Abstract
The impact of returning secondary results from exome sequencing (ES) on patients/participants is important to understand as ES is increasingly utilized in clinical care and research. Participants were recruited from studies using ES and were separated into two arms: 107 who had ES and were offered the choice to learn secondary results (ES group) and 85 who had not yet had ES (No ES group). Questionnaires were administered at baseline and 1 and 12 months, following results disclosure (ES group) or enrollment (No ES group). While the majority (65%) elected to learn all results following pre-test counseling, it was reduced from the 76% who indicated a desire for all results at baseline. Thirty-seven percent received results associated with an increased personal disease risk. There were no differences in changes in any of the psychological and social measures from baseline to post-results disclosure between the ES and No ES groups. Receiving a wide range of secondary findings appeared to have little measurable impact on most participants. The experience of learning secondary results may be related to participants’ previous experiences with genetics, as well as the genetic counseling provided. Future research with a more diverse, genetically naïve group, as well as scalable methods of delivery, is needed.
Keywords: Exome sequencing, secondary findings, incidental findings, psychosocial measures, genetic counseling
INTRODUCTION
The management of secondary or incidental results (results unrelated to the indication for the test) from exome/genome sequencing (ES, GS) has been a growing concern with increasing sequencing use (Biesecker, Burke, Kohane, Plon, & Zimmern, 2012; Clarke, 2014; Wolf et al., 2012). ES is now widely used in both research and clinical care, and the number of people having ES is expected to continue to grow, with nearly 30 clinical laboratories offering ES or GS (concertgenetics.com) and with research studies such as “All of Us” and Genomics England (https://allofus.nih.gov/, https://www.genomicsengland.co.uk/) under way.
When asked, patients/participants usually indicate a desire to learn about secondary results. Attributes of the conditions, including degree of risk, screening options, availability and effectiveness of treatment, severity, confidence in the genetic interpretation, and cost of testing have been identified as influencing participant/patient preferences for results, though whether and how these factors are considered is specific to the person (Bennette et al., 2013; Bollinger, Scott, Dvoskin, & Kaufman, 2012; Fernandez et al., 2014; Regier et al., 2015; Townsend et al., 2012; Yushak et al., 2016). In some cases, participants’ choices appear not to be based on the findings’ relevance to clinical care, such as having a preference to learn about conditions without known treatment (i.e., Alzheimer’s), while not wanting to learn about conditions not associated with a disease risk but that may be relevant to medical care (i.e., pharmacogenetic variants) (Wynn et al., 2016). The basis for preferences is complex and likely influenced by many factors, including personal and family experience or knowledge of a particular condition. Patients/participants have expressed a sense of possessiveness of their genetic data and often feel there is an obligation on the clinician/researcher to share this information (Bollinger et al., 2012). They emphasize their right to choose what information is released to them and are often accepting of possible uncertainty of the findings (Townsend et al., 2012). Patient/participants also express concerns about the privacy of this information and the potential negative impact on insurance coverage (Yushak et al., 2016).
Physicians and researchers, in turn, have concerns about responsibly returning secondary results. These concerns include having the necessary knowledge and support to facilitate informed consent, accurately interpret variants in genes with which they are not familiar, and provide appropriate clinical recommendations. Despite these concerns, many clinicians and researchers agree that patients/participants should, at a minimum, be provided the opportunity to learn about selected secondary results, with options depending on the context (research or clinical) and age of the patient (Green et al., 2012; Wynn et al., 2015). Many clinicians and researchers support allowing the patient/participant to decline secondary results (Jarvik et al., 2014; Klitzman, Appelbaum, Fyer, et al., 2013). A dynamic disclosure process allowing for patients/participants to access results as needed or as desired has also been proposed (Yu, Jamal, Tabor, & Bamshad, 2013). Some caution against genetic exceptionalism and advocate for liberal disclosure policies (Green, Lupski, & Biesecker, 2013; Wilfond, Fernandez, & Green, 2015). For clinical sequencing, the American College of Medical Genetics recommends allowing patients the opportunity to opt out of return of secondary findings from a minimal list of 59 medically actionable genes (Green, Berg, et al., 2013; Kalia et al., 2017). Some clinical laboratories offer return of additional genes beyond the ACMG recommended list, including carrier results and pharmacogenetic variants. While the ACMG guidelines are followed by the majority of clinical labs (e.g., GeneDx, Ambry, Baylor/Miraca), there is currently no consensus policy for return of secondary findings from ES/GS in research. Practices depend on what each researcher views as appropriate, guided by the policies of their institutional review board.
Some of the concern surrounding return of secondary results relates to possible negative impact on the patient/participant. Clinical genetic testing based on personal or family history can cause distress, anxiety, and uncertainty, but generally patients adapt to the information, and these feelings can activate patients to take initiatives to reduce health threats (Bosch et al., 2012; Cella et al., 2002; Krabbenborg et al., 2016; Lumish et al., 2017; Rosell et al., 2016). The experience of receiving secondary results may differ from the results of focused clinical testing because in the latter case patients are likely to have greater familiarity with the condition, have deliberately sought specific genetic information, and have the opportunity to prepare psychologically for the findings. Another concern is that patients/participants may incorrectly assume they have no risk for a condition if secondary results are negative and may forgo recommending screening and preventive behavior (Klitzman, Appelbaum, & Chung, 2013). Early research indicates that participants receiving research results find the information valuable, even when the results are negative, though they do not report changes in their lifestyle behaviors. Participants generally share the information with their health care providers, report the experience to be positive or neutral and not negative, and have no changes in their psychological wellbeing (Lewis et al., 2016; S. C. Sanderson et al., 2017).
We previously examined preferences for receipt of secondary results of participants enrolled in genomic research studies and found that the majority of participants preferred to learn all types of results offered. Participants who reported greater concern about genetic privacy were more likely to indicate a preference to limit the types of results returned (Wynn et al., 2016). Here, we present the experience of this same group after a subset of participants was given the option to receive secondary results. We examine how actual choices differ from initial preferences, and the psychological, medical and social impact of receiving a wide range of secondary results within the context of a research study.
METHODS
Study Sample
Participant recruitment and study sample were previously described (Wynn et al., 2016). Briefly, participants were recruited via invitation letter and follow-up phone call from a population of English-speaking, adult research participants enrolled in studies using ES to identify novel human disease genes at Columbia University Medical Center (CUMC). Written consent was obtained. The Columbia University institutional review board approved this study.
The participants came from three categories of research studies (parent study) utilizing ES: 1) a study of probands with a history of breast cancer, 2) two studies of congenital heart defects (CHD) in which all but one participant, who was an adult affected proband, were unaffected parents of an affected child, and 3) studies of congenital diaphragmatic hernia (CDH), multiple congenital anomalies (MCA) developmental delay (DD), muscle weakness (MW), neurodevelopmental disorders, or familial diabetes in which all but one participant, who was an affected adult proband with diabetes, were unaffected parents of an affected child. Enrolled participants were separated into two study arms: participants who had ES as part of the parent study (ES group) with the potential for secondary findings, and participants who had not yet undergone ES in the parent study (No ES group) and thus had no secondary findings. The decision as to which participants had ES at the time of this study was based on the research goals of the parent study, in general reflecting family history, age of onset, and disease severity. Participants did not receive any results related to the indication for their entry into the parent study (primary results) during the course of this study and were only able to make choices about secondary results for themselves.
