with MRSA.
| Titer of MRSA Processed in NPA (cells/mL) | Avg Cp Without MagVor | Avg Cp With MagVor | Difference between Processing with and without Magvor (Δ Cp) |
|---|---|---|---|
| 108 | 28.86 ± 0.07 | 23.69 ± 0.32 | 5.17 |
| 107 | 28.56 ± 0.97 | 25.5 ± 0.37 | 3.06 |
| 106 | 31.67 ± 0.36 | 29.32 ± 0.13 | 2.36 |
| 105 | 33.98 ± 0.01 | 31.81 ± 0.04 | 2.17 |
Table 2.
The effect on recovery when using MagVor to process NPA spiked with M. tuberculosis.
| Titer of M. tuberculosis processed in NPA (cells/mL) | Avg Cp Without MagVor | Avg Cp With MagVor | Difference between processing with and without MagVor (Δ Cp) |
|---|---|---|---|
| 107 | 20.38 ± 0.28 | 17.31 ± 0.49 | 3.08 |
| 106 | 23.82 ± 0.37 | 21.33 ± 0.16 | 2.49 |
| 105 | 26.50 ± 0.01 | 24.33 ± 0.02 | 2.17 |
| 104 | 29.77 ± 0.16 | 28.79 ± 0.23 | 0.98 |
| 103 | 32.76 ± 0.06 | 31.59 ± 0.01 | 1.17 |
| 102 | 34.52 ± 0.85 | 33.63 ± 0.06 | 0.89 |
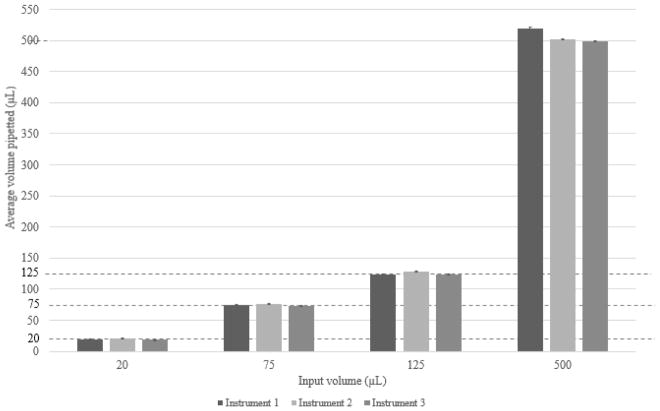
Pipetting precision studies of weighing water transferred by the system demonstrate the reproducibility and precision of the custom designed pipette manifold as shown in Figure 2. The corresponding coefficient of variations (CVs) were 2%, 0.5%, 0.3%, and 0.2% at volumes of 20, 75, and 125, and 500 μL across three workstations. The correlation coefficients were 0.99, 1, and 1 for linear fit for Workstations 1, 2 and 3, respectively.
Figure 2.
Pipetting precision testing across three replicate workstations.
In the silica extraction methods, wash reagents can often carry over into the elution and cause inhibition in the downstream amplification method. Therefore, it is important to achieve sufficient drying after the wash steps to eliminate residual organics, in this case, ethanol. Drying studies showed an ethanol concentration of 0.2% in the eluate after 15 seconds of pressurized airflow through the TruTip matrix; the drying efficiency plateaued after 30 seconds. The average residual concentration of ethanol in the eluate was 10% in the absence of air drying and 0.2%, 0.1%, 0.1%, and 0.1% at 15, 30, 45, and 60 seconds of air drying, respectively. Repeated aspiration and dispense pipetting cycles of air (without pressurized air) did not reduce the ethanol concentration in the eluate; the ethanol was 10% for both conditions of 30 and 60 aspiration-dispense cycles with TruTips not immersed in liquid.
We examined the potential for cross-contamination by processing eight samples in parallel using a checker board study (alternating positive and negative samples). The cross-contamination study results (Table 3) did not show evidence of DNA in the “negative” wells across three different specimen and cell types at high titers (105 copies of M. tuberculosis, 100 μL of blood, and 105 copies of influenza A). The standard deviation of Cp for the replicate “positive” wells was 0.29 for M. tuberculosis, 0.59 for blood, and 0.72 for influenza A.
