Abstract
Thrombospondin-2 (TSP2) is an anti-angiogenic matricellular protein that inhibits tumor growth and angiogenesis. Tumor-associated blood vascular endothelial cells (BECs) were isolated from human invasive bladder cancers and from matched normal bladder tissue by immuno-laser capture microdissection. Exon expression profiling analyses revealed a particularly high expression of a short TSP2 transcript containing only the last 9 (3′) exons of the full-length TSP2 transcript. Using 5′ and 3′ RACE (rapid amplification of cDNA ends) and Sanger sequencing, we confirmed the existence of the shorter transcript of TSP2 (sTSP2) and determined its sequence which completely lacked the anti-angiogenic thrombospondin type 1 repeats domain. The largest open reading frame predicted within the transcript comprises 209 amino acids and matches almost completely the C-terminal lectin domain of full-length TSP2. We produced recombinant sTSP2 and found that unlike the full-length TSP2, sTSP2 did not inhibit vascular endothelial growth factor-A-induced proliferation of cultured human BECs, but in contrast when combined with TSP2 blocked the inhibitory effects of TSP2 on BEC proliferation. In vivo studies with stably transfected A431 squamous cell carcinoma cells revealed that full-length TSP2, but not sTSP2, inhibited tumor growth and angiogenesis. This study reveals that the transcriptional program of tumor stromal cells can change to transcribe a new version of an endogenous angiogenesis inhibitor that has lost its anti-angiogenic activity.
Introduction
Bladder cancer growth is thought to dependent on the activation of angiogenesis [1–3]. However, the therapeutic targeting of vascular endothelial growth factor-A (VEGF-A) or its receptors has shown no or only limited benefit in clinical trials of bladder cancer [4], indicating that other regulators of angiogenesis might play a major role.
Recent discoveries of the ENCODE and FANTOM5 projects [5, 6] and of other RNA-sequencing projects [7, 8] revealed that approximately 90% of human genes undergo alternative splicing or alternative transcription and that there are around 10 to 12 alternative transcripts per gene on average that can code for proteins that show very different protein interactions and perform different biological roles [9]. In order to find new unexplored mediators of tumor angiogenesis in bladder cancer, we isolated tumor-associated blood vascular endothelial cells (tBEC) from invasive human bladder cancers and endothelial cells (ECs) from adjacent normal tissue (nBEC) using immuno-laser capture microdissection (LCM). We then assessed the differences in alternative splicing and transcription using microarray technology. Among other changes, we identified the expression of an alternative transcript of the potent endogenous angiogenesis inhibitor thrombospondin-2 (TSP2) in tBECs.
The alternative TSP2 transcript comprises the 3′-end region of the full-length transcript, spanning from exon 15 to the poly-A tail and does not contain the anti-angiogenic thrombospondin type 1 repeats domain that is located at the N-terminal region of the full-length TSP2 sequence [10–13]. We investigated the potential loss of anti-angiogenic activities of sTSP2 both in vitro and in vivo in mouse models of cancer. Our results demonstrate the first example of the conversion of an endogenous angiogenesis inhibitor into a non-angiogenesis inhibiting molecule by alternative splicing (transcription) in tumor-associated stromal cells, thus revealing a potential new mechanism by which natural anti-angiogenic mechanisms can be inactivated in cancers.
Results
Identification of alternative transcripts of invasive bladder cancer-associated blood vessels
BECs were isolated from frozen sections of invasive bladder cancers (tBECs) and adjacent normal (nBEC) bladder tissue (n = 6) using LCM. The purity of isolates was confirmed by high mRNA expression of PECAM1 (EC marker; among 15% highest expressed genes) and low mRNA expression of KRT7 (urothelial cell marker; among 10% lowest expressed genes).
