Abstract
Ki-67 protein has been widely used as a proliferation marker for human tumor cells for decades. In recent studies, multiple molecular functions of this large protein have become better understood. Ki-67 has roles in both interphase and mitotic cells, and its cellular distribution dramatically changes during cell cycle progression. These localizations correlate with distinct functions. For example, during interphase Ki-67 is required for normal cellular distribution of heterochromatin antigens and for the nucleolar association of heterochromatin. During mitosis, Ki-67 is essential for formation of the perichromosomal layer (PCL), a ribonucleoprotein sheath coating the condensed chromosomes. In this structure, Ki-67 acts to prevent aggregation of mitotic chromosomes. Here, we present an overview of functional roles of Ki-67 across the cell cycle and also describe recent experiments that clarify its role in regulating cell cycle progression in human cells.
Keywords: Ki-67, cell cycle, perichromosomal layer, heterochromatin
1. Introduction
Ki-67 was first identified as an antigen in Hodgkin lymphoma cell nuclei (Gerdes et al. 1983) that is highly expressed in cycling cells but strongly down-regulated in resting G0 cells (Gerdes et al. 1984). This characteristic has made Ki-67 a clinically important proliferation marker for grading multiple types of cancers (Gerdes et al., 1987; Dowsett et al., 2011), with well-established prognostic value in large studies (Luo et al., 2015; Pyo et al., 2015; Pezzilli et al., 2016; Richards-Taylor et al., 2015). Despite this long-standing clinical utility, much less attention has been paid to the molecular functions of Ki-67. Here, we review recent studies that have uncovered roles for Ki-67 in cell cycle regulation, heterochromatin maintenance, and assembly of the perichromosomal layer on mitotic chromosomes (Sobecki et al., 2016; Cuylen et al., 2016; Booth et al., 2014; Sun et al., 2017). We also discuss recent experiments demonstrating how Ki-67 affects cell cycle progression in p21 checkpoint-proficient human cells ( Sun et al. 2017), and how Ki-67 is itself regulated by cell cycle position (Sobecki et al. 2017). Additionally, we discuss new data regarding the protein RepoMan, which shares an important protein interaction domain with Ki-67 (Booth et al., 2014; Kumar et al., 2016) and is critical for important aspects of heterochromatin localization (de Castro et al. 2017).
2. Ki-67 primary structure and interaction partners
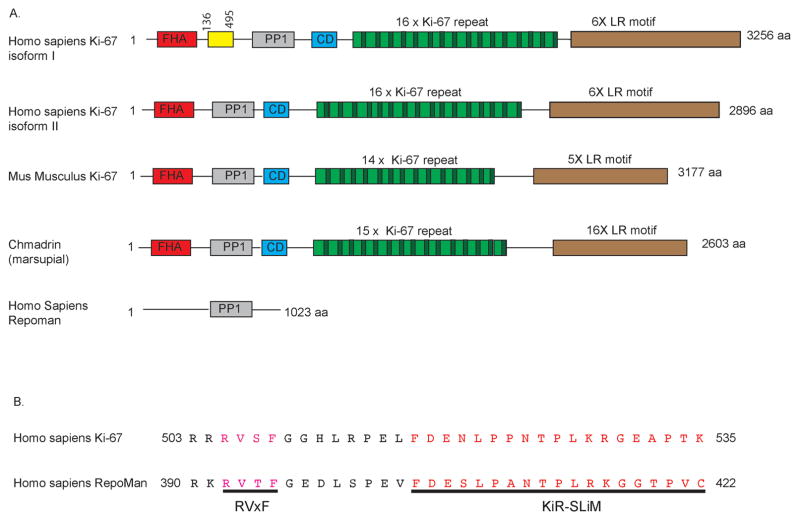
The Ki-67 protein is found in vertebrates (Booth and Earnshaw 2017). In humans, Ki-67 is encoded by the gene MKI67. Although Ki-67 contains several conserved functional regions (Figure 1), the primary sequence conservation outside of these regions is low. Major regions of the Ki-67 protein include an N-terminal forkhead-associated (FHA) domain, a protein Phosphatase 1 (PP1) binding domain, a large central region comprising tandem repeats, and a C-terminal LR (leucine/arginine-rich) chromatin-binding domain (Schluter et al., 1993).
Figure 1. A schematic diagram of human Ki-67 structure.
A: Comparison of evolutionarily conserved regions of Ki-67 (NCBI NP_002408) and RepoMan (NP_689775).
FHA: forkhead-associated domain; PP1: PP1 binding domain; CD: conserved domain with unknown functions, LR: leucine-arginine rich domain
B: Comparison of the primary sequences of PP1-binding domains of human Ki-67 (isoform I) and RepoMan (isoform I).
Additionally, the MKI67 primary transcript is alternatively spliced. Two protein isoforms with molecular weights of 320 kDa and 350 kDa were originally described, encoded by the two major transcript variants which differ by the alternative inclusion of exon 7 (Gerdes et al. 1991). Three additional splicing variants of human Ki-67 have been detected in different tissues and primary and cultured cells, with different patterns of expression in different cell lines (Schmidt et al. 2004). Despite different N-termini, all five Ki-67 variants contain identical central tandem repeats and C-terminal regions (Schmidt et al. 2004). Notably, in stimulated peripheral blood lymphocytes, the splicing pattern changes after stimulation, with the long isoform appearing more slowly (Schmidt et al. 2004). Furthermore, in HeLa cells, overexpression of alternative exon 7, which is present in the major long but not short isoform, reduced proliferation, whereas overexpression of an N-terminal fragment increased cell proliferation (Schmidt et al. 2004). These results suggest that Ki-67 isoforms may differentially impact cell proliferation and cell cycle progression. In addition, expression of either exon 7 or a set of three of the internal repeats as protein fragments causes translocation of cyclin B from the cytoplasm to the nucleolus in HeLa cells (Schmidt et al. 2003). Notably, initiation of mitosis requires active import of cyclin B to the nucleus during the G2/M transition (Moore et al. 1999), and the mitotic localization of Ki-67 is regulated by the balance of CDK/cyclin phosphorylation and PP1 dephosphorylation (Takagi et al. 2014). Thus, Ki-67 is implicated in major nuclear structural transitions during mitotic entry and exit.
FHA domain
The N-terminus of Ki-67 contains a forkhead-associated (FHA) domain (Takagi et al. 2001), a motif that preferentially recognizes phosphorylated protein epitopes (Durocher et al., 1999; Durocher and Jackson, 2002). Via this FHA domain, Ki-67 interacts with two phosphoproteins during mitosis: kinesin-like motor protein Hklp2/Kif15, and nucleolar protein NIFK (Sueishi et al. 2000; Takagi et al., 2001; Durocher and Jackson, 2002).
Hklp2 is a plus-end-directed spindle motor, and is required for the maintenance of spindle bipolarity (Vanneste et al. 2009). Bipolar spindle assembly requires a balance of forces, in which plus-end-directed motors including Eg5 produce outward pushing forces to antagonize the inward pulling forces from minus-end-directed motors (Kapitein et al., 2005; Mitchison et al., 2005; Tanenbaum et al., 2008). In mitotic HeLa cells, Hklp2 localizes either to microtubules through RanGTP-regulated factor TPX2 or to the periphery of chromosomes in a Ki-67-dependent manner (Wittmann et al., 1998; Vanneste et al., 2009). In one study, Hklp2 failed to localize to chromosomes upon Ki-67 depletion, resulting in increased Hklp2 association with microtubules. This generated more spindle-associated Hklp2 motor activity, yielding longer metaphase spindles and more bipolar spindles upon inhibition of Eg5 (Kapitein et al., 2005; Vanneste et al., 2009). However, more recent studies detected no clear anomalies in spindle lengths upon Ki-67 depletion (Cuylen et al. 2016; Takagi et al. 2016; Takagi et al. 2017). The physiological function of the interaction between Ki-67 and Hklp2 therefore remains to be determined.
