Abstract
Jasmonates (JAs) are signalling molecules involved in stress responses, development and secondary metabolism biosynthesis, although their roles in fleshy-fruit development and ripening processes are not well known. In strawberry fruit, it has been proposed that JAs could regulate the early development through the activation of the JAs biosynthesis. Moreover, it has been reported that JA treatment increases anthocyanin content in strawberry fruit involving the bioactive jasmonate biosynthesis. Nevertheless, JA signalling pathway, of which main components are the COI1-JAZ co-receptor and the MYC transcription factors (TFs), has not been characterized in strawberry until now. Here we identified and characterized the woodland strawberry (Fragaria vesca) JAZ and MYC genes as well as studied their expression during development and ripening stages in commercial strawberry (Fragaria × ananassa) fruit. We described twelve putative JAZ proteins and two MYC TFs, which showed high conservation with respect to their orthologs in Arabidopsis thaliana and in other fleshy-fruit species such as Malus × domestica, Vitis vinifera and Solanum lycopersicum as revealed by gene synteny and phylogenetic analyses. Noteworthy, their expression levels exhibited a significant decrease from fruit development to ripening stages in F. × ananassa, along with others of the JA signalling-related genes such as FaNINJA and FaJAMs, encoding for negative regulators of JA responses. Moreover, we found that main JA signalling-related genes such as FaMYC2, and FaJAZ1 are promptly induced by JA treatment at early times in F. × ananassa fruit. These results suggest the conservation of the canonical JA signalling pathway in strawberry and a possible role of this pathway in early strawberry fruit development, which also correlates negatively with the beginning of the ripening process.
Introduction
Jasmonates (JAs) regulate development, metabolism and tolerance against biotic and abiotic stresses [1–3]. Their roles have not been extensively studied in fleshy fruits, although several reports have shown a role as stimulants of the phenylpropanoid pathway and ethylene biosynthesis [4]. Moreover, JAs and related oxylipins could play an early role during strawberry and grape fruit development since 12-oxo-phytodienoic acid (OPDA), methyl jasmonate (MeJA), jasmonic acid (JA), and the bioactive JA jasmonoyl-isoleucine (JA-Ile) accumulate at flowering and immature fruit stages, and then decrease as fruit ripens [5–7]. JAs are also involved in the anthocyanin accumulation in Arabidopsis seedlings [8] and exogenous MeJA application induces anthocyanin biosynthesis with the concomitant upregulation of JA biosynthesis-related genes in Chilean strawberry (Fragaria chiloensis) fruit [9]. In commercial strawberry (Fragaria × ananassa) fruit, after exogenous MeJA application, and coincident with the anthocyanin accumulation, JA-Ile levels increased along with anthocyanin accumulation, while the main ripening-associated hormone abscisic acid (ABA) decreased in developing treated fruit [7]. Recently, we characterized the dynamics of endogenous JAs during F. × ananassa fruit development and ripening [7]. A correlation between reduction of JA-Ile levels and downregulation of FaJAR1, the key gene encoding for the JA-Ile synthesis enzyme, and the JA turnover-related genes (i.e, FaMJE and FaJIH1) was reported [7]. Nevertheless, the molecular characterization of the JA signalling-related components has not been performed in strawberry until now.
The physiological effects mediated by JA-Ile require activation of the signalling pathway, which has been well characterized in Arabidopsis [2,10]. The F-box CORONATINE INSENSITIVE1 protein (COI1) is part of the Skp-Cullin-F-box-type E3 ubiquitin ligase complex (SCFCOI1) and together with JASMONATE ZIM-DOMAIN (JAZ) form the JA-Ile receptor [11–13]. When JA-Ile levels are low, JAZ transcriptional repressors bind to MYC2 and additional transcription factors (TFs) repressing expression of early JA-responsive genes [2,10]. Moreover, Novel Interactor of JAZ (NINJA) adaptor allows the establishment of the co-repressor complex consisting of TOPLESS (TPL) [14] and histone deacetylases (HDAs) [15]. Once JA-Ile level rises, COI1 binds to JAZs that are degraded by the 26S proteasome after ubiquitination [2,10]. Then MYC2 and additional TFs induce the expression of early JA-responsive genes such as JAZs, MYCs and JA biosynthetic ones [10,11,16]. In Arabidopsis, 13 JAZ proteins have been identified until now [10]. JAZ1-12 contain the conserved TIFY and Jas domains where JAZ13 is a non-TIFY JAZ protein [17]. The TIFY domain near N-terminal region contains the TIF[F/Y]XG motif [11,13,18], which mediates homo- and heteromeric interactions between TIFY proteins [19] and the interaction with NINJA [14]. The Jas domain is a conserved sequence at the C-terminal region of JAZ containing a conserved SLX2FX2KRX2RX5PY motif, mediating the hormone-dependent COI1-JAZ complex formation [12]. The Jas domain is also responsible for the interaction of JAZs with MYCs, suppressing JA responses [20]. On the other hand, PEAPOD (PPD) subfamily is part of the TIFY family proteins, which contain a highly conserved TIFY domain, a degenerated Jas domain and an N-terminal PPD domain. However, until now a PPD putative role in JA response has not been described [18,21,22]. MYC2, MYC3, MYC4 and MYC5 are bHLH-like TFs that contain a basic helix-loop-helix (bHLH) domain for binding to G-box-containing promoters [23–25] and the transcription is regulated by MED25 subunit [26]. MYC2, MYC3 and MYC4 interact with JAZs through the JAZ-interacting domain (JID) [27]. Other components of JA signalling pathway are JASMONATE-ASSOCIATED MYC2-like1/2/3 (JAM1, JAM2 and JAM3), which are antagonistic and negative regulators of MYC-like TFs and JA responses [28,29]. JAM1, JAM2 and JAM3 corresponding to bHLH003, bHLH013 and bHLH017 TFs, respectively [30]. Therefore, JAZs together with MYCs are the main regulators for JA-Ile responses [11] including upregulation of JAZ genes [11,24], which encoded JAZ proteins perform a negative feedback to turn off JA-Ile-mediated responses [11]. Recently, in a transcriptome analysis performed in F. × ananassa fruit, the repression of a JAZ from green to white and partially ripe stages was reported [31]. Nevertheless, a deep molecular characterization of the JAZ gene family and JA signalling components has not been reported in Fragaria species.
Thus, in this work we characterized the main JA signalling components in Fragaria species as JAZs and MYCs, using woodland strawberry (Fragaria vesca) to perform the genomic in silico studies and F. × ananassa for transcriptional analysis during fruit development and ripening and in response to JA treatment.
Materials and methods
Identification and characterization of JA signalling-related genes in F. vesca
Arabidopsis sequences of JA signalling-related proteins, including JAZs and MYCs, were used to search ortholog genes in F. vesca genome and transcriptome [32] by tblastn search tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (S1 Table). Sequences with higher coverage and identity, and with lower e-value were selected for further bioinformatics analysis (S1 Table). TIFY5b-like sequence (GenBank accession number XM_011469361.1) was obtained directly from F. vesca transcriptome because it showed lower coverages, identities and higher e-values comparing with Arabidopsis JAZ proteins.
Mapping, duplication and synteny analysis of JAZ and MYC genes in F. vesca and Arabidopsis thaliana
Physical chromosomal locations of F. vesca (Fv) and Arabidopsis thaliana (At) JAZs and MYCs were obtained from Arabidopsis (TAIR10, https://www.arabidopsis.org/) and Genome databases (https://www.ncbi.nlm.nih.gov/genome/). Gene synteny analysis were performed between the F. vesca, Malus × domestica (Md), Vitis vinifera (Vv), Solanum lycopersicum (Sl), Oryza sativa (Os) and A. thaliana genomes (S2 Table) using information available in the Plant Genome Duplication Database (PGDD) [33].
Gene structure, protein sequence and phylogenetic analyses
Exon-intron organizations of JAZ and MYC genes were determined by Gene Display Server 2.0 [34] using the information available at Genome database (https://www.ncbi.nlm.nih.gov/genome). F. vesca and Arabidopsis JAZ and MYC TFs sequences were analyzed for structural and functional domains. F. vesca JAZs and MYCs encoding sequences were obtained from F. vesca transcriptome (https://www.ncbi.nlm.nih.gov/refseq) (S1 Table). TIFY and Jas domains were identified through multiple sequence alignment by Clustal Omega [35] and visualized by Jalview software [36]. Logo sequences of JAZ and MYCs domains were obtained using Weblogo 3 [37]. Unrooted phylogenetic trees were built using full-length amino acidic sequences by the neighbor-joining (NJ) method and a bootstrap of 1,000 replicates. Phylogenetic trees were visualized by Evolview tool [38]. We renamed the annotated F. vesca TIFY and MYC sequences according to their homology degree; length, domain location and clustering in phylogenetic tree with the corresponding Arabidopsis sequences.
Plant material and JA treatment
Strawberry (F. × ananassa cv. Aromas) flowers and fruit were collected at different developmental stages from plants grown in a commercial field at Angol, Araucanía Region, Chile (latitude 37°45’18” S; longitude 72°36’49” W) during three different dates in the 2014 growing season. The owner of the land gave permission to conduct the study on this site. The picked flowers and fruit were transported to the laboratory under refrigerated conditions and classified in six developmental stages corresponding to 0 (flowering, F), 10 (small green, SG), 17 (large green, LG), 20 (white, W), 21 (turning, T), 23 (50% red receptacle, 50%R) and 25 (100% red receptacle, 100%R) days after anthesis (DAA) as previously reported [7].
On the other hand, we performed an experiment to verify JA treatment effects on MYC2 and JAZs gene expression during an in vitro fruit ripening system. Peduncles of three fruit at white stage (W) were trimmed to a uniform length of 50 mm and immersed in sterile tubes (50 ml) with autoclaved distilled water containing 88 mM sucrose and 1 mM hydroxyquinoline hemisulfate (HQS) plus 100 μM MeJA according to previously reported [7,9]. The fruit in solution were incubated in a growth chamber under standard fluorescent lights (16 h photoperiod and 40 μmol/m2.s1 light intensity) at 24°C. Fruit sampling was performed at 15 min, 30 min, 1 h and 6 h of MeJA incubation. At each treatment and time, three biological replicates were used for gene expression analysis.
Gene expression analysis
Total RNA was isolated using the CTAB method [39] and mini-columns for RNA purification (RNeasy Plus Mini Kit, Qiagen, Germany). The cDNA synthesis was performed using the RevertAid H Minus First Strand cDNA Synthesis Kit (ThermoScientific, Finland) according to the manufacturer's instructions. Expression analysis of JA signalling-related genes was performed using reverse transcription-qPCR (RT-qPCR). Primer3 and Primer-BLAST tools using full-length CDS sequences of F. vesca as templates were utilized for primer design for each gene. Primers used for RT-qPCR are described in S3 Table and generated single products. FvJAZ4-1, FvJAZ4-2 and FvJAZ4-3 primers were designed in a shared sequence region by the three CDS. RT-qPCR was performed following the instructions of KAPA SYBR FAST qPCR kit (KAPA Biosystems, USA) according to the manufacturer's instructions in a PikoReal Real-Time PCR System (Thermo Scientific, Finland). The expression levels were calculated according to the 2-ΔΔCT method [40] and expressed in relative arbitrary units, normalized according to housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The PCR conditions were as follows: 95°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 15 s and 72°C for 15 s; and a melting curve of 60°C for 30 s, 95°C for 15 s and 20°C for 10 min. All RT-qPCR reactions were performed with three biological and three technical replicates. The Heatmapper software [41] was used to estimate the fold-change of gene expression.
Determination of anthocyanins and proanthocyanidins (PAs) contents
Five grams of frozen fruit skin tissue without achenes were grounded with liquid nitrogen, homogenized with 15 ml of acetone/water (80/20 v/v) and stored at -20°C until use. Anthocyanins quantification was performed according to previously reported [42]. Briefly, 50 μl of fruit extract and 150 μl of corresponding buffer were dispensed into a 96-well plate. The absorbance was measured at 509 nm and 700 nm, considering ε = 17,330 L/cm1.mol1. The results were expressed as mg of pelargonidin-3-glucoside equivalents per 100 g of fresh weight (FW). PAs content was measured according to previously reported [43]. Briefly, 70 μl of fruit extract diluted (1/10, v/v) and 210 μl of dimethylaminocinnamaldehyde (DMAC) reagent were dispensed into wells of a 96-well plate. The microplate was read for 20 min at 640 nm. The concentration was calculated from a calibration curve, using catechin as standard. The results were expressed as mg of catechin equivalents per 100 g of FW.