Using exon microarrays, alternative splicing was determined by the relative inclusion or skipping rate of an exon in tumor-associated blood vessels and in blood vessels of healthy bladder tissue. Although alternative start/termination of transcription is not an alternative splice event, the exon array technology can also detect differences in the usage of transcription start/stop sites. Since the analysis of exon arrays is challenging, as there are no clear standards of analysis [14] and most of the splicing analysis methods suffer from high false-positive detection rates, we combined three different analysis methods: microarray detection of alternative splicing (MIDAS [15]), analysis of splice variation (ANOSVA [16, 17]), and finding isoforms using robust multichip analysis (FIRMA [18]) to increase the stringency (described in detail in Materials and methods section). We identified, using MIDAS with a cut-off of p < 0.05, 963 genes with alternative splicing events (Fig. 1a). As a second method to analyze alternative splicing, we used ANOSVA with a cut-off of p < 0.01 and found 2,095 genes with alternative splicing events (Fig. 1a). As a third method, we used FIRMA with a cut-off of p < 0.001 and found 1,402 genes with alternative splicing events (Fig. 1a).
Fig. 1.
Identification of a short alternative transcript of TSP2 that is highly expressed in bladder cancer-associated blood vessels. a Blood vascular endothelial cells (BECs) were isolated from frozen sections of invasive bladder cancers (tBEC) and adjacent normal (nBEC) bladder tissue (n = 6). RNA was isolated and hybridized to Human Exon 1.0 ST arrays. a Detection of alternative splicing and transcription was performed using three different bioinformatics analysis tools, namely MIDAS, ANOSVA, and FIRMA. The numbers indicate the number of genes with alternative splicing or alternative transcription identified by each method and their overlap. b Exon array expression levels of the full-length and the short transcript of TSP2 and exon structure as shown in the GenBank database. c qRT-PCR analysis of the expression of the full-length and the short TSP2 transcripts in tumor-associated vessels and in vessels of healthy bladder tissue. For five out of six patients, matched samples of tumor-associated and healthy vessels could be analyzed as highlighted by the connecting dotted lines. The positions of the primers used are indicated by arrows in b. Horizontal lines represent median values. Dots represent patient samples. Relative expression was normalized to expression of vessels from healthy tissues in both groups. *p < 0.05
Combining these three analysis approaches, we found 76 alternatively spliced genes that were identified by all three methods (Fig. 1a, Table 1 and Supplementary Table 1). Alternative transcripts that were found by all three approaches were visually inspected by evaluating the expression of individual exons and comparing them to the deposited alternative transcript sequence in the GenBank database. We focused on the transcripts with alternative transcription; setting the threshold to excision/inclusion of five consecutive exons that could encode for a novel transcript (Table 2). As a validation of our method to identify alternative transcripts, we could validate the existence of alternative transcripts with the capability to encode for proteins (Supplementary Figure 1). In the 3′ end of RABGAP1L, we identified ENST00000478442.5 and ENST00000465412.5 (Supplementary Figure 1A) from the ENSEMBL database with higher expression in tBEC. Furthermore, we detected a higher expression of the alternative transcript ENST00000465413.6 in tBEC, a shorter transcript of ALCAM, previously deposited at ENSEMBL (Supplementary Figure 1B).
Table 1.
Genes with statistically significant splicing identified by all three bioinformatics methods (FIRMA, MIDAS, ANOSVA)
| Splicing of a single exon | Splicing of 2–4 consecutive exons | Splicing of >5 consecutive exons | ||
|---|---|---|---|---|
| RPL11 | ILF2 | SPARC | CCNA2 | IARS2 |
| NEXN | ZBTB41 | SOX4 | SLC17A5 | BUB1 |
| CEP350 | USP34 | STXBP5 | EZH2 | ALCAM |
| PRDX1 | IDH1 | BRAF | RDH10 | APP |
| PDIA6 | VCAN | PRKDC | PTK2 | TNFRSF21 |
| SEC61A1 | SSR1 | SIRT1 | CAPRIN1 | LEMD3 |
| LAT2 | SMC2 | MARS | ESYT1 | CAND1 |
| SPIN1 | TAOK1 | ATP2A2 | GDE1 | TRIM33 |
| TTC17 | MED13 | SLC38A1 | GNAS | MANF |
| PCGF2 | GATA6 | DLGAP5 | RABGAP1L | THBS2 |
| GNA13 | DSG2 | RPN2 | COL12A1 | ADAMTS9 |
| GALNT1 | SLC39A6 | PSMA7 | MTCH2 | TPX2 |
| TCF3 | GNB1 | CCT2 | COL5A2 | |
| YTHDF1 | MATR3 | CLSPN | ||
| HMGXB4 | CCNJ | |||
| RCC2 | ZBTB44 | |||
| DARS | RAP2A | |||
| TFRC | YY1 | |||
| KIF23 | ||||
Genes were separated in three categories based on the number of consective exons spliced
The list of 76 genes identified was divided into 3 categories, based on FIRMA significance analysis: genes with splicing in 1 exon, genes with splicing in 2–4 adjacent exons, and genes with splicing in 5 adjacent exons
MIDAS microarray detection of alternative splicing, FIRMA finding isoforms using robust multichip analysis, ANOSVA analysis of splice variation
Table 2.
