SUMMARY
The processing of triglyceride-rich lipoproteins (TRLs) in capillaries provides lipids for vital tissues, but our understanding of TRL metabolism is limited, in part because TRL processing and lipid movement have never been visualized. To investigate the movement of TRL-derived lipids in the heart, mice were given an injection of [2H]triglyceride-enriched TRLs, and the movement of 2H-labeled lipids across capillaries and into cardiomyocytes was examined by NanoSIMS. TRL processing and lipid movement in tissues was extremely rapid. Within 30 sec, TRL-derived lipids appeared in the subendothelial spaces and in the lipid droplets and mitochondria of cardiomyocytes. 2H enrichment in capillary endothelial cells was not greater than in cardiomyocytes, implying that endothelial cells may not be a control point for lipid movement into cardiomyocytes. Remarkably, a deficiency of the putative fatty acid transport protein CD36, which is expressed highly in capillary endothelial cells, did not impede entry of TRL-derived lipids into cardiomyocytes.
eTOC BLURB
XXX et al used NanoSIMS to visualize the movement of triglyceride-rich lipoproteins (TRLs)-derived lipids in the heart. TRL-derived lipids mitochondria and lipid droplets within seconds. Also, loss of CD36 did not impede entry of TRL-derived lipids into cardiomyocytes.
INTRODUCTION
The lipolytic processing of triglyceride-rich lipoproteins (TRLs) by lipoprotein lipase (LPL) is the central event in plasma lipid metabolism, providing lipid nutrients for adipose tissue, heart, skeletal muscle, and other tissues (Havel, 2010; Havel and Kane, 2001; Kane and Havel, 2001). The processing of TRLs has been studied for decades (Havel and Gordon, 1960; Korn, 1955a; Korn, 1955b), but only recently have key mechanisms fallen into place (Fong et al., 2016). LPL is synthesized by parenchymal cells and secreted into the interstitial spaces, where it is captured by glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) on endothelial cells and transported to the lumen of capillaries (Beigneux et al., 2007; Davies et al., 2010). GPIHBP1 moves bidirectionally across endothelial cells (Davies et al., 2012), with each excursion providing an opportunity to pick up LPL and move it to the capillary lumen. In the setting of GPIHBP1 deficiency, LPL is stranded within the interstitial spaces (Davies et al., 2010), where it is useless for intravascular TRL processing. GPIHBP1 and GPIHBP1-bound LPL are located only on capillary endothelial cells and are absent from endothelial cells of arterioles and venules (Beigneux et al., 2007; Davies et al., 2010).
Aside from shuttling LPL to the capillary lumen, GPIHBP1 plays two additional functions in intravascular lipolysis. First, the GPIHBP1–LPL complex is crucial for the margination of TRLs along capillaries (Goulbourne et al., 2014), making it possible for LPL-mediated TRL processing to proceed. Second, the binding of LPL to GPIHBP1 stabilizes the structural integrity of LPL’s hydrolase domain and preserves its enzymatic activity, even in the presence of physiologic inhibitors (e.g., ANGPTL4), and by doing so GPIHBP1 serves to focus LPL activity in capillaries (Fong et al., 2016; Mysling et al., 2016a; Mysling et al., 2016b).
While recent studies have clarified functions of GPIHBP1 and LPL in TRL processing, other topics in the realm of intravascular lipolysis have remained enigmatic. In particular, no one understands the mechanisms by which the fatty acid products of TRL processing move across endothelial cells and into parenchymal cells (Hagberg et al., 2013). An early electron microscopy (EM) study (Scow et al., 1976) suggested that the products of lipolysis move across endothelial cells in “channels,” but the existence of such channels is questionable and has never been substantiated. In recent years, several groups have proposed that fatty acid movement across endothelial cells is regulated and requires transporter proteins (e.g., CD36, fatty acid transporters) (Hagberg et al., 2013). The movement of fatty acids across capillaries has been proposed to involve fatty acid entry into the endothelial cell cytoplasm and formation of acyl-coenzyme-A derivatives (Hagberg et al., 2013). Also, endothelial cells have been proposed to be a “control point” for regulating fatty acid movement, protecting parenchymal cells from excessive lipid uptake (Hagberg et al., 2013). According to the latter view, one could easily imagine that TRL-derived lipids might accumulate in endothelial cells before moving to parenchymal cells. On the other hand, others have proposed that fatty acids simply diffuse freely across mammalian cells without assistance from protein transporters (Guo et al., 2006; Hamilton et al., 2001; Xu et al., 2013). Another enigma has been the itinerary of TRL-derived fatty acids after entering parenchymal cells. It remains controversial whether fatty acids can enter mitochondria directly or whether they first must enter cytosolic triglyceride droplets.
Understanding how lipids move across capillaries and into surrounding parenchymal cells represents an important issue in lipoprotein physiology, but thus far the insights into this process have been limited and have depended on cell culture models or indirect evidence (e.g., drawing inferences from transcript levels for putative lipid-transport proteins or measuring lipid stores within tissue extracts) (Bharadwaj et al., 2010; Hagberg et al., 2013; Hagberg et al., 2010; Jang et al., 2016). We suspected that it would be possible to gain additional insights into TRL processing and lipid movement by imaging TRL processing within capillaries.
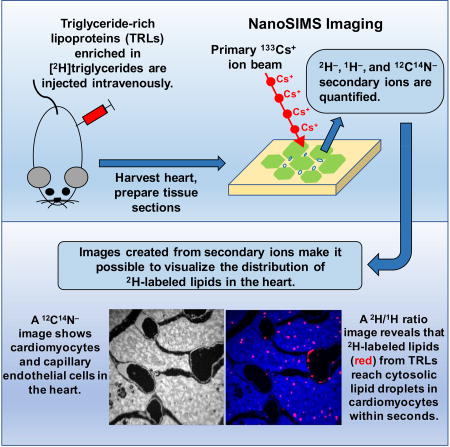
In the current studies, we used nanoscale secondary ion mass spectrometry (NanoSIMS) to visualize both TRL processing in heart capillaries and the fate of TRL-derived lipids in adjacent cardiomyocytes. NanoSIMS imaging uses a Cs+ beam to bombard a tissue section, releasing secondary ions that can be analyzed by mass spectrometry and used to create images based solely on the isotopic content of the tissue (He et al., 2017a; He et al., 2017b; Jiang et al., 2014a; Jiang et al., 2014b). After giving mice an injection of TRLs enriched in [2H]triglycerides, NanoSIMS imaging makes it possible to visualize the 2H-labeled products of TRL processing in endothelial cells, the subendothelial spaces, the mitochondria and/or cytosolic lipid droplets of cardiomyocytes. In NanoSIMS imaging studies, all of the secondary ion data are available for quantitative analyses. In the current studies, we show that the movement of TRL-derived lipids across endothelial cells and into cytosolic lipid droplets and mitochondria of cardiomyocytes is extraordinarily rapid, occurring within seconds. We found no evidence that capillary endothelial cells are a substantial barrier to the movement of TRL-derived lipids into cardiomyocytes. Also, we found no evidence that CD36 deficiency impedes the entry of TRL-derived lipids into cardiomyocytes.
RESULTS
The Lipolytic Processing of Triglyceride-Rich Lipoproteins (TRLs) and Lipid Movement into Cardiomyocytes Is Rapid
2H-enriched TRLs (2H-TRLs) were harvested from the plasma of Gpihbp1−/− mice after administering [2H]fatty acids by gastric gavage. The 2H content of TRLs harvested from Gpihbp1−/− mice was quite high, as judged by the fact that their density exceeded 1.006 g/ml (i.e., virtually all of the 2H-TRLs were on the bottom of the ultracentrifuge tube when spun at d = 1.006 g/ml). Quantitative lipidomics studies confirmed that large percentages of the fatty acyl chains in the 2H-TRLs were deuterated (Table S1). Indeed, the majority of the linoleic acid was deuterated. We observed consistent findings by NanoSIMS. When the 2H-TRL particles were analyzed by NanoSIMS, the 2H/1H ratio in the larger TRLs was ~3,000–6,000-times the natural abundance of 2H, such that about one-third of all hydrogens in the lipoprotein particles were 2H (Figure S1).
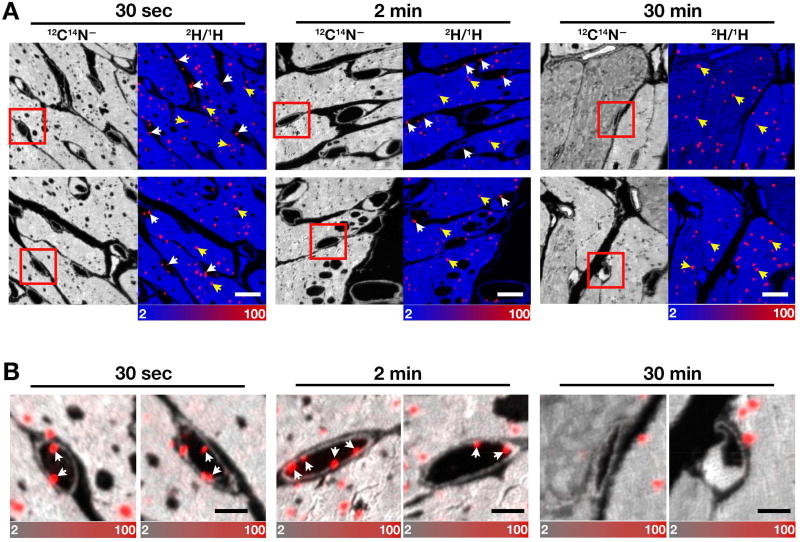
The 2H-TRLs (40 µg triglycerides in a volume of 200 µl) were injected intravenously into wild-type mice. After 30 sec, 2 min, or 30 min, the mice were euthanized; perfused extensively with PBS; and then perfusion-fixed with carbodiimide/glutaraldehyde. [Carbodiimide is effective in fixing carboxylate-containing small molecules in cells or tissues (Dallwig and Deitmer, 2002; Takei et al., 2012; Tymianski et al., 1997), and our studies, in which [3H]oleic acid was added to CHO-K1 cells, revealed 60–75% greater [3H]oleic acid fixation with carbodiimide/glutaraldehyde than with glutaraldehyde alone.] After fixation, the left ventricular apex was embedded in epoxy resin, sectioned, and analyzed by NanoSIMS. Images from 12C14N− ions (a cluster secondary ion that reveals tissue nitrogen content) were useful for visualizing tissue morphology; composite 12C14N− and 2H/1H images identified regions of 2H enrichment (Figure 1). In the composite images, the 2H/1H scale ranges from 0.0002 to 0.01 (from slightly above 2H natural abundance to ~70-times natural abundance). Margination of 2H-TRLs along the luminal surface of capillary endothelial cells was detectable 30 sec and 2 min after the injection of 2H-TRLs (Figures 1A and 1B, white arrows), but “marginated TRLs” were not encountered in capillaries at the 30-min time point. The 2H/1H ratio in the marginated lipoproteins at 30 sec and 2 min ranged from 15–450-times natural abundance, suggesting that processing had removed some of the [2H]triglycerides from the particles. 2H enrichment within cardiomyocyte lipid droplets (40–50-times natural abundance) was observed as early as 30 sec after injecting 2H-TRLs (Figure 1, yellow arrows).
