Abstract
Background/Aims
The development of nonalcoholic fatty liver disease (NAFLD) is associated with multiple genetic and environmental factors.
Methods
We performed a genome-wide association study to identify the genetic factors related to NAFLD in a Korean population-based sample of 1,593 subjects with NAFLD and 2,816 controls. We replicated the data in another sample that included 744 NAFLD patients and 1,137 controls. We investigated single-nucleotide polymorphisms (SNPs) that were related to NAFLD.
Results
After adjusting for age, sex and body mass index, rs738409, rs12483959 and rs2281135, located in the PNPLA3 gene, were validated in our population (p<8.56×10−8) in the same linkage disequilibrium block. Additionally, rs2143571, rs3761472, and rs2073080 in the SAMM50 gene showed significant associations with NAFLD (p<8.56×10−8). Furthermore, these six SNPs showed significant associations with the severity of fatty liver (all p<2.0×10−10 in the discovery set and p<2.0×10−6 in the validation set) and NAFLD, with elevated levels of alanine aminotransferase (all p<2.0×10−10 in the discovery set and p<2.0×10−6 in the validation set).
Conclusions
We demonstrated that the PNPLA3 and SAMM50 genes are significantly associated with the presence and severity of NAFLD in a Korean population. These findings confirm the important roles of genetic factors in the pathogenesis of NAFLD.
Keywords: Non-alcoholic fatty liver disease, Genome-wide association study, Polymorphism, single nucleotide
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) has been recognized as the leading cause of chronic liver disease, with a prevalence up to 20%–30% in the general population.1 Most cases of NAFLD follow a benign clinical course, however, once simple steatosis progresses to nonalcoholic steatohepatitis (NASH), 25% of patients may experience further progression to liver fibrosis and cirrhosis.2,3 Furthermore, patients with NASH have increased liver-related mortality compared with the general population.4 Genetic and environmental factors have important roles in the development of NAFLD.5,6 Single-nucleotide polymorphisms (SNPs) are widely used to identify genetic factors in various common diseases.
The first genome-wide association study (GWAS) of NAFLD found that a SNP (rs738409, encoding I148M) in the patatin-like phospholipase domain-containing protein 3 (PNPLA3) gene on chromosome 22 conferred susceptibility to NAFLD in a population that included Hispanic, African-American and European-American individuals. Subjects homozygous for this allele showed a 2-fold increase in hepatic fat content compared to noncarriers.7 A systematic review showed that rs738409 GG genotype was associated not only with liver fat accumulation but also with susceptibility to more aggressive disease.8 A recent GWAS of a European population identified four additional variants associated with NAFLD including neurocan (NCAN), glucokinase regulatory protein (GCKR), lysophospholipase-like 1 (LYPLAL1) and protein phosphatase 1 regulatory subunit 3b (PPP1R1B).
Although genetic features and their contributions differ among ethnicities, there are few studies incorporating GWAS of Asian populations. In a Japanese study, there was a significant association of the PNPLA3 gene with histologically diagnosed NAFLD.9 Another GWAS in Japan reported the association of two additional genes with the development and progression of NAFLD.10 The aim of this study was to evaluate the associations of genome-wide SNPs with the presence and severity of NAFLD in a Korean population.
MATERIALS AND METHODS
1. Study population
For our study, we analyzed the database from a previously described GENIE cohort.11 Briefly, 8,000 people donated blood samples during a routine health check-up program at Seoul National University Hospital Healthcare System Gangnam Center between January 2014 and December 2014, and their blood samples were stored at a biorepository with their informed consent.
Subjects with significant alcohol intake (>20 g/day for males and >10 g/day for females) were excluded (n=842). We also excluded 248 individuals who were positive for hepatitis B virus and 48 subjects who were positive for hepatitis C virus. Additionally, subjects with missing information were excluded from the study. This study was approved by the Institutional Review Board (No. 1601-010-730) of the Seoul National University Hospital with a waiver of informed consent.
2. Clinical and laboratory assessments
Each subject completed a past medical history questionnaire and underwent anthropometric assessment. The laboratory and radiological tests were performed on the same day. The body weight and height of each subject were measured using a digital scale. Body mass index (BMI) was calculated as the ratio of weight (kg) to area (m2, estimated from height). Waist circumference was measured at the midpoint between the lower costal margin and the anterior superior iliac crest by a well-trained person. Systolic blood pressure and diastolic blood pressure were each measured twice, and their mean values were reported. After an overnight 12 hours period, blood samples were collected from the antecubital vein of each individual. The laboratory evaluations included serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), total cholesterol, triglyceride, high density lipoprotein cholesterol, fasting glucose, hepatitis B surface antigen and antibody to hepatitis C virus. Abnormal liver enzyme levels were defined as elevated serum ALT above the strict cutoff point based on the updated definition (ALT >30 IU/ L for male and >19 IU/L for female) by Prati et al.12 We defined the suspected NASH as presence of ultrasonographic fatty liver with serum ALT elevation. Hypertension was defined as having systolic blood pressure ≥140 mm Hg, having diastolic blood pressure ≥90 mm Hg or using of anti-hypertensive medication. Diabetes mellitus was defined as either a fasting serum glucose level of ≥126 mg/dL or the use of anti-diabetic medication.