Questionnaires
Questionnaires were administered at three time points in the study (Figure S1). The baseline questionnaire was administered following consent to the study. After baseline questionnaire completion, participants were notified if they were in the ES or No ES group. The second questionnaire was administered to the ES group one month following results disclosure and to the No ES group one month after completion of the baseline survey. The third questionnaire was administered to the ES group 12 months following results disclosure and to the No ES group 12 months after completion of the baseline survey. The questionnaires were administered online (21%) or by paper (78%), according to the participants’ preferences.
Educational videos
A 30-minute genetic educational video was produced for this study and mailed to participants in both the ES and No ES groups following consent to the study. Video content was developed by genetic counselors, geneticists, psychologists and a genetic research assistant. The 30-minute video introduced the study, reviewed facts about genetic information and inheritance, and discussed reproductive options. It also discussed types of possible results, including pharmacogenetic results, carrier status and results for Mendelian conditions. The video reviewed one or more examples for each category. The video used a lecture format with basic graphics featuring some animation, with a voiceover explaining the information being presented (https://youtu.be/PlIDMmhfXk4). The video was an optional component of the study and whether the participant watched the video was not formally assessed.
Genetic counseling
Participants in the ES group received education and counseling with the genetic counselor (JW), and in some cases also the medical geneticist (WKC), prior to making a final decision about the types of results they wanted to learn, and had a follow-up appointment at which results were disclosed. The sessions were conducted in person or by video conference, according to participant preference. The pre-test session began with a three generation family history and then a client-centered approach was used to elicit the participant’s choices about the types of secondary results they would like to learn. To facilitate a conversation, the participant was asked about their understanding and expectations of the study. When a specific topic was not raised by the participant, opened questions were used to start a conversation about any specific concerns the participant had about a particular types of results and the degree of risk, severity and treatability of results. Participants were also asked to imagine how they might respond to specific types of results and the potential impact on them and their family and reflect on how they have managed uncertain or unexpected information, both medical and non-medical. The initial vignettes in the baseline survey about which patients made hypothetical choices were chosen because the diseases were associated with varying degrees of risk, severely and treatability; however result choices in the pre-test sessions were not limited to the bins of these categories. Participants were able to allowed to make more granular choices about the results they would like to receive and were not restricted by categories or conditions. Results disclosure sessions were also conducted in a client-centered approach and began with review of the results the participant elected to receive and confirmation of this choice. The participant was then asked what information they would first like to learn about. Open ended questions were asked to assess their understanding of the results and recommendations and to assess their emotional experience. Sessions were audio-recorded and reviewed by JW immediately following the visit. Quotes that highlighted themes raised in the session were extracted. Extracted quotes were then reviewed for common themes by WKC, RJK and PSA.
Instruments
The methology of developing the survey has been previously published (Wynn et al., 2016). Briefly, the surveys were developed to explore the experience of receiving genomic results, including impact on emotions, health, and life behaviors. The surveys were assessed for length and clarity by 15 medical professionals, including clinical geneticists, psychologists, psychiatrists, genetic counselors, and research coordinators. A quality check was completed following the administration of the first 90 baseline surveys and questions, that provided further granularity about family composition, genetic essentialism, burden of medical conditions, planning for the future, and numeracy, were removed to improve flow and shorten the survey.
Survey items included demographics, religiosity, medical history and psychosocial measures. In the baseline questionnaire, participants were asked to review vignettes about 11 types of genetic results and to indicate the likelihood that they would want each result. The results of these preferences have been previously published (Wynn et al., 2016).
In both the ES and the No ES groups the following measures, previously validated in other studies, were assessed at all three time points: the Health Locus of Control Scale (Wallston, Wallston, Kaplan, & Maides, 1976), Beck Anxiety Inventory (BAI) (Gech, Epstein, Brown, & Steer, 1988) and Personal Health Questionnaire (PHQ–9) (depression) (Spitzer, Kroenke, & Williams, 1999). The survey also included a question from the General Sleep Disturbance Scale (Lee, Hicks, & Nino-Murcia, 1991), and questions from the Genetic Knowledge Measure (Erblich et al., 2005) (adapted to assess general genetic knowledge rather than cancer-specific knowledge). Genetic stigma, genetic secrecy, healthy behavior and health worry questions were developed for this study (Wynn et al., 2016). We also asked questions about social supports and life changes (Table SI).
Additional measures and questions were administered to the ES group at 1 and 12 months post-results disclosure to assess the psychological impact of the results and satisfaction with their decision to receive results. The Multidimensional Impact of Cancer Risk (MICRA) scale (Cella et al., 2002), which was developed to evaluate distress, uncertainty and positive experiences associated with receiving results from hereditary breast and ovarian cancer genetic testing, was amended to address all genetic results. Other validated scales were used to address satisfaction with the decision to receive results (Satisfaction with Decision scale (Wills & Holmes-Rovner, 2003)), and actions and behaviors taken to cope with the results (brief COPE scale (Carver & Scheier, 1989; Cooper, Katona, Orrell, & Livingston, 2006)). The ES group was also asked questions about sharing results with medical providers and friends/families, and about distress related to their results.
Statistical analysis
Descriptive statistics are presented in frequencies for categorical variables and means and standard deviations for quantitative variables. The ES group was separated into two sub-groups: those with results associated with an increased personal disease risk (ES w/PDR) and those without (ES w/o PDR) who received only carrier, pharmacogenetics or reduced or average risk for Alzheimer’s disease. Differences in the demographic variables across the three result groups (No ES, ES w/PDR, and ES w/o PDR) were assessed by chi-squared tests and ANOVA (age variable) to evaluate for possible confounders. The mean change in scores (outcome) from baseline to 1 month and baseline to 12 months of the psychological and social measures across the three results groups (predictors) were assessed using linear regression models. Mean scores (outcome) at 1 month and 12 months of the measures assessing experience of receiving ES results, comparing the ES w/PDR and ES w/o PDR groups (predictors), were assessed using linear regression models. These analyses were completed both unadjusted and adjusted for the confounder of parent study type. A p-value of 0.05 was considered relevant for our exploratory analysis. Statistical analysis was completed in SAS and R 3.3.1 (R Core Development Team, 2010; "SAS Institute Inc. SAS 9.4 [computer program]," 2014).