Ki-67’s other FHA binding partner, NIFK, promotes cell proliferation and cancer metastasis (Pan et al., 2015; Lin et al., 2016). Downregulation of NIFK reduced growth in patient-derived lung cancer cell lines H661 and H1299 (Lin et al., 2016). Conversely, in A549 and PC13 lung cancer cell lines which express low NIFK levels, overexpression of wild type NIFK increased cell proliferation (Lin et al. 2016). Notably, the increased proliferation in A549 and PC13 cells requires an intact Ki-67 binding motif on NIFK (Lin et al. 2016), although the functionally relevant NIFK binding partner(s) remains to be definitively identified. Additionally, NIFK enhances the metastatic ability of lung cancer cells by destabilizing the transcription factor RUNX1, which in turn promotes a pro-metastatic Wnt/β-catenin signaling pathway (Hernandez et al., 2012; Li et al., 2012). Depletion of RUNX1 is largely attenuated when NIFK is incapable of Ki-67 binding (Lin et al. 2016). Again, whether RUNX1 destabilization by NIFK requires Ki-67 itself or another protein that shares its binding site remains to be determined.
An additional activity has been mapped to the Ki-67 N-terminus, which includes the FHA domain. In two human cell lines, spontaneously immortalized MCF10A breast epithelial cells and colorectal tumor-derived DLD-1 cells, one study inserted a stop codon into the first coding exon of the MKI67 gene, resulting in loss of expression of Ki-67 protein. This did not affect cell proliferation, but decreased clonogenic survival of cells when plated at low density (Cidado et al. 2016). Reduced growth rates of xenograft tumors derived from these knock-out cell lines were also observed. For comparison, the investigators inserted stop codons into the large exon 13 that encodes the Ki-67 internal repeats, thereby maintaining expression of an N-terminal fragment containing the FHA and PP1-interacting domains (Fig. 1). Cells expression this truncated Ki-67 protein displayed no defects in clongenic survival or xenograft tumor growth (Cidado et al. 2016). It would therefore be of interest to know whether these activities of the Ki-67 N-terminal fragment are also dependent on the NIFK protein. Additionally, it remains to be determined whether distinct subsets of Ki-67 molecules interact with different FHA-binding partners, and whether there is crosstalk between Hklp2 and NIFK regulation.
PP1 interaction domain
All homologues of Ki-67 contain a canonical Protein Phosphatase 1 (PP1) binding motif (RVxF)(Booth et al. 2014). The PP1 family contains three isoforms (α, β, and γ), and is estimated to catalyze approximately one third of all eukaryotic protein dephosphorylation events, spanning a wide variety of cellular functions (Rebelo et al. 2015). The versatility of PP1 functions is largely determined by the binding of its catalytic subunits to different regulatory proteins that define when and where the phosphatase acts (Rebelo et al. 2015).
The PP1 interaction region of Ki-67 displays high similarity to the protein RepoMan (Figure 1). In vivo, both Ki-67 and RepoMan bind specifically to PP1β and PP1γ isoforms, but not to PP1α (Booth et al., 2014; Kumar et al., 2016). Notably, both RepoMan and Ki-67 target PP1γ to anaphase chromosomes (Trinkle-Mulcahy et al. 2006; Takagi et al. 2014), which is a critical step during mitotic exit for the removal of histone phosphorylation (Vagnarelli et al., 2011; Qian et al. 2011; de Castro et al. 2017). Crystal structures of Ki-67:PP1γ and RepoMan:PP1γ holoenzyme complexes identified a supplementary, novel PP1 interaction motif termed KiR-SLiM (Ki-67-RepoMan small linear motif), which is just C-terminal to the canonical PP1 interaction motif RVxF (Kumar et al. 2016). The KiR-SLiM motif contributes to PP1γ binding, because removing it results in five-fold decreased binding affinity (Kumar et al. 2016). Notably, Ki-67 and RepoMan are the only known proteins that utilize this additional motif for PP1 binding (Kumar et al. 2016).
In Ki-67-depleted cells, the accumulation of PP1γ on anaphase chromatin is partially disrupted, but removal of mitotic histone phosphorylation on H3S10 is not affected (Takagi et al., 2014; Booth et al., 2014). These findings suggest functional redundancy among PP1-targeting proteins with regard to histone dephosphorylation at mitosis. A good candidate for this overlapping function is RepoMan, which unlike Ki-67 is essential for cell viability (Trinkle-Mulcahy et al. 2006). RepoMan/PP1 contributes to the the dephosphorylation of histone H3 residues Thr3, Ser10 and Ser28 (Vagnarelli et al., 2011; de Castro et al., 2017). H3K9me3 is a hallmark of constitutive heterochromatin and is recognized by HP1. During mitosis, phosphorylation of the neighboring H3S10 residue prevents HP1 binding to H3K9me3 (Fischle et al. 2005). At anaphase, RepoMan/PP1-mediated dephosphorylation of H3S10P allows for rebinding of HP1 to chromatin, thereby re-establishing heterochromatin in post-mitotic cells (Fischle et al., 2005; Vagnarelli et al., 2011). A similar “phospho-methyl switch” regulation applies to H3K27me3, a modification enriched on facultative heterochromatin and catalyzed by polycomb repressive complex 2 (PRC2) (Margueron and Reinberg 2011). During interphase, phosphorylation of H3S28 occurs upon stress and mitogenic stimulation or upon retinoic acid-induced neuronal differentiation (Gehani et al. 2010). H3S28P in turn promotes dissociation of PRC2 from the adjacent H3K27me3 mark at gene promoters, thus favoring transcriptional derepression (Gehani et al. 2010).
Recent landmark experiments from the Vagnarelli laboratory show that recombinatnt RepoMan protein binds to nucleosomes that contain H3K27 modifications (de Castro et al. 2017). Furthermore, tethering experiments indicate that RepoMan contributes to the formation of H3K27me3-rich chromatin domains. Conversely, cells depleted of RepoMan display increased levels of H3S28 phosphorylation and decreased accumulation of H3K27me3 at Polycomb-regulated genes (de Castro et al. 2017). Additionally, this study found that RepoMan contributed to heterochromatin localization at the nuclear periphery in a manner dependent on nuclear pore protein Nup153. Therefore, this study elegantly outlined how a protein could link specific chromatin regions to specific nuclear localizations, in a manner that can be regulated dynamically, in this case via phospho/methyl switch involving H3K27me3/S28P. The contribution of Ki-67 to such processes is a major outstanding question.