Genes with statistically significant splicing identified by all three bioinformatics methods (FIRMA, MIDAS, ANOSVA) with five and more consecutive exon spliced
| Gene symbol | Number of adjacent exons spliced | FIRMA scores | FIRMAp values | MIDAS scores | MIDASp values | ANOSVA p values |
|---|---|---|---|---|---|---|
| RABGAP1L | 5 | 2.67 | 0.00009 | 2.08 | 0.029123 | 0.00557443 |
| COL5A2 | 11 | −2.69 | 0.0001 | 3.78 | 0.018022 | 5.59315E−12 |
| THBS2 | 8 | −2.13 | 0.00012 | −2.82 | 0.026826 | 1.82292E−10 |
| CAPRIN1 | 4 | −2.71 | 0.00016 | 1.77 | 0.016125 | 0.00401313 |
| CCT2 | 5 | −2.91 | 0.00017 | −5.31 | 0.034639 | 1.66782E−08 |
| PTK2 | 4 | −2.47 | 0.00024 | 1.05 | 0.029415 | 0.000841486 |
| CAND1 | 7 | 2.94 | 0.00036 | 1.72 | 0.036665 | 1.18221E−05 |
| ALCAM | 6 | 1.95 | 0.00036 | 1.49 | 0.042686 | 3.01511E−06 |
| IARS2 | 6 | −2.16 | 0.00043 | 4.17 | 0.02786 | 0.00132895 |
| GNAS | 4 | 2.09 | 0.00043 | 1.26 | 0.048403 | 3.58804E−06 |
| CLSPN | 12 | −1.71 | 0.00045 | −2.78 | 0.018561 | 7.88203E−10 |
| COL12A1 | 5 | −2.16 | 0.00049 | 3.61 | 0.017637 | 1.24805E−11 |
| MANF | 8 | −1.69 | 0.00051 | 4.77 | 0.028571 | 1.57539E−08 |
| ESYT1 | 4 | 1.78 | 0.00052 | −3.86 | 0.024699 | 6.8431E−07 |
| SLC17A5 | 4 | −1.78 | 0.00054 | 3.04 | 0.030499 | 0.000364402 |
| APP | 6 | −1.94 | 0.00064 | 2.25 | 0.044619 | 0.000200504 |
| EZH2 | 4 | 4.95 | 0.00067 | 3.06 | 0.046242 | 2.39594E−10 |
| TPX2 | 10 | −1.54 | 0.0007 | 2.58 | 0.048631 | 5.44606E−10 |
| CCNA2 | 4 | −1.77 | 0.00072 | 2.83 | 0.032232 | 1.44783E−10 |
| TRIM33 | 8 | −2.05 | 0.00073 | 1.60 | 0.021483 | 0.00122048 |
| GDE1 | 4 | −1.61 | 0.00076 | 2.90 | 0.045743 | 7.47662E−08 |
| RDH10 | 4 | 1.95 | 0.00078 | 1.91 | 0.02083 | 0.000160419 |
| ADAMTS9 | 10 | −1.96 | 0.00086 | −2.23 | 0.020655 | 0.000177544 |
| LEMD3 | 7 | 2.09 | 0.0009 | 2.86 | 0.048904 | 0.000502659 |
| BUB1 | 6 | −3.24 | 0.0009 | −2.30 | 0.020682 | 0.00287803 |
| TNFRSF21 | 7 | 2.05 | 0.00098 | 3.26 | 0.030943 | 1.01738E−08 |
| MTCH2 | 5 | −1.68 | 0.00099 | −3.38 | 0.013583 | 0.00801725 |
Scores and significance values for genes that have five or more adjacent exons that are spliced; potential alternative transcripts
MIDAS microarray detection of alternative splicing, FIRMA finding isoforms using robust multichip analysis, ANOSVA analysis of splice variation
Identification of an alternative short TSP2 transcript highly expressed in bladder cancer-associated blood vessels
In the blood vessels of healthy bladder tissue, we detected the expression of the long transcript variant of TSP2 (NM_003247), while in tumor-associated blood vessels, there was a much higher expression of a short TSP2 variant (sTSP2) (sequence almost identical to BX641023, deposited at GenBank) than of the long transcript (Fig. 1b). To confirm the results obtained by the microarrays, we performed quantitative reverse transcription PCR (qRT-PCR) using two different primer pairs. We designed one primer pair that was specific for the long transcript (exon 10 to the exon 10/exon 11 junction) and another primer pair that binds in the 3′ region of both transcripts (exon 20 to exon 21/exon 22 junction). We found that in the vessels of healthy bladder tissue, the expression level of long transcripts was similar to the expression level of the short transcript. In contrast, in tumor-associated blood vessels, expression of the short transcript was much higher than expression of the long transcript (31.5-fold increase, p < 0.05) (Fig. 1c).