Table 3.
Real-time PCR results from cross contamination study using M. tuberculosis (MTB) spiked in NPA; blood; and influenza A spiked in NPS.
| Well Location | Condition | MTB Study Results (Cp) | Blood Study Results (Cp) | Influenza A Study Results (Cp) | |
|---|---|---|---|---|---|
| Columns 1 – 6 | A | + | 23.31 | 29.49 | 31.88 |
| B | − | Negative | Negative | Negative | |
| C | + | 23.38 | 30.58 | 30.32 | |
| D | − | Negative | Negative | Negative | |
| E | + | 23.11 | 30.28 | 29.55 | |
| F | − | Negative | Negative | Negative | |
| G | + | 23.24 | 30.15 | 30.42 | |
| H | − | Negative | Negative | Negative | |
| Columns 7–12 | A | − | Negative | Negative | Negative |
| B | + | 23.35 | 29.51 | 29.84 | |
| C | − | Negative | Negative | Negative | |
| D | + | 23.47 | 30.2 | 29.93 | |
| E | − | Negative | Negative | Negative | |
| F | + | 23.1 | 31.12 | 30.57 | |
| G | − | Negative | Negative | Negative | |
| H | + | 22.55 | 29.44 | 29.95 |
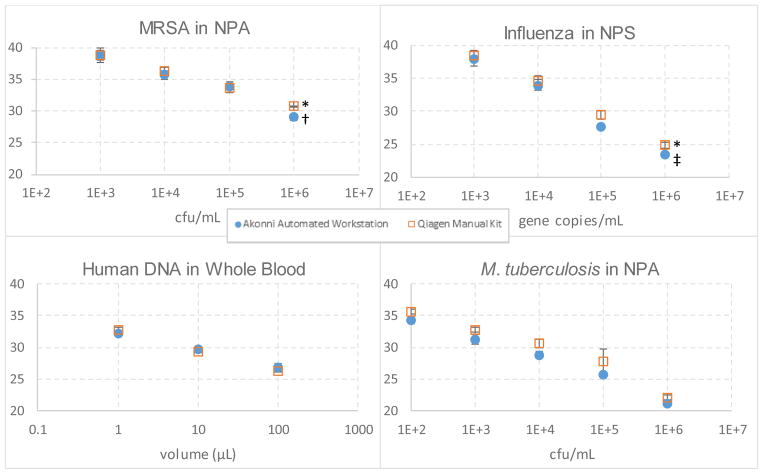
Nucleic acid recovery for the TruTip automated workstation was compared to that of Qiagen’s manual spin column extraction kits, which require multiple centrifugation steps. The results in Table 4 show comparable recoveries between the workstation and the Qiagen Kits. The correlation coefficient of Cp to the log of the concentration was 0.97 for MRSA in NPA, 0.99 for M. tuberculosis in NPA, 0.99 for influenza A in NPS, and 0.99 for blood. For all applications tested, the workstation recovery was statistically equivalent or better than the manual Qiagen method in all cases except for higher volumes of whole blood (10 and 100 μL). For both methods, limits of detection were greater than 103 cells/mL for MRSA and greater than 103 gene copies/mL for influenza (i.e., detection was less than 95% at these levels), and both methods resulted in correct detection for blood and M. tuberculosis at the lowest concentration.
The purity of nucleic acid extracts is typically measured by UV absorbance ratios of 260/230 and 260/280, where 260 nm is the wavelength of nucleic acid absorption and impurities such as salts or proteins absorb at 230 or 280 nm, respectively. Extracts with ratios greater than 1.7 are considered high quality. The overall purity of the isolated nucleic acid from whole blood is consistent with those reported in previous studies (Holmberg et al., 2013a). Absorbance measurements showed high eluate purity; A260/280 ratios were 1.88 ± 0.15, and A260/230 ratios were 1.92 ± 0.09 across two different whole blood specimens with eight replicates each.