The diagnosis of NAFLD was based on the ultrasonographic findings (Aloka a-10, Aloka, Tokyo, Japan) of experienced radiologists who were unaware of the clinical information. The sonographic features of fatty liver were classified based on previously described standard categories as follows;13,14 normal, normal echogenicity; mild, slight diffuse increase in bright homogenous echoes in the liver parenchyma, with normal visualization of the diaphragm and the portal and hepatic vein borders, and with normal hepatorenal contrast if echogenic; moderate, diffuse increase in bright echoes in the liver parenchyma, with slightly impaired visualization of the peripheral portal and hepatic vein borders; severe, marked increase in bright echoes at a shallow depth, with deep attenuation and impaired visualization of the diaphragm and marked vascular blurring.
3. Genome-wide genotyping and quality control
Genomic DNA was isolated from venous blood samples and 200 ng of DNA from each patient was genotyped using Axiom® Customized Biobank Genotyping Arrays (Affymetrix, Santa Clara, CA, USA). The PLINK program version 1.07 (Free Software Foundation Inc., Boston, MA, USA) was used for quality control procedures. Samples meeting any of the following criteria were removed: (1) gender inconsistency, (2) call rate ≤97%, and (3) related and cryptically related individuals (identical by descent >0.185). SNPs were filtered if (1) the call rate was <95%, (2) the minor allele frequency was ≤0.01 (n=172,546), or (3) there was a significant deviation from the Hardy-Weinberg equilibrium permutation test (p<0.0001). After quality control, 584,061 autosomal SNPs remained for the association analysis.
4. Statistical analysis
A total of 584,061 SNPs that passed the quality control were used for the GWAS. We used the multivariable logistic regression model in the PLINK software package (version 1.07) and ordinal logistic regression in the R statistical software package version 3.1.1 (R development Core Team; R Foundation for Statistical Computing, Vienna, Austria) to test the association of NAFLD with SNPs in the genome. Age, gender and BMI were used as covariates. The R statistical software package, was used for the statistical analysis and to draw the Manhattan plot of −log10.
The results were verified using discovery and validation sets. We divided the enrolled population into two groups based on the time of enrollment. Samples from subjects who enrolled from January 2014 to October 2014 were used as the discovery set, and samples from subjects who enrolled in the subsequent months were used as the validation set. Comparisons of continuous variables between the two groups were performed using Student t-test, and categorical variables were compared using a chi-square test or Fisher exact test. Logistic regression analysis was used to analyze the association between NAFLD and control groups while controlling for potential confounders. Ordinal logistic regression analysis was used to analyze the association according to the severity of NAFLD. SNPs that had a p-value of less than 8.56×10−8 (calculated by the Bonferroni correction method) in the discovery set were re-evaluated in the validation set. p-values of less than 0.05 in the validation set were considered significant.
RESULTS
1. Study populations
A total of 6,290 subjects were included. The mean subject age was 50.0±10.6 years, and 53.9% of the subjects were male. Based on the definition described in the methods, there were 4,409 samples in the discovery set and 1,881 samples in the validation set. The characteristics of the study population are described in Table 1. The subjects in the validation set showed greater proportions of male, NAFLD and suspected NASH, and higher diastolic pressure, ALT, BMI and fasting glucose (p<0.05).
Table 1.
Baseline Characteristics of the Study Population
| Characteristic | Discovery set (n=4,409) | Validation set (n=1,881) | p-value |
|---|---|---|---|
| Age, yr | 50.2±10.5 | 50.0±9.9 | 0.509 |
| Male sex | 2,321 (52.6) | 1,067 (56.7) | 0.003 |
| Diabetes mellitus | 286 (6.5) | 111 (5.9) | 0.382 |
| Hypertension | 491 (11.1) | 237 (12.6) | 0.106 |
| Systolic blood pressure, mm Hg | 114.6±13.4 | 115.3±13.1 | 0.077 |
| Diastolic blood pressure, mm Hg | 74.9±10.3 | 76.1±10.0 | <0.001 |
| Body mass index, kg/m2 | 22.9±3.0 | 23.2±3.0 | 0.002 |
| Waist circumference, cm | 82.0±8.8 | 82.0±8.7 | 0.850 |
| AST, IU/L | 22.5±9.4 | 22.9±10.2 | 0.189 |
| ALT, IU/L | 22.2±15.1 | 23.4±16.7 | 0.009 |
| Cholesterol, mg/dL | 192.7±33.7 | 194.5±34.2 | 0.046 |
| Triglycerides, mg/dL | 104.2±69.8 | 107.0±72.1 | 0.142 |
| HDL cholesterol, mg/dL | 53.9±11.9 | 53.7±11.9 | 0.644 |
| LDL cholesterol, mg/dL | 121.1±30.2 | 122.4±30.4 | 0.128 |
| Fasting glucose, mg/dL | 97.1±15.3 | 98.8±18.6 | <0.001 |
| NAFLD | 1,593 (36.1) | 744 (39.6) | 0.011 |
| Severity of NAFLD | 0.049 | ||
| No fatty liver | 2,816 (63.9) | 1,137 (60.5) | |
| Mild | 981 (22.2) | 456 (24.2) | |
| Moderate | 547 (12.4) | 251 (13.3) | |
| Severe | 65 (1.5) | 37 (2.0) | |
| Suspected NASH | 0.026 | ||
| No fatty liver | 2,797 (63.9) | 1,131 (60.5) | |
| Fatty liver with normal ALT | 925 (21.1) | 420 (22.4) | |
| Fatty liver with elevated ALT | 656 (15.0) | 320 (17.1) |
Data are presented as the mean±SD or number (%).