Figure 1. NanoSIMS Images of Triglyceride-rich Lipoproteins (TRLs) Along Heart Capillary Endothelial Cells.
Four-month-old wild-type female mice were fasted for 4 h and then injected with 200 µl of 2H-TRLs containing 40 µg triglycerides (produced in Gpihbp1−/− mice after administering [2H]fatty acids by gavage). After 30 sec, 2 min, or 30 min, mice were euthanized and perfusion-fixed with carbodiimide/glutaraldehyde; sections were prepared for NanoSIMS imaging. Three mice were used in this experiment, one for each time point.
(A) 12C14N− NanoSIMS images were generated to visualize tissue morphology; composite 12C14N− (blue) and 2H/1H ratio (red) images revealed 2H-TRLs that had marginated along capillary endothelial cells (white arrows) and 2H-enriched cytosolic lipid droplets in cardiomyocytes (yellow arrows). The scale shows the 2H/1H ratio multiplied by 10,000. The 2H natural abundance, relative to 1H, is 0.000156. Scale bar, 6 µm.
(B) The boxed areas in panel A images are shown at higher magnification to reveal capillary endothelial cells. Composite 12C14N− (gray) and 2H/1H ratio (red) images show marginated lipoproteins (arrows) in the capillary lumen and 2H-enriched cytosolic lipid droplets in cardiomyocytes. Scale bar, 2 µm. See also Figure S1.
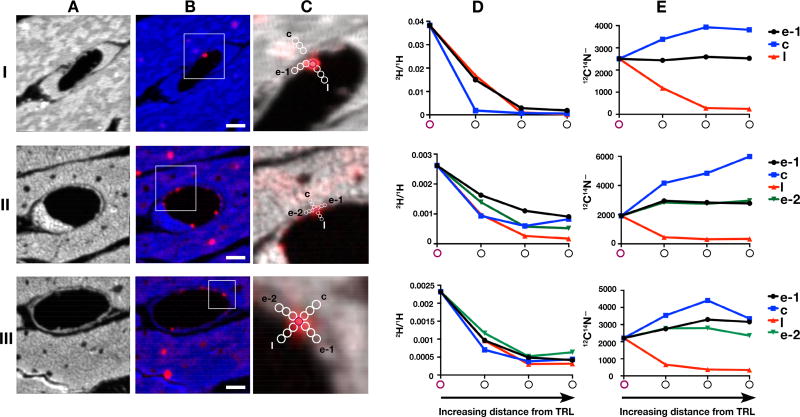
The margination of TRLs along capillaries is mediated by the LPL–GPIHBP1 complex (Goulbourne et al., 2014); thus, there is little doubt that the “marginated TRLs” in Figure 1 were undergoing processing by LPL. Interestingly, we were unable to document streaming of 2H-labeled lipids away from marginated TRLs and into endothelial cells or cardiomyocytes (Figure 2). When we quantified the 2H content of marginated TRLs and the 2H content of immediately adjacent regions of endothelial cells and cardiomyocytes, we observed a “fall-off” in 2H enrichment with increasing distance from the TRL (Figure 2). One might have easily jumped to the conclusion that the “fall-off” in 2H enrichment represented streaming of fatty acids away from the TRL, but we do not believe that this was the case, simply because we observed a similar “fall-off” in 2H enrichment with increasing distance into the capillary lumen. In our tissue sections, the capillary lumen is filled with epoxy resin, which is rich in 12C but devoid of 14N. We suspect that the appearance of 2H enrichment in the lumen of heart capillaries (which had been thoroughly perfused before fixation) was the result of short-distance diffusion of triglycerides into the resin-filled capillary lumen during tissue processing. Because of this observation, we do not believe that we observed bona fide streaming of fatty acids away from TRLs and into endothelial cells or cardiomyocytes. We suspect that the streaming of fatty acids away from TRLs, which obviously must occur physiologically, is so rapid that it was not detectable by NanoSIMS at the 30-sec or 2-min time points. In support of that reasoning, we never observed higher levels of 2H enrichment in segments of cardiomyocytes that were immediately adjacent to capillaries (Figure 1), implying that movement of 2H-labeled lipids into and across cardiomyocytes is quite rapid.
Figure 2. Movement of Lipids from 2H-TRLs to Surrounding Cardiomyocytes.
Four-month-old female wild-type mice were fasted for 4 h and then given an intravenous injection of 200 µl of 2H-TRLs containing 40 µg triglycerides. After 30 sec or 2 min, mice were euthanized and perfusion-fixed with carbodiimide/glutaraldehyde; tissue sections were prepared for NanoSIMS. Images from three regions (I, II, III) of the left ventricular apex were recorded. Two mice were used in this experiment.
(A) 12C14N− NanoSIMS images to show tissue morphology. Section I was prepared from tissue that had been stained with imidazole.
(B and C) Composite 12C14N− (blue in B and grey in C) and 2H/1H ratio (red) images were generated to visualize 2H-enriched TRLs in the capillary lumen and 2H-enriched lipid droplets within cardiomyocytes. The boxed area of the composite image is shown at higher magnification in panel C. 12C14N− ions and the 2H/1H ratio were measured in a TRL particle (purple circle) along the luminal surface of a capillary endothelial cells and in regions (white circles) extending into the endothelial cell (e-1, e-2), an adjacent cardiomyocyte (c), or into the capillary lumen (l). Scale bar, 2 µm.
(D) Graph showing a progressive decrease in the 2H/1H ratio—starting with the TRL particle in the capillary lumen and extending into the endothelial cell, the adjacent cardiomyocyte, and the capillary lumen.
(E) Graph showing 12C14N− ions in the TRL particle and in regions extending into the endothelial cell, the cardiomyocyte, and the capillary lumen.
The Entry of 2H-TRL-Derived Lipids into Cardiomyocytes Depends on Intravascular Processing of 2H-TRLs by LPL
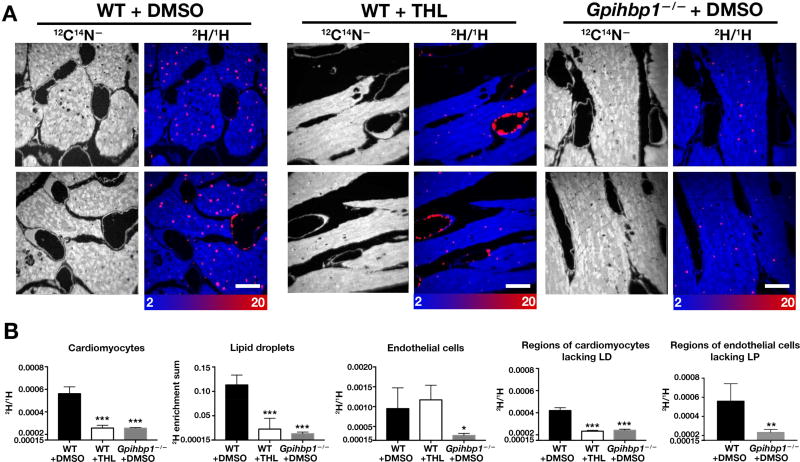
To be confident that the rapid appearance of 2H in cytosolic lipid droplets of cardiomyocytes (Figure 1) was secondary to LPL-mediated 2H-TRL processing, we performed experiments in isolated, perfused mouse hearts, which allowed us to compare 2H entry into cardiomyocytes in the presence or absence of an LPL inhibitor. We also examined 2H uptake into hearts of Gpihbp1−/− mice (where LPL is absent from the luminal surface of capillaries) (Davies et al., 2012; Davies et al., 2010). In perfused hearts of wild-type mice treated with a nonspecific lipase inhibitor (tetrahydrolipistatin), the appearance of 2H into cardiomyocytes was lower than in the absence of the inhibitor (Figures 3A and 3B). 2H enrichment of cardiomyocytes was also lower in the setting of Gpihbp1 deficiency (Figures 3A and 3B). In wild-type hearts that had been treated with the LPL inhibitor, numerous 2H-TRLs were found along endothelial cells, reflecting an accumulation of marginated but unprocessed 2H-TRLs in capillaries (Figure 3A). As expected, 2H enrichment was lower in endothelial cells of Gpihbp1−/− hearts (Figure 3), where TRL margination does not occur (Goulbourne et al., 2014). When we measured the 2H/1H ratio in endothelial cells of wild-type mice (focusing exclusively on segments devoid of marginated TRLs) with the 2H/1H ratio in endothelial cells of Gpihbp1−/− mice, we found a lower 2H/1H ratio in Gpihbp1−/− endothelial cells, reflecting reduced TRL processing (Figure 3B). In both wild-type and Gpihbp1−/− mice, the 2H/1H ratio in endothelial cells was similar to the 2H/1H ratio in surrounding cardiomyocytes (Figure 3B).
Figure 3. The 2H-enriched Lipids That Enter Cardiomyocytes Are Derived from LPL-mediated Intravascular Lipolysis, as Judged by Studies in Isolated, Perfused Hearts.