ES Analysis
ES libraries were prepared using the NimbleGen VCRome targeted capture design (Roche NimbleGen, Madison, WI). Sequencing was performed on the Illumina HiSeq2500 platform using 75bp paired-end reads. We had an average coverage of 150x within the capture regions with an average of 85% bases covered at least 15X. The fastq files were aligned to human reference genome (GRCh37/hg19) using Burrows-Wheeler Alignment tool(Li & Durbin, 2009). GATK software package was used to remove PCR duplicates, recalibrate base quality scores, realign around indels, and call variants. The quality filters were used in accordance with GATK best practices. Variants were annotated with Annovar software (Wang, Li, & Hakonarson, 2010) and custom scripts. Annotations included population frequencies (1000 Genomes Project, ExAC, dbSNP, and the Exome Variant Server), deleteriousness scores (CADD(Kircher et al., 2014), PolyPhen-2(Adzhubei et al., 2010), SIFT(Kumar, Henikoff, & Ng, 2009), MetaSVM), conservation scores (GERP++(Davydov et al., 2010)), ClinVar database, and Cosmic database. After annotations, variants with an allele frequency of >1% in the 1000G or ExAC and variants which occurred in our internal variant database with a frequency of >5% were excluded. Non-synonymous coding and canonical splice site variants were retained. The remaining variants were manually curated with visual review of alignments. Variants were prioritized based on prior annotation as a monogenic disease causing variant in OMIM, HGMD, ClinVar, PubMed, and Google Scholar. Pathogenic/likely pathogenic variants were confirmed by Sanger sequencing (Applied Biosystems; Life Technologies, Darmstadt, Germany). All results that were returned to participants were independently confirmed in a Clinical Laboratory Improvement Amendment (CLIA)-certified laboratory with a new specimen.
RESULTS
Demographics
Results of participant enrollment were published previously (Wynn et al., 2016). The current analysis includes an additional four participants who completed enrollment after the 219 participants describe in the prior publication. There were 85 participants in the No ES group and 107 in the ES group (Figure S1). The other 31 participants were invited to the ES group but did not return up to five follow-up phone calls or declined to participate. The most common reasons for declining, when a participant was reached, were lack of time and the inability to commit to pre- and post-test counseling sessions.
The majority of the participants were female, White, non-Hispanic, married, over 45 years-old, and had a college degree or higher. Participants with ES were more frequently from the breast cancer study or CHD study and male compared to the No ES group (p-value 0.01, 0.04, respectively) (Table 1). Gender and parent study type were correlated, and therefore only parent study type was identified as a confounder.
Table 1.
Demographics of participants by study group (n=192).
| Demographics | ES w/PDR (n=40) | ES no PDR (n=67) | No ES (n=85) | p-value |
|---|---|---|---|---|
| Female | 73% | 66% | 84% | 0.038 |
| Married | 90% | 88% | 83% | 0.530 |
| Age | ||||
| Mean ± SD (range) | 50 ± 13.6 (33–79) | 50 ± 14.6 (22–84) | 48 ± 14.2 (23–88) | 0.600 |
| Ethnicity and Race | ||||
| White, Non-Hispanic | 80% | 88% | 81% | 0.431 |
| Education | ||||
| Up to HS or vocational training | 8% | 11% | 16% | 0.740 |
| Some college/Associate degree | 15% | 22% | 19% | |
| College degree | 33% | 27% | 26% | |
| Advanced degree | 45% | 40% | 39% | |
| Employment | ||||
| Employed (full or part-time) | 68% | 67% | 59% | 0.480 |
| Current Religion (n=191) | ||||
| Christian | 58% | 47% | 41% | 0.473 |
| Jewish | 20% | 26% | 38% | |
| Other (Buddhist, Taoist, Meditation) or >1 religion | 5% | 6% | 5% | |
| None | 18% | 21% | 16% | |
| Parent study type | ||||
| Breast cancer | 28% | 30% | 36% | 0.011 |
| CHD | 45% | 49% | 24% | |
| CDH, DD, D, MCA, MW, ES | 28% | 21% | 40% | |
| Disease status (n=184) | ||||
| Personally affected | 33% | 43% | 46% | 0.404 |
| Children (n=191) | ||||
| Affected children | 70% | 58% | 50% | 0.143 |
| Unaffected children only | 30% | 34% | 39% | |
| No children | 0% | 8% | 11% |
Abbreviations: congenital heart disease (CHD), congenital diaphragmatic hernia (CDH), developmental delay (DD), diabetes (D), multiple congenital anomalies (MCA), muscle weakness (MW), exome sequencing study (ES), personal disease risk (PDR).
p-values were calculated by Chi-squared test or ANOVA (age variable)
The 31 participants who did not continue with the study were more frequently from the CHD study (p-value 0.01), not personally affected (p-value 0.04), and on average 8 years younger (p-value 0.009) as compared to the 192 who proceeded with the study (Table S2).
There was participant attrition throughout the course of the study for both the ES and no ES groups. Two participants in the ES group who had a pre-test counseling session did not return to receive results despite five follow-up calls. One indicated that she did not have time, while the other did not return any follow-up phone calls or emails from the study coordinator and genetic counselor. At one month, participant retention for the ES w/PDR, ES w/o PDR and No ES groups was 83%, 94% and 95%, respectively, and at 12 months it was 78%, 84%, and 84%, respectively (Figure S1).
Actual versus hypothetical choices to learn results
Among participants in the ES group, following the pre-test counseling session, participants chose to learn fewer results compared to their baseline preferences. On the baseline questionnaire, 76% (77/101) of the ES group indicated a preference to learn all types of results, while following pre-test genetic counseling only 65% (66/101) did so (p-value <0.0001). Of those who changed their choice, 14 who initially indicated a preference to learn all results elected not to receive some results and three participants elected to learn all results though they had indicated a preference for more limited results at baseline. Hypothetical preferences were unknown for six particiants who did not provide these choices prior to the genetic cousenling session because they did not have time (n=4) or did not feel comfortable making these choices without consultation with a genetic counselor (n=2).
Several themes were identified in the audio recordings of the pre-test counseling sessions. Some participants expressed a desire to learn information to help them take care of their health or plan for the future, even if there was no known treatment or prevention.
“I want to know regardless of the fact that I cannot do anything because I do want to plan.” RoR285, elected all results.
Other felt some obligation to learn their results to help their family members.
“That is like some of the benefits. If they [children] want to be exposed to that information, they can be. And they can have it. That may be something they want to know and maybe not. That will be their choice when they are old enough.” RoR283, elected all results.
Many also reflected on their curiosity about their genetic information.
“I have to die at some point so why not know more about it? I’m also just intellectually curious.” RoR269, elected all results.
Several participants reflected on an experience with a particular condition and how this affected their desire to learn or not learn a result. Some participants had discordant choices despite similar experiences with a condition.
I have had life experience with friends’ and families’ Alzheimer’s who were diagnosed and I have seen what happens, and I just would not want to know.” RoR360, elected limited results; no results associated with dementia.
“I would like to know about Alzheimer’s since I suspect I’m at risk of it …But it would make me feel more empowered to be able to do something about it if I knew about it rather than just wonder if I was at a high risk. Again, it could work in the reverse if it came back that you don’t have the genetic make up to make you more likely. Then it would be a relief but again it wouldn’t be a huge change to how I live.” RoR333, elected limited results, but elected results related to dementia because she cared for her grandmother with dementia.
Others also described their life experiences of having difficulty with their own or their child’s health and how those experiences affected their decisions.
“We already have a dark cloud–exactly–this one won’t affect us.” RoR277, elected all results; has a child with neurocognitive disabilities
“I think if we hadn’t gone through that, then all this talk would be a little bit more scary.” “I have all that to deal with this, I don’t want to say it is nothing, but it [ the study] doesn’t seem all that scary.” RoR338, elected all results; has a child with a congenital heart defect.