Results
Identification, chromosomal location and synteny of JAZ and MYC genes in F. vesca
In order to identify ortholog JAZ and MYC genes in F. vesca, a tblastn search was performed using Arabidopsis JAZs and MYCs protein sequences as queries (S1 Table). In F. vesca, we identified and named 12 non-redundant members (FvJAZ1, FvJAZ4-1, FvJAZ4-2, FvJAZ4-3, FvJAZ5, FvJAZ7, FvJAZ8.1, FvJAZ8.2, FvJAZ9, FvJAZ10, FvJAZ11 and FvJAZ12), which are annotated as TIFY proteins in F. vesca database (Table 1, S1 and S4 Tables). Due to the similar number of JAZs in F. vesca and Arabidopsis, we decided to name the F. vesca JAZ according to their well-studied Arabidopsis ortholog genes. Moreover, we identified two MYC transcription factors, FvMYC2 and FvMYC2-like, which are annotated as MYC2 and MYC2-like in F. vesca databases (Table 1, S1 and S5 Tables). However, Arabidopsis MYC3, MYC4 and MYC5 orthologs were not identified in F. vesca databases.
Table 1. Genomic data of JAZ and MYC gene family in woodland strawberry (Fragaria vesca) and Arabidopsis, and their corresponding CDS and protein lengths.
| Gene name a | Gene ID | Chromosome | Start | End | Gene (bp) |
CDS (bp) | ORF (aa) |
|---|---|---|---|---|---|---|---|
| FvJAZ1 / FvTIFY10A | 101302102 | 1 | 6964291 | 6965689 | 1349 | 909 | 302 |
| FvJAZ4-1 / FvTIFY6B.1 | 101298700 | 4 | 19330995 | 19333777 | 1934 | 1158 | 385 |
| FvJAZ4-2 / FvTIFY6B.2 | 101298700 | 4 | 19330995 | 19333777 | 1960 | 1155 | 384 |
| FvJAZ4-3 / FvTIFY6B.3 | 101298700 | 4 | 19330995 | 19333777 | 1701 | 1083 | 360 |
| FvJAZ5 / FvTIFY11A-like | 101305492 | 6 | 24774113 | 24774773 | 1016 | 558 | 185 |
| FvJAZ7 / FvTIFY5B | 105352369 | 6 | 27798266 | 27799489 | 814 | 372 | 123 |
| FvJAZ8.1 / FvTIFY5A | 101295112 | 3 | 2612521 | 2613468 | 862 | 393 | 130 |
| FvJAZ8.2 / FvTIFY5B-like | 105350256 | 3 | 2616234 | 2616765 | 794 | 420 | 139 |
| FvJAZ9 / FvTIFY6B | 101303423 | 5 | 9961254 | 9964696 | 1678 | 1116 | 371 |
| FvJAZ10 / FvTIFY9 | 101299545 | 1 | 1660803 | 1661807 | 1489 | 576 | 191 |
| FvJAZ11 / FvTIFY3A-like | 105349490 | Unknown | 159768 | 161085 | 561 | 561 | 186 |
| FvJAZ12 / FvTIFY3B | 101312185 | 1 | 7358182 | 7359713 | 1172 | 609 | 202 |
| FvMYC2 | 101299702 | 7 | 2955350 | 2956825 | 1955 | 1476 | 491 |
| FvMYC2-like | 101308180 | 5 | 21462454 | 21464502 | 2690 | 2049 | 682 |
| AtJAZ1 / AtTIFY10A | AT1G19180 | 1 | 6622312 | 6623271 | 1892 | 762 | 253 |
| AtJAZ2 / AtTIFY10B | AT1G74950 | 1 | 28148919 | 28150258 | 1873 | 750 | 249 |
| AtJAZ3 / AtTIFY6B | AT3G17860 | 3 | 6119968 | 6122691 | 3338 | 1059 | 352 |
| AtJAZ4 / AtTIFY6A | AT1G48500 | 1 | 17931658 | 17934255 | 3273 | 834 | 310 |
| AtJAZ5 / AtTIFY11A | AT1G17380 | 1 | 5955654 | 5957070 | 2357 | 825 | 274 |
| AtJAZ6 / AtTIFY11B | AT1G72450 | 1 | 27274336 | 27276136 | 2595 | 810 | 269 |
| AtJAZ7 / AtTIFY5B | AT2G34600 | 2 | 14573172 | 14573718 | 923 | 447 | 148 |
| AtJAZ8 / AtTIFY5A | AT1G30135 | 1 | 10596516 | 10597095 | 990 | 396 | 131 |
| AtJAZ9 / AtTIFY7 | AT1G70700 | 1 | 26654951 | 26656804 | 2822 | 804 | 243 |
| AtJAZ10 / AtTIFY9 | AT5G13220 | 5 | 4219001 | 4220502 | 2292 | 504 | 197 |
| AtJAZ11 / AtTIFY3A | AT3G43440 | 3 | 15367670 | 15369774 | 2612 | 717 | 238 |
| AtJAZ12 / AtTIFY3B | AT5G20900 | 5 | 7090883 | 7092201 | 1836 | 564 | 187 |
| AtJAZ13 | AT3G22275 | 3 | 7878807 | 7879810 | 827 | 378 | 125 |
| AtMYC2 | AT1G32640 | 1 | 11799042 | 11800913 | 3289 | 1872 | 623 |
| AtMYC3 | AT5G46760 | 5 | 18974231 | 18976009 | 2569 | 1779 | 592 |
| AtMYC4 | AT4G17880 | 4 | 9933702 | 9935471 | 2360 | 1770 | 589 |
| AtMYC5 | AT5G46830 | 5 | 19002719 | 19004254 | 1821 | 1536 | 511 |
a F. vesca (Fv) and Arabidopsis (At) gene sequences were obtained from the National Center for Biotechnology Information NCBI, https://www.ncbi.nlm.nih.gov/genome) and Arabidopsis database (TAIR10, http://www.arabidopsis.org), respectively.
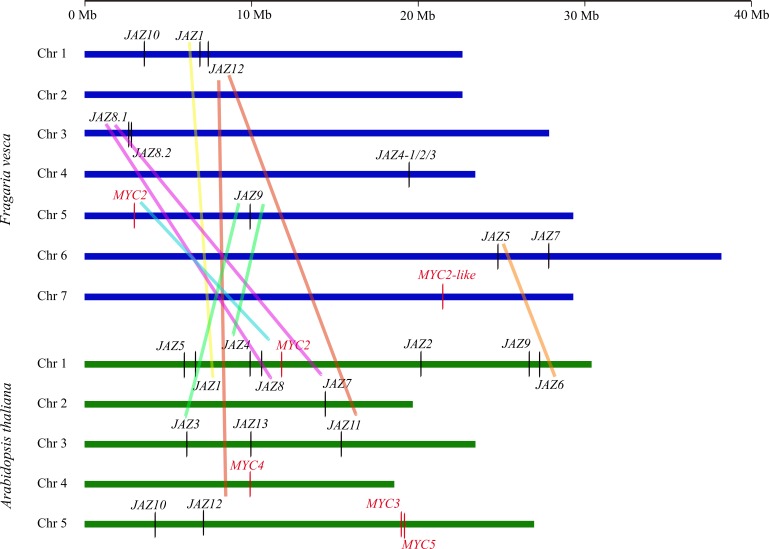
Next, we compared chromosomal locations of JAZ and MYC genes and constructed a schematic representation of their distribution (Fig 1) based on information available from F. vesca and Arabidopsis genome databases (Table 1 and S2 Table). The FvJAZ genes were located on chromosomes 1, 3, 4, 5 and 6, while FvMYC2 and FvMYC2-like did on chromosomes 5 and 7, respectively (Fig 1, Table 1 and S2 Table). However, FvJAZ11 could not be located in F. vesca genome (Fig 1, Table 1 and S2 Table), likely because of F. vesca genome is not completely assembled (https://www.ncbi.nlm.nih.gov/genome/3314). Additionally, we searched and detected conserved syntenic regions corresponding to segmental duplications between JAZ and MYC genes of F. vesca and Arabidopsis using the Plant Genome Duplication Database (PGDD). FvJAZ1, FvJAZ8.1 and FvJAZ12 exhibited synteny with their Arabidopsis ortholog genes and FvJAZ8.1 and FvJAZ12 showed an additional synteny with near ortholog genes AtJAZ7 and AtJAZ11, respectively (Fig 1 and S1 Table). We found a tandemly duplication in F. vesca genome corresponding to FvJAZ8.1 and FvJAZ8.2 genes (Fig 1 and S1 Table). FvJAZ5 and FvJAZ9 showed synteny with AtJAZ6 and AtJAZ3 and AtJAZ4, respectively (Fig 1). Other genes such as FvJAZ4-1, FvJAZ4-2, FvJAZ4-3, FvJAZ7, FvJAZ8.2, FvJAZ10 and FvMYC2-like could not be mapped to syntenic regions into Arabidopsis genome (Fig 1). FvMYC2 showed synteny with its AtMYC2 ortholog gene (Fig 1 and S1 Table). Moreover, chromosome location and synteny analysis of M. × domestica, S. lycopersicum, V. vinifera and O. sativa JAZ and MYC-like genes was performed respect A. thaliana orthologs, using information available in PGDD and species-genomic databases (S1 Fig and S2 Table). V. vinifera displayed higher number of JAZ and MYC-like syntenic genes respect to A. thaliana genes (S1 Fig). An additional non-annotated VvMYC2-like gene showed synteny with AtMYC2 (S1 Fig). In the case of M. × domestica, S. lycopersicum and O. sativa, only four syntenic regions were detected for each one (S1 Fig). In summary, FvJAZ and FvMYC are orthologs and syntenic genes respect to A. thaliana ones, similarly to M. × domestica, V. vinifera, S. lycopersicum and O. sativa.
Fig 1. Genome distribution and synteny of JAZ and MYC genes in Fragaria vesca and Arabidopsis chromosomes.
Chromosomes are indicated as horizontal blue and green bars. JAZs are indicated by black letters and vertical lines and MYCs-like are indicated by red letters and vertical lines. Thick colored lines denote syntenic regions. FvJAZ11 has unknown location. JAZ, jasmonate ZIM-domain.
Exon-intron structure analysis for JAZ and MYC genes in F. vesca
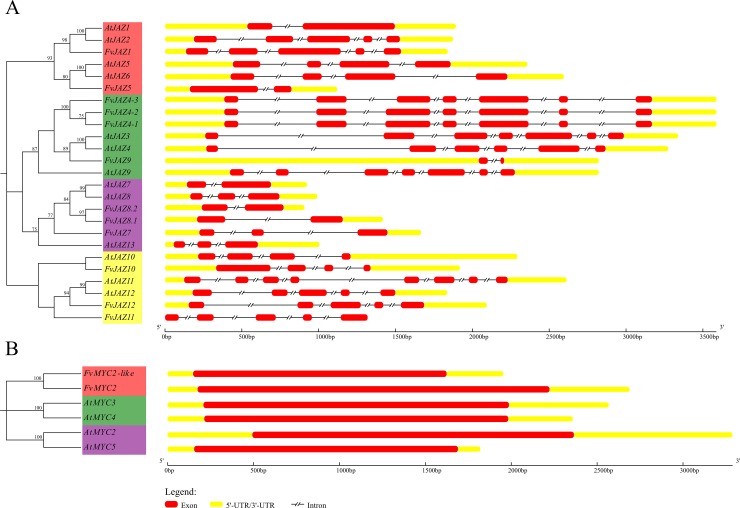
In order to gain insights into the diversification of the JAZs and MYCs, we compared exon-intron organization among all genes of F. vesca and Arabidopsis (Fig 2). We observed that FvJAZs and AtJAZs contain a variable intron numbers (Fig 2A); in contrast, all FvMYC and AtMYC genes lack introns (Fig 2B). We detected variations in length and number of introns in FvJAZ genes. FvJAZ10 and FvJAZ12 maintained the number of introns, although with length variations, with respect to Arabidopsis orthologs (Fig 2A). Other FvJAZ genes such as FvJAZ4s, FvJAZ5, FvJAZ7, FvJAZ8.1, FvJAZ8.2 and FvJAZ9 showed variable numbers of introns from one to six (Fig 2A). On the other hand, FvJAZ1 gained three introns and FvJAZ4s and FvJAZ7 genes gained one intron each one; while FvJAZ5, FvJAZ8.1, FvJAZ9 and FvJAZ11 lost two, one, five and three introns, respectively, regarding their Arabidopsis ortholog genes (Fig 2A). Moreover, FvMYC2 and FvMYC2-like lack introns (Fig 2B) as well as A. thaliana orthologs. Overall, some FvJAZ genes display a variable exon-intron organization, while FvMYC2 and FvMYC2-like genes keep the lack of introns.