Ki-67 internal repeats
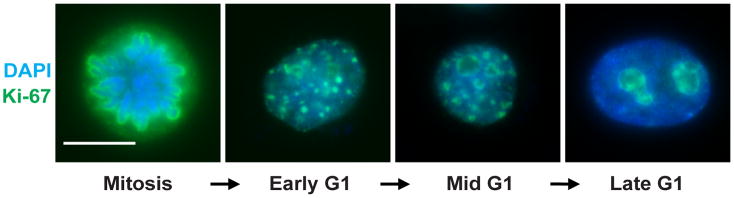
Ki-67 has distinct nuclear localization patterns, which appear at distinct of cell-cycle periods (Gerdes et al. 1983; Gerdes et al. 1984; Verheijen et al. 1989a; Verheijen et al. 1989b; Isola et al., 1990; Scholzen and Gerdes 2000; Figure 2). The central region of Ki-67 is comprised of tandem repeats that contain residues phosphorylated by CDK1 during mitosis (Schluter et al. 1993; MacCallum and Hall 1999; Endl and Gerdes 2000; Takagi et al. 2014). Notably, Ki-67’s mitotic localization and in vitro DNA binding affinity are affected by phosphorylation (MacCallum and Hall, 1999; Endl and Gerdes, 2000). In interphase, dephosphorylated Ki-67 forms fibre-like structures surrounding nucleoli, overlapping the perinucleolar heterochromatin (Kill 1996; Cheutin et al 2003). This localization appears to be functionally important, because depletion of Ki-67 reduces the association of heterochromatin around nucleoli (Booth et al., 2014; Sobecki et al. 2016; Matheson and Kaufman 2017). At the onset of mitosis, Ki-67 becomes hyperphosphorylated and thereby binds less avidly to DNA, and is highly mobile on the chromosome periphery until anaphase (MacCallum and Hall, 1999; Endl and Gerdes, 2000; Saiwaki et al., 2005). Dephosphorylation of Ki-67 during mitotic exit stimulates its dissociation from the perichromosomal layer (MacCallum and Hall, 1999; Takagi et al. 2014). On the mitotic chromosome surface, the highly positive electrostatic charge of Ki-67 serves as an electrostatic barrier important for prevention of hyperaggregation of chromosome arms (Cuylen et al. 2016; Takagi et al. 2016). This will be discussed in more detail in Section 4.
Figure 2. Ki-67 localization throughout the cell cycle.
HeLa S3 cells were stained with anti-Ki67 antibodies (green) and DAPI to visualize DNA (blue), illustrating different Ki-67 localizations across the cell cycle. In mitotic cells, Ki-67 coats the condensed chromosomes as the foundation of the perichromosomal layer. As cells exit mitosis and enter early G1 phase, small puncta of Ki-67 leave the decondensing chromosomes. These then coalesce at the periphery of the reformed nucleoli as G1 phase progresses. Scale bar, 10 μm.
This figure is reprinted from Matheson and Kaufman, Mol. Biol. Cell (2017), with permissions from the authors.
C-terminal LR domain
Ki-67 has a weakly conserved leucine/arginine-rich C-terminal domain (LR domain) which can bind to DNA in vitro (Takagi et al., 1999; Scholzen et al., 2002) and is required for association with chromosomes in living cells (Saiwaki et al., 2005; Cuylen et al., 2016). This C-terminal domain of human Ki-67 binds all three isoforms of heterochromatin protein 1 (HP1) in vitro and in vivo (Kametaka et al. 2002; Scholzen et al., 2002). Via its methyllysine-binding chromodomain, HP1 binds to the hallmark of constitutive heterochromatin, di- and tri-methylated histone H3K9 residues (Jacobs and Sepideh, 2002; Nielsen et al., 2002). Through its chromoshadow domain, HP1 interacts with a large number of proteins involved in heterochromatin formation, including DNA methyltransferases DNMT1 and DNMT3a, histone methyltransferase SUV39H1, and the p150 subunit of the chromatin assembly factor-1 complex (CAF-1) (Murzina et al. 1999; Lachner et al., 2001; Fuks et al., 2003; Hiragami-Hamada et al., 2016). Therefore, by reading H3K9-methyl marks and recruiting heterochromatin assembly proteins, HP1 contributes to formation of the constitutive heterochromatin enriched at centromeres and telomeres in every cell type (Saksouk et al. 2015). Furthermore, overexpression of human, marsupial or Xenopus Ki-67 results in accumulation of HP1 and H3K9me3 at sites of high Ki-67 concentration and induces the appearance of more compacted chromatin as measured by the intensity of DAPI staining (Takagi et al., 1999; Scholzen et al., 2002; Sobecki et al., 2016). Other examples of LR domain function will be discussed in Section 6.
3. Ki-67 expression during the cell cycle
The initial characterization of Ki-67 localization detected a nuclear protein in proliferating human cells (Gerdes et al. 1983). Additionally, studies of phytohemagglutinin (PHA)-stimulated peripheral mononuclear blood leukocytes (PBL) showed that unstimulated (G0 phase) cells were negative for the Ki-67 antigen (Gerdes et al. 1984). We now know that the MKI67 gene promoter itself is cell cycle-regulated, containing binding sites for the canonical G1-regulatory E2F family of transcription factors (Ishida et al. 2001), and that Ki-67 mRNA levels increase during G1 (Sobecki et al., 2016; Sobecki et al., 2017). Ki-67 protein degradation also occurs during G1 via ubiquitin proteasome complex APC/C-Cdh1 (Chierico et al., 2017; Sobecki et al., 2017). Therefore, Ki-67 protein concentrations in G1 are controlled by two opposing mechanisms. In addition, unlike deeply quiescent or senescent cells, cells in early stages of cell cycle arrest have low levels of Ki-67, which can carry over after re-entering the cell cycle (Sobecki et al. 2017). Therefore, slight variation in the degree of quiescence can result in variable Ki-67 levels in the first G1 phase after cell cycle re-entry, likely contributing to variable observations of in different experiments (Gerdes et al., 1984; Lopez et al., 1991; Sobecki et al., 2017).
4. Ki-67 coats mitotic chromosomes
In the past few years, several studies have greatly increased our understanding of Ki-67’s mitotic functions. As cells enter mitosis, chromosomes undergo a remarkable series of structural transformations known as chromosome condensation. A proteinaceous sheath termed the perichromosomal layer (PCL) exists at the outer surfaces of individual chromosomes (Van Hooser et al. 2005), and comprises approximately one-third of the protein mass of mitotic chromosomes (Booth et al., 2016; Booth and Earnshaw, 2017). Ki-67 is one of the earliest proteins associated with this structure and remains on it until telophase (Van Hooser et al. 2005). Several studies have found that Ki-67 is required for the formation of the human PCL. Acute depletion of Ki-67 in human cells caused dispersal of all other PCL components tested, including nucleolar proteins nucleolin, nucleophosmin, NIFK, PES1, cPERP-B, cPERP-C, cPERP-D, cPERP-F and pre-ribosomal RNAs; conversely, depletion of these components did not alter perichromosomal localization of Ki-67 (Booth et al., 2014; Sobecki et al. 2016; Hayashi et al., 2017). These conventional immunofluorescence studies were bolstered by elegant 3D correlative light and electron microscopy studies (“CLEM”) that indicated a loss of approximately one-third the volume of mitotic chromosomes upon depletion of Ki-67 (Booth et al. 2016).
Therefore, Ki-67 is the essential foundational component of the perichromosomal layer. What is the function of this structure? Two major findings have been made recently. First, disruption of the PCL upon Ki-67 depletion delocalizes nucleolar components during mitosis, which in turn leads to their asymmetric distribution in daughter cells (Booth et al. 2014). Second, Ki-67 prevents the aggregation of mitotic chromosomes (Cuylen et al. 2016; Takagi et al. 2016). Indeed, molecular dissection of Ki-67 showed that its large size and high density of positively charged amino acids causes it to act as a “surfactant” on the mitotic chromosome surface (Cuylen et al. 2016). During prophase, Ki-67 is not required for the initial individualization of chromosomes as they condense (Cuylen et al. 2016). However, after nuclear envelope breakdown, cells depleted of Ki-67 display increased mitotic chromosome associations, resulting in impaired spindle assembly and metaphase plate formation, thereby prolonging progression from prometaphase to anaphase (Cuylen et al. 2016). Another study reports aberrant mitotic chromosome structure upon rapid removal of Ki-67 via the mAID degron system, which results in aberrant swollen mitotic chromosome structures (Takagi et al. 2016). Structural contributions of Ki-67 are most dramatically demonstrated in a new preprint, which shows that Ki-67 and condensin protein complexes make independent contributions to mitotic chromosome structure. Notably, in this study acute depletion of both these factors results in more severe morphological defects than upon depletion of either factor alone (Takagi et al. 2017).