AST, aspartate aminotransferase; ALT, alanine aminotransferase; HDL, high-density lipoprotein; LDL, low-density lipoprotein; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis.
2. Genome-wide association analysis of NAFLD
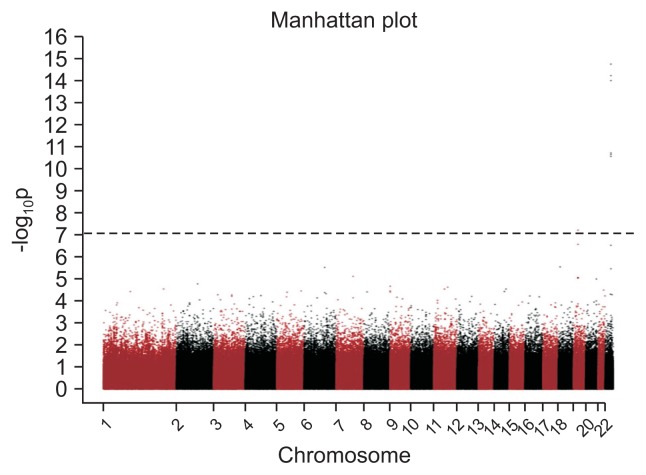
We first evaluated genome-wide associations in the discovery set, with a significance threshold of p<8.56×10−8 as the threshold after adjusting for age, gender and BMI. Fig. 1 shows a Manhattan plot with one peak, located on chromosome 22, that was associated with NAFLD. In the discovery set of the GWAS, seven SNPs on two chromosomes were significantly associated with NAFLD and we performed a validation test with the validation set, all remained significant (Table 2). Among them, three SNPs, namely, rs738409, rs12483959, and rs2281135 located in the PNPLA3 gene showed strong associations with NAFLD (discovery set: p=1.74×10−15, p=5.79×10−15, and p=9.72×10−15; validation set: p=2.56×10−8, p=1.34×10−8, and p=1.29×10−7, respectively). All the three SNPs were in strong linkage disequilibrium (LD; r2>0.9). An additional three SNPs, including rs3761472, rs2143571, and rs2073080 located in the SAMM50 gene, were in strong LD (r2>0.9) showed significant associations with NAFLD (discovery set: p=2.22×10−11, p=1.88×10−11, and p=2.72×10−11; validation set: p=3.28×10−7, p=5.86×10−7, and p=7.83×10−7, respectively). In addition, rs58542926, located in the TM6SF2 gene showed a weak association with NAFLD (discovery set, p=6.08×10−8; validation set, p=0.01385).
Fig. 1.
Manhattan plot of the association (−log10 [p-values], Y-axis) between single-nucleotide polymorphisms (X-axis, chromosome and chromosomal position) and nonalcoholic fatty liver disease in the genome-wide association study. The horizontal line indicates the Bonferroni-adjusted significance threshold. Threshold line: 8.56×10−8.
Table 2.
SNPs Associated with NAFLD in a Korean Population
| SNP | Chromosome | Position | Nearest genes | Risk allele | Discovery | Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Risk allele frequency | OR (95% CI) | p-value* | Risk allele frequency | OR (95% CI) | p-value* | |||||||
|
|
|
|||||||||||
| NAFLD | Control | NAFLD | Control | |||||||||
| rs58542926 | 19 | 19379549 | TM6SF2 | T | 0.094 | 0.066 | 1.671 (1.387–2.012) | 6.08E-8 | 0.086 | 0.070 | 1.419 (1.074–1.875) | 0.01385 |
| rs738409 | 22 | 44324727 | PNPLA3 | G | 0.466 | 0.398 | 1.537 (1.383–1.709) | 1.74E-15 | 0.467 | 0.382 | 1.542 (1.324–1.797) | 2.56E-08 |
| rs12483959 | 22 | 44325996 | PNPLA3 | A | 0.465 | 0.397 | 1.525 (1.365–1.686) | 5.79E-15 | 0.467 | 0.3382 | 1.556 (1.336–1.812) | 1.34E-08 |
| rs2281135 | 22 | 44332570 | PNPLA3 | A | 0.464 | 0.396 | 1.517 (1.365–1.686) | 9.72E-15 | 0.459 | 0.379 | 1.509 (1.295–1.757) | 1.29E-07 |
| rs3761472 | 22 | 44368122 | SAMM50 | G | 0.450 | 0.390 | 1.431 (1.288–1.590) | 2.22E-11 | 0.451 | 0.378 | 1.489 (1.278–1.735) | 3.28E-07 |
| rs2143571 | 22 | 44391686 | SAMM50 | A | 0.453 | 0.395 | 1.437 (1.292–1.597) | 1.88E-11 | 0.448 | 0.380 | 1.475 (1.266–1.717) | 5.86E-07 |
| rs2073080 | 22 | 44394402 | SAMM50 | T | 0.454 | 0.395 | 1.432 (1.288–1.591) | 2.72E-11 | 0.449 | 0.380 | 1.467 (1.260–1.708) | 7.83E-07 |
An additive genetic model was used.