4. DISCUSSION
The TruTip Automated Workstation, which includes a software interface that allows the user to write custom scripts, is amenable to at least four different specimen/cell types. The Workstation protocols can be customized with respect to: homogenization and lysis by varying the MagVor duration and rotational speed, TruTip pipetting protocols including volumes and flow rates, or TruTip drying using a pressurized air drying system. The scripts can be tailored to the application needs and may include the implementation of MagVor when the specimens are heterogeneous and viscous or the organisms are difficult to lyse. Alternatively, MagVor can also be used without beads to simply mix reagents (i.e., protocols for human gDNA in whole blood and influenza A in NPS).
The intent of the TruTip design is to allow for ease of interfacing to liquid handling systems. One distinction of the TruTip protocol compared to many liquid handling operations, however, is that most steps of the TruTip procedure do not include liquid transport. Rather, the procedure consists of cycling liquid back and forth through the matrix while the TruTip resides in the well of the reagent plate; the discarded liquid is then dispensed back into the well from which it came. This approach of confining bound nucleic acid to the matrix inside the pipette tip, therefore does not require the transport of liquids from well to well across the reagent plate. Minimizing liquid transfer steps has an inherently low susceptibility to contamination, as evidenced by the cross-contamination studies.
The TruTip protocol is designed to ensure that the entire aliquot of sample/bind buffer travels through and beyond the matrix in the tip so that all nucleic acid in the aliquot is exposed to the DNA-binding silica surface; this also applies to the elution buffer. The distance that the liquid travels beyond the matrix, however, can be arbitrary, provided that the entire aliquot flows through the matrix. The pipetting tolerance requirement for the TruTip protocol is, therefore, less stringent than many liquid handling applications. The preparation of the reagent plate, however, does require tighter pipetting tolerance than operations on the Workstation to ensure that the aliquoted volumes are properly proportioned when combining reagents. Precise metering of the liquid can therefore be imposed upon the preparation (manufacturing) of the reagent plate – as opposed to the operation of the Workstation – using conventional liquid handling systems.
Nevertheless, the Workstation pipetting experiments demonstrated high precision: 0.2 to 2% CV for a single 1.2 mL tip type across volumes from 20 μL to 500 μL. These CVs are consistent with the Hamilton STAR Liquid Handling System, which is marketed to achieve 3.5% CV at 10 μL and 0.75% at 100 and 1000 μL. The use of this single 1.2 mL pipette tip type for all pipetting functions reduces the instrument and consumable complexity (i.e., multiple pipette heads and tips are not required).
Bi-directional flow across the TruTip matrix allows the user to tailor the number of aspirate and dispense pipetting cycles for each of the steps (i.e., bind, wash, dry, and elute). This approach offers an ability to increase the liquid residence time in the matrix as compared to centrifugation methods that rely on only a single pass of unidirectional flow. Increased residence time is particularly important for viscous samples, which reduce molecular diffusivity. As an illustration of the ability to customize pipetting cycles, the bind steps were 20, 10, 7, and 20 pipetting cycles for the M. tuberculosis, blood, influenza, and MRSA protocols, respectively. Thus, rather than making changes to the porosity or thickness of the TruTip matrix, we made changes to the number of pipetting cycles in the protocol.
We find that removal of residual organic wash reagents is improved nearly 100-fold with pressurized air drying as compared to bi-directional air flow using the aspiration and dispense strokes of the pipette plunger. In these studies, the 25 μL real-time PCR reactions consist of only 2 or 5 μL eluate volumes, thus the fraction of eluate to the reaction volume is low compared to assays that require large eluate volumes such as those that rely entirely on reconstituting lyophilized reagents with the eluate liquid. Thus, pressurized air drying may not be necessary for all applications (Griesemer et al., 2013; Homberg et al., 2013a; Holmberg et al., 2013b), but is likely to be important for assays that are inhibited by organics and require large eluate volumes.