Because of these important interphase and mitotic roles, a major question is how Ki-67 localization is specified and modulated across the cell cycle. Recent studies show that the p150 subunit of histone chaperone Chromatin Assembly Factor-1 (CAF-1) regulates Ki-67 localization at all cell cycle points. Specifically, interphase nucleolar localization, mitotic PCL localization, and the appearance of numerous small foci in early G1 cells were all perturbed upon p150 depletion (Smith et al. 2014; Matheson and Kaufman 2017). Each of these localizations depends on the sumoylation interacting motif (SIM) within p150. Because sumoylated proteins are implicated in liquid demixing reactions (Banani et al. 2016), these results raise the possibility that p150 can modulate the phase transition properties of Ki-67.
5. Contributions of Ki-67 to human cell cycle progression
5a. Ki-67 perturbation experiments
Although Ki-67 expression is tightly correlated with proliferation, differing results have been obtained in functional tests for contributions of Ki-67 to cell cycle progression. In early studies, microinjection of an anti-Ki-67 antibody inhibited proliferation of mouse Swiss 3T3 cells (Starborg et al. 1996). Likewise, human IM-9 multiple myeoloma and RT-4 bladder carcinoma cell lines displayed reduced proliferation rates upon treatment with Ki-67-targeted anti-sense oligonucleotides or si-RNAs (Schluter et al., 1993; Kausch et al. 2003). Together with the clinical use of Ki-67 as a tumor categorization marker, these types of data were consistent with the idea that Ki-67 could affect cell cycle progression.
However, some recent studies challenged this view. For example, one study found that mouse NIH-3T3 cells lacking Ki-67 proliferate normally without apparent cell cycle delays (Sobecki et al. 2016). Furthermore, depletion of Ki-67 in human HeLa and U2OS cells did not alter cell cycle distribution, ribosomal RNA synthesis, or transcription of cell cycle-related genes (Sobecki et al. 2016). In a different study, proliferation of human MCF-10A epithelial breast or DLD-1 colon cancer cells were not affected by loss of Ki-67, although clonogenic growth of highly diluted cell populations were decreased (Cidado et al. 2016). These studies raised the possibility that the contributions of Ki-67 to cell cycle progression could be cell type specific, at least in human cells.
Indeed, recent studies have shown that differing effects of Ki-67 depletion on human cell cycle progression are correlated with the status of G1/S checkpoints. Human primary fibroblasts (WI-38, IMR90 and HFF) as well as non-tumor derived diploid cell lines (hTERT-RPE1 and hTERT-BJ) are able to induce the cyclin-dependent kinase inhibitor checkpoint protein p21 upon Ki-67 depletion. Thus, these cells have been termed “Ki-67-sensitive” (Sun et al. 2017). p21 is required for transcriptional repression by the Rb/E2F and DREAM complexes, both of which repress genes required for cell cycle progression during G1/S phases (Fischer et al. 2016). p21 is also a direct inhibitor of DNA synthesis via its interaction with the DNA polymerase processivity clamp protein, PCNA (Waga and Stillman 1998). In p21 checkpoint-proficient cell lines, siRNA-mediated Ki-67 depletion delays S phase entry and coordinately downregulates DNA replication-related mRNA levels. To control for possible indirect effects of the siRNA treatment, CRISPR/Cas9-mediated gene editing was used to generate homozygous siRNA-insensitive MKI67 alleles in the hTERT-RPE1 cell line. In these cells, siKi-67 no longer downregulated S phase RNA levels or induced p21 (Sun et al. 2017). Therefore, the observed effects are attributed to a function of the Ki-67 protein.
In contrast to “Ki-67 sensitive” cell types, tumor-derived cell lines (HeLa, U2OS and 293T cells) do not induce p21 or display altered S phase populations upon Ki-67 depletion (Sun et al. 2017). Notably, although p21 is a direct transcriptional target of p53 (El-Deiry et al. 1993), p53 status cannot always predict a cell’s sensitivity to Ki-67 depletion. For example, both MCF7 breast adenocarcinoma and HCT116 colorectal carcinoma cells express wild-type p53. Upon Ki-67 depletion, MCF-7 cells induce p21 expression and repress DNA replication genes while HCT116 cells do not (Sun et al. 2017). It remains to be determined what mechanisms thwart p21 induction and thereby prevent Ki-67 sensitivity in a subset of p53-positive cell lines.
Several additional issues remain unresolved regarding the role of Ki-67 in cell cycle progression. In one study, Ki-67 depletion delayed S phase entry in multiple p21 checkpoint-proficient cells including hTERT-BJ cells (Sun et al. 2017). However, a second study observed no clear difference in the numbers of S phase cells between control and Ki-67-depleted hTERT-BJ cells (Sobecki et al. 2016). These differences likely result at least in part from the specific assays used. For example, one dimensional FACS analysis of propidium iodide-stained cells, or 3-hour EdU labeling experiments are unable to detect short and transient delays (~2 hr) in S phase entry. Instead, brief (20 min) EdU pulse labeling of HU-synchronized cells or two-dimensional BrdU/propidium iodide measurement of S phase populations allowed for detection of the effects of Ki-67 depletion (Sun et al. 2017).
A more mysterious paradox arises in the comparison of the mouse and human data. Notably, mice with greatly reduced Ki-67 expression are viable, develop normally, and are fertile; furthermore, cell cycle length in Ki-67 wt and mutant mouse embryonic fibroblasts is indistinguishable (Sobecki et al. 2016). Perhaps organismal development is largely insensitive to short cell cycle progression perturbations that may not be recapitulated in immortalized fibroblasts. More generally, it is possible that the role of Ki-67 in preventing p21-mediated checkpoint activation is present in human but not in mouse cells. Future experiments will be required to explore the idea that the role of Ki-67 in protecting from G1/S checkpoint activation is species-specific.
5b. CDK inhibitor experiments
Notably, the phenotypes of Ki-67 sensitive cells often correlate with the response to the small molecule CDK4/6 inhibitor (CDKi) termed palbociclib (otherwise known as PD0332991, (Fry et al. 2004)). Palbociclib blocks Rb phosphorylation by CDKs (Fry et al. 2004), thereby disfavoring S phase entry because unphosphorylated Rb is the form able to bind and inhibit the S phase-promoting transcription factor E2F (Chellappan et al., 1991; Weintraub et al., 1992). One of the E2F target genes is MKI67, encoding Ki-67 (Ishida et al., 2001; Ren et al., 2002; Tian et al., 2011). In an important study supporting the cancer relevance of Ki-67, the laboratory of Daniel Fisher found that in some cell types, palbociclib treatment depleted Ki-67 and cyclin A protein levels, because constitutive proteasome degradation was no longer balanced by E2F-driven transcription (Sobecki et al. 2017). Furthermore, these protein depletions correlated with G1 arrest upon drug treatment, as would be expected for cells lacking cyclin A. Thus, cell types arrested in G1 by palbociclib also become depleted of Ki-67 and cyclin A, and can be termed “CDKi-sensitive”.