Overall, 64% (68/107) of the ES group elected to receive all results. For the 39 participants who choose not to learn about some results, these choices were often unique. One participant chose not to learn about carrier results and elected to learn about all other results. Other participants had specific disease types they elected or declined to learn about. For example, several participants declined to learn about dementia but elected to learn all other types of results. Other participants had more granular choices; one declined to learn about myopathies and dementias and another elected to learn about conditions associated with CHD, diabetes, breast cancer as well as pharmacogenetic results and declined all other results. Other participants’ choices reflected the penetrance or treatability of the condition. One participant declined to learn about dementia as well as any condition that was associated with a less than 60% penetrance and another declined to learn about any condition where the penetrance was uncertain. Some participants declined to learn about any results without available treatment while others specified specific conditions they would or would not like to learn depending on the available treatment. Some participants had different criteria depending on the type of condition. For example, one participant declined to learn about results associated with young onset dementia and any type of myopathies but elected to learn about all other types of results.
Personal Disease Risk
Over a third (40/107) of the participants in the ES group received results associated with an increased personal disease risk (Table 2). None were aware of their status prior to the study. With the exception of one participant with a MYH7 pathogenic variant who had a history of mild left ventricular hypertrophy with normal function, the participants did not have a known diagnosis of the condition associated with the risk identified at the time of the results disclosure. Some, but not all, had a family history of the condition.
Table 2.
Personal disease risk (PDR) identified in the 40 participants.
| Gene | Condition | N* |
|---|---|---|
| ABCA4 | Age Related Macular Degeneration | 4 |
| APO E4/E3* | Alzheimer's Disease | 19 |
| APO E4/E4 | Alzheimer's Disease | 1 |
| DSP | Arrythmogenic Right Ventricular Cardiomyopathy | 1 |
| G6PD** | Glucose 6 Phosphate Dehydrogenase | 1 |
| GCKR | Hypertriglyceridemia | 1 |
| GJB1** | Charcot-Marie-Tooth Neuropathy X type 1 | 1 |
| HFE*** | Hereditary Hemochromatosis | 1 |
| KCNE2 | Arrhythmia | 1 |
| LHX4 | Pituitary hormone deficiency | 1 |
| MC4R | Obesity | 1 |
| MEFV*** | Familial Mediterranean Fever | 1 |
| MYBPC3 | Cardiomyopathy | 1 |
| MSH6 | Hereditary Nonpolyposis Colorectal Cancer | 1 |
| MYH7 | Cardiomyopathy | 1 |
| SCN5A | Arrhythmia | 3 |
| SERPINA1 S/Z | Alpha-1-antitrypsin deficiency | 1 |
| SERPINA10 | Venous thromboembolic disease | 1 |
| VWD | Von Willebrand Disease | 1 |
| ZEB1 | Fuchs' corneal dystrophy | 2 |
| Total * | 44 |
4 people had APO E4/E3 and an additional pathogenic variant
X-linked condition, participant was male or a phenotype was also reported in females
Participant has compound heterozygous pathogenic variants
Psychological Measures
As previously reported, in the study sample as a whole, the frequency of anxiety measured by a score of > 10 on the BAI, indicating moderate to severe anxiety, was somewhat greater (28.2%) than the general population (18.1%), whereas depression measured by a score of > 10 on the PHQ-9 (7.4%), indicating moderate to severe depression, was similar to the general population (7.6%) (Kessler, Chiu, Demler, Merikangas, & Walters, 2005; Wynn et al., 2016). In this analysis, the levels were somewhat different among the study groups: 38% of the No ES group, 20% of the PDR w/ES and 21% of the ES w/o PDR were anxious (p-value 0.016) and 12% of the No ES group, 3% of the ES w/PDR and 2% of the ES w/o PDR were depressed (p-value 0.05).
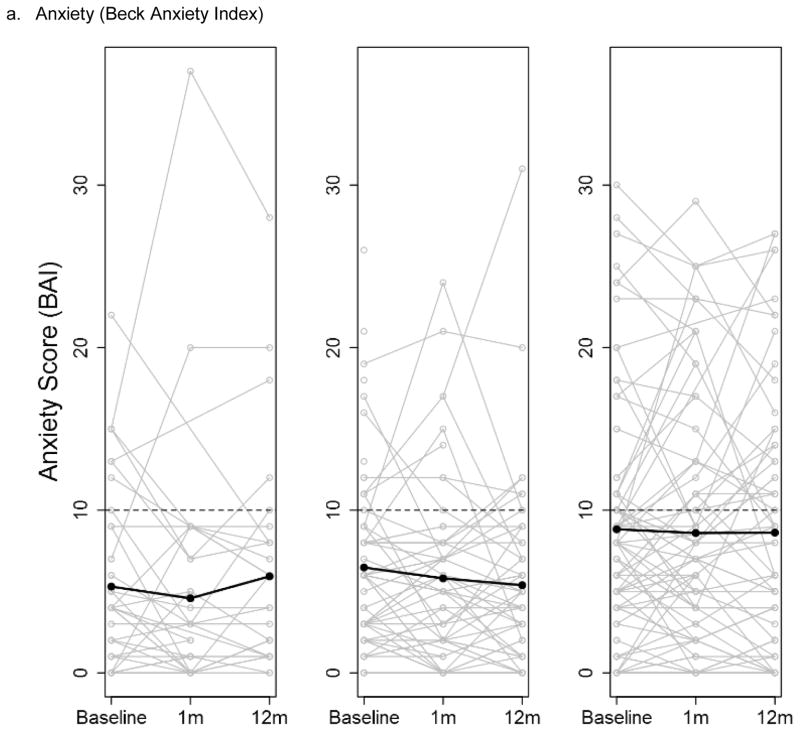
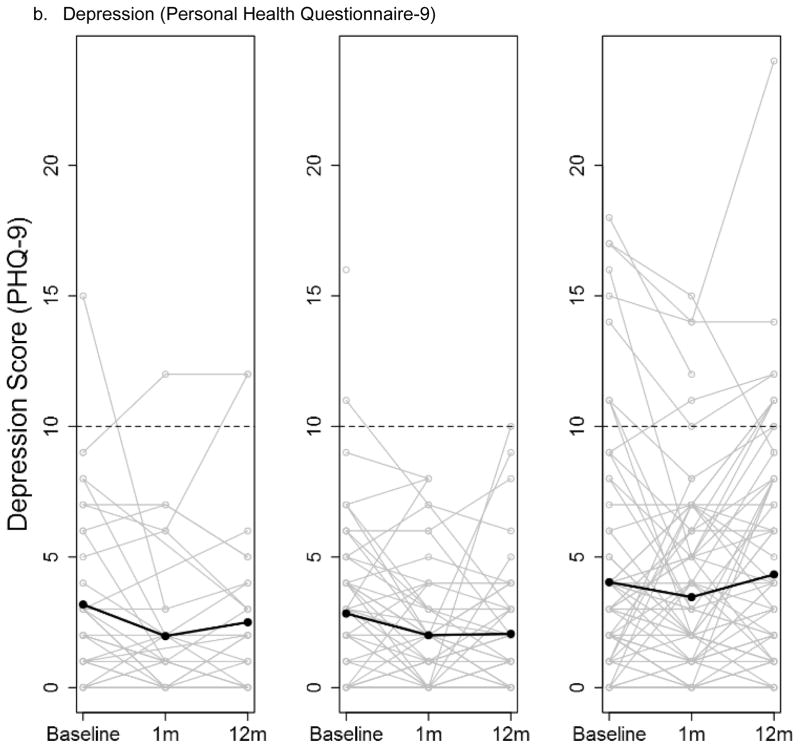
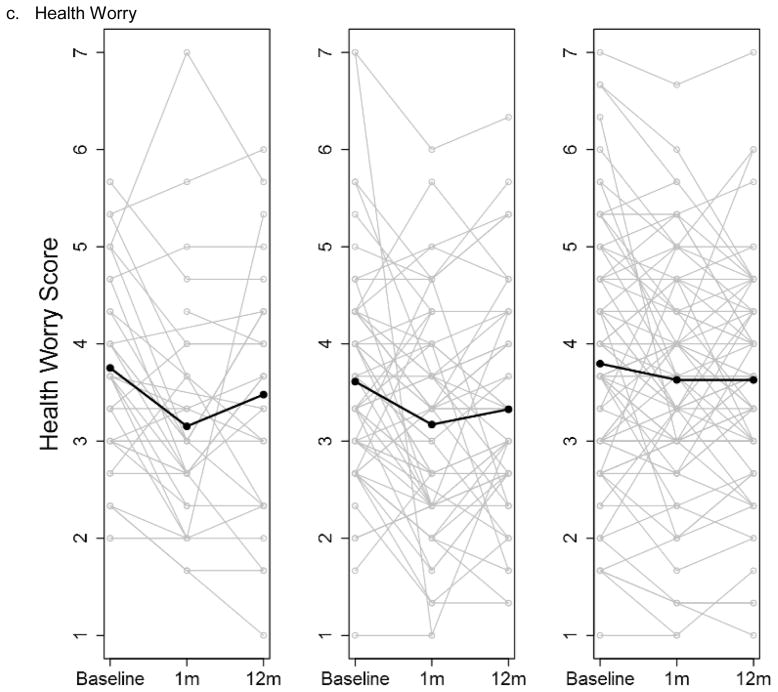
There were few differences in changes over time in the psychological and social measures for the three groups throughout the study; analyses adjusted and unadjusted for the parent study type were similar. At baseline, the mean scores of the measures were similar, with the exception of anxiety, which was higher in the no ES group (Table S3a). There were no differences in mean change in any of the measures across the three groups from baseline to 1 month post-results disclosure (ES group)/ post-enrollment (No ES group) or baseline to 12 months post-disclosure (ES group)/ post-baseline (No ES group) (Table S3b–c). The depression and anxiety scores displayed considerable variability among the participants, and that variability was maintained over the duration of the study in all three study groups (Figure 1a–b). The degree of health worry was also variable across participants; there was a non-significant trend of a decrease in health worry at 1 month and then return to near baseline at 12 months for the two ES groups that was not seen in the No ES group (p-value 0.11) (Figure 1c).