SNPs, single-nucleotide polymorphisms; NAFLD, nonalcoholic fatty liver disease; OR, odds ratio; CI, confidence interval.
p-values are adjusted for age, sex and body mass index.
3. Impact of SNPs on the severity of NAFLD
We next investigated the associations of the SNPs found in the GWAS with NAFLD severity based on ultrasonographic grade, namely, normal versus mild versus moderate versus severe. The ordinal logistic regression analysis after adjusting for age, gender and BMI showed that that 10 SNPs showed significant associations with the severity of NAFLD (Table 3). Among them, three SNPs (rs738409, rs12483959, and rs2281135, located in the PNPLA3 gene) that had shown strong associations with the presence of NAFLD, also showed strong associations with the severity of NAFLD (discovery set: p=2.02×10−19, p=1.42×10−18, and p=2.21×10−18; validation set: p=1.70×10−8, p=5.14×10−9, and p=4.92×10−8, respectively). An additional three SNPs (rs3761472, rs2143571, and rs2073080 located in the SAMM50 gene), showed significant associations with the severity of NAFLD (discovery set: p=1.03×10−14, p=1.08 ×10−14, and p=1.97×10−14; validation set: p=2.07×10−7, p=9.01×10−7, and p=1.03×10−6, respectively).
Table 3.
SNPs Associated with the Severity of NAFLD in a Korean Population
| SNP | Chromosome | Position | Nearest genes | Risk allele | Discovery | Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Risk allele frequency | B (SE) | p-value* | Risk allele frequency | B (SE) | p-value* | |||||||
|
|
|
|||||||||||
| NAFLD | Control | NAFLD | Control | |||||||||
| rs58542926 | 19 | 19379549 | TM6SF2 | T | 0.094 | 0.066 | 0.489 (0.086) | 1.15E-08 | 0.086 | 0.069 | 0.335 (0.128) | 0.008 |
| rs2965185 | 19 | 19525792 | GATAD2A | C | 0.177 | 0.142 | 0.347 (0.064) | 7.54E-08 | 0.181 | 0.151 | 0.248 (0.095) | 0.009 |
| rs139051 | 22 | 44324676 | PNPLA3 | G | 0.342 | 0.382 | −0.302 (0.060) | 1.83E-09 | 0.343 | 0.394 | −0.269 (0.074) | 0.0002 |
| rs738409 | 22 | 44324727 | PNPLA3 | G | 0.466 | 0.398 | 0.444 (0.049) | 2.02E-19 | 0.467 | 0.382 | 0.399 (0.071) | 1.70E-08 |
| rs12483959 | 22 | 44325996 | PNPLA3 | A | 0.465 | 0.397 | 0.433 (0.049) | 1.42E-18 | 0.467 | 0.382 | 0.414 (0.071) | 5.14E-09 |
| rs2281135 | 22 | 44332570 | PNPLA3 | A | 0.464 | 0.396 | 0.429 (0.049) | 2.21E-18 | 0.459 | 0.379 | 0.386 (0.071) | 4.92E-08 |
| 5s3761472 | 22 | 44368122 | SAMM50 | G | 0.45 | 0.390 | 0.378 (0.049) | 1.03E-14 | 0.451 | 0.378 | 0.369 (0.071) | 2.07E-07 |
| rs2143571 | 22 | 44391686 | SAMM50 | A | 0.453 | 0.395 | 0.380 (0.049) | 1.08E-14 | 0.448 | 0.380 | 0.348 (0.071) | 9.01E-07 |
| rs2073080 | 22 | 44394402 | SAMM50 | T | 0.454 | 0.395 | 0.376 (0.049) | 1.97E-14 | 0.449 | 0.380 | 0.345 (0.071) | 1.03E-06 |
| rs1007863 | 22 | 44395451 | PARVB | T | 0.466 | 0.500 | −0.277 (0.049) | 1.69E-08 | 0.458 | 0.491 | −0.290 (0.070) | 3.17E-05 |
Genomic position is based on NCBI build 37.
SNPs, single-nucleotide polymorphisms; NAFLD, nonalcoholic fatty liver disease; SE, standard error.
p-values are adjusted for age, sex and body mass index.