Full-length sequence of the short TSP2 transcript
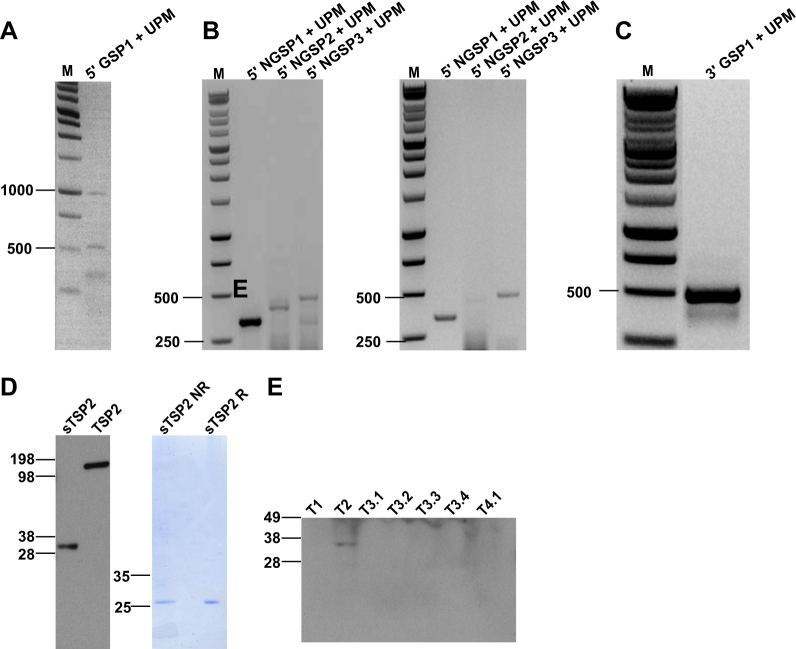
To obtain the full-length sequence of the short TSP2 RNA transcript, we performed rapid amplification of both the 5′ and 3′ cDNA ends (5′ and 3′ RACE (rapid amplification of cDNA ends) [19]). Using the RNA isolated from a sample of tumor-associated vasculature (RNA not previously amplified or hybridized to exon arrays), we first generated cDNA using reverse transcription and template switching with incorporation of a SMART II A oligo. Then, using a gene-specific primer (5′ GSP1) and a universal primer mix (binds to SMART II A oligo), we obtained two weak bands of around 0.5 and 1 kb (Fig. 2a). A second round of PCR was performed using nested primers on the two products from the 5′ RACE PCR in order to increase specificity and yield. Primers were designed in reverse direction and specific to TSP2. We obtained bands of approximately 300 bp (5′ NGSP1) and 400 bp (5′ NGSP2 and 5′ NGSP3) from both bands (left—0.5 kb, right—1 kb; Fig. 2b). We isolated the bands and Sanger sequenced them and thus could assemble the 5′ end sequence. For determining the 3′ end sequence, we performed 3′ RACE using a gene-specific primer (3′ GSP1) and universal primer mix to obtain a 500 nucleotide band (Fig. 2c). Sanger sequencing determined the 3′ sequence.