Figure 1.
Participant anxiety (a), depression (b) and health worry (c) scores at baseline, 1 month and 12 months post-results disclosure (ES groups) and post-enrollment (No ES group) stratified by study group (Exome sequencing with personal disease risk (ES w/PDR), ES w/o PDR and No ES). Individual participant scores are shown by grey lines with overlaid means scores at each time point shown by a black line. The dotted line indicates the threshold for diagnosis of anxiety or depression.
Abbreviations: exome sequencing (ES) personal disease risk (PDR)
Other impact of ES results
Overall, the impacts of results for the ES w/PDR and ES w/o PDR groups were similar (Table S4a–b). There was high satisfaction (1=complete satisfaction) with participants’ choices to learn about results in both groups at both 1 and 12 months (1 month means: 1.03 ES w/PDR, 1.04 ES w/o PDR). When asked about specific actions and behaviors to cope with their results (Brief COPE), on average, participants were not engaging in these actions or behaviors (score=1) or were only doing so a little bit (score=2) at both time points. There was a trend for ES w/PDR participants to report using slightly more of all types of coping methods compared to those without PDR results, and this was significant for emotion-based coping (acceptance, seeking emotional support, using humor, religion and positive reframing) at 1 month (means: 1.52 ES w/PDR, 1.22 ES w/o PDR, p-value 0.01) and 12 months (means: 1.76 ES w/PDR, 1.23 ES w/o PDR, p-value 0.001), and problem-based coping (active coping, instrumental support and making a plan) at 12 months (means: 1.75 ES w/PDR, 1.29 ES w/o PDR, p-value 0.002) (Table S4a–b). The impact of the results, as measured by the aMICRA, was similarly modest for both groups, with a mean total aMICRA score at 1 month for the ES w/PDR of 12.8 and for the EWS w/o PDR of 13.3 (p-value 0.73), though the range of aMICRA scores was wider for the ES w/ PDR group (2–31) compared to the ES w/o PDR group (0–23). Both groups had low levels of distress and uncertainty and modest positivity related to the results on the aMICRA subdomains. Few in either PDR group described being upset with any of the specific types of results. No one in either group regretted being part of the study (data not shown).
In review of the audio recordings, the participants’ reactions in the disclosure sessions reflected the modest impact of the results. The participants without PDR often expressed disappointment about the lack of results to help guide their health care.
“I thought the results would be more specific and I am disappointed it is not more comprehensive.” ROR180, elected all results; did not have PDR.
While some expected to learn more than they did, they were relieved not to have any risks identified.
“Basically I was more worried about having some genetic markers for cancer or heart [disease] and feel like coming away with a reasonably clean slate there. I'm always tending to assume the worst case, I expected more than I actually did have.” ROR281, elected selected results; did not have PDR.
Other participants who did not have PDR but did have other results, such as carrier or pharmacogenetics, acknowledged the importance of sharing their results with family members.
“Most important thing is that I'm able to let my daughters know to get testing. It is not a usual test so the normal genetic screen would not identify it. It’s really difficult to have a child with special needs” [when discussing her carrier results]. ROR454, elected all results
Those participants who received PDR results often expressed feelings of empowerment from having the knowledge.
“What I was worried about I’m not worried about because now I know that I can be screened and never get it.” RoR270, elected selected results; received PDR (cancer risk).
Some participants who received PDR results recognized that to have the results helps them to seek targeted medical care to reduce their risk or enable early diagnosis.
“Relieved that I have the test results - give me a starting point. I'm going to see a cardiologist.” ROR357, elected all results; received PDR (Arrhythmia)
Many were also relieved despite having PDR, and often felt that the results could have been more severe or have had a greater health consequence.
“I was all prepared just in case there was bad news, but I was very relieved there was no really bad news” RoR340 elected selected results, received PDR (Arrhythmogenic Right Ventricular Cardiomyopathy).