Fig 2. Exon-intron structures of the Fragaria vesca and Arabidopsis JAZ and MYC genes.
Exon-intron organization of JAZ genes (A) and MYC genes (B) in F. vesca (Fv) and Arabidopsis thaliana (At) grouped according their gene orthology. Yellow and red bars indicate untranslated (UTR) regions and exons, respectively. Black interrupted lines indicate introns. JAZ, jasmonate ZIM-domain.
Conserved domains of JAZ and MYC proteins in F. vesca
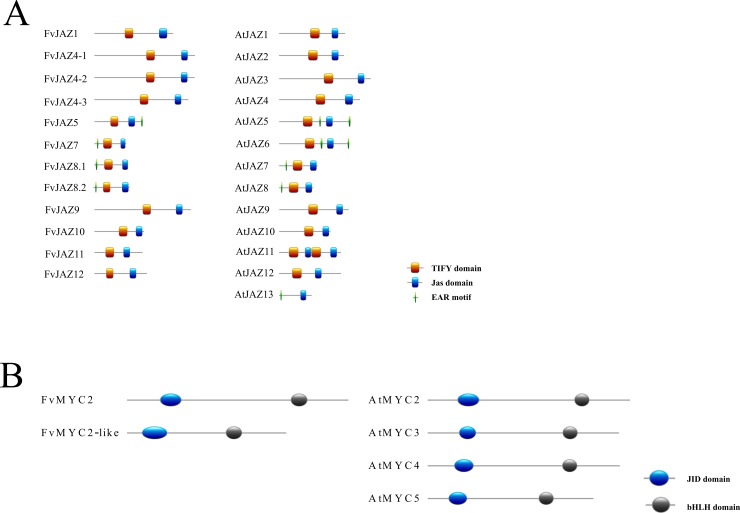
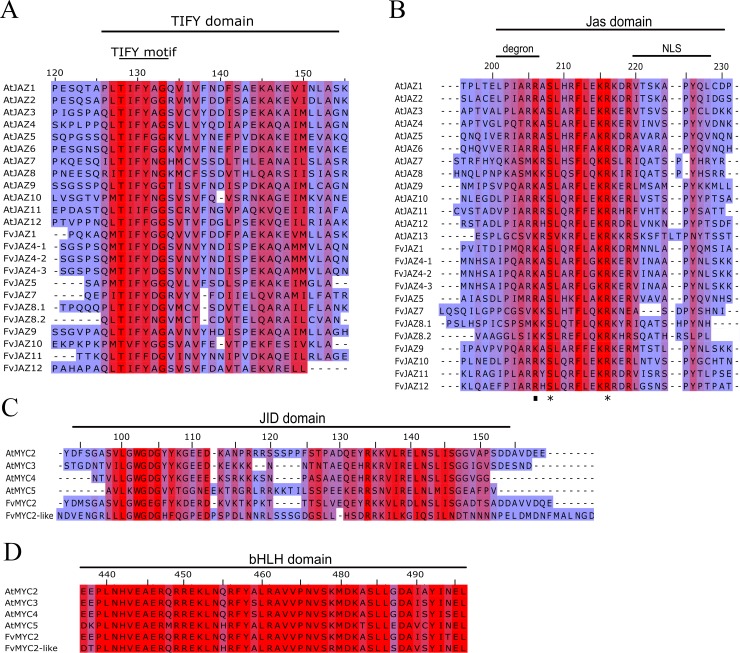
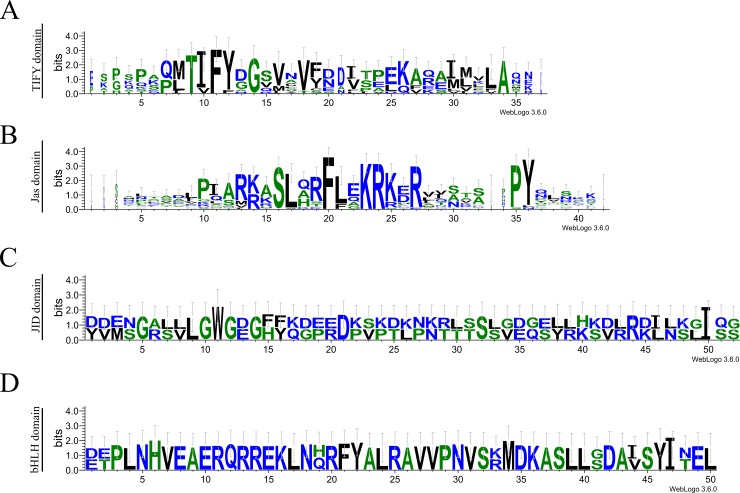
To confirm evolutionary relationships and gain further insights in the primary protein structure between JAZ proteins, the domain and motifs positions were evaluated (Figs 3 and 4). FvJAZ proteins showed a length range from 123 to 385 amino acid residues according to that observed in Arabidopsis (Table 1 and S6 Table). The distribution of domains is similar between protein sequences of F. vesca and their orthologs in Arabidopsis (Fig 3A). Analysis of the deduced amino acid sequence of F. vesca JAZ showed the conservation of the TIFY/ZIM and Jas domains (Fig 4A and 4B). In F. vesca, the TIFY domain displayed the highly conserved TIFY[F/Y]XG motif (Fig 4A), although with TVFYXG and TIFFXG variants present in FvJAZ10 and FvJAZ11, respectively (Fig 4A). Jas domain displayed conserved sequence SLX2FLXKR[K/R]X[R/E] like consensus sequence in F. vesca (Fig 4B). Although some proteins like FvJAZ7, FvJAZ8.1 and FvJAZ8.2 showed a variant Jas sequence (Fig 4B and S3 Fig). FvJAZ10 exhibited the canonical degron LPIARK whereas other F. vesca JAZ proteins exhibited variations in degron amino acid residues such as IPMQRK in FvJAZ1, IPQARK in FvJAZ4s, LPIMRR in FvJAZ5, VPQARK in FvJAZ9, IPLARR in FvJAZ11 and FPIARR in FvJAZ12 (Fig 4B). FvJAZ11 did not show the duplicated TIFY and Jas domains that are present in AtJAZ11 (Fig 3A). On the other hand, C-terminal X5PYX2 region, which may act as Nuclear Localization Signal (NLS) exhibited conservation among JAZ proteins of F. vesca (Fig 4B). In this regard, FvJAZ5 showed an EAR-motif at the C-terminus, while FvJAZ7, FvJAZ8.1 and FvJAZ8.2 displayed this motif at the N-terminus (Fig 3A and S2 Fig). In the case of FvJAZ5, we noticed that it lacks the DLNEPT motif or similar sequence (Panel A in S2 Fig). TIFY and Jas domain logo sequences showed a highly residue conservation in FvJAZ proteins (Fig 5A and 5B). On the other hand, domain structure and sequences analysis of FvMYC TFs were evaluated and compared with their Arabidopsis orthologs (Figs 3B, 4C and 4D). FvMYC2 and FvMYC2-like showed two conserved domains corresponding to JID and bHLH and displayed conserved position and different domain lengths (Figs 3B, 4C and 4D). Deduced consensus sequences of FvMYC2 and FvMYC2-like JID domain showed conservation between some amino acidic residues (Fig 4C), while bHLH domain displayed highly similarity between MYC-like proteins (Fig 4D). FvMYC2-like contain amino acidic residues more similar to AtMYC5 (Fig 4D). The logo sequences of JID and bHLH domains indicated high conservation of residues in FvMYC proteins (Fig 5C and 5D). Globally, FvJAZs and FvMYCs contain domains highly conserved, with similar domain locations and protein lengths.
Fig 3. Distribution of JAZ and MYC protein domains and motifs in Fragaria vesca and Arabidopsis.
Comparative distribution of TIFY, Jas and EAR domains and motifs in JAZ proteins (A) and comparative distributions of JID and bHLH domains in MYC-like proteins (B) of Fragaria vesca (Fv) and Arabidopsis thaliana (At). The relative position of each domain within each protein are displayed in colors. bHLH, basic helix-loop-helix; EAR, ethylene-responsive element binding factor-associated amphiphilic repression; JAZ, jasmonate ZIM-domain; JID, JAZ-interacting domain.
Fig 4. Multiple alignment of JAZ and MYC protein domains in Fragaria vesca and Arabidopsis.
Multiple sequence alignment of TIFY (A), Jas (B), JID (C) and bHLH (D) domains of Arabidopsis and putative F. vesca JAZ and MYC sequences. Red and blue colors indicate higher and lower amino acidic residues conservation, respectively. Dot and asterisks indicate conserved residues involved in interaction with COI1 and MYCs, respectively. bHLH, basic helix-loop-helix; JAZ, jasmonate ZIM-domain; JID, JAZ-interacting domain; NLS, nuclear localization signal.
Fig 5. Logo sequences for FvJAZ and FvMYC proteins.
Logo sequences for TIFY (A), and Jas (B) domains of FvJAZ proteins, and JID (C) and bHLH (D) domains of FvMYC proteins. bHLH, basic helix-loop-helix; JID, JAZ-interacting domain; JAZ, jasmonate ZIM-domain.
Phylogenetic analysis of F. vesca JAZ and MYC2-like proteins
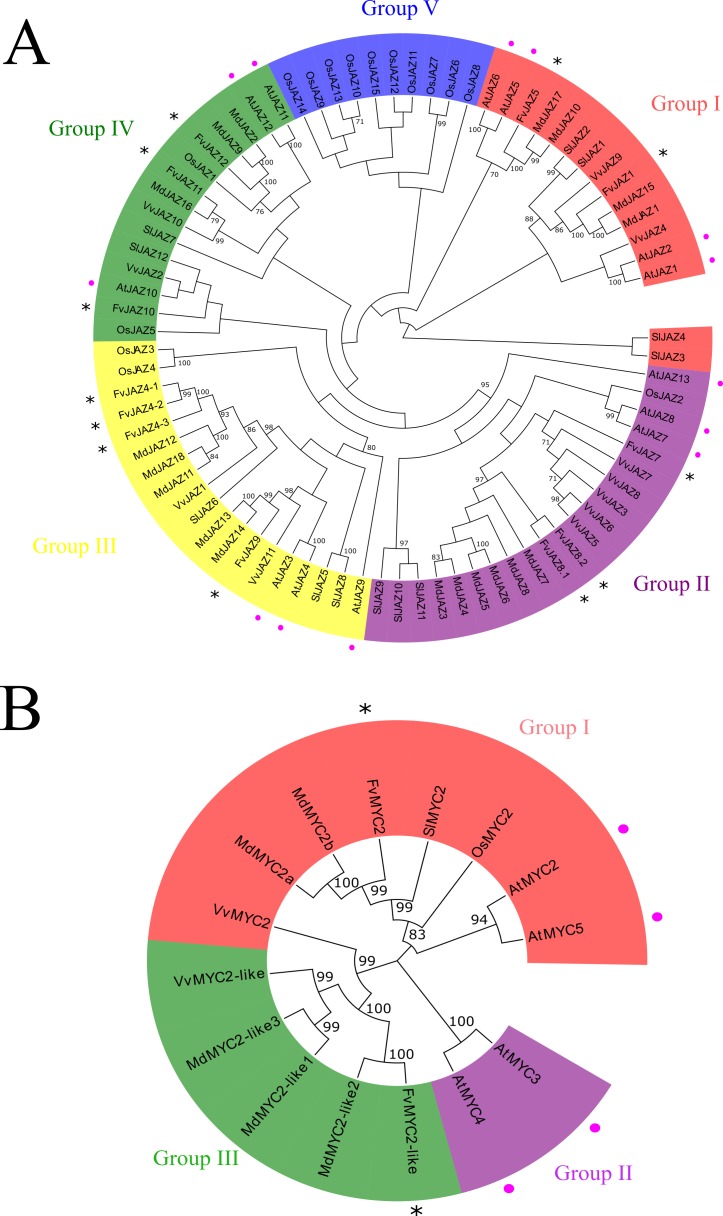
Unrooted phylogenetic trees were generated by neighbor-joining (NJ) algorithm by bootstrap of 1,000 replicates, showing evolutionary relationships between the JAZ and MYC2-like proteins of the dicot plants such as A. thaliana, F. vesca, M. × domestica, V. vinifera, S. lycopersicum and the monocot O. sativa. JAZ proteins were clustered in five groups (Fig 6). Regarding to the JAZ protein subfamilies, FvJAZ1 and FvJAZ5 were clustered together with AtJAZ1, AtJAZ2, AtJAZ5 and AtJAZ6 proteins into group I together some M. × domestica, V. vinifera and S. lycopersicum ortholog proteins (Fig 6A) and showed 40.1–43.9% identity with the A. thaliana orthologs AtJAZ1 and AtJAZ5, respectively (S4 Table). Otherwise, in group II, FvJAZ7, FvJAZ8.2 and FvJAZ8.1 clustered together (Fig 6A) and showed between 36.8–47% identity with their corresponding A. thaliana orthologs (S4 Table), and clustered in the same group of the non-TIFY protein AtJAZ13 and M. × domestica, V. vinifera, S. lycopersicum JAZs and OsJAZ2 proteins (Fig 6A). On the other hand, FvJAZ4s and FvJAZ9 were grouped into group III together with AtJAZ3, AtJAZ4 and AtJAZ9 including JAZ proteins from other species (Fig 6A). Finally, A. thaliana and F. vesca JAZ10, JAZ11 and JAZ12 proteins were clustered in the group IV (Fig 6A). Nevertheless, FvJAZ10 and FvJAZ11 showed higher similarity with AtJAZ4 on the sequence identity matrix (S4 Table), which was not reproduced in the phylogenetic tree position (Fig 6A). Additionally, some O. sativa JAZ proteins were clustered together in group V displaying divergence with dicots-associated JAZs (Fig 6A). Bootstrap values showed (> 70%) indicated high reliability in most clusters (Fig 6A), and values ≥ 95% represented groupings with the higher confidence.