In most cases, cells that are CDKi-sensitive are also Ki-67-sensitive (relevant checkpoint features of different cell lines examined are listed in Table 1). For example, Rb- and p53-positive cells like primary fibroblasts, diploid fibroblast lines (e.g. IMR-90, WI-38), hTERT-RPE1 epithelial and MCF7 breast adenocarcinoma cells induce p21 expression in response to Ki-67 depletion and are also CDKi-sensitive. Conversely, cells lacking p53-driven induction of p21 (e.g. virally transformed cells like HeLa) do not induce p21 upon Ki-67 depletion, nor do they arrest in G1 upon CDKi treatment. A counter-example to this correlation is the MDA-MB-231 breast adenocarcinoma line. These cells are CDKi-sensitive, yet do not induce p21 or downregulate S phase target genes upon Ki-67 depletion. Notably, the gain of function p53 R280K allele in MDA-MB-231 cells dominantly blocks transcriptional induction of p21 (Junk et al. 2008). Therefore, these cells do not downregulate S phase genes upon siKi-67 mediated depletion. A simple interpretation of these data would be that Ki-67 depletion is a downstream effect of CDKi treatment, explaining the frequent overlap of responses to these treatments. CDKi inhibition however has additional consequences, so that some cell types unable to induce p21 upon Ki-67 depletion are still sensitive to other effects of the CDKi (e.g. via inhibition of Rb). Importantly, CDKi sensitivity in culture is correlated with sensitivity to growth inhibition by palbociclib in a xenograft tumor model (Sobecki et al. 2017). Therefore, it will be of great interest to see future experiments that extend the analysis of the CDKi-sensitivity phenotype and the contribution of Ki-67 depletion in the suppression of tumorigenesis.
Table 1.
Summary of p53, p21 and Rb status of CDKi and Ki-67 sensitive cells
| Cell line | Rb status (Sobecki et al. 2017) | p53 status (Leroy et al. 2014) | p21 induction upon Ki-67 KD (Sun et al., 2017) | CDKi sensitivity (Sobecki et al. 2017) | Ki-67 sensitivity (Sun et al., 2017) |
|---|---|---|---|---|---|
| IMR-90 | + | + | + | + | + |
| MCF-7 | + | + | + | + | + |
| HDF | + | + | + | + | + |
| hTERT-RPE1 | + | + | + | ND | + |
| MDA-MB-231 | + | R280K mutant, dominantly defective for transcriptional activation | − | + | − |
| MDA-MB-436 | − | − | ND | − | ND |
| HCT116 | + | + | − | − | − |
| HeLa | Inactivated by HPV | Inactivated by HPV | − | − | − |
| U2OS | − | + | − | − | − |
HDF, Human diploid fibroblast; ND, not determined
6. Interphase Ki-67 organizes heterochromatin
Early studies showed that Ki-67 colocalizes with Hoechst 33258-stained chromocenters in mouse Swiss 3T3 fibroblasts during the S and G2 cell cycle phases, suggesting that Ki-67 may play a general role in heterochromatin organization (Starborg et al. 1996). Consistent with this idea, Ki-67 association with heterochromatic regions was also detected in early G1 phase cells. In this case, immuno-FISH experiments revealed that Ki-67 localizes to hundreds of distinct foci within two hours after mitotic exit (Lopez et al., 1991; Isola et al., 1990). These early G1 foci co-localize with several different classes of heterochromatic repetitive elements, including centromeric alpha satellite, telomeric repeats and Satellite III (Bridger et al. 1998). Notably, these repeats frequently localize to the nucleolar periphery during the rest of interphase (Koningsbruggen et al., 2010; Nemeth et al., 2010).
These observations are underpinned by several recent studies showing that Ki-67 promotes association of multiple heterochromatic regions with the nucleolar periphery. Nucleoli are surrounded by a subset of the genome termed nucleolar-associated DNA sequences (NADs) (Norton et al., 2009; Koningsbruggen et al., 2010; Nemeth et al., 2010; Dillinger et al. 2017). Multiple studies showed that Ki-67 depletion decreased the nucleolar assocation of NADs, including a LacO array proximal to the rDNA repeats on chromosome 13 (Booth et al. 2014), and chromosome 17 alpha satellite repeats (Matheson and Kaufman 2017). Additionally, Ki-67 depletion delocalized centromeric histone variant CENP-A away from nucleoli, indicating reduced centromeric chromatin association with nucleoli (Sobecki et al. 2016). An additional prominent example of nucleolar association is the inactive X chromosome in female mammalian cells (Barr and Bertram, 1949; Zhang et al., 2007). In diploid female hTERT-RPE1 cells, depletion of Ki-67 altered the S phase-dependent nucleolar localization of the inactive X (Xi) chromosome, and perturbed several characteristic features of Xi facultative heterochromatin (Sun et al. 2017).
Ki-67 affects nucleolar localization of heterochromatin in multiple ways. The first of these appears to be restricted to G1/S checkpoint-proficient cells. For example, Ki-67 depletion delays S phase entry in p21 checkpoint-proficient cell lines such as hTERT-RPE1 cells. In these female cells, Ki-67 depletion also delays the S phase-dependent association of the Xi chromosome with nucleoli. This is accompanied by increased transcription of Cot1-hybridizing DNA repetitive elements and reduced H3K27me3 and H4K20me1 enrichment on the Xi in a subset of cells in which the Xi was localized away from the nuclear periphery (Sun et al. 2017). In contrast to checkpoint-proficient hTERT-RPE1 cells, no alteration of Xi localization or erosion of heterochromatin features is observed in 293T cells which enter S phase without delay and do not induce p21 upon Ki-67 depletion (Sobecki et al., 2016; Sun et al., 2017). Therefore, the impaired maintenance of epigenetic features in hTERT-RPE1 cells on non-laminar Xi chromosomes could indicate that laminar associations protect from this type of erosion. Alternatively, these observations could indicate that movement away from the lamina is required for downstream alterations.
However, not all heterochromatin effects of Ki-67 depletion are dependent on cell cycle checkpoints. For example, depletion of Ki-67 in HeLa cells does not alter cell cycle distribution (Sobecki et al., 2016; Sun et al., 2017), but the nucleolar association frequency of alpha satellite DNA becomes significantly reduced (Matheson and Kaufman 2017). Therefore, Ki-67 promotes NAD localization even in checkpoint-deficient cells. How does Ki-67 promote these interactions? There are several possibilities that are not mutually exclusive. These include recognition of primary sequences or chromatin features of NADs (Bridger et al., 1998; Kreitz et al., 2000), via direct interaction (Takagi et al., 1999; Scholzen et al., 2002), or via binding of other nucleolar–targeting proteins. Furthermore, RNAs are important for several higher order chromosome interactions (Hacisuleyman et al., 2014; Yang et al., 2015), including examples at nucleoli (Mondal et al. 2008; Fedoriw et al. 2012), and these have not been investigated in this regard. Ki-67 may also form a charged surface coating on nucleoli in analogy to its role on mitotic chromosomes (Cuylen et al. 2016) and thereby attract heterochromatin (Larson et al. 2017; Strom et al. 2017). It is possible that phase separation of heterochromatin, such as that recently demonstrated for HP1 (Larson et al. 2017; Strom et al. 2017), could contribute to such a mechanism.
Besides mediating long-range chromosome interactions, Ki-67 also promotes chromatin compaction. Upon Ki-67 depletion, HeLa cells displayed reduced fluorescence lifetime imaging-fluorescence resonance energy transfer (FLIM-FRET) signals between H2B-eGFP and H2B-mCherry (Sobecki et al. 2016), which report on inter-nucleosomal interactions on the scale of ~1–10 nm (Lleres et al. 2009). Thus, the elimination of high FRET signals emanating from perinuclear and perinucleolar heterochromatic regions indicates reduced heterochromatin compaction (Sobecki et al. 2016). Because this has been observed in HeLa cells, this activity is not dependent on G1 checkpoint events.