The extraction method comparison study shows equivalent, and in some cases improved, recoveries compared to Qiagen for all applications. Yet, the Qiagen workflow was manual whereas the Workstation automated all steps including lysis of difficult-to-disrupt cells such as M. tuberculosis. The stepwise linear decrease in Cp value with a logarithmic increase in concentration across all four applications suggests that the binding capacity of the matrix has not been exceeded up to the maximum titers.
The purity of the resulting nucleic acid is important for optimal detection by downstream molecular methods. Typical contaminating substances that adversely affect PCR efficiency include residual organics such as ethanol, salts from wash reagents, heme in blood, and/or specimen debris such as proteins and cells. While more targeted studies are required to determine assay compatibility with eluates from this automated Workstation, the data reported here indicate extracted DNA and RNA are of sufficient purity for real-time PCR detection. Furthermore, the high A260/A280 and A260/230 ratios following extraction of human gDNA from blood suggest low protein and salt content in the final eluate. And, the ethanol concentration of 0.1% in the eluate after pressurized air drying suggests low residual ethanol concentration. Thus, we expect that the eluate will be of sufficient purity for many types of amplification assays in future applications.
Automated nucleic acid extraction from the sample types reported here expand upon those described in our previous publication (Griesemer et al., 2013). We address these additional sample types by implementing new features in an Akonni-developed workstation. For example, the TruTip Automated Workstation reported in this work includes MagVor and forced air drying. Another distinction for the influenza in NPS sample type, compared to Griesemer et al. (2013), is the use of different TruTips (1.2 mL SPT tips) as compared with the epMotion TruTips (1 mL LPT tips). (SPT has a smaller porosity than LPT). In this study we include MagVor to improve the homogeneity and reduce the viscosity of the sample, so that the sample can permeate the smaller SPT pores, which reduces the diffusion path length of the nucleic acid to the surface of the binding matrix.
These data demonstrate the feasibility and flexibility of the Workstation for isolating DNA from difficult-to-disrupt organisms as well as RNA from more labile viral capsids in clinical samples. Specifically, the flexibility of this platform allows users to define their own automated protocols to address, potentially unique, nucleic acid isolation applications. Additional work to evaluate the TruTip Automated Workstation is underway at a number of collaborator sites to test the system with challenging specimen types such as sputum and stool. Future work will also expand these analytical studies (clinical matrices supplemented with known titers of target cells) to additionally include clinical studies (retrospective and prospective sample testing).
Figure 3.
Real-time PCR results comparing the automated workstation (n=24) to the manual Qiagen extraction kit (n=8) as a function of Cp, which is inversely proportional to nucleic acid recovery. No template controls were not detected.
* 6 of 8 replicates detected in this data set.
† 17 of 24 replicates detected in this data set.
‡ 21 of 24 replicates detected in this data set.
Highlights.
A new lysis and homogenization method is combined with nucleic acid purification.
Nucleic acid purification and concentration occur in a pipette tip.
Both blood and nasopharyngeal samples are processed on a new Workstation.
Nucleic acid extraction from bacteria and viruses are automated on the Workstation.
The automated Workstation shows comparable recoveries to standard manual methods.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) under the Center of Excellence in Translational Research (CETR) grant U19 AI109755, R44 EB011274, R01 AI111435, and R44 GM103053. The authors would also like to acknowledge Dr. Kirsten St. George from Wadsworth Center and the Little Company of Mary for providing de-identified specimens and samples.
Footnotes
6. CONFLICTS OF INTEREST
All Akonni-affiliated authors are employees of Akonni Biosystems Inc. NT, DPC, and CGC are also Akonni shareholders.