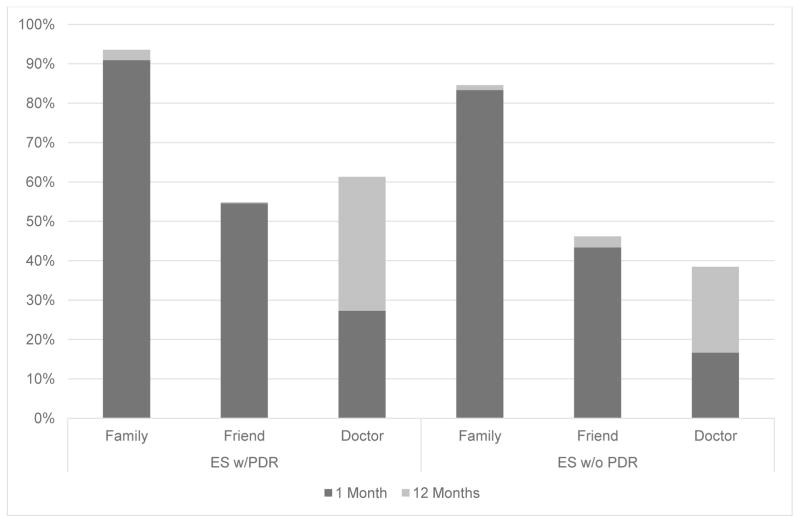
The majority of participants shared their results with family members and over half had shared their results with friends by 12 months (Figure 2). Only 61% of participants with PDR reported sharing their results with a doctor and even fewer (38%) did so if they did not have PDR (Figure 2). Nearly all participants in the ES groups would recommend this type of genetic analysis to others (88% PDR, 87% without PDR).
Figure 2.
Percentage of participants who had shared results with different types of people at 1 month and 12 months post-results disclosure stratified by those with PDR and those without. (chi squared test: no significant difference for any category)
Abbreviations: exome sequencing (ES) personal disease risk (PDR)
DISCUSSION
This study found that most genomic research participants choose to learn some or all secondary results offered when given a hypothetical option, though fewer actually want to learn all results when the choice is real. Often-expressed concerns about returning secondary findings are that the results will cause undue stress or anxiety, and increase genetic stigmatization, or that individuals with no findings will pursue more risky health behaviors or forego routine screening (Klitzman, Appelbaum, & Chung, 2013; Townsend et al., 2012). Our exploration of the psychological and health behavior impacts of returning secondary results did not identify any measurable changes in depression, anxiety, perceived genetic stigma, or desire for secrecy. Nor did participants have changes in health behaviors or perceptions of control over their health. The majority of the participants shared their results with friends and family members. However, fewer shared their results with their doctors, suggesting that participants did not perceive utility of results for their current medical care or had not yet seen a relevant healthcare provider within the 12 months after disclosure. Participants with PDR results reported using slightly more emotional and problem-based coping than those without PDR but coping behaviors and actions were minimal in both groups. Overall, learning about a wide range of secondary findings (well beyond the ACMG list of secondary findings) appeared to have little measurable impact on most participants, which is consistent with previous reports (Lewis et al., 2016; S. C. Sanderson et al., 2017). Although a direct comparison is not possible, it is notable that the values of the adjusted MICRA scale were lower than typically found in women having hereditary breast and ovarian cancer genetic testing (Cella et al., 2002; Lumish et al., 2017), suggesting less psychological impact of secondary results.
Several aspects of the study should be considered when examining the experiences reported by the participants. Participants had the option to view an educational video about the genetic testing and had a formal genetic counseling session before making the final decision about what results to receive. They were given the option to make granular choices about their preferences for results based on the disease and the medical impact (with or without effective medical intervention, carrier status and pharmacogenetics). Prior to making a decision, participants were encouraged to reflect on their experiences with disease and genomics and their comfort with uncertainty and unexpected health information, and were guided in the genetic counseling session on how to consider these factors when making their choices.
When participants chose to learn only selected results, the specific choices were often unique to the participant, and reflective of their experiences with certain diseases and comfort with uncertainty, and were not necessarily consistent with traditional clinical categories (e.g. severity, actionability). Some elected not to learn about a specific health condition for individual reasons (e.g., having had a friend who had the condition, or being someone who would never consider having an implantable cardiac defibrillator), others had a threshold of penetrance that guided their decisions, and some considered the availability of treatment and prevention options for health conditions. Notably, the number of participants who wanted all results offered decreased from 76% after pretest education to 65% after completing pre-test counseling. The experience of the genetic counselor and her ability to guide the participants to consider all relevant variables and to make informed decisions may have influenced participants’ choices and generally positive post-test satisfaction with the results they received. Our findings confirm that hypothetical choices are not equivalent to actual choices, a phenomenon that has been observed in other studies and is likely influenced by factors including confronting the reality of proceeding with testing and the potential implications of the results and the logistics of having the test (Ropka, Wenzel, Phillips, Siadaty, & Philbrick, 2006; S. Sanderson, O’Neill, Bastian, Bepler, & McBride, 2009; Sawyer et al., 2006).
The observation of a decrease in health worry (for those with and without PDR) at the 1-month post-disclosure time point and return to baseline at 12 months may be a reflection of the nature of the results disclosed, the preparedness of the participants and the framing of the results in the larger health maintenance context. Results were disclosed by the same geneticist and genetic counselor who completed the pre-test counseling session. The previously established, trusting relationship with genomic professionals and confidence in the information about the degree of risk associated with the results, as well as the frequent availability of preventive measures, may have evoked feelings of empowerment and control over their own health.
While not directly measured in this study, it is important to note that one of the most common reactions identified in the review of the audio recordings of the disclosure sessions was a feeling of disappointment when there were no PDR results. Participants displayed a tendency to overestimate the ability of genomics to predict disease risk and subsequent disappointment about the lack of overall results or of results related to a specific disease or trait, which has been documented in other genomic studies, suggesting a need to anticipate and manage unrealistic expectations of GS (Amendola et al., 2015; Lewis et al., 2016; S. C. Sanderson et al., 2017).
Limitations
There are several limitations to the generalizability of our results. Our study participants were mostly white, non-Hispanic, with college or higher levels of education, and more than half were beyond reproductive age. The 31 who did not proceed with the study after completion of the baseline survey were more likely be younger and from the CHD study, and therefore parents of affected children, than those who proceeded with the study. The burden of committing to two genetic counselling sessions while taking care of a sick child may have been too great. The experiences of those who dropped out after the baseline survey may have been different from the other participants in this study. All of the participants were part of a primary genomic research study and had previous exposure to information about genetics and its potential impact on health. Many had previously had genetic counseling and clinical genetic testing. A large proportion had experienced and were continuing to experience significant personal and family health issues and were coping with these challenges. These issues, and other unmeasured effects, may have influenced their depression and anxiety during the study period (as evidenced by the baseline elevation and variability of these measures across time points in the No ES), which in turn may have limited our ability to fully detect the psychological impact of ES results in their lives.
Only approximately one-third of the ES group received PDR results and many of them were incompletely penetrant and associated with a low-to-moderate risk; people who receive secondary findings associated with more penetrant conditions might have a different experience. The sample size of the study was limited; it is possible that a small proportion of people will have experiences we did not observe on returning secondary findings, and that the current study did not have sufficient power to detect these rare events. The measures used were validated for different adult patient populations and in most cases this is the first time they have been used to assess the experience of genetic testing. Therefore, our negative findings may relate to the failure of the measures to detect differences that are present. In particular, the mean decisional satisfaction and coping measure scores were close to the limit of the scales (upper for satisfaction and lower for coping) and therefore these measures were unable to assess an differences due to the type of results received.