Fig 6. Phylogenetic analysis of Fragaria vesca JAZ and MYC proteins.
Phylogenetic tree of JAZ (A) and MYC-like proteins sequences (B) from Fragaria vesca (Fv), Arabidopsis thaliana (At), Malus × domestica (Md), Vitis vinifera (Vv), Solanum lycopersicum (Sl) and Oryza sativa (Os). The phylogenetic analysis was performed using full-length JAZ and MYC protein sequences. FvJAZ and FvMYC2-like, and AtJAZ and AtMYC-like proteins are indicated by asterisk and pink dots, respectively. Nodes with bootstrap values > 70% are labelled and bootstrap values ≥ 95% show highlight bootstrap. JAZ, jasmonate ZIM-domain.
To analyze the evolutionary relationships between MYC-like TFs of F. vesca, A. thaliana, M. × domestica, V. vinifera, S. lycopersicum and O. sativa, an unrooted phylogenetic tree was performed (Fig 6B). FvMYC2 showed the highest identity (58.5%) with AtMYC2 (S5 Table) and it was clustered in the group I close to AtMYC2 in the phylogenetic tree (Fig 6B). Other MYC2 TFs corresponding to M. × domestica, S. lycopersicum and O. sativa were also clustered in this group (Fig 6B). On the other hand, FvMYC2-like was grouped into group II along with VvMYC2-like TFs (Fig 6B). AtMYC3 and AtMYC4 were clustered together in a group III (Fig 6B). Phylogenetic tree nodes showed bootstrap values ≥ 95%, except for OsMYC2, exhibiting high confidence levels (Fig 6B). In summary, FvJAZ, FvMYC2 and FvMYC2-like proteins share high similarity and show closer evolutionary relationships within dicots (F. vesca, A. thaliana, M. × domestica, S. lycopersicum and V. vinifera).
Expression of FaJAZ and FaMYC genes during development and ripening of F. × ananassa fruit
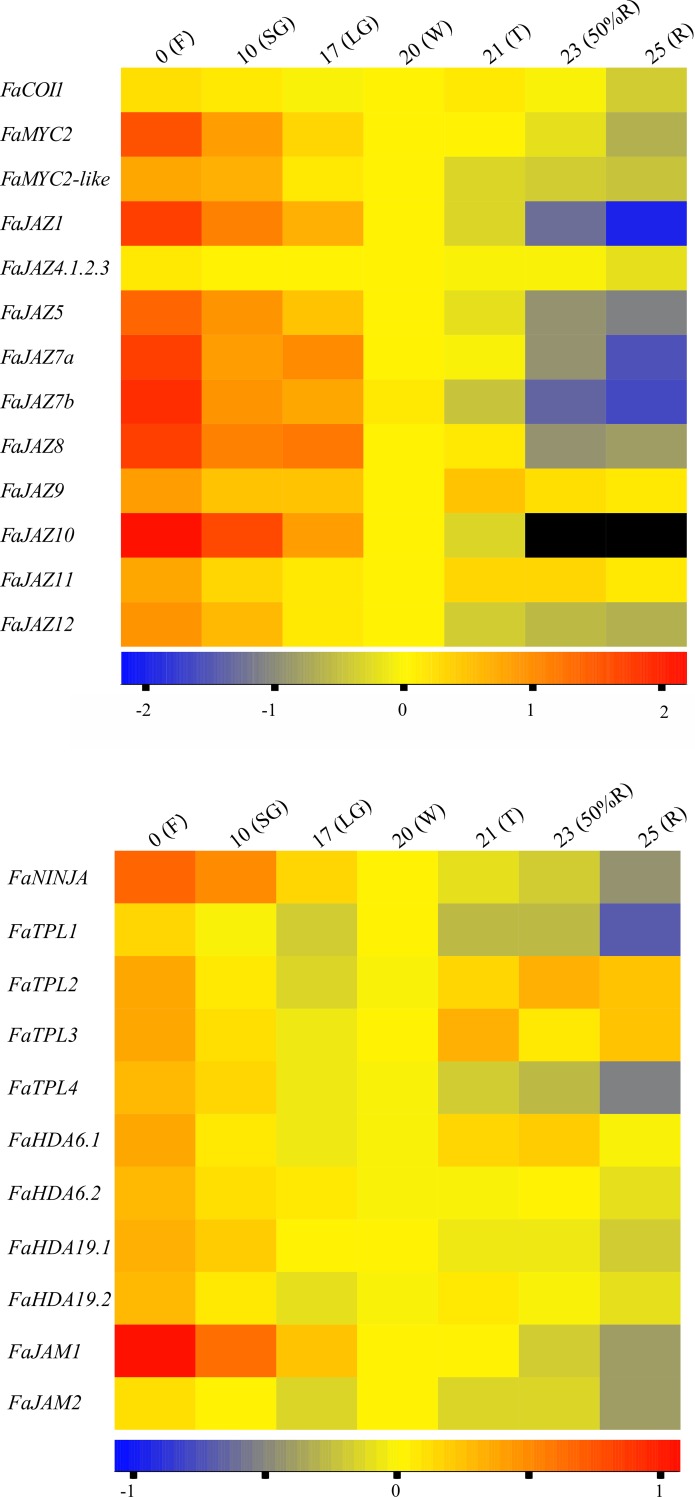
To determine the expression dynamics of FvJAZ and FvMYC genes during fruit development and ripening, RT-qPCR assays were performed, and expression changes were represented by heatmaps (Fig 7A, S7 and S8 Tables). We included in the analysis the JA-Ile co-receptor COI1-encoding gene (FaCOI1), which showed two peaks in expression levels at F and T stages during fruit development and ripening (Fig 7A). Overall, F. × ananassa JAZ encoding genes (FaJAZs) were downregulated from 0 to 25 DAA (F to R stages) (Fig 7A). Notably, the expression of FaJAZ1, FaJAZ5 and FaJAZ8.1 exhibited the highest reduction by 1043, 125 and 157-fold (p-value ≤ 0.05), respectively, from 0 to 25 DAA stages (F to R stages) (Fig 7A and S7 Table). These genes showed a similar expression pattern consisting in a reduction by 3.6, 2.3 and 3.4-fold (p-value ≤ 0.05) from 0 to 10 DAA (F to SG stages), a steady level from 10 to 21 DAA (SG to T stages), and then a reduction by 6.2, 4.3 and 6.8-fold (p-value ≤ 0.05), respectively, to 23 DAA (50%R stage) (Fig 7A and S7 Table). In contrast, the expression of FaJAZ10 and FaJAZ12 presented a similar reduction pattern from 0 to 25 DAA stages (F to R stages), being this reduction more pronounced for FaJAZ10 with undetected levels at 23 and 25 DAA (50%R and R stages) (Fig 7A and S7 Table). Other JAZ genes (e.g., FaJAZ4s, FaJAZ8.2 FaJAZ9, FaJAZ11) also exhibited a higher expression at F stage and then lower constant levels during fruit development and ripening (Fig 7A). FaMYC2 and FaMYC2-like showed an expression reduction during fruit development in a similar way to that observed for FaJAZs (Fig 7A) although a greater expression decline was observed for FaMYC2 than FaMYC2-like between 0 and 10 DAA (F and SG stages) (Fig 7A and S7 Table). Higher relative expression levels were observed for FaMYC2 and FaMYC2-like in 0 and 10 DAA (F and SG stages) and a constant decrease was registered through fruit development and ripening (Fig 7A and S7 Table).
Fig 7. Expression heatmaps of Fragaria × ananassa JA signalling-related genes during fruit development and ripening.
Expression heatmaps of FaCOI1, FaMYCs, FaJAZs (A) and FaNINJA, FaTPLs, FaHDAs, FaJAMs (B). The Log-transformed values of relative expression levels based on RT-qPCR assays were used to perform heatmaps. The color scale represents relative expression levels with red and blue colors as high and low values, respectively. Black means no detection. The expression level of FaGAPDH was used as reference gene to normalize each reaction. The data was from three biological and three technical replicates. Developmental stages correspond to 0 (flowering, F), 10 (small green, SG), 17 (large green, LG), 20 (white, W), 21 (turning, T), 23 (50% red receptacle, 50%R) and 25 (100% red receptacle, 100%R) days after anthesis (DAA) in F. × ananassa cv. Aromas. JAZ, jasmonate ZIM-Domain.
Expression of other JA signalling-related genes during development and ripening in F. × ananassa fruit
To characterize temporal expression of additional components of the JA-related repressor machinery during F. × ananassa fruit development, we analyzed the expression levels of the NINJA adaptor-, TPL-, HDA co-repressors- and JAM-encoding genes (Fig 7B, S7 and S8 Tables). FaNINJA showing a higher level at 0 and 10 DAA (F and SG stages) a then progressively declined to 25 DAA (R stage) (Fig 7B and S7 Table). During F. × ananassa fruit development, different expression patterns were detected for FaTPL1, FaTPL2, FaTPL3, FaTPL4, FaHDA6.1, FaHDA6.2, FaHDA19.1 and FaHDA19.2 (Fig 7B and S7 Table). We also observed a constant reduction of FaJAM1 and FaJAM2 transcript accumulation through fruit development and ripening (Fig 7B) in accordance to the expression pattern of FaMYCs (Fig 7A). Overall, F. × ananassa JAZs, MYC2, MYC2-like and other JA-signalling related genes are downregulated during fruit development and ripening.
Expression of FaJAZ1, FaJAZ8.1 and FaMYC2 under JA treatment
To gain insights into the expression response of F. × ananassa JAZ and MYC genes to JA treatment, we evaluated the expression profiles for FaMYC2, FaJAZ1 and FaJAZ8.1 in MeJA-treated W stage fruit (Fig 8 and S9 Table). We selected FaJAZ1 and FaJAZ8.1 because they showed the highest reduction in expression levels from 21 to 25 DAA (T and R stages) (Fig 7A), when F. × ananassa (cv. Aromas) starts the anthocyanin accumulation (S4 Fig).
Fig 8. Expression of FaJAZ1, FaJAZ8.1 and FaMYC2 under MeJA treatment in Fragaria × ananassa fruit.