Publication in JMM does not imply that the journal or publisher is endorsing the product and method.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Belgrader P. Apparatus, system, and method for purifying nucleic acids. 7759112 US Patent. 2010
- Belgrader P, Hindson B. Magnetic lysis method and device. 8399190 US Patent. 2013
- Boom RCJA, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, Van der Noordaa JPME. Rapid and simple method for purification of nucleic acids. Journal of Clinical Microbiology. 1990;28(3):495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevnov M, Mundt J, Benfield J, Treat-Clemons L, Kalusche G, Meredith J, Porter G, Furtado MR, Shewale JG. Automated extraction of DNA from forensic sample types using the PrepFiler automated forensic DNA extraction kit. JALA: Journal of the Association for Laboratory Automation. 2009;14(5):294–302. [Google Scholar]
- Buser JR, Wollen A, Heiniger EK, Byrnes SA, Kauffman PC, Ladd PD, Yager P. Electromechanical cell lysis using a portable audio device: enabling challenging sample preparation at the point-of-care. Lab on a Chip. 2015;15(9):1994–1997. doi: 10.1039/c5lc00080g. [DOI] [PubMed] [Google Scholar]
- Byrnes SA, Bishop JD, Lafleur L, Buser JR, Lutz B, Yager P. Onestep purification and concentration of DNA in porous membranes for point-of-care applications. Lab on a Chip. 2015;15(12):2647–2659. doi: 10.1039/c5lc00317b. [DOI] [PubMed] [Google Scholar]
- Casavant BP, Guckenberger DJ, Beebe DJ, Berry SM. Efficient sample preparation from complex biological samples using a sliding lid for immobilized droplet extractions. Analytical Chemistry. 2014;86(13):6355–6362. doi: 10.1021/ac500574t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler DP, Griesemer SB, Cooney CG, Holmberg R, Thakore N, Mokhiber B, Belgrader P, Knickerbocker C, Schied J, George KS. Rapid, simple influenza RNA extraction from nasopharyngeal samples. Journal of Virological Methods. 2012;183(1):8–13. doi: 10.1016/j.jviromet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KD, Nacham O, Yu H, Li T, Yamsek MM, Ronning DR, Anderson JL. Extraction of DNA by magnetic ionic liquids: tunable solvents for rapid and selective DNA analysis. Analytical Chemistry. 2015;87(3):1552–1559. doi: 10.1021/ac504260t. [DOI] [PubMed] [Google Scholar]
- Cooney CG, Belgrader P. Sample preparation device. 8574923 US Patent. 2013
- Cui F, Rhee M, Singh A, Tripathi A. Microfluidic sample preparation for medical diagnostics. Annual Review of Biomedical Engineering. 2015;17:267–286. doi: 10.1146/annurev-bioeng-071114-040538. [DOI] [PubMed] [Google Scholar]
- de-Paris F, Beck C, Machado ABMP, Paiva RM, da Silva Menezes D, de Souza Nunes L, Kuchenbecker R, Barth AL. Optimization of one-step duplex real-time RT-PCR for detection of influenza and respiratory syncytial virus in nasopharyngeal aspirates. Journal of Virological Methods. 2012;186(1–2):189–192. doi: 10.1016/j.jviromet.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Dineva MA, Mahilum-Tapay L, Lee H. Sample preparation: a challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings. Analyst. 2007;132(12):1193–1199. doi: 10.1039/b705672a. [DOI] [PubMed] [Google Scholar]
- Doebler RW, Erwin B, Hickerson A, Irvine B, Woyski D, Nadim A, Sterling JD. Continuous-flow, rapid lysis devices for biodefense nucleic acid diagnostic systems. JALA: Journal of the Association for Laboratory Automation. 2009;14(3):119–125. [Google Scholar]
- Govindarajan AV, Ramachandran S, Vigil GD, Yager P, Böhringer KF. A low cost point-of-care viscous sample preparation device for molecular diagnosis in the developing world; an example of microfluidic origami. Lab on a Chip. 2012;12(1):174–181. doi: 10.1039/c1lc20622b. [DOI] [PubMed] [Google Scholar]