Hearts from 4-month-old female wild-type or Gpihbp1−/− mice were isolated and cannulated. Beating hearts were perfused with Tyrode’s buffer containing CaCl2 and either an LPL inhibitor (1 mM tetrahydrolipistatin) or vehicle (DMSO) alone. Next, hearts were perfused with 2H-TRLs. After 5 min, the hearts were perfused with PBS (to remove non-marginated TRLs) and then perfusion-fixed with carbodiimide/glutaraldehyde. Sections of hearts were prepared for NanoSIMS. Three mice were used in this experiment, one for each time point.
(A) 12C14N− images were created to visualize morphology (left panels); composite 12C14N− (blue) and 2H/1H ratio (red) images (right panels) were used to visualize 2H-TRLs in capillaries and 2H-enriched lipid droplets in cardiomyocytes. The scale shows the 2H/1H ratio multiplied by 10,000. Scale bar, 2 µm.
(B) Bar graphs show 2H/1H ratios in cardiomyocytes; the sum of 2H/1H ratios in cytosolic lipid droplets; 2H/1H ratios in capillary endothelial cells; 2H/1H ratios in regions of cardiomyocytes devoid of lipid droplets (LD); and 2H/1H ratios in regions of endothelial cells devoid of marginated lipoproteins (LP). In each graph, the y axis starts at 0.00015 (natural abundance of 2H). *p < 0.05; **p < 0.01; ***p < 0.0005. Each graph represent quantification of four 30 × 30-µm images. Data are presented as mean ± SD and were analyzed with one-way ANOVA with multiple comparisons.
Investigating the Fate of TRL-derived Lipids Within Cardiomyocytes
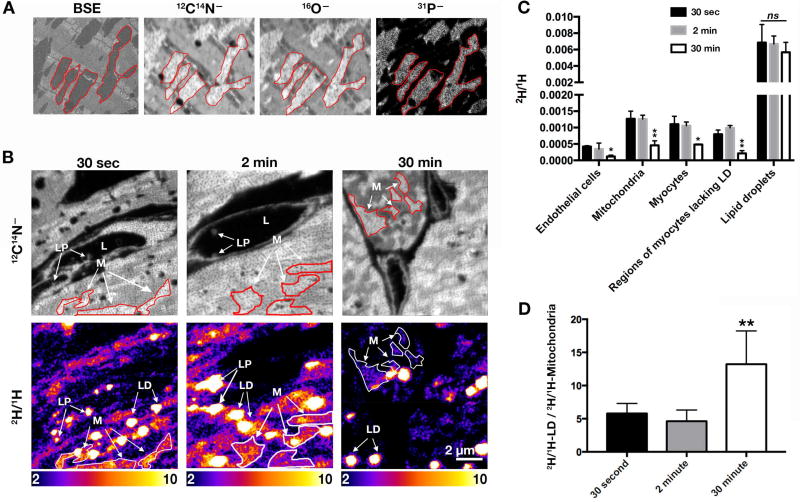
The mitochondria of cardiomyocytes could be identified in backscattered electron (BSE) images and in 12C14N−, 16O−, and 31P− NanoSIMS images (Figure 4A). Visualizing mitochondria with 12C14N− images was particularly helpful because the NanoSIMS 50L instrument is capable of recording 12C14N−, 2H−, and 1H− ions simultaneously. In Figure 4B, we show high-resolution 12C14N− and 2H/1H NanoSIMS images of the heart after injecting 2H-TRLs. The 2H/1H scale in these images differs from those in Figure 1, ranging from slightly above 2H natural abundance to ~7-times natural abundance. When the mice were sacrificed 30 sec or 2 min after the injection of 2H-TRLs, marginated TRLs could be identified in the capillary lumen, and there was 2H enrichment in both mitochondria and cytosolic lipid droplets of cardiomyocytes (Figure 4C). At the 30-min time point, we did not find marginated TRLs in capillaries; 2H enrichment persisted in cardiomyocyte mitochondria and lipid droplets, but the amount of 2H enrichment in mitochondria was lower in the 30-min images than in the 30-sec or 2-min images (Figures 4C and 4D), almost certainly reflecting mitochondrial catabolism of 2H–fatty acids in the 30-min time span after the injection of 2H-TRLs. We found no evidence, at any of the time points, that 2H enrichment in endothelial cells was greater than in adjacent cardiomyocytes (Figures 1–3; data not shown), implying that TRL–derived lipids do not accumulate in endothelial cells before moving into parenchymal cells.
Figure 4. NanoSIMS Imaging to Visualize Lipids Released from 2H-enriched TRLs.
Three-month-old female wild-type mice were fasted for 4 h and then given an intravenous injection of 200 µl of 2H-TRLs (40 µg triglycerides). After 30 sec, 2 min, or 30 min, the mice were euthanized and perfusion-fixed with carbodiimide/glutaraldehyde. Tissue sections were prepared for NanoSIMS and backscattered electron (BSE) imaging. Three mice were used in this experiment, one for each time point.
(A) NanoSIMS images (12C14N−, 16O−, 31P−) from tissue not stained with imidazole. A BSE image of the same region was useful for morphology. Several mitochondria are outlined in red.
(B) NanoSIMS images of heart sections 30 sec, 2 min, or 30 min after the injection of 2H-TRLs. 12C14N− images to visualize cell and tissue morphology (top panels); 2H/1H ratio images (bottom panels) to visualize the fate of lipids released from 2H-TRLs. LP, lipoproteins; L, capillary lumen; M, mitochondria; LD, lipid droplets. The scale shows the 2H/1H ratio multiplied by 10,000.
C) Bar graph showing the 2H/1H ratio in endothelial cells, mitochondria, myocytes, regions of myocytes lacking lipid droplets (LD), and lipid droplets at different time points. Three 30 × 30-µm images (each containing a minimum of 2–3 cardiomyocytes) were analyzed at each time point (endothelial cells, n = 8–13; mitochondria, n = 24–73; cardiomyocytes, n = 7–18; regions of myocytes lacking lipid droplets, n = 40; lipid droplets, n = 40–90). Data are presented as mean ± SD. Differences were assessed with one-way ANOVA. *p < 0.05; **p < 0.01; ns, nonsignificant.
(D) Bar graph depicting 2H/1H in lipid droplets divided by 2H/1H in mitochondria at each time point (mean ± SD). Each graph represent data from two 10 × 10-µm images and three 30 × 30-µm images. Data are presented as mean ± SD and were analyzed with one-way ANOVA with multiple comparisons. **p < 0.01. See also Figure S2.
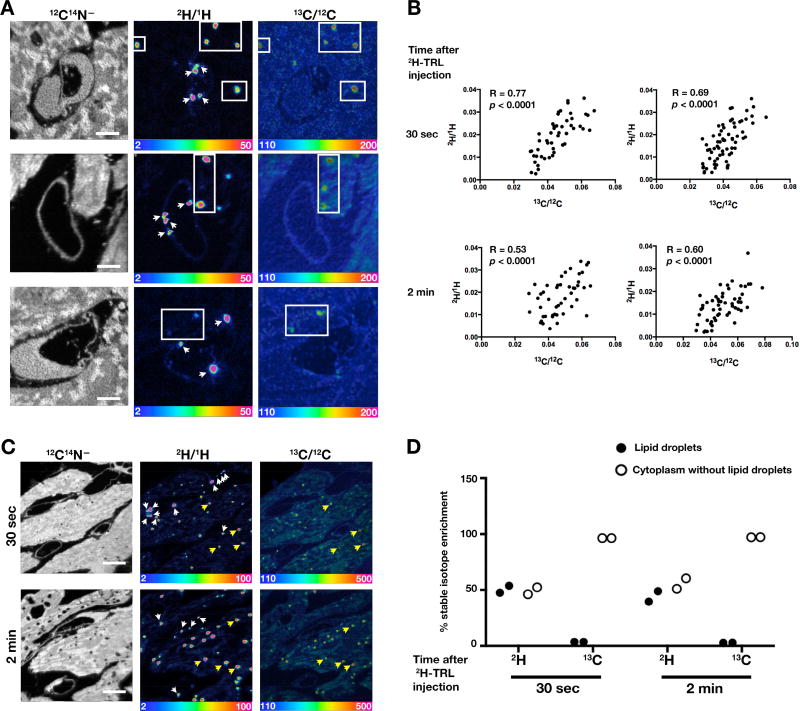
In some experiments, 13C-labeled fatty acids were given to mice by gastric gavage for several days before administering the intravenous injection of 2H-TRLs. In these studies, we once again observed 2H-TRLs margination in endothelial cells and 2H enrichment in cardiomyocyte lipid droplets 30 sec after injecting 2H-TRLs (Figure 5A). 2H-labeled lipids invariably entered cytosolic lipid droplets that were already labeled with 13C (Figure 5A), and the levels of 2H and 13C enrichment in lipid droplets were positively correlated (p < 0.001) (Figure 5B). Of note, however, the percentages of the labels in cytosolic lipid droplets were different. Approximately 50% of cardiomyocyte 2H was located in cytosolic lipid droplets, whereas only ~5% of the 13C was in lipid droplets (Figures 5C and 5D). This difference was not particularly surprising because 13C-labeled fatty acids (administered a day earlier) would be expected to contribute to membrane phospholipids. Also, fatty acid metabolites are used for synthesis of nonessential amino acids (Sidossis et al., 1995). Indeed, the fact that metabolites from [13C]fatty acids contribute to protein synthesis was illustrated by NanoSIMS images of brown adipose tissue. As expected, the cytosolic lipid droplets in brown adipocytes were enriched in 13C but devoid of nitrogen (i.e., as judged by the 12C14N− images) (Figure S2). Interestingly, the hemoglobin in one erythrocyte (almost certainly a newly released erythrocyte) was enriched in 13C, but 13C enrichment was negligible in older blood cells (two leukocytes and one erythrocyte) (Figure S2). The nitrogen-rich cytoplasm of brown adipocytes was also enriched in 13C, but to a lesser degree than cytosolic lipid droplets (Figure S2).