Finally we further stratified the subjects with NAFLD by serum ALT levels, as a possible proxy for NASH comparing; normal versus NAFLD with normal ALT versus NAFLD with elevated ALT (>30 for male and 19 for female, IU/L), which may reflect the presence of suspected NASH, and examined the association between SNPs and suspected NASH. We found that eight SNPs that had significant relationships with the severity of NAFLD also showed significant associations fatty liver with elevated ALT. Among them, rs58542926, located in the TM6SF gene, showed significant associations with suspected NASH (discovery set, p=2.85×10−8; validation set, p=0.0018) (Table 4).
Table 4.
SNPs Associated with NAFLD and Elevated ALT
| SNP | Chromosome | Position | Nearest genes | Risk allele | Discovery | Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Risk allele frequency | B (SE) | p-value* | Risk allele frequency | B (SE) | p-value* | |||||||
|
|
|
|||||||||||
| NAFLD | Control | NAFLD | Control | |||||||||
| rs58542926 | 19 | 19379549 | TM6SF2 | T | 0.066 | 0.093 | 0.480 (0.086) | 2.85E-08 | 0.086 | 0.070 | 0.400 (0.128) | 0.00180 |
| rs139051 | 22 | 44324676 | PNPLA3 | G | 0.382 | 0.342 | −0.280 (0.050) | 2.44E-08 | 0.341 | 0.392 | −0.233 (0.074) | 0.00170 |
| rs738409 | 22 | 44324727 | PNPLA3 | G | 0.397 | 0.466 | 0.410 (0.049) | 8.44E-17 | 0.468 | 0.384 | 0.375 (0.071) | 1.42E-07 |
| rs12483959 | 22 | 44325996 | PNPLA3 | A | 0.397 | 0.464 | 0.403 (0.049) | 3.29E-16 | 0.468 | 0.383 | 0.383 (0.071) | 8.89E-08 |
| rs2281135 | 22 | 44332570 | PNPLA3 | A | 0.395 | 0.463 | 0.398 (0.049) | 5.91E-16 | 0.460 | 0.381 | 0.346 (0.071) | 1.08E-06 |
| rs3761472 | 22 | 44368122 | SAMM50 | G | 0.390 | 0.450 | 0.349 (0.049) | 9.66E-13 | 0.452 | 0.379 | 0.348 (0.072) | 1.14E-06 |
| rs2143571 | 22 | 44391686 | SAMM50 | A | 0.396 | 0.453 | 0.347 (0.049) | 1.65E-12 | 0.450 | 0.380 | 0.339 (0.072) | 1.72E-06 |
| rs2073080 | 22 | 44394402 | SAMM50 | T | 0.395 | 0.453 | 0.344 (0.049) | 2.31E-12 | 0.451 | 0.381 | 0.341 (0.071) | 1.46E-06 |
An additive genetic model was used.
SNPs, single-nucleotide polymorphisms; NAFLD, nonalcoholic fatty liver disease; ALT, alanine aminotransferase; SE, standard error.
p-values are adjusted for age, sex and body mass index.
When diabetes was added as an adjustment factor in both analysis, there was no change in the significant SNPs (data were not shown).
4. Analysis of metabolic traits and NAFLD-suspected SNPs
Because NAFLD is recognized as a hepatic manifestation of metabolic syndrome, we investigated the associations between metabolic traits and NAFLD-suspected SNPs. Six SNPs were associated with increased serum ALT and AST levels in the NAFLD group, but not in the control group (p-values <0.0001) (Table 5). Other metabolic traits were not significantly associated with the six SNPs.
Table 5.
Associations between Significant SNPs and Metabolic Factors
| SNP | Subjects | Fasting glucose | Triglycerides | HDL | Cholesterol | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| B (SE) | p-value | B (SE) | p-value | B (SE) | p-value | B (SE) | p-value | ||
| rs58542926 | Control | −0.081 (0.006) | 0.002 | −0.070 (0.024) | 0.003 | 0.560 (0.600) | 0.350 | 1.742 (1.745) | 0.318 |
| NAFLD | 0.042 (0.009) | 0.636 | −0.104 (0.03) | 5.5×E-4 | 1.357 (0.582) | 0.012 | −2.002 (2.175) | 0.357 | |
| rs738409 | Control | −0.018 (0.006) | 7.4×E-4 | −0.003 (0.012) | 0.019 | 0.080 (0.309) | 0.797 | −0.988 (0.896) | 0.270 |
| NAFLD | −0.001 (0.005) | 0.848 | −0.054 (0.002) | 0.002 | 0.614 (0.341) | 0.072 | 0.393 (1.276) | 0.758 | |
| rs12483959 | Control | −0.01 (0.003) | 9.6×E-4 | −0.027 (0.012) | 0.033 | 0.038 (0.310) | 0.903 | −1.004 (0.900) | 0.265 |