How does Ki-67 regulate heterochromatin compaction? One clue comes from the observation that in the absence of Ki-67, H3K9me3 become less focally clustered (Sobecki et al. 2016). H3K9me3 is particularly relevent here because in most eukaryotes chromatin compaction depends on core histone deacetylation and histone H3K9 di- and tri-methylation (Eissenberg and Elgin 2014). HP1 recognizes methylated H3K9 and also directly binds Ki-67 with high affinity (Jacobs and Sepideh 2002; Nielsen et al. 2002). This interaction has consequences in cells, because ectopic expression of HP1 relocalizes Ki-67 to sites of high HP1 concentration (Scholzen et al. 2002). The interaction between Ki-67 and HP1 may allow Ki-67 to link H3K9me2/3-enriched regions with Ki-67 interacting proteins involved in chromatin modifications (Sobecki et al. 2016). Notably, many such candidate interactors were recently identified via proteomics (Sobecki et al. 2016). The associated proteins include UHRF1, which binds H3K9me3 and DNA methyltransferase DNMT1 (Bostick et al. 2007); SUZ12, a component of H3K27 methyltransferase complex PRC2 (Pasini et al. 2004); and TIP5, the major component of the nucleolar remodeling complex (NoRC) which contributes to the silent state of inactive rDNA repeats (Strohner et al. 2004). Thus, many potential molecular links between Ki-67 and heterochromatin remain to be explored in detail.
Additionally, the existing data provide an apparantly paradoxical scenario: although Ki-67 is required for the 3D-organization of H3K9me3 foci, all three HP1 isoforms maintained their cellular localization in Ki-67-depleted cells (Sobecki et al. 2016). One possibile explanation is that the localization of HP1 proteins can be regulated at least in part in a histone modification-independent manner, as has been demonstrated in C. elegans (Garrigues et al. 2015). Additionally, it remains to be determined how DNA association by HP1 proteins changes upon loss of Ki-67, and whether HP1 contributes to heterochromatin changes detected upon Ki-67 depletion.
7. Concluding remarks
The growing interest in Ki-67 protein functions has led to several recent important findings. Multiple molecular functions of Ki-67 display cell type-specific variations and are correlated with distinct stages of cell cycle. During mitosis, Ki-67 coats the surface of chromosomes and is required for the formation of the perichromosomal layer, which constitutes approximately one-third of the mass of mitotic chromosomes (Booth et al. 2014; Booth et al. 2016); reviewed in (Booth and Earnshaw 2017). As part of the PCL, Ki-67’s high net positive charge enables it to prevent mitotic chromosomes from sticking together (Cuylen et al. 2016). Additionally, recent data indicate that Ki-67 and cohesin complexes make distinct contributions to the stuctural integrity of mitotic chromosomes, such that co-depletion of Ki-67 and condensin causes chromosomes to form an amorphous “slime ball” (Takagi et al. 2017). Therefore, there is great interest in understanding the molecular details of how Ki-67, as the keystone of forming the PCL, contributes to mitotic chromosome structure.
After mitosis, Ki-67 relocalizes to the nucleolar periphery, overlapping perinucleolar heterochromatin. Nucleoli are the largest non-membrane-bound subnuclear structures. Analyses in Xenopus oocyte nuclei and Drosophila embryos reveal that the internal organization of nucleoli depends on liquid-liquid phase separation (Feric et al. 2016). This raises the question of whether Ki-67 has a surfactant role in nucleolus organization, and whether this may contribute to Ki-67’s role in organizing heterochromatin around the nucleolus (Sobecki et al., 2016; Matheson and Kaufman, 2015; Matheson and Kaufman, 2017). Additionally, there has been no systematic analysis of NAD relocalization in response to Ki-67 depletion, so it is not known if there are unforeseen specificities regarding this activity. It will be particularly interesting to know if any NAD associations are senstive to the G1/S checkpoint status of the cells examined.
Ki-67 forms a functional holoenzyme with PP1 in a similar manner as RepoMan (Trinkle-Mulcahy et al., 2006; Takagi et al., 2014; Booth et al., 2014; Kumar et al., 2016), but the contribution of the Ki-67/PP1 holoenzyme to histone dephosphorylation remains to be characterized. It will be of great interest to determine whether the Ki-67/PP1 holoenzyme complex makes its contributions to heterochromatin localization via similar phospho/methyl switch mechanisms.
Notably, PP1 has 200 interacting/regulator proteins, meaning that therapeutic targeting of this enzyme would likely cause broad pleotropic effects. However, the KiR-SLiM:PP1 interaction surface could be a candidate drug target that would be predicted to inhibit only the Ki-67:PP1 and RepoMan:PP1 holoenzymes (Rebelo et al. 2015). Such tools would be invaluable biological probes of events during mitotic exit, and would be interesting candidate therapeutics.
Acknowledgments
We thank Timothy Matheson for the use of Figure 2.
Funding:
This study was funded by National Institutes of Health (grant number 1U01 DA040588).
Footnotes
Conflict of Interest:
Xiaoming Sun declares that she has no conflict of interest.
Paul D. Kaufman declares that he has no conflict of interest.
Ethical approval:
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Banani SF, Rice AM, Peeples WB, et al. Composition control of Phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- Booth DG, Beckett AJ, Molina O, et al. 3D-CLEM Reveals that a Major Portion of Mitotic Chromosomes Is Not Chromatin. Mol Cell. 2016;64:790–802. doi: 10.1016/j.molcel.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth DG, Earnshaw WC. Ki-67 and the Chromosome Periphery Compartment in Mitosis. Trends Cell Biol S0962-8924. 2017:30136–8. doi: 10.1016/j.tcb.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Booth DG, Takagi M, Sanchez-Pulido L, et al. Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. Elife. 2014 doi: 10.7554/eLife.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kyong KJ, Esteve P-O, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science (80- ) 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Bridger JM, Kill IR, Lichter P. Association of pKi-67 with satellite DNA of the human genome in early G 1 cells. Chromosom Res. 1998;6:13–24. doi: 10.1023/a:1009210206855. [DOI] [PubMed] [Google Scholar]
- Chellappan SP, Hiebert S, Mudryj M, et al. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-F. [DOI] [PubMed] [Google Scholar]
- Chierico L, Rizzello L, Guan L, et al. The role of the two splice variants and extranuclear pathway on Ki-67 regulation in non-cancer and cancer cells. PLoS One. 2017;12:e0171815. doi: 10.1371/journal.pone.0171815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cidado J, Wong HY, Rosen DM, et al. Ki-67 is required for maintenance of cancer stem cells but not cell proliferation. Oncotarget. 2016;7:6281–6293. doi: 10.18632/oncotarget.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Blaukopf C, Politi AZ, et al. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 2016;535:308–312. doi: 10.1038/nature18610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro IJ, Budzak J, Di Giacinto ML, et al. Repo-Man/PP1 regulates heterochromatin formation in interphase. Nat Commun. 2017;8:14048. doi: 10.1038/ncomms14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillinger S, Straub T, Nemeth A. Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear reorganisation. PLoS One. 2017;12:e0178821. doi: 10.1371/journal.pone.0178821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowsett M, Nielsen TO, A’Hern R, et al. Assessment of Ki67 in Breast Cancer: Recommendations from the international Ki67 in breast cancer working Group. J Natl Cancer Inst. 2011;103:1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Henckel J, Fersht AR, et al. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4:387–94. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513:58–66. doi: 10.1016/S0014-5793(01)03294-X. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin S. HP1a: A structural chromsomal protein regulating transcription. Trends Genet. 2014;30:103–110. doi: 10.1016/j.tig.