Figure 5. NanoSIMS Images of Heart Tissue from Mice That Had Been Given [13C]Fatty Acids by Gastric Gavage and on the Following Day an Intravenous Injection of 2H-TRLs.
[13C]Fatty acids were administered to 3–4-month-old female wild-type mice by gastric gavage (two 80-mg doses 12 h apart). Twenty-two h after the second dose, mice were fasted for 4 h and then injected with 200 µl of 2H-TRLs containing 40 µg triglycerides. Thirty sec or 2 min after the injection, the mice were euthanized and perfusion-fixed with carbodiimide/glutaraldehyde; sections of heart were prepared for NanoSIMS imaging. Two mice were used in this experiment, one for each time point.
(A) NanoSIMS images of heart tissues 30 sec after the injection of 2H-TRLs. 12C14N− images were useful for morphology; 2H/1H ratio images revealed marginated 2H-TRLs in capillaries (white arrows) and 2H-enriched lipids in lipid droplets of cardiomyocytes (boxed regions). 13C/12C ratio images show 13C distribution. 2H/1H and 13C/12C ratio scales are multiplied by 10,000. Scale bar, 2 µm.
(B) Scatter plots comparing 2H/1H and 13C/12C ratios in the center of cardiomyocyte lipid droplets. Data were obtained from lipid droplets (n = 50–60) in two NanoSIMS images (31 × 31 µm).
(C) NanoSIMS images showing 2H enrichment in marginated lipoproteins in capillaries (white arrows) and cardiomyocyte lipid droplets (several indicated by yellow arrows). 12C14N− images show tissue morphology; 2H/1H ratio images show the distribution of 2H 30 sec or 2 min after the injection of 2H-TRLs; 13C/12C ratio images show 13C distribution one day after giving 13C-labeled fatty acids by gastric gavage. The same lipid droplets were identified in both 2H/1H and 13C/12C images. 2H/1H and 13C/12C ratio scales are multiplied by 10,000. Scale bar, 5 µm.
(D) Plot depicting the percentage of cellular 2H and 13C in cytosolic lipid droplets, relative to 1H and 12C, respectively (two 35 × 35-µm NanoSIMS images analyzed at each time point). See also Figure S3.
The adult mouse brain primarily uses glucose for fuel, and TRL margination is absent in the capillaries of the brain (Goulbourne et al., 2014). For those reasons, we were skeptical that we would detect significant amounts of 13C or 2H enrichment in the brain after administering 13C-labeled fatty acids or 2H-TRLs. Indeed, after administering 13C-labeled fatty acids by gastric gavage and subsequently administering 2H-TRLs intravenously, we were unable to detect 13C or 2H enrichment in the cerebral cortex (Figure S3A).
NanoSIMS instruments have extraordinarily accurate mass resolution, making it nearly impossible to mistake one isotope for another. However, to be confident that we identified 2H− and 13C− ions correctly, we created NanoSIMS images from the heart of a mouse that had not been given 13C-labeled fatty acids or 2H-TRLs. We had no difficulty in generating 12C14N− NanoSIMS images of heart tissue, but as expected there was no enrichment in 13C or 2H (Figure S3B).
Detecting 2H-labeled Lipids in the Subendothelial Spaces
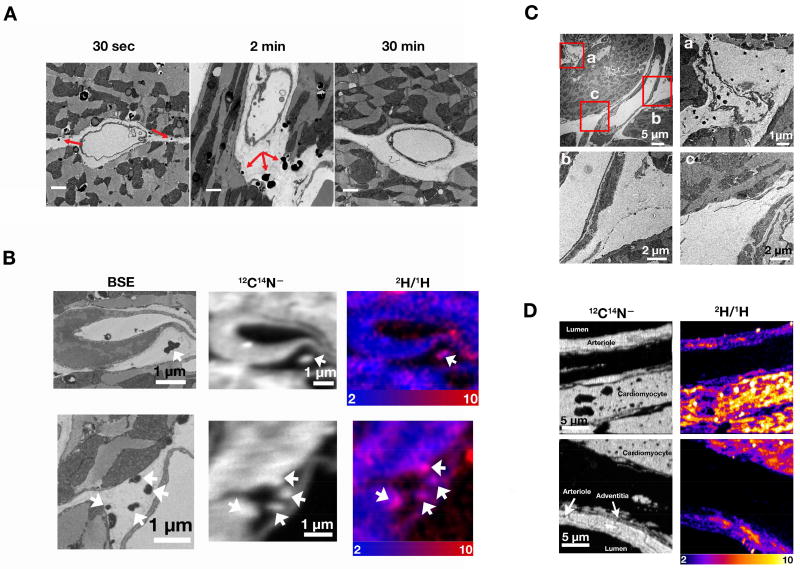
The TRL-derived lipids that enter cardiomyocytes obviously must have traversed the subendothelial spaces around capillaries. After administering an intravenous injection of TRLs and then staining heart tissue with OsO4 and imidazole, we observed irregular, darkly-stained structures in the subendothelial spaces around capillaries. [Imidazole is a nitrogen-rich mordant that increases OsO4 binding to lipids (Angermüller and Fahimi, 1982).] The irregular darkly staining structures were frequent at the 30-sec and 2-min time points but were uncommon at the 30-min time point (Figure 6A). To determine if the darkly-staining subendothelial structures contained TRL-derived lipids, we performed correlative BSE/NanoSIMS imaging on OsO4/imidazole–stained heart tissue harvested 30 sec after an intravenous injection of 2H-TRLs. BSE images revealed irregular, darkly stained structures in the subendothelial spaces (Figures 6B and S4A); NanoSIMS imaging of the same sections revealed that these structures were enriched in 2H (Figures 6B and S4A) from 2H-TRL processing. Because these structures were stained with imidazole, they were enriched in nitrogen, as judged by 12C14N− images (Figures 6B and S4A).
Figure 6. The Lipid Products of TRL Processing Can Be Identified in the Subendothelial Spaces.
(A) Transmission electron micrographs of the left ventricular apex from 4-month-old female wild-type mice 30 sec, 2 min, or 30 min after an intravenous injection of 200 µl of TRLs. Irregular, darkly stained structures were observed in the subendothelial spaces (red arrows) in OsO4/imidazole–stained tissue; these structures were more common at the 30-sec and 2-min time points. Scale bar, 1 µm. 2H/1H ratio scales were multiplied by 10,000. Three mice were used in this experiment, one for each time point.
(B) Visualizing irregular, darkly stained structures in the subendothelial spaces by NanoSIMS and BSE imaging. A 4-month-old female wild-type mouse was fasted for 4 h, injected with 200 µl of 2H-TRLs and sacrificed 30 sec later. Heart tissue was stained with OsO4/imidazole and processed for NanoSIMS and BSE imaging. The irregular, darkly staining subendothelial structures in the BSE images (left panel) were enriched in nitrogen (12C14N− ions, middle panels), reflecting the high nitrogen content of imidazole, and were enriched in 2H judged by composite 12C14N− (blue) and 2H/1H ratio (red) images (right panels).
(C) The irregular, darkly staining subendothelial structures were found in the subendothelial spaces around capillaries (a) but not in the vicinity of a large blood vessel (b and c).
(D) NanoSIMS images of an arteriole and an adjacent cardiomyocyte 2 min after an injection of 2H-TRLs. 2H enrichment was observed in mitochondria and cytosolic lipid droplets of cardiomyocytes and in the mitochondria of arteriolar smooth muscle cells, but 2H enrichment was not observed in the interstitial spaces around the arteriole. See also Figure S4.
Staining of lipids by imidazole was also evident in NanoSIMS images of cardiomyocytes from mice that had been given [13C]fatty acids by gastric gavage (Figure S4B). Normally, 13C-enriched cytosolic lipid droplets in cardiomyocytes are devoid of nitrogen and appear “black” in a 12C14N− NanoSIMS image. When the same tissue was stained with imidazole, the cytosolic lipid droplets became nitrogen-rich and were “white” in a 12C14N− image (Figure S4B).
The irregular OsO4/imidazole–stained structures were less common around larger blood vessels, where GPIHBP1 and LPL are absent (Davies et al., 2010; Fong et al., 2016). In the electron micrograph shown in Figure 6C, many irregular, darkly stained structures were found in and around a capillary, but not in the vicinity of an arteriole (Figure 6C and S4C). By NanoSIMS, we could not detect 2H enrichment in the subendothelial spaces surrounding arterioles, even though 2H-labeled lipids from TRL processing were present in adjacent cardiomyocytes and in the smooth muscle cells of nearby arterioles (Figures 6D).