| NAFLD | −9E04 (0.005) | 0.862 | −0.054 (0.018) | 0.002 | 0.614 (0.341) | 0.072 | 0.429 (1.276) | 0.737 | |
| rs2281135 | Control | −0.010 (0.003) | 0.001 | −0.02 (0.012) | 0.088 | −0.039 (0.309) | 0.900 | −0.812 (0.896) | 0.365 |
| NAFLD | 0.001 (0.005) | 0.791 | −0.048 (0.018) | 0.006 | 0.592 (0.340) | 0.081 | 0.414 (1.273) | 0.745 | |
| rs3761472 | Control | −0.009 (0.003) | 0.002 | −0.020 (0.012) | 0.107 | 0.017 (0.306) | 0.956 | −0.351 (0.890) | 0.693 |
| NAFLD | 0.005 (0.005) | 0.321 | −0.031 (0.018) | 0.080 | 0.864 (0.342) | 0.011 | 2.116 (1.278) | 0.098 | |
| rs2143571 | Control | 0.001 (0.003) | 6.3×E-4 | −0.012 (0.012) | 0.108 | 0.007 (0.306) | 0.956 | −0.819 (0.894) | 0.360 |
| NAFLD | 0.004 (0.005) | 0.493 | −0.003 (0.018) | 0.137 | 0.878 (0.341) | 0.010 | 2.560 (1.278) | 0.045 | |
| rs2073080 | Control | −0.001 (0.003) | 6.6×E-4 | −0.019 (0.012) | 0.121 | −0.017 (0.309) | 0.956 | −0.871 (0.896) | 0.331 |
| NAFLD | 0.004 (0.005) | 0.466 | −0.027 (0.018) | 0.121 | 0.934 (0.341) | 0.006 | 2.467 (1.275) | 0.038 | |
|
| |||||||||
| SBP | DBP | ALT | AST | ||||||
|
| |||||||||
| rs58542926 | Control | −0.352 (0.496) | 0.478 | −0.575 (0.656) | 0.380 | 0.006 (0.022) | 0.790 | 0.002 (0.015) | 0.868 |
| NAFLD | 0.265 (0.559) | 0.636 | 0.522 (0.572) | 0.488 | 0.102 (0.030) | 5.7×E-4 | 0.523 (0.019) | 0.007 | |
| rs738409 | Control | −0.316 (0.255) | 0.215 | −0.509 (0.337) | 0.131 | −0.018 (0.011) | 0.113 | −0.003 (0.008) | 0.673 |
| NAFLD | −0.181 (0.331) | 0.585 | −0.106 (0.445) | 0.811 | 0.084 (0.017) | 6.0×E-7 | 0.051 (0.011) | 7.6×E-6 | |
| rs12483959 | Control | −0.308 (0.255) | 0.228 | −0.581 (0.338) | 0.086 | −0.021 (0.012) | 0.069 | −0.004 (0.008) | 0.644 |
| NAFLD | −0.227 (0.330) | 0.493 | −0.257 (0.443) | 0.561 | 0.086 (0.017) | 6.0×E-7 | 0.051 (0.011) | 7.6×E-6 | |
| rs2281135 | Control | −0.257 (0.255) | 0.314 | −0.562 (0.337) | 0.095 | −0.020 (0.011) | 0.083 | −0.005 (0.008) | 0.546 |
| NAFLD | −0.114 (0.330) | 0.729 | −0.093 (0.443) | 0.835 | 0.087 (0.017) | 6.0×E-7 | 0.050 (0.011) | 9.7×E-6 | |
| rs3761472 | Control | 0.144 (0.253) | 0.560 | −0.353 (0.334) | 0.292 | −0.192 (0.011) | 0.901 | −0.005 (0.007) | 0.493 |
| NAFLD | 0.228 (0.332) | 0.492 | 0.082 (0.445) | 0.853 | 0.084 (0.017) | 1.3×E-6 | 0.049 (0.011) | 1.8×E-5 | |
| rs2143571 | Control | −0.142 (0.254) | 0.577 | −0.413 (0.336) | 0.220 | −0.021 (0.011) | 0.062 | −0.007 (0.008) | 0.367 |
| NAFLD | 0.256 (0.332) | 0.441 | 0.156 (0.446) | 0.727 | 0.081 (0.017) | 3.5×E-6 | 0.049 (0.011) | 1.8×E-5 | |
| rs2073080 | Control | −0.173 (0.255) | 0.497 | −0.449 (0.337) | 0.183 | −0.020 (0.011) | 0.075 | −0.007 (0.008) | 0.383 |
| NAFLD | 0.312 (0.331) | 0.346 | 0.236 (0.444) | 0.596 | 0.080 (0.017) | 3.7×E-6 | 0.049 (0.011) | 2.1×E-5 | |
Data were derived from a linear regression analysis. Each metabolic factor was adjusted for age, gender and body mass index. Genomic position is based on NCBI build 37.
SNPs, single-nucleotide polymorphisms; HDL, high-density lipoprotein; SE, standard error; NAFLD, nonalcoholic fatty liver disease; SBP, systolic blood pressure; DBP, diastolic blood pressure; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
DISCUSSION
The significant associations between the PNPLA3 gene and NAFLD are well-known. However, there has been no study based on GWAS in a Korean population. In this study, we investigated the significance of association between the PNPLA3 gene and ultrasonography-diagnosed NAFLD or NAFLD severity. Moreover, we demonstrated that SAMM50 and TM6SF2 are most likely involved in NAFLD in the Korean population.