Fig. 2.
Characterization of the short TSP2 transcript. 5′-RACE and 3′-RACE reactions were performed on cDNA synthesized from bladder cancer-associated blood vessels. Representative ethidium bromide-stained 1% agarose gels are shown. a 5′-RACE with UPM (forward primer) and GSP (reverse primer) yielded two products. b The 0.5 kb (left panel) and the 1 kb product (right panel) were used as templates for nested PCR using UPM (forward primer) and NGSPs (reverse primers). c 3′-RACE with UPM and GSP yielded one distinct product. d Recombinant TSP2 and sTSP2 proteins were expressed in CHO cells and analyzed by SDS-PAGE and Western blot (left panel). Recombinant sTSP2 was electrophoresed under non-reducing (NR) and reducing (R) conditions and stained with Coomassie blue (right panel). e IBC tissue lysates (of seven tissue samples from four patients) were subjected to Western blot analysis using an antibody directed against the 3′ end of TSP2
Based on the determined sequence of TSP2 (Supplementary Figure 2), we next determined the longest open reading frame of the short TSP2 variant (Supplementary Figure 3) and found a coding sequence that codes for a 209 amino acid-long protein sequence (Supplementary Figure 4) corresponding to the C-terminal lectin domain (213 amino acids, first 4 amino acids are missing in sTSP2). Next, we investigated whether the transcript is indeed translated into protein. The sTSP2 protein obtained was of the predicted size and was secreted as a monomer (Fig. 2d). Due to the limited amount of protein that can be isolated from blood vessels by LCM, we isolated protein from total tumor biopsies. We detected sTSP2 protein in one of tested seven invasive bladder cancer (IBC) samples (Fig. 2e; seven tumor lysates from four IBC patients, these samples were from different tumors than those used for LCM where only limited remaining material was available).
Full-length TSP2 but not sTSP2 inhibits proliferation and migration of BECs in vitro
We next used CHO-S cells to recombinantly produce the sTSP2 protein. Previously, it was found that TSP2 inhibits proliferation of ECs in vitro [20–22]. We therefore investigated how sTSP2 may affect BEC proliferation. BECs were incubated with recombinant full-length (TSP2) or short TSP2 proteins (sTSP2) for 4, 24, or 48 h, followed by treatment with VEGF-A until measurement of cell numbers at 72 h. Pre-treatment with TSP2 for 4, 24, or 48 h significantly inhibited VEGF-A-induced BEC proliferation (Fig. 3a). In contrast, pre-treatment with sTSP2 did not result in any inhibition of VEGF-A-induced BEC proliferation (Fig. 3b). Pre-treatment with a combination of TSP2 and sTSP2 in equal amounts for 4 h resulted in a loss of the inhibitory effect of TSP2 on BEC proliferation (Fig. 3c). Treatment with TSP2, but not sTSP2, also inhibited the migration of BECs, as studied in a “Scratch” wound healing assay (Fig. 3d).
Fig. 3.

Full-length TSP2 but not the short TSP2 variant inhibits VEGF-A-induced proliferation and migration of blood vascular endothelial cells in vitro. a Proliferation of BECs incubated with recombinant full-length TSP2 with or without VEGF-A. b Proliferation of BECs incubated with the recombinant short TSP2 variant with or without VEGF-A. Cells were treated with full-length TSP2 or sTSP2 (100 ng/mL) for 4, 24 and 48 h and then VEGF-A was added to the cells (20 ng/mL). Relative cell number was measured after 72 h using the MUH assay. c Cells were treated with a full-length TSP2 and/or sTSP2 (100 ng/mL) for 4 h and then VEGF-A was added to the cells (20 ng/mL). Relative cell number was measured after 72 h using the MUH assay. d Migration of BECs with pre-treatment with a full-length TSP2 and/or sTSP2 (100 ng/mL) for 4 h with a subsequent addition of VEGF-A (20 ng/mL). Migration was assessed using a scratch wound healing closure assay. Control treated cells were treated with PBS. Data are presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001
Stable overexpression of full-length TSP2 but not of short TSP2 in A431 cells inhibits tumor growth and angiogenesis
To assess the effects of sTSP2 on tumor growth, we next used our previously established experimental A431 tumor model in which we had identified TSP1 and TSP2 as inhibitors of tumor angiogenesis [12, 23–26]. The A431 cell line does not express TSP2 or sTSP2 and therefore can serve as an appropriate model for overexpressing TSP2. Due to the lack of specific antibodies for the 3′ end of TSP2, we overexpressed sTSP2 with a 6× His-tag (Supplementary Fig. 5A). A431 empty plasmid control cells and A431 cells overexpressing full-length TSP2 or sTSP2 showed comparable proliferation rates in vitro (Supplementary Fig. 5B).