Assessing the Impact of CD36 Deficiency on the Entry of TRL-derived Lipids into Cardiomyocytes
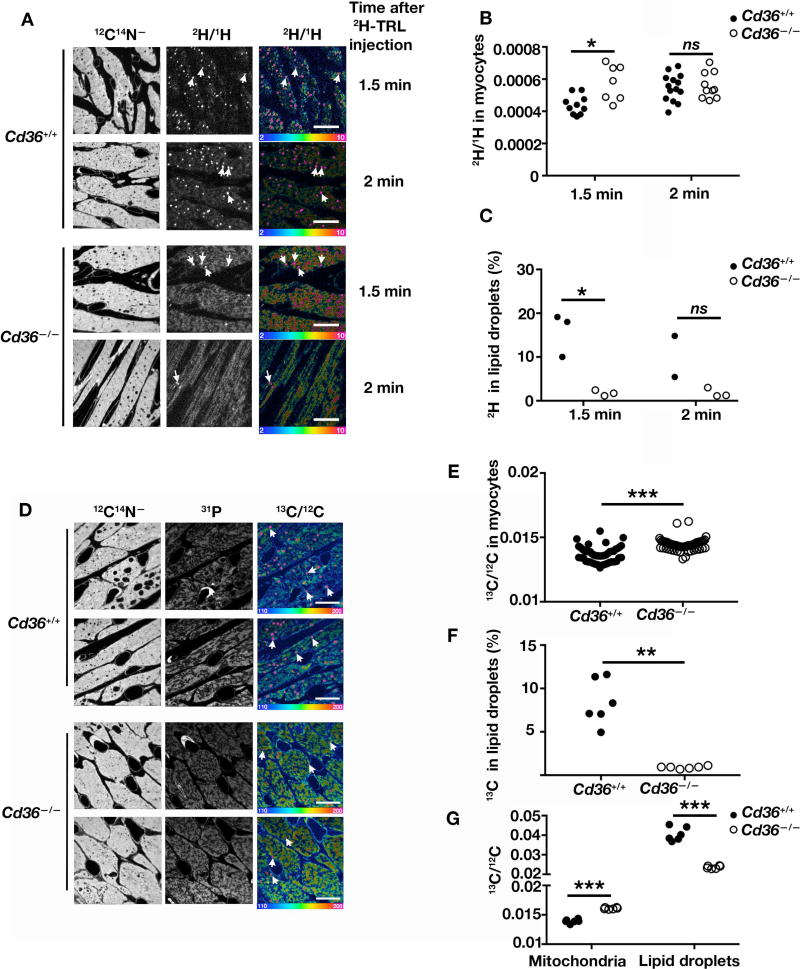
NanoSIMS imaging opens the door to examining the functional relevance of specific proteins in the movement of TRL-derived lipids into and across capillaries. CD36, a putative fatty acid transporter, is expressed at high levels in heart capillary endothelial cells (Greenwalt et al., 1995) (Figure S5) and has been hypothesized to play a role in the movement of fatty acids into cardiomyocytes (Bharadwaj et al., 2010; Hagberg et al., 2013). In the current study, we focused on testing whether a deficiency of CD36 might impede the movement of 2H-TRL–derived lipids into cardiomyocytes. Wild-type and Cd36−/− mice were injected intravenously with equivalent amounts of 2H-TRLs, and heart sections were prepared for NanoSIMS imaging. At 1.5- and 2-min time points, 2H enrichment in cardiomyocytes was equivalent in wild-type and Cd36−/− mouse hearts (Figures 7A and 7B), but the amount of 2H in cytosolic lipid droplets was lower in Cd36−/− hearts (as a percentage of total 2H in cells) (Figures 7A and 7C). When we administered [13C]fatty acids to wild-type and Cd36−/− mice by gastric gavage, we observed consistent findings—similar amounts of 13C in wild-type and Cd36−/− cardiomyocytes (Figures 7D, 7E) but reduced amounts of 13C in cytosolic lipid droplets of Cd36−/− cardiomyocytes (as a percentage of total 13C in cells) (Figures 7D and 7F). In addition to reduced amounts of total 13C in lipid droplets, the 13C/12C ratio in lipid droplets was lower in Cd36−/− cardiomyocytes (Figure 7G). Interestingly, 13C/12C ratio was slightly higher in Cd36−/− mitochondria (Figure 7G). In another series of NanoSIMS studies, we once again found similar amounts of 13C enrichment in wild-type and Cd36−/− cardiomyocytes (Figure S6A and S6B), but the percentage of total 13C in lipid droplets was lower in Cd36−/− cardiomyocytes (Figure S6C). CD36 deficiency has been reported to be associated with lower levels of acyl-CoA in cells (Bharadwaj et al., 2010), but whether that finding is due to the reduced amounts of 2H in cytosolic lipid droplets of Cd36−/− mice is unknown. We considered the possibility that the lipid droplets in Cd36−/− cardiomyocytes were simply too small to be visualized by NanoSIMS (where the lateral resolution is ~40–50 nm). By electron microscopy, the average area of cytosolic lipid droplets in Cd36−/− cardiomyocytes was lower than in wild-type cardiomyocytes (Figure S7), but this difference was small, probably too small to explain the paucity of lipid droplets in the NanoSIMS images of Cd36−/− cardiomyocytes.
Figure 7. TRL-derived Lipids Move Across Endothelial Cells and Enter Cardiomyocytes in Both Wild-type and Cd36-deficient Mice.
NanoSIMS analyses of 2H− ions in the heart after administering an intravenous injection of 200 µl of 2H-TRLs to 4-month-old female wild-type (Cd36+/+) and Cd36−/− mice. 1.5 or 2 min after the injection, hearts were harvested and sections were prepared for NanoSIMS. Differences in quantitative NanoSIMS data were analyzed with a Student’s t test with Welch’s correction. Four mice were used in this experiment, one for each time point.
(A) 12C14N− images (left) were used to define tissue morphology; the 2H/1H ratio images (middle) and the 2H/1H hue saturation images (right) show sites of 2H enrichment. White arrows indicate examples of cytosolic lipid droplets. Ratio scales were multiplied by 10,000. Scale bar, 10 µm.
(B) 2H/1H ratios in cardiomyocytes of Cd36+/+ and Cd36−/− mice. Each data point represents one cardiomyocyte in two or three 30 × 30-µm NanoSIMS images. *p < 0.05; ns, nonsignificant.
(C) 2H in cytosolic lipid droplets as a percentage of total 2H in cardiomyocytes. Graphs were generated by quantifying two or three 30 × 30-µm NanoSIMS images for each time point. NanoSIMS analyses of 13C− ions in the heart after feeding [13C]fatty acids by gavage (three 80-mg doses 12 h apart). 24 h after the last dose, mice were fasted for 4 h, and hearts were harvested and prepared for NanoSIMS.
(D) 12C14N− NanoSIMS images were used to visualize morphology; 31P images were used to visualize mitochondria; 13C/12C ratio images show sites of 13C enrichment. Arrows indicate examples of cytosolic lipid droplets. Ratio scales were multiplied by 10,000. Scale bar, 10 µm. *p < 0.05; ns, nonsignificant.
(E) 13C/12C ratio in cardiomyocytes of Cd36+/+ and Cd36−/− mice. Each data point represents one cardiomyocyte in six 30 × 30-µm NanoSIMS images. ***p < 0.0005.
(F) 13C enrichment in cardiomyocyte lipid droplets as a percentage of total 13C in cardiomyocytes. Graphs were generated by quantifying six 30 × 30-µm images for each sample. **p < 0.01. See also Figure S5, S6, and S7.
(G) 13C/12C ratio in mitochondria and lipid droplets of Cd36+/+ and Cd36−/− mice. Six 30 × 30-µm NanoSIMS images were analyzed. Each data point shows the mean 13C/12C ratio in the mitochondria and lipid droplets in each image (>50 mitochondria and 5–50 lipid droplets were analyzed per image). ***p < 0.0005.
DISCUSSION
We used NanoSIMS imaging along with electron microscopy to visualize intravascular TRL processing and the fate of TRL-derived lipids in cardiomyocytes. We gleaned three insights from our studies. First, TRL processing and the uptake of TRL lipids by cardiomyocytes is extremely rapid—occurring within seconds after TRLs enter the bloodstream. Second, during active TRL processing, we observed no evidence that 2H-TRL–derived lipids accumulate to higher levels in capillary endothelial cells than in adjacent cardiomyocytes. Third, we found no evidence that CD36 deficiency substantially impedes entry of TRL-derived lipids into cardiomyocytes.
For decades, the lipid metabolism field has recognized that the half-life of TRLs is measured in minutes (Havel and Kane, 2001), but we were nevertheless quite surprised by how rapidly TRL-derived lipids traverse capillary endothelial cells and enter cardiomyocytes. NanoSIMS imaging showed that 2H-TRL–derived lipids appear in cardiomyocytes within seconds (preferentially in cytosolic lipid droplets but also in mitochondria). Other features of the NanoSIMS images were consistent with extremely rapid movement of fatty acids into and across tissues. For example, we never observed more 2H enrichment in capillary endothelial cells than in cardiomyocytes, nor did we observe greater 2H enrichment in regions of cardiomyocytes adjacent to capillaries. Also, 2H rapidly entered arteriolar smooth muscle cells, despite the fact that TRL processing in arterioles is almost certainly negligible, owing to an absence of GPIHBP1 and LPL (Davies et al., 2010). All of these observations appear to be consistent with the proposal that fatty acids diffuse rapidly across cells (Guo et al., 2006; Hamilton et al., 2001; Xu et al., 2013) or alternatively that there is a yet-to-be-identified abundant high-capacity transport system for fatty acid movement across endothelial cells. Before reaching cardiomyocytes, the products of lipolysis obviously must traverse subendothelial spaces. In imidazole-stained heart tissue, we observed darkly-staining structures in the subendothelial spaces, and those structures contained 2H-labeled lipids from 2H-TRL processing.
An attractive feature of NanoSIMS analysis is that the secondary ion data are available for quantitative analyses, pixel by pixel, with >260,000 pixels/image. In our studies, quantitative analyses revealed that an LPL inhibitor drug or a deficiency of GPIHBP1 is accompanied by reduced uptake of 2H-TRL–derived lipids into cardiomyocytes. Quantitative analyses also allowed us to quantify the distribution of 2H-TRL–derived lipids within cardiomyocytes. 30 sec or 2 min after an injection of 2H-TRLs, ~50% of the 2H in cardiomyocytes was located in cytosolic lipid droplets—4–6-fold more than in mitochondria. After 30 min, 2H enrichment was ~12-fold greater in lipid droplets than in mitochondria, likely reflecting intervening oxidation of 2H-labeled fatty acids in mitochondria. We also examined 13C distribution in cardiomyocytes one day after administering [13C]fatty acids by gavage. Only ~5% of the 13C in cardiomyocytes was located in cytosolic lipid droplets, likely reflecting the use of fatty acids for membrane phospholipids and the use of fatty acid metabolites in the synthesis of nonessential amino acids.