Romeo et al.,7 previously found that rs738409 is associated with increased hepatic fat, as measured by magnetic resonance spectroscopy in Hispanic, African American and European populations. Other groups have replicated significant associations between the SNP and fatty liver.8,15,16 In addition, many subsequent studies were performed to identify new SNPs with roles in the development and progression of NAFLD.17,18 However, previous studies has been primarily based on European or American populations. Because there are genetic variations among different ethnicities, it is valuable to evaluate the genetic background of NAFLD in Asian populations. In a GWAS using a Japanese population, SNPs in the PNPLA3 and SAMM50 genes were significantly associated with NAFLD, in accord with our results.9,10,19 We observed that the SNPs that were associated with the presence of NAFLD also showed significant associations with the severity of NAFLD.
Although ultrasonography is widely used as a first-line method to detect NAFLD according to practical clinical guidelines,20,21 ultrasonography has limitations when grading the severity of NAFLD.22 A recent prospective study reported that serum ALT level elevation was associated with the degree of hepatic steatosis.23 Thus, in our study, we further stratified NAFLD subjects according to their ALT levels. Using criteria for NAFLD categorization that were similar those in our study, Sung et al.24 showed that overall cardiovascular risk was significantly greater in NASH, defined as steatosis accompanied by increased serum ALT levels, than in either simple steatosis or elevated ALT alone, suggesting that the NAFLD categories used in this study are useful for predicting the prognosis of NAFLD. We found significant associations of the PNPLA3 gene with suspected NASH in our study, consistent with previous studies showing association of rs738409 with serum ALT levels in Japanese18 and U.S. populations.25
The PNPLA3 gene is involved in lipid metabolism and modulates hepatic triglyceride accumulation. Recently, rs738409 was associated with decreased serum levels of triglycerides in type 2 diabetes.10,26 In addition, homozygous carriers of the PNPLA3 risk allele showed higher levels of fasting glucose than heterozygous carriers.27 However, inconsistencies remain, there are no significant associations between rs738409 and NAFLD-related metabolic traits28 in consistent with our results. These findings suggest the possibility of disconnect between metabolic risk factors and the risk of NAFLD conferred by rs738409. Metabolic syndrome may be related to obesity, dyslipidemia, hypertension and glucose intolerance while NAFLD may be more related to endogenous genetic predispositions toward fat accumulation in the liver. The role of SAMM50 and TM6SF2 in lipid metabolism and hepatic triglycerides accumulation have been reported. However, controversies remain that SAMM50 gene were associated with reduced or increased levels of serum triglycerides.10,29 TM6SF2 was known to be a regulator of hepatic fat metabolism.30,31 Further studies are needed to confirm the mechanism by which metabolic traits and NAFLD are associated.
The strength of our study is that this is the first genetic analysis based on GWAS regarding NAFLD in Korean population, reflecting ethnic characteristics. Moreover, because of the relatively large sample size, the results of our study might have adequate statistical power.
There are several limitations in this study. First, we selected the discovery set and the validation set from the same population (those who had regular health check-ups at our institute). In the future, a validation study should be performed using a different population set. Second, because the study population was based on subjects who visited a single health screening center in Korea for health check-ups, it may not be representative of the general population, and there might be a selection bias. Third, it is impossible to discriminate normal, NAFLD and NASH using single ultrasonographic exam and serum ALT level. Fourth, hepatic sonography cannot diagnose a small fatty infiltration below 30% and it is not so much accurate as Fibroscan or liver biopsy to measure the degree of fatty liver, thus there may be the possible measurement bias. Finally, we cannot identify the effect of insulin resistance as a metabolic trait because there was no data regarding insulin levels in this study.