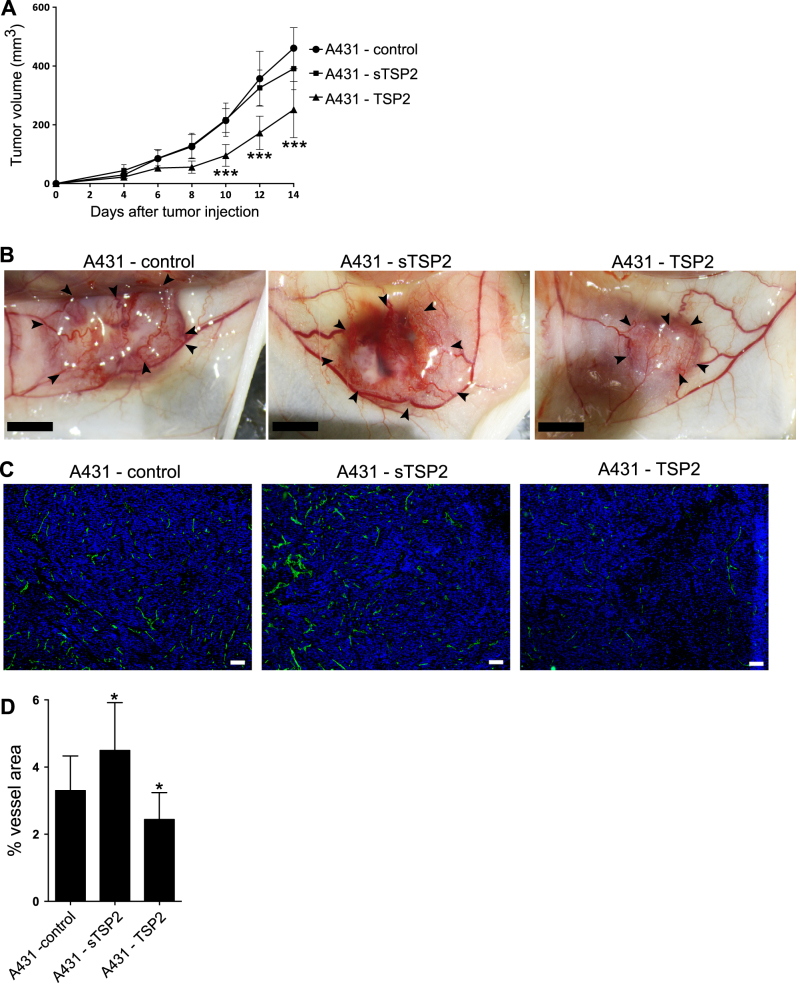
We next injected A431 cells intradermally into mice as this model was previously used to evaluate the effects of TSP1 and TSP2 on tumor angiogenesis [12, 23, 24]. After 10 days, there was a significant decrease in tumor size in A431 tumors expressing full-length TSP2 compared to control tumors expressing empty plasmid; this effect was seen throughout day 14 when animals were sacrificed (day 10: 55%, p < 0.001; day 12: 52%, p < 0.001; day 14: 45%, p < 0.001; Fig. 4a). No significant differences in animal weights were detected (Supplementary Fig. 5C). Already by visual observation on the day of sacrifice, a significant reduction and a more regular arrangement of tumor-supplying blood vessels was found in TSP2-overexpressing squamous cell carcinomas, whereas tumors overexpressing sTSP2 showed increased vasculature and immature blood vessels similar to control tumors (Fig. 4b). Quantitative image analysis of tumor sections stained for the vascular marker MECA-32 (Fig. 4c, d) revealed a significant decrease in the area covered by blood vessels in tumors expressing full-length TSP2 (−26%; p = 0.04) and a significant increase in tumors expressing sTSP2 (+36%; p = 0.03).