CD36 is expressed at high levels in capillary endothelial cells (Greenwalt et al., 1995) and is often assumed to play a role in uptake of fatty acids by cells and tissues (Tanaka et al., 1997; Yang et al., 2007). When radioiodinated fatty acid derivatives are injected intravenously into humans, they are taken up by the heart. Uptake of those radiopharmaceuticals by the heart is reduced in subjects with CD36 deficiency (Hwang et al., 1998; Tanaka et al., 1997). Also, when Cd36−/− mice are fasted, fatty acids accumulate in the plasma and triglyceride accumulation in the heart is reduced (Bharadwaj et al., 2010). These observations suggest that CD36 could be important for the uptake of free fatty acids in the plasma compartment, but it is unclear whether CD36 deficiency influences uptake of TRL-derived nutrients by cardiomyocytes. Bharadwaj et al. (Bharadwaj et al., 2010) loaded human VLDL with [14C]triglycerides in vitro and then injected the TRLs into wild-type and Cd36−/− mice; they found reduced amounts of 14C in heart tissue extracts from Cd36−/− mice. On the other hand, Cd36 deficiency appeared to have little effect on the uptake of lipids from TRLs that had been endogenously labeled with 14C (14C-TRLs prepared in Lpl-deficient mice). In the current study, we used NanoSIMS to assess the effect of Cd36 deficiency on the fate of 2H-TRL–derived lipids in the heart. After 1.5 or 2 min, 2H entry into wild-type and Cd36−/− cardiomyocytes was nearly identical, indicating that CD36 deficiency has little effect on the movement of TRL-derived lipids into cardiomyocytes. [This observation seems consistent with an absence of impaired cardiac performance in Cd36−/− mice (Koonen et al., 2007).] However, we observed reduced entry of 2H into cardiomyocyte lipid droplets in Cd36−/− mice. The cytosolic lipid droplets were slightly smaller in Cd36−/− mice, but we are skeptical that their smaller size prevented us from detecting them by NanoSIMS. Cd36 deficiency has complex effects on cardiomyocytes, including effects on insulin and calcium signaling, fatty acyl-CoA levels, and fatty acid esterification (Abumrad and Goldberg, 2016; Bharadwaj et al., 2010; Xu et al., 2013), and those effects presumably could contribute, directly or indirectly, to changes in the distribution of 2H-TRL–derived lipids within cardiomyocytes.
NanoSIMS analyses are particularly helpful when, as in the current study, there are anatomical questions to address (i.e., an interest in defining the distribution of an isotope in different cell types or different subcellular compartments). NanoSIMS imaging will continue to be a useful approach in sorting out factors that regulate lipolysis and lipid movement within tissues. In the past, others have concluded, based largely on perturbations of lipid stores in knockout mice, that vascular endothelial growth factor-B (VEGF-B) is crucial for regulating fatty acid transport across capillary endothelial cells (Hagberg et al., 2010). Fatty acid binding proteins and fatty acyl-CoA synthetases have also been proposed to play a role in the delivery of lipids to myocytes (Hagberg et al., 2013; Hagberg et al., 2010). Recently, a valine metabolite, 3-hydroxyisobutyrate, was proposed to regulate fatty acid transport across endothelial cells (Jang et al., 2016), based largely on in vitro models and measurements of lipid stores in tissues. NanoSIMS imaging could be useful for defining the impact of both specific genes and small molecules in lipid movement into and across capillaries. The utility of NanoSIMS imaging for metabolism research will extend well beyond TRL metabolism. For example, NanoSIMS could simultaneously quantify the uptake and subcellular distribution of 2H-labeled glucose (or 2-deoxyglucose), 15N-labeled glutamine, and 13C-labeled fatty acids in solid tumors, making it possible to define metabolic heterogeneity in tumor cells, stromal cells, and endothelial cells.
There are certain limitations to our study. One is that the number of mice that we studied was low, and the number of images generated per mouse was rather limited. NanoSIMS imaging is very expensive, slow, and technically demanding. In a 24-h period, only 8–12 images could be obtained from one very small region of the heart. While each image is generated from an extraordinary amount of quantitative data on secondary ions, it is simply not feasible to image many mice per group, many different lines of mice, or many different tissues. That is why we believe that NanoSIMS is most suitable for addressing an anatomical issue and less suitable when scientific questions can be addressed by measuring tritium-labeled lipids in tissue extracts. Another limitation of our study is that it is conceivable that there was some fatty acid diffusion within tissues during the PBS perfusion (prior to perfusion-fixation with carbodiimide and glutaraldehyde) or some fatty acid diffusion after fixation during the preparation of tissues for NanoSIMS imaging. However, this type of lipid diffusion could not explain the rapid entry of deuterium into neutral lipids of cytosolic lipid droplets of cardiomyocytes. Our NanoSIMS images uncovered deuterium-labeled lipids in the subendothelial spaces of the heart, but it is likely that the patchy localization of those lipids was due to imidazole and osmium binding. Also, even though the deuterium-enriched, imidazole-staining patches were primarily located around small capillaries, we cannot fully exclude the possibility that some of the deuterium-labeled lipids moved into the subendothelial spaces during tissue processing. Finally, we would point out that the 13C NanoSIMS images (obtained at least 22 h after the administration of 13C-labeled fatty acids) should not be interpreted as exclusively showing lipid distribution, given that some of the 13C-labeled fatty acids were metabolized and used to generate amino acid substrates for protein synthesis.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to the Lead Contacts, Stephen G. Young (sgyoungmednet.ucla.edu) and Haibo Jiang (haibo.jianguwa.edu.au).
EXPERIMENTAL MODEL DETAILS
Mice
Cd36-deficient mice (Cd36−/−) (Febbraio et al., 1999) and wild-type mice were obtained from The Jackson Laboratory (strain C57BL/6). Gpihbp1-deficient mice (Gpihbp1−/−) (strain C57BL/6) have been described previously (Beigneux et al., 2007). Mice were housed in a barrier facility with a 12-h light-dark cycle and unless otherwise noted were fed a standard chow diet. All studies were approved by UCLA’s Animal Research Committee.
METHOD DETAILS
2H-labeled Triglyceride-rich Lipoproteins
Gpihbp1−/− mice (6–12-months-old) were placed on a fat-free (62% sucrose) diet (Envigo). After 10 days, 67 µl of 2H-labeled mixed fatty acids (2H-FA; 1.0 mg/µl; Cambridge Isotope Laboratories) and 13 µl sunflower oil were administered by gastric gavage every 12 h for 4.5 days. The deuterated mixed fatty acids from Cambridge Isotope Laboratories contain 35–55% palmitic acid, 10–15% palmitoleic acid, 20–35% oleic acid, and 10–20% linoleic acid. This acyl chain distribution is similar to the composition of the acyl chains in the lipids of mouse plasma lipoproteins (~20% palmitic acid, ~3% palmitoleic acid, 18% oleic acid, and 27% linoleic acid) (Yao G., 2000). Four hours after the last dose, blood was withdrawn from the inferior vena cava into a syringe containing EDTA. The plasma was adjusted to a density of 1.02 g/ml and spun at 539,511 × g in a Beckman TLA-100.3 rotor for 2 h at 10°C. The 2H-TRLs were collected, added to 2.5 ml of a 1.02 g/ml density solution, and spun again at 539,511 × g at 10°C for 2 h. 2H-TRLs were collected and stored at 4°C and used promptly. The size of TRLs isolated from Gpihbp1−/− mice is 50–150 nm (Weinstein et al., 2010). The percentages of fatty acyl chains in 2H-TRLs that were deuterated was determined by quantitative proteomics. 2H-TRLs in suspension (10 µl) were subjected to Bligh-Dyer extraction (Bligh and Dyer, 1959). Extracted lipid was then subjected to either base hydrolysis or thin-layer chromatography using silica gel G as solid phase and petroleum ether/ethyl ether/acetic acid (80/20/1, v/v/v) to resolve total phospholipid (origin) from triglyceride (Rf = 0.6). Silica associated with phospholipid and triglyceride from thin-layer chromatography was subsequently extracted and subjected to base hydrolysis. Base hydrolysis products (free fatty acids) were then converted to pentafluorobenzyl esters and quantified by gas chromatography in negative-ion chemical ionization mode with methane (Shao and Ford, 2014; Quehenberger et al., 2010).
Administration of [13C]Fatty Acids and 2H-TRLs to Mice
Wild-type and Cd36−/− mice were given 80 µl of [13C]fatty acids (about 1 mg/µl; Cambridge Isotope Laboratories) by gastric gavage. In other experiments, mice were anesthetized with ketamine/xylazine and given an intravenous injection of 200 µl of 2H-TRLs (40 µg triglycerides) through the inferior vena cava (for the 30-sec, 1.5-min, and 2-min time points) or the tail vein (for the 30-min time point). After the injection of 2H-TRLs, the diaphragm was cut to stop the heart at the indicated time points. Immediately thereafter, the mice were perfused with 10 ml of ice-cold PBS at 3 ml/min (5 ml though the right ventricle and 5 ml through the left ventricle). Next, mice were perfusion-fixed through the left ventricle with ice-cold 4% N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (“carbodiimide;” Sigma-Aldrich) (mass/vol) and 0.4% glutaraldehyde (Electron Microscopy Sciences) (vol/vol) in a 0.1 M phosphate buffer. The apex of the left ventricle was collected and cut into 1-mm3 pieces. The tissue was then placed in a 0.1 M phosphate buffer containing 4% carbodiimide and 2.5% glutaraldehyde overnight at 4°C.
Isolated Mouse Heart Experiments
Wild-type and Gpihbp1−/− mice were euthanized and perfused with 10 ml of Tyrode’s buffer [136 mM NaCl, 5.4 mM KCl, 0.33 mM NaH2PO4, 1 mM MgCl2, 10 mM Hepes (pH 7.4), 10 mM glucose] containing 1 mM CaCl2 (5 ml though the right ventricle and 5 ml through the left ventricle). The hearts were removed, cannulated, and submerged in PBS (with Ca2+ and Mg2+). Next, hearts were perfused with 3 ml of Tyrode’s buffer containing 1 mM CaCl2, 1% bovine serum albumin (BSA), and either 1 mM tetrahydrolipistatin (THL) or vehicle (DMSO) alone. Hearts were perfused with 1 ml of Tyrode’s buffer containing 1 mM CaCl2, 1% BSA, and 2H-TRLs (45 µg of triglyceride) and incubated for 5 min at room temperature. The hearts were then perfused with 3 ml of PBS (without Ca2+ and Mg2+) to remove unbound 2H-TRLs. Hearts were then perfusion-fixed with 3 ml of ice-cold 4% carbodiimide and 0.4% glutaraldehyde in 0.1 M phosphate buffer. The apex of the left ventricle was collected, cut into 1-mm3 pieces, and placed in 4% carbodiimide and 2.5% glutaraldehyde in 0.1 M phosphate buffer overnight at 4°C.