In conclusion, we demonstrated that the PNPLA3 and SAMM50 genes are significantly associated with the presence and severity of NAFLD in a Korean population. These findings confirm the important roles of genetic factors in the pathogenesis of NAFLD.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther. 2011;34:274–285. doi: 10.1111/j.1365-2036.2011.04724.x. [DOI] [PubMed] [Google Scholar]
- 2.Ekstedt M, Franzén LE, Mathiesen UL, et al. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006;44:865–873. doi: 10.1002/hep.21327. [DOI] [PubMed] [Google Scholar]
- 3.Adams LA, Lymp JF, St Sauver J, et al. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–121. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Rafiq N, Bai C, Fang Y, et al. Long-term follow-up of patients with nonalcoholic fatty liver. Clin Gastroenterol Hepatol. 2009;7:234–238. doi: 10.1016/j.cgh.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Wilfred de Alwis NM, Day CP. Genes and nonalcoholic fatty liver disease. Curr Diab Rep. 2008;8:156–163. doi: 10.1007/s11892-008-0027-9. [DOI] [PubMed] [Google Scholar]
- 6.Gambino R, Cassader M, Pagano G, Durazzo M, Musso G. Polymorphism in microsomal triglyceride transfer protein: a link between liver disease and atherogenic postprandial lipid profile in NASH? Hepatology. 2007;45:1097–1107. doi: 10.1002/hep.21631. [DOI] [PubMed] [Google Scholar]
- 7.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonal-coholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 9.Kawaguchi T, Sumida Y, Umemura A, et al. Genetic polymorphisms of the human PNPLA3 gene are strongly associated with severity of non-alcoholic fatty liver disease in Japanese. PLoS One. 2012;7:e38322. doi: 10.1371/journal.pone.0038322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitamoto T, Kitamoto A, Yoneda M, et al. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of non-alcoholic fatty liver disease in Japan. Hum Genet. 2013;132:783–792. doi: 10.1007/s00439-013-1294-3. [DOI] [PubMed] [Google Scholar]
- 11.Choe EK, Lee Y, Cho JY, et al. Search for genetic factor association with cancer-free prostate-specific antigen level elevation on the basis of a genome-wide association study in the Korean population. Eur J Cancer Prev. doi: 10.1097/CEJ.0000000000000359. Epub 2017 May 3. https://doi.org/10.1097/CEJ.0000000000000359. [DOI] [PubMed]
- 12.Prati D, Taioli E, Zanella A, et al. Updated definitions of healthy ranges for serum alanine aminotransferase levels. Ann Intern Med. 2002;137:1–10. doi: 10.7326/0003-4819-137-1-200207020-00006. [DOI] [PubMed] [Google Scholar]
- 13.Saadeh S, Younossi ZM, Remer EM, et al. The utility of radiological imaging in nonalcoholic fatty liver disease. Gastroenterology. 2002;123:745–750. doi: 10.1053/gast.2002.35354. [DOI] [PubMed] [Google Scholar]
- 14.Saverymuttu SH, Joseph AE, Maxwell JD. Ultrasound scanning in the detection of hepatic fibrosis and steatosis. Br Med J (Clin Res Ed) 1986;292:13–15. doi: 10.1136/bmj.292.6512.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan X, Waterworth D, Perry JR, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150:1219–1230.e6. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers JC, Zhang W, Sehmi J, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hotta K, Yoneda M, Hyogo H, et al. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet. 2010;11:172. doi: 10.1186/1471-2350-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korean Association for the Study of the Liver (KASL) KASL clinical practice guidelines: management of nonalcoholic fatty liver disease. Clin Mol Hepatol. 2013;19:325–348. doi: 10.3350/cmh.2013.19.4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology. 2012;142:1592–1609. doi: 10.1053/j.gastro.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Schwenzer NF, Springer F, Schraml C, Stefan N, Machann J, Schick F. Non-invasive assessment and quantification of liver steatosis by ultrasound, computed tomography and magnetic resonance. J Hepatol. 2009;51:433–445. doi: 10.1016/j.jhep.2009.05.023. [DOI] [PubMed] [Google Scholar]
- 23.Cruz MA, Cruz JF, Macena LB, et al. Association of the nonalcoholic hepatic steatosis and its degrees with the values of Liver Enzymes and Homeostasis Model Assessment-Insulin Resistance index. Gastroenterology Res. 2015;8:260–264. doi: 10.14740/gr685w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung KC, Ryan MC, Wilson AM. The severity of nonalcoholic fatty liver disease is associated with increased cardiovascular risk in a large cohort of non-obese Asian subjects. Atherosclerosis. 2009;203:581–586. doi: 10.1016/j.atherosclerosis.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Hernaez R, McLean J, Lazo M, et al. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third National Health and Nutrition Examination Survey. Clin Gastroenterol Hepatol. 2013;11:1183–1190.e2. doi: 10.1016/j.cgh.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer CN, Maglio C, Pirazzi C, et al. Paradoxical lower serum triglyceride levels and higher type 2 diabetes mellitus susceptibility in obese individuals with the PNPLA3 148M variant. PLoS One. 2012;7:e39362. doi: 10.1371/journal.pone.0039362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krawczyk M, Gruenhage F, Mahler M, Tirziu S, Acalovschi M, Lammert F. The common adiponutrin variant p.I148M does not confer gallstone risk but affects fasting glucose and triglyceride levels. J Physiol Pharmacol. 2011;62:369–375. [PubMed] [Google Scholar]
- 28.Speliotes EK, Butler JL, Palmer CD, et al. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Lin Z, Jiang M, et al. Genetic variants in the SAMM50 gene create susceptibility to nonalcoholic fatty liver disease in a Chinese Han population. Hepat Mon. 2015;15:e31076. doi: 10.5812/hepatmon.31076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen LZ, Xia HH, Xin YN, Lin ZH, Xuan SY. TM6SF2 E167K variant, a novel genetic susceptibility variant, contributing to nonal-coholic fatty liver disease. J Clin Transl Hepatol. 2015;3:265–270. doi: 10.14218/JCTH.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahdessian H, Taxiarchis A, Popov S, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A. 2014;111:8913–8918. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]