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- Endl E, Gerdes J. Posttranslational modifications of the Ki-67 protein coincide with two major checkpoints during mitosis. J Cell Physiol. 2000;182:371–380. doi: 10.1002/(SICI)1097-4652(200003)182:3<371::AID-JCP8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Fedoriw AM, Calabrese JM, Mu W, et al. Differentiation-Driven Nucleolar Association of the Mouse Imprinted Kcnq1 Locus. G3. 2012;2:1521–1528. doi: 10.1534/g3.112.004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, et al. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016;44:6070–6086. doi: 10.1093/nar/gkw523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Fry DW, Harvey PJ, Keller PR, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–38. 3/11/1427. [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305– 2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues JM, Sidoli S, Garcia Ba, Strome S. Defining heterochromatin in C. elegans through genome-wide analysis of the heterochromatin protein 1 homolog HPL-2. Genome Res. 2015;25:76–88. doi: 10.1101/gr.180489.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehani SS, Agrawal-Singh S, Dietrich N, et al. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell. 2010;39:886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Gerdes J, Lemke H, Baisch H, et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- Gerdes J, Li L, Schlueter C, et al. Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am J Pathol. 1991;138:867–873. [PMC free article] [PubMed] [Google Scholar]
- Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- Gerdes J, Stein H, Pileri S, et al. Prognostic relevance of tumour-cell growth fraction in malignant non-Hodgkin’s lymphomas. Lancet. 1987;2:448–449. doi: 10.1016/S0140-6736(87)90977-9. [DOI] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Kato K, Kimura K. The hierarchical structure of the perichromosomal layer comprises Ki67, ribosomal RNAs, and nucleolar proteins. Biochem Biophys Res Commun. 2017;493:1043–1049. doi: 10.1016/j.bbrc.2017.09.092. [DOI] [PubMed] [Google Scholar]
- Hernandez AR, Klein AM, Kirschner MW. Kinetic Responses of beta-catenin specify the sites of wnt control. Science (80- ) 2012;338:1337–1340. doi: 10.1126/science.1228734. [DOI] [PubMed] [Google Scholar]
- Hiragami-Hamada K, Soeroes S, Nikolov M, et al. Dynamic and flexible H3K9me3 bridging via HP1β dimerization establishes a plastic state of condensed chromatin. Nat Commun. 2016;7:11310. doi: 10.1038/ncomms11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, et al. Role for E2F in Control of Both DNA Replication and Mitotic Functions as Revealed from DNA Microarray Analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isola J, Helin H, Kallioniemi OP. Immunoelectron-microscopic localization of a proliferation-associated antigen Ki-67 in MCF-7 cells. Histochem J. 1990;22:498–506. doi: 10.1007/BF01007235. [DOI] [PubMed] [Google Scholar]
- Jacobs S, Sepideh K. Structure of the HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Nature. 2002;295:2080–3. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Junk DJ, Vrba L, Watts GS, et al. Different mutant/wild-type p53 combinations cause a spectrum of increased invasive potential in nonmalignant immortalized human mammary epithelial cells. Neoplasia. 2008;10:450–61. doi: 10.1593/neo.08120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka A, Takagi M, Hayakawa T, et al. Interaction of the chromatin compaction-inducing domain (LR domain) of Ki-67 antigen with HP1 proteins. Genes to Cells. 2002;7:1231–1242. doi: 10.1046/j.1365-2443.2002.00596.x. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJG, Kwok BH, et al. The biplolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–8. doi: 10.1038/nature03493.Published. [DOI] [PubMed] [Google Scholar]
- Kausch I, Lingnau A, Endl E, et al. Antisense treatment against Ki-67 mRNA inhibits proliferation and tumor growth in vitro and in vivo. Int J Cancer. 2003;105:710–716. doi: 10.1002/ijc.11111. [DOI] [PubMed] [Google Scholar]
- van Koningsbruggen S, Gierlinski M, Schofield P, et al. High-Resolution Whole-Genome Sequencing Reveals That Specific Chromatin Domains from Most Human Chromosomes Associate with Nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz S, Fackelmayer FO, Gerdes J, Knippers R. The proliferation-specific human Ki-67 protein is a constituent of compact chromatin. Exp Cell Res. 2000;261:284–292. doi: 10.1006/excr.2000.5064. [DOI] [PubMed] [Google Scholar]
- Kumar GS, Gokhan E, De Munter S, et al. The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism. Elife. 2016;5:e16539. doi: 10.7554/eLife.16539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, Carroll D, Rea S, et al. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy B, Girard L, Hollestelle A, et al. Analysis of TP53 mutation status in human cancer cell lines: A reassessment. Hum Mutat. 2014;35:756–765. doi: 10.1002/humu.22556.Analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li VSW, Ng SS, Boersema PJ, et al. Wnt Signaling through Inhibition of β-Catenin Degradation in an Intact Axin1 Complex. Cell. 2012;149:1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Lin TC, Su CY, Wu PY, et al. The nucleolar protein NIFK promotes cancer progression via ck1α/β-catenin in metastasis and ki-67-dependent cell proliferation. Elife. 2016;5:e11288. doi: 10.7554/eLife.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llères D, James J, Swift S, et al. Quantitative analysis of chromatin compaction in living cells using FLIM-FRET. J Cell Biol. 2009;187:481–496. doi: 10.1083/jcb.200907029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez F, Belloc F, Lacombe F, et al. Modalities of synthesis of Ki67 antigen during the stimulation of lymphocytes. Cytometry. 1991;12:42–49. doi: 10.1002/cyto.990120107. [DOI] [PubMed] [Google Scholar]
- Luo Y, Ren F, Liu Y, et al. Clinicopathological and prognostic significance of high Ki-67 labeling index in hepatocellular carcinoma patients: A meta-analysis. Int J Clin Exp Med. 2015;8:10235–10247. [PMC free article] [PubMed] [Google Scholar]
- MacCallum DE, Hall PA. Biochemical characterization of pKi67 with the identification of a mitotic-specific form associated with hyperphosphorylation and altered DNA binding. Exp Cell Res. 1999;252:186–98. doi: 10.1006/excr.1999.4600. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson TD, Kaufman PD. Grabbing the genome by the NADs. Chromosoma. 2015;125:361–371. doi: 10.1007/s00412-015-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson TD, Kaufman PD. The p150N domain of chromatin assembly factor-1 regulates Ki-67 accumulation on the mitotic perichromosomal layer. Mol Biol Cell. 2017;28:21–29. doi: 10.1091/mbc.E16-09-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Maddox P, Gaetz J, et al. Role of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus Extract meitotic spindles. Mol Biol Cell. 2005;16:3064–76. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal T, Fraser P, Kanduri C. Kcnq1ot1/Lit1 Noncoding RNA Mediates Transcriptional Silencing by Targeting to the Perinucleolar Region †. 2008;28:3713–3728. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Yang J, Truant R, Kornbluth S. Nuclear import of Cdk/cyclin complexes: Identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J Cell Biol. 1999;144:213–224. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: Interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/S1097-2765(00)80204-X. [DOI] [PubMed] [Google Scholar]
- Németh A, Conesa A, Santoyo-Lopez J, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- Norton JT, Wang C, Gjidoda A, et al. The perinucleolar compartment is directly associated with DNA. J Biol Chem. 2009;284:4090–4101. doi: 10.1074/jbc.M807255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Tsai H, Wang S, et al. The RNA recognition motif of NIFK is required for rRNA maturation during cell cycle progression. RNA Biol. 2015;12:255–267. doi: 10.1080/15476286.2015.1017221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, et al. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzilli R, Partelli S, Cannizzaro R, et al. Ki-67 prognostic and therapeutic decision driven marker for pancreatic neuroendocrine neoplasms (PNENs): A systematic review. Adv Med Sci. 2016;61:147–153. doi: 10.1016/j.advms.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Pyo JS, Kang G, Sohn JH. Ki-67 labeling index can be used as a prognostic marker in gastrointestinal stromal tumor: a systematic review and meta-analysis. Int J Biol Markers. 2015;31:0. doi: 10.5301/jbm.5000183. [DOI] [PubMed] [Google Scholar]