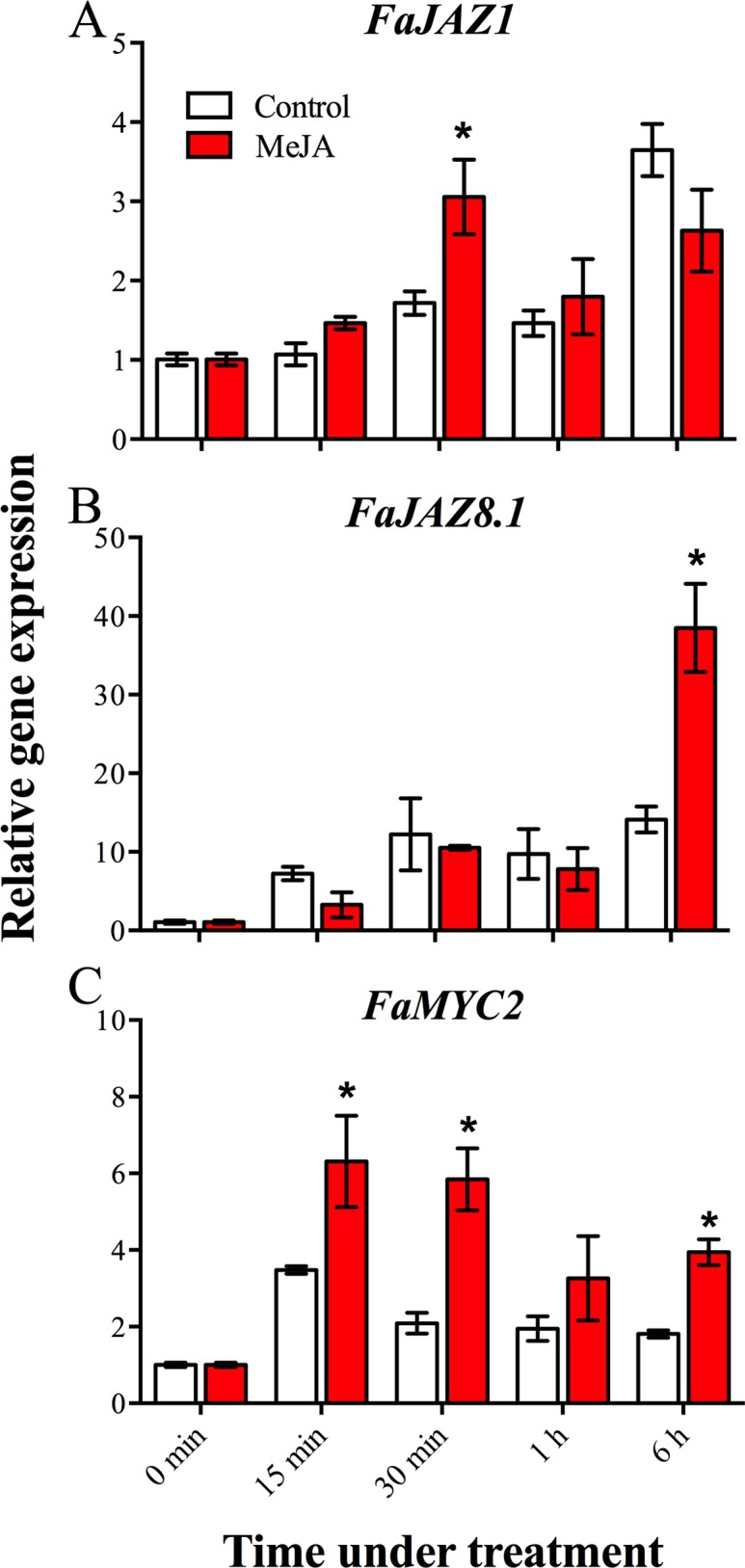
Changes in relative expression of FaJAZ1 (A), FaJAZ8.1 (B) and FaMYC2 (C) genes at 15 min, 30 min, 1 h and 6 h under 100 μM MeJA treatment. The expression level of FaGAPDH was used as reference gene to normalize each reaction. The data are from three biological and three technical replicates. Data were subjected to one-way ANOVA test, differences among means ± SE (n = 3) were determined using LSD test. Different letters indicate significant differences between developmental stages (p ≤ 0.05). JAZ, jasmonate ZIM-domain.
FaJAZ1 increased its expression 1.4-fold (p-value ≤ 0.05) respect to control after 30 min of 100 μM MeJA application in fruit (Fig 8A and S9 Table). FaJAZ8.1 showed an upregulation of 2.7-fold (p-value ≤ 0.05) at 6 h after MeJA treatment with respect to a control (Fig 8B and S9 Table). On the other hand, FaMYC2 exhibited a significant upregulation of 1.8, 2.8 and 2.2-fold (p-value ≤ 0.05) at 15 min, 30 min and 6 h after MeJA treatment, respectively (Fig 8C and S9 Table). These results demonstrate that FaJAZ1, FaJAZ8.1 and FaMYC2 are JA-responsive genes in F. × ananassa fruit. Moreover, FaMYC2 responds earlier than FaJAZ1 and FaJAZ8.1 (Fig 8).
Molecular characterization and expression of PPDs in strawberry
To gain insights into the existence of non-JAZ TIFY proteins in strawberry, we characterized inferred amino acid sequences containing conserved domains for TIFY and Jas. In this sense, we identified two PEAPOD (PPD) genes in F. vesca genome using AtPPD ortholog protein sequences as queries (S1 Table), named as FvPPD1-1 and FvPPD1-2 according to the gene nomenclature proposed for the Rosaceae [44]. We compared exon-intron structures between F. vesca and A. thaliana PPD genes and observed that FvPPD1-1 and FvPPD1-2 have an extra intron and longer introns in comparison to those of A. thaliana orthologs (Panel A in S5 Fig). FvPPDs proteins showed conserved position of PPD, TIFY and degenerated Jas domains respect to those present in AtPPD proteins (Panel B in S5 Fig) along with a high identity sequence as we observed from multiple sequence alignment analysis (Panel C in S5 Fig). Additionally, we constructed a phylogenetic tree that showed evolutionary relationships between FvPPDs and A. thaliana, M. x domestica, V. vinifera and S. lycopersicum orthologs (Panel D in S5 Fig). TIFY family was characterized in O. sativa, however PPDs were not reported [45]. FvPPD1-1 and FvPPD1-2 were clustered along with MdPPDs and VvPPD1 in group II (Panel D in S5 Fig). Finally, we determined the relative expression levels of FaPPD1-1, displaying a decreasing pattern during fruit development and ripening (Panel E in S5 Fig) as we observed for most of the JA signalling-related genes in strawberry.
Discussion
FvJAZ, FvMYC2 and FvMYC2-like genes conserve synteny in F. vesca genome
JAZ and MYC along with COI1 co-receptor establish the core of JA signalling pathway [10,46]. Eleven to eighteen JAZ genes have been identified in the genome of higher plant species: thirteen JAZ protein members belonging to TIFY family in Arabidopsis [10,17], 15 members in rice [45], 14 members in wheat [47], 13 members in tomato [48], 18 members in apple and bamboo [49,50] and 11 members in grape [51]. We identified 12 non-redundant JAZ genes in F. vesca genome, FvJAZ1, FvJAZ4-1, FvJAZ4-2, FvJAZ4-3, FvJAZ5, FvJAZ7, FvJAZ8.1, FvJAZ8.2, FvJAZ9, FvJAZ10, FvJAZ11 and FvJAZ12 (Table 1). On the other hand, there are four MYC members involved in the JA signalling pathway in Arabidopsis: MYC2, MYC3, MYC4 and MYC5 [23–25]. Nevertheless, we only found two genes encoding for MYC TFs in F. vesca genome: MYC2 and MYC2-like (Table 1). The number of MYC2-like genes is variable between species, for instance M. × domestica contains five MYC2–like TFs (S2 Table) [52,53], but others species such as V. vinifera, Nicotiana tabacum, Nicotiana attenuata, Salvia miltiorrhiza, and S. lycopersicum contain two MYC2-like TFs [51,54–56] like in F. vesca (Table 1 and S2 Table). In the case of apple, which belongs to Rosaceae family and is evolutionary related to strawberry, contains five MYC2-like encoding genes defined as MdMYC2a [52], MdMYC2b, MdMYC2-like1, MdMYC2-like2 and MdMYC2-like3 [53].
Tandem, segmental and whole duplication are key processes in the expansion of gene families [57,58] and genome comparisons provide information about roles and evolutionary relationships between genes [59]. Tandemly duplicated genes were considered as adjacent homologous in the same chromosome according to observed in rice and apple [45,50]. Specifically, gene duplications play an important role in expansion of the TIFY family [21], to which belongs JAZ subfamily as observed for JAZ7, JAZ8 and JAZ9, JAZ10 in V. vinifera and S. lycopersicum genomes, respectively [48,51], similar to that observed in apple JAZ genes [50]. These results indicate that FvJAZ, AtJAZ, FvMYC2 and AtMYC2 syntenic genes share likely a common ancestor and tandemly and segmental gene duplications were important for the expansion of JAZ subfamily [50]. In the case of FvPPD genes, they did not show syntenic regions within A. thaliana genome.
Exon-intron organization plays a role in diversification and evolution of gene families through gain/loss and insertion/deletions [58]. FvJAZ genes showed variable lengths and number of introns with their respective Arabidopsis orthologs (Fig 2A). These differences could be a consequence of rearrangements and fusions similar to that observed in apple TIFY gene family [50]. The presence of introns allows expanding the repertoire of some JAZ proteins, as reported for the different splice variants for AtJAZ10 with different stability in their encoded proteins and roles in JA responses [60]. In contrast to FvJAZ genes, FvMYC2 and FvMYC2-like genes lack introns as Arabidopsis MYC-like orthologs (Fig 2B). In some cases, introns could have additional functions related to gene expression regulation [61], and this could be related with a key role in JAs responses [62–64], because their absence could be related with faster and efficient expression [65].
FvJAZ proteins show conserved TIFY and Jas domains
To gain further insights in the primary protein structure and evolutionary relationships in JAZ and MYC protein families, multiple sequence alignment and phylogenetic analyses were performed (Figs 3–6, S2 and S3 Figs). FvJAZ proteins exhibited similar length and conserved structure according to that observed in Arabidopsis (Table 1) and previously reported in V. vinifera [51]. Analysis of the deduced amino acid sequence of F. vesca JAZs showed the conservation of the TIFY/ZIM domain that characterize this family (Figs 4A and 5A) [18,21] and a Jas domain (Figs 4B and 5B), which is specific of JAZ subfamily [10,20,21]. In F. vesca, the TIFY domain contains the highly conserved TIFY[F/Y]XG motif [18] (Figs 4A and 5A) as consensus sequence, however, some alternative sequences like TVFYXG and TIFFXG were found in FvJAZ10 and FvJAZ11, respectively (Fig 4A), as previously described for TIFY proteins in other species [21]. Jas domain maintained the conserved central amino acidic residues SLX2FLXKR[K/R]X[R/E] according to consensus sequence observed in F. vesca (Figs 4B and 5B) and similar to the previously reported SLX2FX2KRX2R sequence in Arabidopsis [20]. Besides, FvJAZ7, FvJAZ8.1 and FvJAZ8.2 displayed a variant Jas sequence (Fig 4B and S3 Fig) similar to that observed in Arabidopsis for JAZ7 and JAZ8 proteins [66]. Most of Arabidopsis JAZs contain a degron sequence at the Jas domain that is necessary for interaction with COI1 and JA-Ile [12], but only FvJAZ10 showed the canonical degron LPIAR(R/K) previously described for AtJAZ1, AtJAZ2, AtJAZ10 and AtJAZ12 proteins [66]. Other FvJAZ proteins displayed variations in degron sequences such as IPMQRK in FvJAZ1 (Fig 4B). Moreover, the degron sequence displayed the conserved residues R(K), and S and R (Fig 4B) interacting with COI1 [12] and MYC3 [67], respectively. Therefore, NLS located in C-terminal region [68] showed amino acidic residues conserved among JAZ proteins of F. vesca (Figs 4B and 5B). In some Arabidopsis JAZs, the N-terminal LxLxL type of EAR motif allows recruitment of TPL co-repressors to repress JA signalling pathway through a NINJA independent molecular mechanism [66]. In this regard, FvJAZ5 displayed the LxLxL type of EAR-motif at C-terminus, and FvJAZ7, FvJAZ8.1 and FvJAZ8.2 presented it at the N-terminus (S2 Fig) similar to that reported for AtJAZ5 and AtJAZ6, and AtJAZ7 and AtJAZ8 proteins, respectively (Fig 3) [66,69]. FvJAZ5 showed the lack of DLNEPT type of EAR-motif (Panel A in S2 Fig) in N-terminal region previously described in AtJAZ5 and AtJAZ6 proteins [66,69]. Phylogenetic analysis displayed evolutionary relationships between F. vesca, A. thaliana, M. × domestica, V. vinifera, S. lycopersicum and O. sativa JAZ proteins (Fig 6A). Protein grouping of FvJAZ proteins are related with high sequence identity respect to AtJAZ proteins (Fig 3A and S4 Table). The FvJAZ proteins clustered together with their Arabidopsis orthologs in a similar fashion than observed in several plant species [45,48,50,51], however, some phylogenetic trees contain lower bootstrap values [51]. At the same time, protein positions of FvJAZs in tree groups (Fig 6A) is in agreement with the identity and synteny analyses (Fig 1 and S1 Fig).