Practice Implications
This study provides important insight for clinicians and researchers disclosing secondary findings from genomic sequencing. Our results indicate that genomic research participants, when provided with appropriate pretest education, pre- and post-test counseling, and the option to choose the information to be received, are minimally impacted—either positively or negatively—by receipt of a wide range of secondary findings. Our results also highlight the unique and individual choices participants make about what types of results to receive and how their choices change following genetic counseling, suggesting that the current model of all or none may not be appropriate for all participants and patients. Clinicians and researchers need to continue to provide appropriate pre-test education and counseling to help guide participants and patients to make individualized choices. Finally, more scalable models of education and counseling will need to be developed to meet the growing population of people having genomic sequencing.
Research Recommendations
Future studies should examine participants’ experiences of receiving secondary findings using alternative methods of delivery (e.g., online portals), with larger sample sizes, in more diverse populations. The medical and personal utility of returning secondary results should be examined further, with studies that include longer periods of follow-up to determine what actions were taken by participants in response to genetic information in the long run and the impact of these actions.
Supplementary Material
Study flow and retention of participants throughout the course of the study
Table S1. Measures on the study questionnaires
Table S2. Demographics stratified by those with ES who proceeded with the study post-baseline (n=107) and those who did not (n=31).
Table S3. Mean score of measures at baseline (a) and mean change in measures from baseline to 1 month (b) and 12 months (c) following results disclosure (ES groups) or study enrollment (No ES group) stratified by ES w/PDR, ES w/o PDR and No ES (type of results). Associations of type of results (primary predictor) and the mean change of the measure overtime (outcome) were assessed by linear regression both unadjusted and adjusted for the confounder of parent study type.
Table S4. Means scores of measures to assess impact of receiving results stratified by ES w/PDR and ES w/o PDR at 1 month (a) and 12 months (b) following results disclosure. Associations of type of results (primary predictor) and the mean score of the measures (outcome) were assessed by linear regression both unadjusted and adjusted for the confounder of parent study type.
Acknowledgments
We gratefully acknowledge the contribution of the research participants. This work was funded by grants from the National Human Genome Research Institute through Grant Number R21 HG006596 (Dr. Appelbaum, PI) and Grant Number R01 HG006600 (Drs. Chung and Phelan, PIs), and Grant Number P50 HG007257 (Dr. Appelbaum, PI), as well as grants from the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number UL1 TR000040, formerly the National Center for Research Resources, Grant Number UL1 RR024156. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
COMPLIANCE WITH ETHICAL STANDARDS CONFLICT OF INTEREST
Julia Wynn, Josue Martinez, Jessica Bulafka, Jimmy Duong, Yuan Zheng, Codruta Chiuzan, Preti Jain, Maria L Cremona, Vaidehi Jobanputra, Abby J Fyer, Robert L Klitzman, Paul S Appelbaum, and Wendy K. Chung have no conflict of interests.
HUMAN STUDIES AND INFORMED CONSENT STATEMENT
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and nation) and with the Helsinski Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients for being included in the study.
ANIMAL STUDIES
No animal studies were carried out by the authors for this article.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, … Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola LM, Lautenbach D, Scollon S, Bernhardt B, Biswas S, East K, … Jarvik GP. Illustrative case studies in the return of exome and genome sequencing results. Per Med. 2015;12(3):283–295. doi: 10.2217/pme.14.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennette CS, Trinidad SB, Fullerton SM, Patrick D, Amendola L, Burke W, … Veenstra DL. Return of incidental findings in genomic medicine: measuring what patients value--development of an instrument to measure preferences for information from next-generation testing (IMPRINT) Genet Med. 2013;15(11):873–881. doi: 10.1038/gim.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Burke W, Kohane I, Plon SE, Zimmern R. Next-generation sequencing in the clinic: are we ready? Nat Rev Genet. 2012;13(11):818–824. doi: 10.1038/nrg3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger JM, Scott J, Dvoskin R, Kaufman D. Public preferences regarding the return of individual genetic research results: findings from a qualitative focus group study. Genet Med. 2012;14(4):451–457. doi: 10.1038/gim.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch N, Junyent N, Gadea N, Brunet J, Ramon y Cajal T, Torres A, … Balmana J. What factors may influence psychological well being at three months and one year post BRCA genetic result disclosure? Breast. 2012;21(6):755–760. doi: 10.1016/j.breast.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Carver CS, Scheier MF. Assessing coping strategies: A theoretically based approach. Journal of Personality and Social Psychology. 1989;56(2):267–283. doi: 10.1037//0022-3514.56.2.267. [DOI] [PubMed] [Google Scholar]
- Cella D, Hughes C, Peterman A, Chang CH, Peshkin BN, Schwartz MD, … Lerman C. A brief assessment of concerns associated with genetic testing for cancer: the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychol. 2002;21(6):564–572. [PubMed] [Google Scholar]
- Clarke AJ. Managing the ethical challenges of next-generation sequencing in genomic medicine. Br Med Bull. 2014;111(1):17–30. doi: 10.1093/bmb/ldu017. [DOI] [PubMed] [Google Scholar]
- Cooper C, Katona C, Orrell M, Livingston G. Coping strategies and anxiety in caregivers of people with Alzheimer's disease: the LASER-AD study. J Affect Disord. 2006;90(1):15–20. doi: 10.1016/j.jad.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erblich J, Brown K, Kim Y, Valdimarsdottir HB, Livingston BE, Bovbjerg DH. Development and validation of a Breast Cancer Genetic Counseling Knowledge Questionnaire. Patient Educ Couns. 2005;56(2):182–191. doi: 10.1016/j.pec.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Fernandez CV, Bouffet E, Malkin D, Jabado N, O'Connell C, Avard D, … McMaster CR. Attitudes of parents toward the return of targeted and incidental genomic research findings in children. Genet Med. 2014;16(8):633–640. doi: 10.1038/gim.2013.201. [DOI] [PubMed] [Google Scholar]
- Gech A, Epstein N, Brown G, Steer R. An inventory for measuring clinical anxiety: psychometric properties. Jouranl of Consulting and Clinical Psychology. 1988;56:5. doi: 10.1037//0022-006x.56.6.893. [DOI] [PubMed] [Google Scholar]