- Qian J, Lesage B, Beullens M, et al. PP1/repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr Biol. 2011;21:766–773. doi: 10.1016/j.cub.2011.03.047. [DOI] [PubMed] [Google Scholar]
- Quivy J-P, Le Roche D, Kirschner D, et al. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. doi:10.1038/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo S, Santos M, Martins F, et al. Protein phosphatase 1 is a key player in nuclear events. Cell Signal. 2015;27:2589–2598. doi: 10.1016/j.cellsig.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, et al. E2F integrates cell cycle progression with DNA repair, replication, and E2F integrates cell cycle progression with DNA repair, replication, and G 2/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards-Taylor S, Ewings SM, Jaynes E, et al. The assessment of Ki-67 as a prognostic marker in neuroendocrine tumours: a systematic review and meta-analysis. J Clin Pathol. 2015 doi: 10.1136/jclinpath-2015-203340. jclinpath-2015-203340. [DOI] [PubMed] [Google Scholar]
- Saiwaki T, Kotera I, Sasaki M, et al. In vivo dynamics and kinetics of pKi-67: Transition from a mobile to an immobile form at the onset of anaphase. Exp Cell Res. 2005;308:123–134. doi: 10.1016/j.yexcr.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Saksouk N, Simboeck E, Déjardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin. 2015;8:3. doi: 10.1186/1756-8935-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter C, Duchrow M, Wohlenberg C, et al. The cell proliferation-associated antigen of antibody Ki-67: A very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J Cell Biol. 1993;123:513–522. doi: 10.1083/jcb.123.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MHH, Broll R, Bruch HP, et al. Proliferation marker pKi-67 occurs in different isoforms with various cellular effects. J Cell Biochem. 2004;91:1280–1292. doi: 10.1002/jcb.20016. [DOI] [PubMed] [Google Scholar]
- Schmidt MHH, Broll R, Bruch HP, et al. The proliferation marker pKi-67 organizes the nucleolus during the cell cycle depending on Ran and cyclin B. J Pathol. 2003;199:18–27. doi: 10.1002/path.1221. [DOI] [PubMed] [Google Scholar]
- Scholzen T, Endl E, Wohlenberg C, et al. The Ki-67 protein interacts with members of the heterochromatin protein 1 (HP1) family: a potential role in the regulation of higher-order chromatin structure. J Pathol. 2002;196:135–144. doi: 10.1002/path.1016. [DOI] [PubMed] [Google Scholar]
- Scholzen T, Gerdes J. The Ki-67 protein: From the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Smith CL, Matheson TD, Trombly DJ, et al. A separable domain of the p150 subunit of human chromatin assembly factor-1 promotes protein and chromosome associations with nucleoli. Mol Biol Cell. 2014;25:2866–81. doi: 10.1091/mbc.E14-05-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobecki M, Mrouj K, Camasses A, et al. The cell proliferation antigen Ki-67 organises heterochromatin. Elife. 2016;5:e13722. doi: 10.7554/eLife.13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobecki M, Mrouj K, Colinge J, et al. Cell cycle regulation accounts for variability in Ki-67 expression levels. Cancer Res. 2017;77:2722–2734. doi: 10.1158/0008-5472.CAN-16-0707. [DOI] [PubMed] [Google Scholar]
- Starborg M, Gell K, Brundell E, Höög C. The murine Ki-67 cell proliferation antigen accumulates in the nucleolar and heterochromatic regions of interphase cells and at the periphery of the mitotic chromosomes in a process essential for cell cycle progression. J Cell Sci. 1996;109(Pt1):143–53. doi: 10.1242/jcs.109.1.143. [DOI] [PubMed] [Google Scholar]
- Strohner R, Németh A, Nightingale KP, et al. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol Cell Biol. 2004;24:1791–8. doi: 10.1128/MCB.24.4.1791-1798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, et al. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueishi M, Takagi M, Yoneda Y. The forkhead-associated domain of Ki-67 antigen interacts with the novel kinesin-like protein Hklp2. J Biol Chem. 2000;275:28888–28892. doi: 10.1074/jbc.M003879200. [DOI] [PubMed] [Google Scholar]
- Sun X, Bizhanova A, Matheson TD, et al. Ki-67 contributes to normal cell cycle progression and inactive X heterochromatin in p21 checkpoint-proficient human cells. Mol Cell Biol. 2017;37:e00569–16. doi: 10.1128/MCB.00569-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Matsuoka Y, Kurihara T, Yoneda Y. Chmadrin: a novel Ki-67 antigen-related perichromosomal protein possibly implicated in higher order chromatin structure. J Cell Sci. 1999;112(Pt 15):2463–2472. doi: 10.1242/jcs.112.15.2463. [DOI] [PubMed] [Google Scholar]
- Takagi M, Natsume T, Kanemaki MT, Imamoto N. Perichromosomal protein Ki67 supports mitotic chromosome architecture. Genes to Cells. 2016;21:1113–1124. doi: 10.1111/gtc.12420. [DOI] [PubMed] [Google Scholar]
- Takagi M, Nishiyama Y, Taguchi A, Imamoto N. Ki67 antigen contributes to the timely accumulation of protein phosphatase 1γ on anaphase chromosomes. J Biol Chem. 2014;289:22877–22887. doi: 10.1074/jbc.M114.556647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Ono T, Natsume T, et al. Ki-67 and condensins support the integrity of mitotic chromosomes through distinct mechanisms. bioRxiv. 2017 doi: 10.1242/jcs.212092. doi: org/10.1101/202390. [DOI] [PubMed]
- Takagi M, Sueishi M, Saiwaki T, et al. A Novel Nucleolar Protein, NIFK, Interacts with the Forkhead Associated Domain of Ki-67 Antigen in Mitosis. J Biol Chem. 2001;276:25386–25391. doi: 10.1074/jbc.M102227200. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Macůrek L, Galjart N, Medema RH. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 2008;27:3235–3245. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Qian GW, Li W, et al. A critical role of Sp1 transcription factor in regulating the human Ki-67 gene expression. Tumor Biol. 2011;32:273–283. doi: 10.1007/s13277-010-0119-4. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Andersen J, Yun WL, et al. Repo-Man recruits PP1 to chromatin and is essential for cell viability. J Cell Biol. 2006;172:679–692. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P, Ribeiro S, Sennels L, et al. Repo-Man Coordinates Chromosomal Reorganization with Nuclear Envelope Reassembly during Mitotic Exit. Dev Cell. 2011;21:328–342. doi: 10.1016/j.devcel.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Yuh P, Heald R. The perichromosomal layer. Chromosoma. 2005;114:377–388. doi: 10.1007/s00412-005-0021-9. [DOI] [PubMed] [Google Scholar]
- Vanneste D, Takagi M, Imamoto N, Vernos I. The Role of Hklp2 in the Stabilization and Maintenance of Spindle Bipolarity. Curr Biol. 2009;19:1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Verheijen R, Kuijpers HJ, Schlingemann RO, et al. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. I. Intracellular localization during interphase. J Cell Sci. 1989a;92( Pt 1):123–30. doi: 10.1242/jcs.92.1.123. [DOI] [PubMed] [Google Scholar]
- Verheijen R, Kuijpers HJ, van Driel R, et al. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. II. Localization in mitotic cells and association with chromosomes. J Cell Sci. 1989b;92( Pt 4):531–540. doi: 10.1242/jcs.92.4.531. [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B. Cyclin-dependent kinase inhibitor p21 modulates the DNA primer-template recognition complex. Mol Cell Biol. 1998;18:4177–87. doi: 10.1128/mcb.18.7.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub SJ, Prater CA, Dean DC. Retinoblastoma protein swithes the E2F site from positive to negative element. Nature. 1992;358:259–61. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Boleti H, Antony C, et al. Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J Cell Biol. 1998;143:673–685. doi: 10.1083/jcb.143.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Deng X, Ma W, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015 doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. Perinucleolar Targeting of the Inactive X during S Phase: Evidence for a Role in the Maintenance of Silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]