FvMYC2 and FvMYC2-like proteins showed high conservation in JID and bHLH domains
In Arabidopsis, MYC2 [64], MYC3, MYC4 [23,70] and MYC5 [25] are master regulators of JA responses. We analyzed conservation domain in FvMYC2 and FvMYC2-like proteins and compared with AtMYC TFs (Fig 3B and S6 Table). The JID domain, which interacts with Jas domain to regulate JA responses [23,27], was present and conserved in FvMYC2 and FvMYC2-like proteins (Figs 3B, 4C and 5C). Both proteins exhibited conserved bHLH domains (Figs 3B, 4D and 5D), which bind to G-box cis elements in JA-response promoters [23–25]. The observed close evolutionary distances, the higher similarity in the identity matrix (S5 Table), and similar sequence lengths and domain positions (Fig 3B and S6 Table) suggest that FvMYC2 is the ortholog TF to AtMYC2. On the other hand, FvMYC2-like is phylogenetically closer to other MYC2-like TFs such as VvMYC2-like or MdMYC2-like TFs (Fig 6B). Furthermore, AtMYC3 and AtMYC4 were clustered as an independent group without ortholog sequences observed in F. vesca genome (Fig 6B) as previously reported in N. attenuata and S. miltiorrhiza, which MYC2 has been grouped in different clusters respect to MYC3 and MYC4 proteins [55,56].
FaMYC2, FaJAZ1 and FaJAZ8.1 are downregulated during development and ripening in F. × ananassa fruit
We analyzed the expression profiles of JAZs, MYC2, MYC2-like and other JA signalling-related genes during fruit development and ripening in F. × ananassa, the worldwide cultivated strawberry species. Most molecular studies on this species have been performed based on F. vesca reference genome [32] which is a subgenome of F. × ananassa [71]. Expression of the JA-Ile co-receptor FaCOI1 (Fig 7A) was similar to that previously reported in F. × ananassa cv. Elsanta fruit [72]. In general, FaJAZ genes displayed a constant reduction pattern from flowering to ripe fruits, and some genes like FaJAZ1, FaJAZ5 and FaJAZ8.1 showed a pronounced reduction at ripe stages (Fig 7A), according to proanthocyanidins (PAs) reduction and opposite to anthocyanin accumulation during fruit development and ripening of cv. Aromas (S4 Fig). On the other hand, FaJAZ10 was not detected in full ripe fruit (Fig 7A). Recently, similar expression profiles for JAZ genes were reported for a JAZ gene from large green to partial red fruit stages in F. × ananassa cv. Hongyan [31]. In this sense, Sánchez-Sevilla et al. also reported expression reduction for the most JAZ, named like TIFY genes, from RNAseq experiments in achene and receptacle during strawberry (F. × ananassa cv. Camarosa) fruit development and ripening (Panel A in S6 Fig and S10 Table) [73]. Moreover, FaMYC2 and FaMYC2-like genes exhibiting an expression reduction from flowering to ripe stages, similar to that observed for FaJAZs (Fig 7A) and to that previously reported both in achene and receptacle (Panel A in S6 Fig and S10 Table) [73]. MYC2-like genes have been reported in tomato and grape, although their expression patterns in fruit stages are unknown [54,74]. Globally, downregulation of FaJAZs and FaMYC2 genes (Fig 7A) matches with previously reported JA-Ile endogenous levels and expression of JA metabolism-related genes during F. × ananassa fruit development and ripening [7]. This suggests that JA metabolism and signalling are in coordination and JA pathway could have a key role in anthesis and physiological events like PAs biosynthesis that occur during early fruit development in strawberry (S4 Fig).
Other JA-signalling genes are downregulated during development and ripening of F. × ananassa fruits
Other components of JA-signalling pathway like NINJA, TPLs and HDAs are key for JAs responses [10]. NINJA is necessary for transcriptional repression by interaction with JAZ proteins and the recruitment of TPL proteins [14,20]. FaNINJA showed the same expression pattern as FaJAZ, FaMYC2 and FaMYC2-like genes during fruit development and ripening (Fig 7), like to previously observed for this gene in RNAseq assays (Panel B in S6 Fig and S10 Table) [73]. Otherwise, different patterns of expression were detected for TPL genes (Fig 7B) and these patterns were similar to those reported for TPL genes in S. lycopersicum fruit [75]. However, TPL gene expression seems to depend of the fruit tissue (Panel B in S6 Fig and S10 Table) [73]. Moreover, HDAs are recruited by TPLs during JAZ-mediated transcriptional repression [76], and FaHDA6 and FaHDA19 expression levels were similar to FaTPL genes (Fig 7B), suggesting a possible transcriptional coordination between both gene families. Finally, FaJAM1 and FaJAM2 exhibited a reduction expression during fruit development and ripening (Fig 7B), according to observed for FaMYC2 and FvMYC2-like genes (Fig 7A) and expression levels previously reported for F. × ananassa JAM1 and JAM2 genes in receptacle by RNAseq assays (Panel B in S6 Fig and S10 Table) [73]. This fact is related with the antagonistic effect of JAMs on MYC2 TFs and the negative regulation of JA responses [28,29]. These results indicate that JA signalling pathway is turned off during development and ripening of F. × ananassa fruit according to the reduction of JA-Ile endogenous levels [7].
FaJAZ1, FaJAZ8.1 and FaMYC2 are JA-responsive genes in F. × ananassa fruit
JAZ and MYC2 genes are JA-responsive genes in Arabidopsis leaves [14]. We evaluated FaJAZ1, FaJAZ8.1 and FaMYC2 expression under 6 h of MeJA treatment. We selected FaJAZ1 and FaJAZ8.1 because they showed the highest reduction in expression levels from turning to ripe stages (Fig 7A), when F. × ananassa cv. Aromas starts the anthocyanin accumulation (S4 Fig). These genes also showed an expression pattern concomitant with the PAs accumulation pattern of developing fruit (S4 Fig). Moreover, we selected FaJAZ1 and FaJAZ8.1 since their deduced amino acid sequences are structurally different: FaJAZ1 contains a variation of the canonical degron sequence (IPMQRK) in contrast to FaJAZ8.1, which lacks canonical degron (Fig 4B) as well as AtJAZ8 [66]. FaJAZ1 raised its expression levels after 30 min under MeJA treatment in fruit at white stage (Fig 8A), according to the previously observed with the ortholog genes in Arabidopsis leaves [14]. FaJAZ8.1 displayed an increase of expression at 6 h after MeJA treatment, later than FaJAZ1 (Fig 8B). On the other hand, FaMYC2 showed higher expression levels at 15, 30 min and 6 h after MeJA application (Fig 8C), similar to observed for AtMYC2 expression in Arabidopsis leaves [14]. In summary, FaMYC2 responds earlier than FaJAZ1 and FaJAZ8.1 (Fig 8) suggesting a previous transcriptional activation according to the master role in JA responses regulation [10,11]. Furthermore, the early induction of FaJAZ1, FaJAZ8.1 and FaMYC2 genes could be related with JA-Ile accumulation under JA treatment, as previously reported [7].
Conclusions
Overall, we identified and characterized 12 JAZ and two MYC genes in F. vesca genome, encoding for key components in the regulation of JA responses in plants [10,46]. Nevertheless, the number of JAZ and MYC genes could be higher to that reported in the present research, since F. x ananassa is an octoploid and hybrid species [71], similar to the findings reported for the triploid M. × domestica species [50]. Synteny analysis using Arabidopsis genome and exon-intron organization indicates that FvJAZ subfamily and FvMYC2-like genes have a common ancestor with Arabidopsis. Besides, protein sequences are highly conserved through domains and position into JAZ and MYC2-like proteins. Finally, we evaluated temporal expression pattern of key JA signalling components during development and ripening of F. × ananassa fruit, indicating that JA signalling pathway is downregulated along with PAs decrease accumulation during fruit development and ripening processes in agreement with previously reported temporal JA-Ile reduction [7]. In addition, we demonstrated that FaMYC2, FaJAZ1 and FaJAZ8.1 are JA-responsive genes in F. × ananassa fruit which could related with the activation of JA-Ile biosynthesis detected in MeJA-treated fruit recently reported [7].
The JA signalling pathway could trigger PAs biosynthesis at early stages of strawberry fruit development, which shows a similar reduction profile with JA pathway during development and ripening. Thus, the present research opens the gates to further studies to decipher specific JA-Ile roles and its signalling pathway-associated components during early development of strawberry and other non-climacteric fruits.
Supporting information
Grey and blue horizontal lines indicate position of JAZ and MYC-like genes along chromosome, respectively. Orange, pink, purple and yellow lines indicate syntenic regions between A. thaliana and M. × domestica, S. lycopersicum, V. vinifera and O. sativa JAZ and MYC-like genes, respectively.
(TIF)
DLNPT (A) and EAR LxLxL (B) motifs of AtJAZ5, AtJAZ6, FvJAZ5, and EAR LxLxL motif of AtJAZ7, AtJAZ8, FvJAZ7, FvJAZ8.1 and FvJAZ8.2 (C). Red and blue colors indicate higher and lower amino acidic residues conservation, respectively. EAR, ethylene-responsive element binding factor-associated amphiphilic repression domain; JAZ, jasmonate ZIM-domain.
(TIF)
Asterisks (*) indicate conserved residues involved for JAZ-MYC interaction (Zhang et al. 2015). Red and blue colors indicate higher and lower amino acidic residues conservation, respectively. JAZ, jasmonate ZIM-domain; NLS, nuclear localization signalling.
(TIF)
Data are from three biological and three technical replicates. Developmental stages correspond to 0 (flowering, F), 10 (small green, SG), 17 (large green, LG), 20 (white, W), 21 (turning, T), 23 (50% red receptacle, 50%R) and 25 (100% red receptacle, 100%R) days after anthesis (DAA) in F. × ananassa cv. Aromas fruit. Data were analysed by one-way ANOVA test, and differences among means ± SE (n = 3) were determined using LSD test. Different letters indicate significant differences between developmental stages (p ≤ 0.05) for each gene.
(TIF)
Exon-intron organization of Arabidopsis thaliana (At) and F. vesca (Fv) PPD genes (A). Yellow and red bars indicate UTR regions and exons, respectively. Black interrupted lines indicate introns. Domain structure of A. thaliana and F. vesca PPD proteins (B). The relative position of each domain within each protein are displayed in colors. Multiple alignment sequences of A. thaliana and F. vesca PPD proteins (C). Red and blue colors indicate higher and lower amino acidic residues conservation, respectively. Phylogenetic analysis between PPDs proteins of A. thaliana, F. vesca, M. × domestica, V. vinifera and S. lycopersicum (D). The phylogenetic analysis was performed using full-length JAZ and MYC protein sequences. Nodes with bootstrap values > 70% are labelled and bootstrap values ≥ 95% show highlight bootstrap. Relative gene expression of F. × ananassa PPD1-1 during fruit development and ripening of F. × ananassa cv. Aromas (E). Developmental stages correspond to 0 (flowering, F), 10 (small green, SG), 17 (large green, LG), 20 (white, W), 21 (turning, T), 23 (50% red receptacle, 50%R), and 25 (100% red receptacle, R) days after anthesis (DAA). Data were analyzed by one-way ANOVA test, and differences among means ± SE (n = 3) were determined using LSD test. Different letters indicate significant differences between developmental stages (p ≤ 0.05).
(TIFF)
Gene expression patterns of FaCOI1, FaMYCs, FaJAZs (A) and FaNINJA, FaJAMs, FaTPLs, FaHDAs (B). Expression data were extracted from accession numbers of F. vesca (Sanchez-Sevilla et al. 2017) and we renamed according to the present research gene nomenclature. FaJAZ12 gene was not found in RNAseq experiments from Sanchez-Sevilla et al. (2017). Developmental stages correspond to GA (green achene), GR (green receptacle), RA (ripe achene), RR (ripe receptacle), TA (turning achene), TR (turning receptacle), WA (white achene), WR (white receptacle). COI1, coronatine insensitive 1; HDA, histone deacetylase; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate-ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS.