Preparation of Tissue Sections for Electron Microscopy and NanoSIMS Imaging
After fixation, 1-mm3 pieces of heart tissue were rinsed three times in 0.1 M sodium cacodylate buffer, pH 7.4 (10 min each) and placed in 1% OsO4 (Electron Microscopy Sciences) at room temperature for 45 min. The samples were then rinsed three times with distilled water (10 min each) and stained with 2% uranyl acetate (SPI-Chem) at 4°C overnight. On the following day, the samples were processed and embedded in Spurr’s resin as described previously (Goulbourne et al., 2014). For imidazole staining, tissue samples were rinsed twice with 0.1 M sodium cacodylate (10 min each) and twice with 0.2 M imidazole (Sigma) (15 min each) after fixation. The samples were then post-fixed with 2% OsO4 in 0.1 M imidazole for 40 min at room temperature. The samples were washed, stained with uranyl acetate, processed, and embedded in Spurr’s resin (Goulbourne et al., 2014).
NanoSIMS and Backscattered Electron (BSE) Imaging
Resin-embedded tissues were trimmed and sectioned (500-nm thick) and placed on silicon wafers. The sections were coated with 5 nm of platinum and analyzed with NanoSIMS 50L or NanoSIMS 50 instruments (CAMECA). A 16-KeV 133Cs+ beam was used to bombard the sample, and secondary electrons (SEs) and secondary ions (1H−, 2H−, 12C−, 13C−, 16O−, 12C14N−, 31P−) (not all secondary ions were analyzed simultaneously) were collected. A 40 × 40-µm region of the section was presputtered with a ~1.2-nA beam current (primary aperture D1=1) for ~3 min to remove the platinum coating and implant 133Cs+ to achieve a steady state of secondary ion release. For low-magnification images, a ~30 × 30-µm region was imaged with a ~1.6-pA beam current (primary aperture D1=2) and a dwell time of 10 ms/pixel per frame. Both 256 × 256- and 512 × 512-pixel images were obtained. To obtain high-magnification images, a ~10 × 10-µm region was imaged with a ~0.3 pA-0.65 pA beam current (primary aperture D1=3) and a dwell time of 20 ms/pixel per frame. Multiple frames and 512 × 512-pixel images were also obtained. The best lateral resolution for NanoSIMS images is approximately 40 nm. The Cs+ beam gradually vaporizes tissues, releasing secondary ions that are collected by the NanoSIMS instrument. The depth of tissue that is vaporized is only a few nm (less than the thickness of a lipid bilayer). Images were adjusted for contrast with the OpenMIMS plugin in ImageJ. For image quantification, 12C14N− ions and 2H/1H and 13C/12C ratios in regions-of-interest were measured with the OpenMIMS plugin and processed by GraphPad Prism 7.0.
For correlative NanoSIMS and backscattered electron (BSE) imaging, sections mounted on silicon wafers were initially examined with an FEI Verios scanning electron microscope. BSE images were recorded with a 1–2-KeV incident beam with a current of ~100 pA (Jiang et al., 2014a; Jiang et al., 2014b). The sections were then transferred to the NanoSIMS instrument, and the same regions were analyzed by NanoSIMS (Jiang et al., 2014a; Jiang et al., 2014b).
Transmission Electron Microscopy
Resin-embedded tissues were trimmed and sectioned to 65 nm with a Leica UC6 ultramicrotome. Sections were picked up on 100-mesh copper grids that were coated sequentially with formvar and carbon and then glow-discharged. The sections on grids were then stained with Reynold’s lead citrate for 9 min and imaged at 200 kV with an FEI T20 iCorr microscope equipped with an Eagle 2K CCD camera. To visualize lipid droplets in cardiomyocytes, Cd36+/+ and Cd36−/− mice were fasted for 16 h or fed an ad libitum diet. Mice were sacrificed, perfused with 10 ml of PBS, and then perfusion-fixed with 5 ml of 2% glutaraldehyde (Electron Microscopy Sciences) (vol/vol), 3.7% paraformaldehyde (Electron Microscopy Sciences) (vol/vol), and 2.1% sucrose in 0.1 M phosphate buffer. The left ventricular apex was collected and cut into 1-mm3 pieces. The tissue pieces were then incubated in the same fixation buffer for 2 h at room temperature. Next, the tissue was rinsed twice in 0.1 M sodium cacodylate (10 min each) and twice in 0.2 M imidazole (Sigma) (15 min each). The samples were then post-fixed with 2% OsO4 and 0.1 M imidazole for 40 min at room temperature. The samples were then washed, stained with uranyl acetate, processed, and embedded in Spurr’s resin (Goulbourne et al., 2014).
Transmission Electron Microscopy of Negatively Stained Lipoproteins
Staining was carried out under 100% humidity. 5 µl aliquots of TRLs were applied to a glow-discharged carbon-coated copper grid for 1 min; excess liquid was removed with a Whatman filter paper. Next, 5 µl of 2% uranyl acetate was added and blotted off; another 5 µl of 2% uranyl acetate was added for 1 min and again blotted off. The grids were allowed to air-dry before being imaged with an FEI T20 transmission electron microscope at 200 kV equipped with an Eagle 2K CCD camera.
Immunohistochemistry
Mice were anesthetized with isoflurane and perfused with PBS followed by 3% paraformaldehyde. The heart was harvested and embedded in OCT medium on dry ice. Tissue sections (7-µm) were fixed with 3% paraformaldehyde at room temperature for 15 min, permeabilized with 0.2% Triton X-100 for 5 min, and blocked at room temperature with 5% donkey serum and 0.2% BSA in PBS/Mg/Ca. Tissues were incubated overnight at 4°C with a rabbit polyclonal antibody against mouse CD36 (Abcam; 5 µg/ml), followed by a 45-min incubation at room temperature with Alexa647-conjugated anti-GPIHBP1 antibody 11A12 [2 µg/ml; (Beigneux et al., 2009)] and Alexa568-conjugated donkey anti–rabbit IgG (ThermoFisher Scientific; 1:200). After washing, the tissues were fixed in 3% paraformaldehyde for 5 min and stained with DAPI to visualize DNA. Images were recorded with an Axiovert 200M microscope and processed with Zen 2010 software (all from Zeiss). Within each experiment, the exposure conditions were identical.
Statistics
Differences were assessed by a Student’s t test or by ANOVA with multiple comparisons (GraphPad Prism software).
Supplementary Material
HIGHLIGHTS.
-
□
Intravascular lipolysis can be visualized by NanoSIMS.
-
□
Lipoprotein-derived lipids enter cardiomyocytes within seconds.
-
□
Lipoprotein-derived lipids do not accumulate in endothelial cells.
-
□
CD36 deficiency does not prevent entry of TRL-derived lipids into cardiomyocytes.
Acknowledgments
We are grateful for the NanoSIMS facilities at California Institute of Technology (Pasadena, CA) and the University of Western Australia (Perth). We acknowledge support from the Leducq Foundation Transatlantic Network Grant (12CVD04) (S.G.Y.), the National Institutes of Health [P01 HL090553 (S.G.Y.), R01 HL087228 (S.G.Y.), HL125335 (S.G.Y.), and a Ruth L. Kirschstein National Research Service Award, F32 HL132471 (C.H.)]. We also acknowledge support from the Australian Microscopy & Microanalysis Research Facility and the Science and Industry Endowment Fund for supporting the Ion Probe Facility at the Centre for Microscopy, Characterisation and Analysis at the University of Western Australia. We thank Dr. Marianne Cilluffo for help in preparing tissue sections.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
C.H. designed, performed, and analyzed experiments, prepared figures and the initial draft of the manuscript; T.A.W. performed electron microscopy and prepared tissue sections for NanoSIMS; R.S.J. performed experiments and analyzed NanoSIMS data; P.H. managed the mouse colony and performed mouse experiments; M.L. performed isolated heart studies; X.H. performed NanoSIMS studies; C.M.A. performed immunohistochemistry studies; P.T., K.R., M.P., and A.P.B. advised on experimental design and data analysis; A.H., M.K., and P.G. assisted with NanoSIMS; D.A.F. performed lipidomics studies; L.G.F. and S.G.Y. analyzed data and prepared the manuscript; H.J. performed NanoSIMS and BSE imaging, analyzed data, and assisted with figures and the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Abumrad NA, Goldberg IJ. CD36 actions in the heart: Lipids, calcium, inflammation, repair and more? Biochim. Biophys. Acta. 2016;1861:1442–1449. doi: 10.1016/j.bbalip.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermüller S, Fahimi HD. Imidazole-buffered osmium tetroxide: An excellent stain for visualization of lipids in transmission electron microscopy. Histochem. J. 1982;14:823–835. doi: 10.1007/BF01033631. [DOI] [PubMed] [Google Scholar]
- Beigneux AP, Davies B, Gin P, Weinstein MM, Farber E, Qiao X, Peale P, Bunting S, Walzem RL, Wong JS, et al. Glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007;5:279–291. doi: 10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigneux AP, Gin P, Davies BSJ, Weinstein MM, Bensadoun A, Fong LG, Young SG. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J. Biol. Chem. 2009;284:30240–30247. doi: 10.1074/jbc.M109.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj KG, Hiyama Y, Hu Y, Huggins LA, Ramakrishnan R, Abumrad NA, Shulman GI, Blaner WS, Goldberg IJ. Chylomicron- and VLDL-derived lipids enter the heart through different pathways: in vivo evidence for receptor- and non-receptor-mediated fatty acid uptake. J. Biol. Chem. 2010;285:37976–37986. doi: 10.1074/jbc.M110.174458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Dallwig R, Deitmer JW. Cell-type specific calcium responses in acute rat hippocampal slices. J. Neurosci. Methods. 2002;116:77–87. doi: 10.1016/s0165-0270(02)00030-4. [DOI] [PubMed] [Google Scholar]