- Green RC, Berg JS, Berry GT, Biesecker LG, Dimmock DP, Evans JP, … Jacob HJ. Exploring concordance and discordance for return of incidental findings from clinical sequencing. Genet Med. 2012;14(4):405–410. doi: 10.1038/gim.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL … Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Lupski JR, Biesecker LG. Reporting genomic sequencing results to ordering clinicians: incidental, but not exceptional. JAMA. 2013;310(4):365–366. doi: 10.1001/jama.2013.41703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik GP, Amendola LM, Berg JS, Brothers K, Clayton EW, Chung W, … Burke W. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet. 2014;94(6):818–826. doi: 10.1016/j.ajhg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, … Miller DT. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62(6):617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitzman R, Appelbaum PS, Chung W. Return of secondary genomic findings vs patient autonomy: implications for medical care. JAMA. 2013;310(4):369–370. doi: 10.1001/jama.2013.41709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitzman R, Appelbaum PS, Fyer A, Martinez J, Buquez B, Wynn J, … Chung WK. Researchers' views on return of incidental genomic research results: qualitative and quantitative findings. Genet Med. 2013;15(11):888–895. doi: 10.1038/gim.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krabbenborg L, Vissers LE, Schieving J, Kleefstra T, Kamsteeg EJ, Veltman JA, … Van der Burg S. Understanding the psychosocial effects of WES test results on parents of children with rare diseases. J Genet Couns. 2016;25(6):1207–1214. doi: 10.1007/s10897-016-9958-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lee KA, Hicks G, Nino-Murcia G. Validity and reliability of a scale to assess fatigue. Psychiatry Res. 1991;36(3):291–298. doi: 10.1016/0165-1781(91)90027-m. [DOI] [PubMed] [Google Scholar]
- Lewis KL, Hooker GW, Connors PD, Hyams TC, Wright MF, Caldwell S, … Biesecker BB. Participant use and communication of findings from exome sequencing: a mixed-methods study. Genet Med. 2016;18(6):577–583. doi: 10.1038/gim.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumish HS, Steinfeld H, Koval C, Russo D, Levinson E, Wynn J, … Chung WK. Impact of panel gene testing for hereditary breast and ovarian cancer on patients. J Genet Couns. 2017 doi: 10.1007/s10897-017-0090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. Retrieved from http://www.R-project.org. [Google Scholar]
- Regier DA, Peacock SJ, Pataky R, van der Hoek K, Jarvik GP, Hoch J, Veenstra D. Societal preferences for the return of incidental findings from clinical genomic sequencing: a discrete-choice experiment. Cmaj. 2015;187(6):E190–197. doi: 10.1503/cmaj.140697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropka ME, Wenzel J, Phillips EK, Siadaty M, Philbrick JT. Uptake rates for breast cancer genetic testing: a systematic review. Cancer Epidemiol Biomarkers Prev. 2006;15(5):840–855. doi: 10.1158/1055-9965.epi-05-0002. [DOI] [PubMed] [Google Scholar]
- Rosell AM, Pena LD, Schoch K, Spillmann R, Sullivan J, Hooper SR, … Shashi V. Not the end of the odyssey: parental perceptions of whole exome sequencing (WES) in pediatric undiagnosed disorders. J Genet Couns. 2016;25(5):1019–1031. doi: 10.1007/s10897-016-9933-1. [DOI] [PubMed] [Google Scholar]
- Sanderson S, O’Neill S, Bastian L, Bepler G, McBride C. What Can Interest Tell Us about Uptake of Genetic Testing? Intention and Behavior amongst Smokers Related to Patients with Lung Cancer. Public Health Genomics. 2009;13(2):116–124. doi: 10.1159/000226595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson SC, Linderman MD, Suckiel SA, Zinberg R, Wasserstein M, Kasarskis A, … Schadt EE. Psychological and behavioural impact of returning personal results from whole-genome sequencing: the HealthSeq project. Eur J Hum Genet. 2017;25(3):280–292. doi: 10.1038/ejhg.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc. SAS 9.4 [computer program] Cary, NC: SAS Institute Inc; 2014. [Google Scholar]
- Sawyer SM, Cerritelli B, Carter LS, Cooke M, Glazner JA, Massie J. Changing their minds with time: a comparison of hypothetical and actual reproductive behaviors in parents of children with cystic fibrosis. Pediatrics. 2006;118(3):e649–656. doi: 10.1542/peds.2005-2551. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Kroenke K, Williams JB. Validation and utility of a self-report version of PRIME-MD: the PHQ primary care study. Primary Care Evaluation of Mental Disorders. Patient Health Questionnaire. JAMA. 1999;282(18):1737–1744. doi: 10.1001/jama.282.18.1737. [DOI] [PubMed] [Google Scholar]
- Townsend A, Adam S, Birch PH, Lohn Z, Rousseau F, Friedman JM. "I want to know what's in Pandora's Box": comparing stakeholder perspectives on incidental findings in clinical whole genomic sequencing. Am J Med Genet A. 2012;158a(10):2519–2525. doi: 10.1002/ajmg.a.35554. [DOI] [PubMed] [Google Scholar]
- Wallston BS, Wallston KA, Kaplan GD, Maides SA. Development and validation of the health locus of control (HLC) scale. J Consult Clin Psychol. 1976;44(4):580–585. doi: 10.1037//0022-006x.44.4.580. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfond BS, Fernandez CV, Green RC. Disclosing secondary findings from pediatric sequencing to families: considering the "Benefit to Families". J Law Med Ethics. 2015;43(3):552–558. doi: 10.1111/jlme.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills CE, Holmes-Rovner M. Preliminary validation of the Satisfaction With Decision scale with depressed primary care patients. Health Expect. 2003;6(2):149–159. doi: 10.1046/j.1369-6513.2003.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SM, Crock BN, Van Ness B, Lawrenz F, Kahn JP, Beskow LM, … Wolf WA. Managing incidental findings and research results in genomic research involving biobanks and archived data sets. Genet Med. 2012;14(4):361–384. doi: 10.1038/gim.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn J, Martinez J, Duong J, Chiuzan C, Phelan JC, Fyer A, … Chung WK. Research participants' preferences for hypothetical secondary results from genomic research. J Genet Couns. 2016 doi: 10.1007/s10897-016-0059-2. [DOI] [PubMed] [Google Scholar]
- Wynn J, Martinez J, Duong J, Zhang Y, Phelan J, Fyer A, … Chung WK. Association of Researcher Characteristics with Views on Return of Incidental Findings from Genomic Research. J Genet Couns. 2015 doi: 10.1007/s10897-014-9817-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Jamal SM, Tabor HK, Bamshad MJ. Self-guided management of exome and whole-genome sequencing results: changing the results return model. Genet Med. 2013;15(9):684–690. doi: 10.1038/gim.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushak ML, Han G, Bouberhan S, Epstein L, DiGiovanna MP, Mougalian SS, … Hofstatter EW. Patient preferences regarding incidental genomic findings discovered during tumor profiling. Cancer. 2016;122(10):1588–1597. doi: 10.1002/cncr.29951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Study flow and retention of participants throughout the course of the study
Table S1. Measures on the study questionnaires
Table S2. Demographics stratified by those with ES who proceeded with the study post-baseline (n=107) and those who did not (n=31).
Table S3. Mean score of measures at baseline (a) and mean change in measures from baseline to 1 month (b) and 12 months (c) following results disclosure (ES groups) or study enrollment (No ES group) stratified by ES w/PDR, ES w/o PDR and No ES (type of results). Associations of type of results (primary predictor) and the mean change of the measure overtime (outcome) were assessed by linear regression both unadjusted and adjusted for the confounder of parent study type.
Table S4. Means scores of measures to assess impact of receiving results stratified by ES w/PDR and ES w/o PDR at 1 month (a) and 12 months (b) following results disclosure. Associations of type of results (primary predictor) and the mean score of the measures (outcome) were assessed by linear regression both unadjusted and adjusted for the confounder of parent study type.