- Griesemer SB, Holmberg R, Cooney CG, Thakore N, Gindlesperger A, Knickerbocker C, Chandler DP, George KS. Automated, simple, and efficient influenza RNA extraction from clinical respiratory swabs using TruTip and epMotion. Journal of Clinical Virology. 2013;58(1):138–143. doi: 10.1016/j.jcv.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiniger EK, Buser JR, Mireles L, Zhang X, Ladd PD, Lutz BR, Yager P. Comparison of point-of-care-compatible lysis methods for bacteria and viruses. Journal of Microbiological Methods. 2016;128:80–87. doi: 10.1016/j.mimet.2016.07.007. [DOI] [PubMed] [Google Scholar]
- Holmberg RC, Gindlesperger A, Stokes T, Brady D, Thakore N, Belgrader P, Cooney CG, Chandler DP. High-throughput, automated extraction of DNA and RNA from clinical samples using TruTip technology on common liquid handling robots. Journal of Visualized Experiments: JoVE. 2013a;(76) doi: 10.3791/50356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg RC, Gindlesperger A, Stokes T, Lopez D, Hyman L, Freed M, Belgrader P, Harvey J, Li Z. Akonni TruTip® and Qiagen® methods for extraction of fetal circulating DNA-evaluation by real-time and digital PCR. PloS one. 2013b;8(8):e73068. doi: 10.1371/journal.pone.0073068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayamohan H, Romanov V, Li H, Son J, Samuel R, Nelson J, Gale BK. Advances in Microfluidics and Lab-on-a-Chip Technologies. Molecular Diagnostics (Third Edition) 2017:197–217. [Google Scholar]
- May M. Automated sample preparation. Science. 2016;351(6270):300–302. [Google Scholar]
- Nacham O, Clark KD, Anderson JL. Extraction and purification of DNA from complex biological sample matrices using solid-phase microextraction coupled with real-time PCR. Analytical Chemistry. 2016;88(15):7813–7820. doi: 10.1021/acs.analchem.6b01861. [DOI] [PubMed] [Google Scholar]
- Ritzi-Lehnert M. Development of chip-compatible sample preparation for diagnosis of infectious diseases. Expert Review of Molecular Diagnostics. 2012;12(2):189–206. doi: 10.1586/erm.11.98. [DOI] [PubMed] [Google Scholar]
- Savelkoul PH, Catsburg A, Mulder S, Oostendorp L, Schirm J, Wilke H, van der Zanden AG, Noordhoek GT. Detection of Mycobacterium tuberculosis complex with real time PCR: comparison of different primer-probe sets based on the IS6110 element. Journal of Microbiological Methods. 2006;66(1):177–180. doi: 10.1016/j.mimet.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sin ML, Mach KE, Wong PK, Liao JC. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Review of Molecular Diagnostics. 2014;14(2):225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmeier O, Keil S, Kanat B, Patel P, Niedrig M, Weidmann M, Hufert F, Drexler J, Zengerle R, von Stetten F. Automated nucleic acid extraction from whole blood, B. subtilis, E. coli, and Rift Valley fever virus on a centrifugal microfluidic LabDisk. RSC Advances. 2015;5(41):32144–32150. [Google Scholar]
- Van Heirstraeten L, Spang P, Schwind C, Drese KS, Ritzi-Lehnert M, Nieto B, Camps M, Landgraf B, Guasch F, Corbera AH, Samitier J. Integrated DNA and RNA extraction and purification on an automated microfluidic cassette from bacterial and viral pathogens causing community-acquired lower respiratory tract infections. Lab on a Chip. 2014;14(9):1519–1526. doi: 10.1039/c3lc51339d. [DOI] [PubMed] [Google Scholar]
- Zimmerman CE, Stamper PD, Bryant L, Farley J, Golova J, Holmberg R, Howard T, Linger Y, Meyers K, Perov A, Rudy GB. Development of a simple, low-density array to detect methicillin-resistant Staphylococcus aureus and mecA dropouts in nasal swabs. Journal of Microbiological Methods. 2012;91(3):366–376. doi: 10.1016/j.mimet.2012.09.010. [DOI] [PubMed] [Google Scholar]