- Davies BS, Goulbourne CN, Barnes RH, 2nd, Turlo KA, Gin P, Vaughan S, Vaux DJ, Bensadoun A, Beigneux AP, Fong LG, et al. Assessing mechanisms of GPIHBP1 and lipoprotein lipase movement across endothelial cells. J. Lipid Res. 2012;53:2690–2697. doi: 10.1194/jlr.M031559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BSJ, Beigneux AP, Barnes RH, II, Tu Y, Gin P, Weinstein MM, Nobumori C, Nyrén R, Goldberg IJ, Olivecrona G, et al. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 2010;12:42–52. doi: 10.1016/j.cmet.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febbraio M, Abumrad NA, Hajjar DP, Sharma K, Cheng W, Pearce SFA, Silverstein RL. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J. Biol. Chem. 1999;274:19055–19062. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- Fong LG, Young SG, Beigneux AP, Bensadoun A, Oberer M, Jiang H, Ploug M. GPIHBP1 and plasma triglyceride metabolism. Trends in Endocrinology and Metabolism: TEM. 2016;27:455–469. doi: 10.1016/j.tem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulbourne CN, Gin P, Tatar A, Nobumori C, Hoenger A, Jiang H, Grovenor CR, Adeyo O, Esko JD, Goldberg IJ, et al. The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 2014;19:849–860. doi: 10.1016/j.cmet.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwalt DE, Scheck SH, Rhinehart-Jones T. Heart CD36 expression is increased in murine models of diabetes and in mice fed a high fat diet. J. Clin. Invest. 1995;96:1382–1388. doi: 10.1172/JCI118173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Huang N, Cai J, Xie W, Hamilton JA. Fatty acid transport and metabolism in HepG2 cells. American journal of physiology. Gastrointestinal and Liver Physiology. 2006;290:G528–534. doi: 10.1152/ajpgi.00386.2005. [DOI] [PubMed] [Google Scholar]
- Hagberg C, Mehlem A, Falkevall A, Muhl L, Eriksson U. Endothelial fatty acid transport: role of vascular endothelial growth factor B. Physiology (Bethesda) 2013;28:125–134. doi: 10.1152/physiol.00042.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg CE, Falkevall A, Wang X, Larsson E, Huusko J, Nilsson I, van Meeteren LA, Samen E, Lu L, Vanwildemeersch M, et al. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature. 2010;464:917–921. doi: 10.1038/nature08945. [DOI] [PubMed] [Google Scholar]
- Hamilton JA, Johnson RA, Corkey B, Kamp F. Fatty acid transport: the diffusion mechanism in model and biological membranes. J. Mol. Neurosci. 2001;16:99–108. doi: 10.1385/JMN:16:2-3:99. discussion 151–107. [DOI] [PubMed] [Google Scholar]
- Havel RJ. Triglyceride-rich lipoproteins and plasma lipid transport. Arterioscler Thromb Vasc. Biol. 2010;30:9–19. doi: 10.1161/ATVBAHA.108.178756. [DOI] [PubMed] [Google Scholar]
- Havel RJ, Gordon RS., Jr Idiopathic hyperlipemia: metabolic studies in an affected family. J. Clin. Invest. 1960;39:1777–1790. doi: 10.1172/JCI104202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havel RJ, Kane JP. Introduction: Structure and metabolism of plasma lipoproteins. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 2705–2716. [Google Scholar]
- He C, Fong LG, Young SG, Jiang H. NanoSIMS imaging: an approach for visualizing and quantifying lipids in cells and tissues. J. Investig. Med. 2017a;65:669–672. doi: 10.1136/jim-2016-000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Hu X, Jung RS, Weston TA, Sandoval NP, Tontonoz P, Kilburn MR, Fong LG, Young SG, Jiang H. High-resolution imaging and quantification of plasma membrane cholesterol by NanoSIMS. Proc. Natl. Acad. Sci. U S A. 2017b;114:2000–2005. doi: 10.1073/pnas.1621432114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang EH, Taki J, Yasue S, Fujimoto M, Taniguchi M, Matsunari I, Nakajima K, Shiobara S, Ikeda T, Tonami N. Absent myocardial iodine-123-BMIPP uptake and platelet/monocyte CD36 deficiency. J. Nucl. Med. 1998;39:1681–1684. [PubMed] [Google Scholar]
- Jang C, Oh SF, Wada S, Rowe GC, Liu L, Chan MC, Rhee J, Hoshino A, Kim B, Ibrahim A, et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med. 2016;22:421–426. doi: 10.1038/nm.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Favaro E, Goulbourne CN, Rakowska PD, Hughes GM, Ryadnov MG, Fong LG, Young SG, Ferguson DJ, Harris AL, et al. Stable isotope imaging of biological samples with high resolution secondary ion mass spectrometry and complementary techniques. Methods. 2014a;68:317–324. doi: 10.1016/j.ymeth.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Goulbourne CN, Tatar A, Turlo K, Wu D, Beigneux AP, Grovenor CR, Fong LG, Young SG. High-resolution imaging of dietary lipids in cells and tissues by NanoSIMS analysis. J. Lipid Res. 2014b;55:2156–2166. doi: 10.1194/jlr.M053363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane JP, Havel RJ. Disorders of the biogenesis and secretion of lipoproteins containing the B apolipoproteins. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 2717–2752. [Google Scholar]
- Koonen DP, Febbraio M, Bonnet S, Nagendran J, Young ME, Michelakis ED, Dyck JR. CD36 expression contributes to age-induced cardiomyopathy in mice. Circulation. 2007;116:2139–2147. doi: 10.1161/CIRCULATIONAHA.107.712901. [DOI] [PubMed] [Google Scholar]
- Korn ED. Clearing factor, a heparin-activated lipoprotein lipase. I. Isolation and characterization of the enzyme from normal rat heart. J. Biol. Chem. 1955a;215:1–14. [PubMed] [Google Scholar]
- Korn ED. Clearing factor, a heparin-activated lipoprotein lipase. II. Substrate specificity and activation of coconut oil. J. Biol. Chem. 1955b;215:15–26. [PubMed] [Google Scholar]
- Mysling S, Kristensen KK, Larsson M, Beigneux AP, Gardsvoll H, Fong LG, Bensadouen A, Jorgensen TJ, Young SG, Ploug M. The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. eLife. 2016a;5:e12095. doi: 10.7554/eLife.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysling S, Kristensen KK, Larsson M, Kovrov O, Bensadouen A, Jorgensen TJ, Olivecrona G, Young SG, Ploug M. The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. eLife. 2016b;5:e20958. doi: 10.7554/eLife.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CR, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scow RO, Blanchette-Mackie EJ, Smith LC. Role of capillary endothelium in the clearance of chylomicrons. A model for lipid transport from blood by lateral diffusion in cell membranes. Circ. Res. 1976;39:149–162. doi: 10.1161/01.res.39.2.149. [DOI] [PubMed] [Google Scholar]
- Shao F, Ford DA. Elaidic acid increases hepatic lipogenesis by mediating sterol regulatory element binding protein-1c activity in huh-7 cells. Lipids. 2014;49:403–413. doi: 10.1007/s11745-014-3883-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidossis LS, Coggan AR, Gastaldelli A, Wolfe RR. Pathway of free fatty acid oxidation in human subjects. Implications for tracer studies. J. Clin. Invest. 1995;95:278–284. doi: 10.1172/JCI117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei S, Hasegawa-Ishii S, Uekawa A, Chiba Y, Umegaki H, Hosokawa M, Woodward DF, Watanabe K, Shimada A. Immunohistochemical demonstration of increased prostaglandin F(2)alpha levels in the rat hippocampus following kainic acid-induced seizures. Neuroscience. 2012;218:295–304. doi: 10.1016/j.neuroscience.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Okamoto F, Sohmiya K, Kawamura K. Lack of myocardial iodine-123 15-(p-iodiphenyl)-3-R,S-methylpentadecanoic acid (BMIPP) uptake and CD36 abnormality--CD36 deficiency and hypertrophic cardiomyopathy. Japanese Circulation Journal. 1997;61:724–725. doi: 10.1253/jcj.61.724. [DOI] [PubMed] [Google Scholar]
- Tymianski M, Bernstein GM, Abdel-Hamid KM, Sattler R, Velumian A, Carlen PL, Razavi H, Jones OT. A novel use for a carbodiimide compound for the fixation of fluorescent and non-fluorescent calcium indicators in situ following physiological experiments. Cell Calcium. 1997;21:175–183. doi: 10.1016/s0143-4160(97)90042-7. [DOI] [PubMed] [Google Scholar]
- Weinstein MM, Yin L, Tu Y, Wang X, Wu X, Castellani LW, Walzem RL, Lusis AJ, Fong LG, Beigneux AP, et al. Chylomicronemia elicits atherosclerosis in mice. Arterioscler. Thromb. Vasc. Biol. 2010;30:20–23. doi: 10.1161/ATVBAHA.109.196329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Jay A, Brunaldi K, Huang N, Hamilton JA. CD36 enhances fatty acid uptake by increasing the rate of intracellular esterification but not transport across the plasma membrane. Biochemistry. 2013;52:7254–7261. doi: 10.1021/bi400914c. [DOI] [PubMed] [Google Scholar]
- Yang J, Sambandam N, Han X, Gross RW, Courtois M, Kovacs A, Febbraio M, Finck BN, Kelly DP. CD36 deficiency rescues lipotoxic cardiomyopathy. Circulation Research. 2007;100:1208–1217. doi: 10.1161/01.RES.0000264104.25265.b6. [DOI] [PubMed] [Google Scholar]
- Yao G. Thèse présentée & la Faculté des tiîudes supérieures de l'université Lavai pour 1'0 btention du grade de maître ès sciences (M. Sc.) Medech Expérimentale. FACULTÉ DE &DECINE, Université Laval. National Library of Canada; 395 Wellington Street, Ottawa, Canada: 2000. Effect of LPL Activity on Fatty acid Composition in Mouse Plasma, Liver, Skeletal and Cardiac Muscles, Adipose and Brain Tissues. http://www.collectionscanada.gc.ca/obj/s4/f2/dsk1/tape2/PQDD_0019/MQ53989.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.