(TIFF)
COI1, coronatine insensitive 1; HDA, histone deacetylase; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate-ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS; PPD, PEAPOD.
(PDF)
JAZ, jasmonate ZIM-domain.
(PDF)
The primers were designed from full-length cDNA sequences of Fragaria vesca. The primers for COI1 and GAPDH genes were obtained from Preuss et al. (2014). COI1, coronatine insensitive 1; GAPDH, glyceraldehyde-3-phospate dehydrogenase; HDA, histone deacetylase; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate-ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS; PPD, PEAPOD.
(PDF)
Bold numbers indicate the highest identity of F. vesca TIFY and JAZ proteins comparing to Arabidopsis. JAZ, jasmonate ZIM-domain.
(PDF)
Bold numbers indicate the highest identity of F. vesca MYCs comparing to Arabidopsis.
(PDF)
Arabidopsis and F. vesca TIFY, JAZ and MYCs proteins were obtained from GenPept database (NCBI).
(PDF)
Developmental stages correspond to 0 (flowering, F), 10 (small green, SG), 17 (large green, LG), 20 (white, W), 21 (turning, T), 23 (50% red receptacle, 50%R), and 25 (100% red receptacle, R) days after anthesis (DAA). Data were analyzed by one-way ANOVA test, and differences among means ± SE (n = 3) were determined using LSD test. Different letters indicate significant differences between developmental stages (p ≤ 0.05) for each gene. nd = no detection. COI1, coronatine insensitive 1; HDA, histone deacetylases; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS.
(PDF)
Developmental stages correspond to 0 (flowering, F), 10 (small green, SG), 17 (large green, LG), 20 (white, W), 21 (turning, T), 23 (50% red receptacle, 50%R), and 25 (100% red receptacle, R) days after anthesis (DAA). nd = no detection. COI1, coronatine insensitive 1; HDA, histone deacetylases; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS.
(PDF)
Expression levels were measured at 0, 15, 30 min, 1 and 6 h under MeJA treatment. At each treatment and time, three biological replicates were used for the different analysis. Data were analyzed by one-way ANOVA test, and differences among means ± SE (n = 3) were determined using LSD test. Asterisks (*) indicate significant differences between control and MeJA treatment (p ≤ 0.05) for each gene. JAZ, jasmonate ZIM-domain.
(PDF)
Expression data were extracted from accession numbers of F. vesca (Sanchez-Sevilla et al. 2017) and we renamed according to the present research gene nomenclature. FaJAZ12 gene was not found in RNAseq experiments from Sanchez-Sevilla et al. (2017). Developmental stages correspond to GA (green achene), GR (green receptacle), RA (ripe achene), RR (ripe receptacle), TA (turning achene), TR (turning receptacle), WA (white achene), WR (white receptacle). COI1, coronatine insensitive 1; HDA, histone deacetylases; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS.
(PDF)
Acknowledgments
We thankfully acknowledge Dr. Andrea Chini (Department of Plant Molecular Genetics, Centro Nacional de Biotecnología-CSIC, Madrid, Spain) for his critical reading and important suggestions on this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was funded by the National Commission for Scientific and Technological Research (CONICYT, Chile, http://www.conicyt.cl/), grant CONICYT, FONDECYT/Regular 1140663 to C.R.F. A.G.-B. acknowledges the support by CONICYT through 'Beca Doctorado Nacional 2015 No. 21151411.' The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ahmad P, Rasool S, Gul A, Sheikh SA, Akram NA, Ashraf M, et al. Jasmonates: Multifunctional Roles in Stress Tolerance. Front Plant Sci. 2016;7: 813 doi: 10.3389/fpls.2016.00813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111: 1021–1058. doi: 10.1093/aob/mct067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wasternack C, Feussner I. The Oxylipin Pathways: Biochemistry and Function. Annu Rev Plant Biol. 2018;69: null. doi: 10.1146/annurev-arplant-042817-040440 [DOI] [PubMed] [Google Scholar]
- 4.Cherian S, Figueroa CR, Nair H. “Movers and shakers” in the regulation of fruit ripening: a cross-dissection of climacteric versus non-climacteric fruit. J Exp Bot. 2014;65: 4705–4722. doi: 10.1093/jxb/eru280 [DOI] [PubMed] [Google Scholar]
- 5.Böttcher C, Burbidge CA, di Rienzo V, Boss PK, Davies C. Jasmonic acid-isoleucine formation in grapevine (Vitis vinifera L.) by two enzymes with distinct transcription profiles. J Integr Plant Biol. 2015;57: 618–627. doi: 10.1111/jipb.12321 [DOI] [PubMed] [Google Scholar]
- 6.Gansser D, Latza S, Berger RG. Methyl Jasmonates in Developing Strawberry Fruit (Fragaria ananassa Duch. Cv. Kent). J Agric Food Chem. 1997;45: 2477–2480. doi: 10.1021/jf9608940 [Google Scholar]
- 7.Garrido-Bigotes A, Figueroa PM, Figueroa CR. Jasmonate Metabolism and Its Relationship with Abscisic Acid During Strawberry Fruit Development and Ripening. J Plant Growth Regul. 2018;37: 101–113. doi: 10.1007/s00344-017-9710-x [Google Scholar]
- 8.Shan X, Zhang Y, Peng W, Wang Z, Xie D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot. 2009;60: 3849–3860. doi: 10.1093/jxb/erp223 [DOI] [PubMed] [Google Scholar]
- 9.Concha CM, Figueroa NE, Poblete LA, Oñate FA, Schwab W, Figueroa CR. Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in Fragaria chiloensis fruit. Plant Physiol Biochem. 2013;70: 433–444. doi: 10.1016/j.plaphy.2013.06.008 [DOI] [PubMed] [Google Scholar]
- 10.Chini A, Gimenez-Ibanez S, Goossens A, Solano R. Redundancy and specificity in jasmonate signalling. Curr Opin Plant Biol. 2016;33: 147–156. doi: 10.1016/j.pbi.2016.07.005 [DOI] [PubMed] [Google Scholar]
- 11.Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448: 666–671. doi: 10.1038/nature06006 [DOI] [PubMed] [Google Scholar]
- 12.Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468: 400–405. doi: 10.1038/nature09430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448: 661–665. doi: 10.1038/nature05960 [DOI] [PubMed] [Google Scholar]
- 14.Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464: 788–791. doi: 10.1038/nature08854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Geyter N, Gholami A, Goormachtig S, Goossens A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012;17: 349–359. doi: 10.1016/j.tplants.2012.03.001 [DOI] [PubMed] [Google Scholar]
- 16.Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, et al. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008;146: 952–964. doi: 10.1104/pp.107.115691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thireault C, Shyu C, Yoshida Y, St Aubin B, Campos ML, Howe GA. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J Cell Mol Biol. 2015;82: 669–679. doi: 10.1111/tpj.12841 [DOI] [PubMed] [Google Scholar]
- 18.Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. The tify family previously known as ZIM. Trends Plant Sci. 2007;12: 239–244. doi: 10.1016/j.tplants.2007.04.004 [DOI] [PubMed] [Google Scholar]
- 19.Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J Cell Mol Biol. 2009;59: 77–87. doi: 10.1111/j.1365-313X.2009.03852.x [DOI] [PubMed] [Google Scholar]
- 20.Pauwels L, Goossens A. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell. 2011;23: 3089–3100. doi: 10.1105/tpc.111.089300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Y, Meng Y, Huang D, Qi Y, Chen M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics. 2011;98: 128–136. doi: 10.1016/j.ygeno.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 22.White DWR. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103: 13238–13243. doi: 10.1073/pnas.0604349103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico J-M, Gimenez-Ibanez S, Geerinck J, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23: 701–715. doi: 10.1105/tpc.110.080788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueroa P, Browse J. The Arabidopsis JAZ2 promoter contains a G-Box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol. 2012;53: 330–343. doi: 10.1093/pcp/pcr178 [DOI] [PubMed] [Google Scholar]
- 25.Figueroa P, Browse J. Male sterility in Arabidopsis induced by overexpression of a MYC5-SRDX chimeric repressor. Plant J Cell Mol Biol. 2015;81: 849–860. doi: 10.1111/tpj.12776 [DOI] [PubMed] [Google Scholar]
- 26.Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, et al. The Arabidopsis Mediator Subunit MED25 Differentially Regulates Jasmonate and Abscisic Acid Signaling through Interacting with the MYC2 and ABI5 Transcription Factors. Plant Cell Online. 2012; tpc.112.098277. doi: 10.1105/tpc.112.098277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, et al. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011;23: 1795–1814. doi: 10.1105/tpc.111.083261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakata M, Mitsuda N, Herde M, Koo AJK, Moreno JE, Suzuki K, et al. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell. 2013;25: 1641–1656. doi: 10.1105/tpc.113.111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, et al. Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol. 2013;163: 291–304. doi: 10.1104/pp.113.220129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fonseca S, Fernández-Calvo P, Fernández GM, Díez-Díaz M, Gimenez-Ibanez S, López-Vidriero I, et al. bHLH003, bHLH013 and bHLH017 Are New Targets of JAZ Repressors Negatively Regulating JA Responses. PLOS ONE. 2014;9: e86182 doi: 10.1371/journal.pone.0086182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q-H, Zhao C, Zhang M, Li Y-Z, Shen Y-Y, Guo J-X. Transcriptome analysis around the onset of strawberry fruit ripening uncovers an important role of oxidative phosphorylation in ripening. Sci Rep. 2017;7: 41477 doi: 10.1038/srep41477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, et al. The genome of woodland strawberry (Fragaria vesca). Nat Genet. 2011;43: 109–116. doi: 10.1038/ng.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee T-H, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41: D1152–D1158. doi: 10.1093/nar/gks1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31: 1296–1297. doi: 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sievers F, Higgins DG. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol Clifton NJ. 2014;1079: 105–116. doi: 10.1007/978-1-62703-646-7_6 [DOI] [PubMed] [Google Scholar]
- 36.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinforma Oxf Engl. 2009;25: 1189–1191. doi: 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14: 1188–1190. doi: 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Z, Zhang H, Gao S, Lercher MJ, Chen W-H, Hu S. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016;44: W236–W241. doi: 10.1093/nar/gkw370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasic K, Hernandez A, Korban SS. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Report. 2004;22: 437–438. doi: 10.1007/BF02772687 [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif. 2001;25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 41.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44: W147–153. doi: 10.1093/nar/gkw419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giusti MM, Wrolstad RE. Characterization and Measurement of Anthocyanins by UV-Visible Spectroscopy Current Protocols in Food Analytical Chemistry. John Wiley & Sons, Inc; 2001. Available: http://onlinelibrary.wiley.com/doi/10.1002/0471142913.faf0102s00/abstract [Google Scholar]
- 43.Prior RL, Fan E, Ji H, Howell A, Nio C, Payne MJ, et al. Multi-laboratory validation of a standard method for quantifying proanthocyanidins in cranberry powders. J Sci Food Agric. 2010;90: 1473–1478. doi: 10.1002/jsfa.3966 [DOI] [PubMed] [Google Scholar]
- 44.Jung S, Bassett C, Bielenberg DG, Cheng C-H, Dardick C, Main D, et al. A standard nomenclature for gene designation in the Rosaceae. Tree Genet Genomes. 2015;11: 108 doi: 10.1007/s11295-015-0931-5 [Google Scholar]
- 45.Ye H, Du H, Tang N, Li X, Xiong L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol. 2009;71: 291–305. doi: 10.1007/s11103-009-9524-8 [DOI] [PubMed] [Google Scholar]
- 46.Fonseca S, Chico JM, Solano R. The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol. 2009;12: 539–547. doi: 10.1016/j.pbi.2009.07.013 [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Qiao L, Bai J, Wang P, Duan W, Yuan S, et al. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). BMC Genomics. 2017;18: 152 doi: 10.1186/s12864-017-3582-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chini A, Ben-Romdhane W, Hassairi A, Aboul-Soud MAM. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLOS ONE. 2017;12: e0177381 doi: 10.1371/journal.pone.0177381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Z, Jin S-H, Guo H-D, Zhong X-J, He J, Li X, et al. Genome-wide identification and characterization of TIFY family genes in Moso Bamboo (Phyllostachys edulis) and expression profiling analysis under dehydration and cold stresses. PeerJ. 2016;4: e2620 doi: 10.7717/peerj.2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Yin X, Wang H, Li J, Guo C, Gao H, et al. Genome-wide identification and analysis of the apple (Malus × domestica Borkh.) TIFY gene family. Tree Genet Genomes. 2015;11: 808 doi: 10.1007/s11295-014-0808-z [Google Scholar]
- 51.Zhang Y, Gao M, Singer SD, Fei Z, Wang H, Wang X. Genome-wide identification and analysis of the TIFY gene family in grape. PloS One. 2012;7: e44465 doi: 10.1371/journal.pone.0044465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An J-P, Li H-H, Song L-Q, Su L, Liu X, You C-X, et al. The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol Biochem. 2016;108: 24–31. doi: 10.1016/j.plaphy.2016.06.032 [DOI] [PubMed] [Google Scholar]
- 53.Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, et al. The Jasmonate-Activated Transcription Factor MdMYC2 Regulates ETHYLENE RESPONSE FACTOR and Ethylene Biosynthetic Genes to Promote Ethylene Biosynthesis during Apple Fruit Ripening. Plant Cell Online. 2017; tpc.00349.2017. doi: 10.1105/tpc.17.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta N, Prasad VBR, Chattopadhyay S. LeMYC2 acts as a negative regulator of blue light mediated photomorphogenic growth, and promotes the growth of adult tomato plants. BMC Plant Biol. 2014;14: 38 doi: 10.1186/1471-2229-14-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woldemariam MG, Dinh ST, Oh Y, Gaquerel E, Baldwin IT, Galis I. NaMYC2 transcription factor regulates a subset of plant defense responses in Nicotiana attenuata. BMC Plant Biol. 2013;13: 73 doi: 10.1186/1471-2229-13-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Sun W, Chen J, Tan H, Xiao Y, Li Q, et al. SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci Rep. 2016;6: 22852 doi: 10.1038/srep22852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4: 10 doi: 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu G, Guo C, Shan H, Kong H. Divergence of duplicate genes in exon–intron structure. Proc Natl Acad Sci U S A. 2012;109: 1187–1192. doi: 10.1073/pnas.1109047109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang H, et al. Finding and Comparing Syntenic Regions among Arabidopsis and the Outgroups Papaya, Poplar, and Grape: CoGe with Rosids. Plant Physiol. 2008;148: 1772–1781. doi: 10.1104/pp.108.124867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21: 131–145. doi: 10.1105/tpc.108.064097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chorev M, Carmel L. The Function of Introns. Front Genet. 2012;3 doi: 10.3389/fgene.2012.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boter M, Ruíz-Rivero O, Abdeen A, Prat S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004;18: 1577–1591. doi: 10.1101/gad.297704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007;19: 2225–2245. doi: 10.1105/tpc.106.048017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 Encodes a MYC Transcription Factor Essential to Discriminate between Different Jasmonate-Regulated Defense Responses in Arabidopsis. Plant Cell Online. 2004;16: 1938–1950. doi: 10.1105/tpc.022319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oswald A, Oates AC. Control of endogenous gene expression timing by introns. Genome Biol. 2011;12: 107 doi: 10.1186/gb-2011-12-3-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shyu C, Figueroa P, Depew CL, Cooke TF, Sheard LB, Moreno JE, et al. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell. 2012;24: 536–550. doi: 10.1105/tpc.111.093005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang F, Yao J, Ke J, Zhang L, Lam VQ, Xin X-F, et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signaling. Nature. 2015;525: 269–273. doi: 10.1038/nature14661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grunewald W, Vanholme B, Pauwels L, Plovie E, Inzé D, Gheysen G, et al. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 2009;10: 923–928. doi: 10.1038/embor.2009.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thatcher LF, Cevik V, Grant M, Zhai B, Jones JDG, Manners JM, et al. Characterization of a JAZ7 activation-tagged Arabidopsis mutant with increased susceptibility to the fungal pathogen Fusarium oxysporum. J Exp Bot. 2016;67: 2367–2386. doi: 10.1093/jxb/erw040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot. 2011; erq408 doi: 10.1093/jxb/erq408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu B, Poulsen EG, Davis TM. Insight into octoploid strawberry (Fragaria) subgenome composition revealed by GISH analysis of pentaploid hybrids. Genome. 2016;59: 79–86. doi: 10.1139/gen-2015-0116 [DOI] [PubMed] [Google Scholar]
- 72.Preuß A, Augustin C, Figueroa CR, Hoffmann T, Valpuesta V, Sevilla JF, et al. Expression of a functional jasmonic acid carboxyl methyltransferase is negatively correlated with strawberry fruit development. J Plant Physiol. 2014;171: 1315–1324. doi: 10.1016/j.jplph.2014.06.004 [DOI] [PubMed] [Google Scholar]
- 73.Sánchez-Sevilla JF, Vallarino JG, Osorio S, Bombarely A, Posé D, Merchante C, et al. Gene expression atlas of fruit ripening and transcriptome assembly from RNA-seq data in octoploid strawberry (Fragaria × ananassa). Sci Rep. 2017;7: 13737 doi: 10.1038/s41598-017-14239-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paolacci AR, Catarcione G, Ederli L, Zadra C, Pasqualini S, Badiani M, et al. Jasmonate-mediated defence responses, unlike salicylate-mediated responses, are involved in the recovery of grapevine from bois noir disease. BMC Plant Biol. 2017;17: 118 doi: 10.1186/s12870-017-1069-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hao Y, Wang X, Li X, Bassa C, Mila I, Audran C, et al. Genome-wide identification, phylogenetic analysis, expression profiling, and protein–protein interaction properties of TOPLESS gene family members in tomato. J Exp Bot. 2014;65: 1013–1023. doi: 10.1093/jxb/ert440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pérez AC, Goossens A. Jasmonate signalling: a copycat of auxin signalling? Plant Cell Environ. 2013;36: 2071–2084. doi: 10.1111/pce.12121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Grey and blue horizontal lines indicate position of JAZ and MYC-like genes along chromosome, respectively. Orange, pink, purple and yellow lines indicate syntenic regions between A. thaliana and M. × domestica, S. lycopersicum, V. vinifera and O. sativa JAZ and MYC-like genes, respectively.
(TIF)
DLNPT (A) and EAR LxLxL (B) motifs of AtJAZ5, AtJAZ6, FvJAZ5, and EAR LxLxL motif of AtJAZ7, AtJAZ8, FvJAZ7, FvJAZ8.1 and FvJAZ8.2 (C). Red and blue colors indicate higher and lower amino acidic residues conservation, respectively. EAR, ethylene-responsive element binding factor-associated amphiphilic repression domain; JAZ, jasmonate ZIM-domain.
(TIF)
Asterisks (*) indicate conserved residues involved for JAZ-MYC interaction (Zhang et al. 2015). Red and blue colors indicate higher and lower amino acidic residues conservation, respectively. JAZ, jasmonate ZIM-domain; NLS, nuclear localization signalling.
(TIF)
Data are from three biological and three technical replicates. Developmental stages correspond to 0 (flowering, F), 10 (small green, SG), 17 (large green, LG), 20 (white, W), 21 (turning, T), 23 (50% red receptacle, 50%R) and 25 (100% red receptacle, 100%R) days after anthesis (DAA) in F. × ananassa cv. Aromas fruit. Data were analysed by one-way ANOVA test, and differences among means ± SE (n = 3) were determined using LSD test. Different letters indicate significant differences between developmental stages (p ≤ 0.05) for each gene.
(TIF)
Exon-intron organization of Arabidopsis thaliana (At) and F. vesca (Fv) PPD genes (A). Yellow and red bars indicate UTR regions and exons, respectively. Black interrupted lines indicate introns. Domain structure of A. thaliana and F. vesca PPD proteins (B). The relative position of each domain within each protein are displayed in colors. Multiple alignment sequences of A. thaliana and F. vesca PPD proteins (C). Red and blue colors indicate higher and lower amino acidic residues conservation, respectively. Phylogenetic analysis between PPDs proteins of A. thaliana, F. vesca, M. × domestica, V. vinifera and S. lycopersicum (D). The phylogenetic analysis was performed using full-length JAZ and MYC protein sequences. Nodes with bootstrap values > 70% are labelled and bootstrap values ≥ 95% show highlight bootstrap. Relative gene expression of F. × ananassa PPD1-1 during fruit development and ripening of F. × ananassa cv. Aromas (E). Developmental stages correspond to 0 (flowering, F), 10 (small green, SG), 17 (large green, LG), 20 (white, W), 21 (turning, T), 23 (50% red receptacle, 50%R), and 25 (100% red receptacle, R) days after anthesis (DAA). Data were analyzed by one-way ANOVA test, and differences among means ± SE (n = 3) were determined using LSD test. Different letters indicate significant differences between developmental stages (p ≤ 0.05).
(TIFF)
Gene expression patterns of FaCOI1, FaMYCs, FaJAZs (A) and FaNINJA, FaJAMs, FaTPLs, FaHDAs (B). Expression data were extracted from accession numbers of F. vesca (Sanchez-Sevilla et al. 2017) and we renamed according to the present research gene nomenclature. FaJAZ12 gene was not found in RNAseq experiments from Sanchez-Sevilla et al. (2017). Developmental stages correspond to GA (green achene), GR (green receptacle), RA (ripe achene), RR (ripe receptacle), TA (turning achene), TR (turning receptacle), WA (white achene), WR (white receptacle). COI1, coronatine insensitive 1; HDA, histone deacetylase; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate-ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS.
(TIFF)
COI1, coronatine insensitive 1; HDA, histone deacetylase; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate-ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS; PPD, PEAPOD.
(PDF)
JAZ, jasmonate ZIM-domain.
(PDF)
The primers were designed from full-length cDNA sequences of Fragaria vesca. The primers for COI1 and GAPDH genes were obtained from Preuss et al. (2014). COI1, coronatine insensitive 1; GAPDH, glyceraldehyde-3-phospate dehydrogenase; HDA, histone deacetylase; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate-ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS; PPD, PEAPOD.
(PDF)
Bold numbers indicate the highest identity of F. vesca TIFY and JAZ proteins comparing to Arabidopsis. JAZ, jasmonate ZIM-domain.
(PDF)
Bold numbers indicate the highest identity of F. vesca MYCs comparing to Arabidopsis.
(PDF)
Arabidopsis and F. vesca TIFY, JAZ and MYCs proteins were obtained from GenPept database (NCBI).
(PDF)
Developmental stages correspond to 0 (flowering, F), 10 (small green, SG), 17 (large green, LG), 20 (white, W), 21 (turning, T), 23 (50% red receptacle, 50%R), and 25 (100% red receptacle, R) days after anthesis (DAA). Data were analyzed by one-way ANOVA test, and differences among means ± SE (n = 3) were determined using LSD test. Different letters indicate significant differences between developmental stages (p ≤ 0.05) for each gene. nd = no detection. COI1, coronatine insensitive 1; HDA, histone deacetylases; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS.
(PDF)
Developmental stages correspond to 0 (flowering, F), 10 (small green, SG), 17 (large green, LG), 20 (white, W), 21 (turning, T), 23 (50% red receptacle, 50%R), and 25 (100% red receptacle, R) days after anthesis (DAA). nd = no detection. COI1, coronatine insensitive 1; HDA, histone deacetylases; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS.
(PDF)
Expression levels were measured at 0, 15, 30 min, 1 and 6 h under MeJA treatment. At each treatment and time, three biological replicates were used for the different analysis. Data were analyzed by one-way ANOVA test, and differences among means ± SE (n = 3) were determined using LSD test. Asterisks (*) indicate significant differences between control and MeJA treatment (p ≤ 0.05) for each gene. JAZ, jasmonate ZIM-domain.
(PDF)
Expression data were extracted from accession numbers of F. vesca (Sanchez-Sevilla et al. 2017) and we renamed according to the present research gene nomenclature. FaJAZ12 gene was not found in RNAseq experiments from Sanchez-Sevilla et al. (2017). Developmental stages correspond to GA (green achene), GR (green receptacle), RA (ripe achene), RR (ripe receptacle), TA (turning achene), TR (turning receptacle), WA (white achene), WR (white receptacle). COI1, coronatine insensitive 1; HDA, histone deacetylases; JAM, jasmonate-associated MYC2-like; JAZ, jasmonate ZIM-domain; NINJA, novel interactor of JAZ; TPL, TOPLESS.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.