Fig. 4.
Stable overexpression of full-length TSP2, but not of the short TSP2 variant, in A431 cells inhibits intradermal tumor growth and angiogenesis. a Tumor growth was measured in mice with A431 tumors expressing either TSP2 or sTSP2 (n = 12 per group). b Representative pictures of NOD-SCID mouse skin flaps containing intradermally injected A431 tumors transfected with control vector, full-length TSP2 (TSP2), or the short TSP2 variant (sTSP2). Arrows indicate tumor borders. Scale bars = 5 mm. c Tumor sections were stained with MECA-32 (vascular marker; green) and with Hoechst (nuclear marker; blue). d Quantification of tumor area covered by blood vessels for tumors of similar size (n = 12 for control and sTSP2, n = 10 for TSP2; 200–600 mm3). Data are presented as mean ± SD. All comparisons for testing for significance were compared to A431-control. *p < 0.05, ***p < 0.001. Scale bars = 100 μm
Discussion
Even though there are numerous studies that have investigated the transcriptional profile of tumor-associated endothelial cells [27–29], there has been no systematic investigation of the alternative transcripts in tumor-associated blood vessels on a global whole transcriptome approach. This is surprising since it is well known that tumor-associated stroma, similar to wound healing-associated stroma, can express alternative splice forms of matrix molecules such as embryonic variants of fibronectin 1 (extra domains A and B) [30–32], and these alternative splice variants have been used to develop targeted anti-tumor therapies [33, 34]. Here, we globally analyzed the splicing changes and alternative transcription of tumor-associated blood vessels in comparison to normal blood vessels, using immuno-LCM of human bladder cancers and of healthy bladder tissue combined with human exon array analysis. There was relatively little overlap between the results of three different analysis methods for exon arrays. This is likely due to the fact that we could not perform a multiple hypothesis testing correction, because the necessary assumption that tests are independent is not valid as the majority of exons belonging to the same gene are clearly correlating. Nonetheless, the combination analysis identified 76 candidate alternative splicing events in tumor-associated endothelial cells revealed by all three methods of analysis. Out of these, we selected a short transcriptional variant of TSP2 for further analyses since we had previously identified TSP2 as a potent endogenous inhibitor of tumor angiogenesis [12, 23, 35, 36].
We found a shorter TSP2 transcript that was specifically expressed in the tumor-associated vasculature (comprising the 3′ end of the full-length mRNA sequence). The existence of this alternative TSP2 transcript in human bladder cancer-associated blood vessels could be confirmed through 5′ and 3′ RACE and Sanger sequencing. The short TSP2 transcript is, with regard to the exon number and sequence, almost identical to a mRNA sequence deposited in the GenBank (NCBI accession number BX641023) that was previously detected in esophageal cancer [37]. Indeed, we could confirm translation of sTSP2 mRNA into a protein, by detecting the sTSP2 in one of the whole tumor lysates. Only detecting the protein in one out of seven IBC tumor lysates is not surprising as vessels represent a very minor part by amount in the whole tumor tissue and abundance of vascular proteins is very low in protein of total tissue.
Under physiological conditions, vascular quiescence is maintained by the tight balance between endogenous angiogenic inhibitors and angiogenic factors [38, 39]. In most human cancers, the balance is however thought to be shifted towards angiogenesis through the increased production of angiogenic factors (e.g. VEGF-A, FGF2, PDGF, PlGF) and reduced production of endogenous inhibitors of angiogenesis (most prominently, TSP1 and TSP2 [40]). Whereas some endogenous angiogenesis inhibitors need to be created by cleavage of extracellular matrix molecules such as collagen XVIII (endostatin [41]) and plasminogen (angiostatin [42]), TSP1 and TSP2 are already secreted in their active form without the need for any further proteolytic cleavage. TSP2-deficient mice have an increased vascularization of several organs [43–45] and show an earlier angiogenic switch and increased tumor formation and angiogenesis when subjected to a multistep skin carcinogenesis regimen [35], revealing TSP2 as a potent endogenous inhibitor of tumor angiogenesis. Importantly, overexpression of TSP2 in cancer cells resulted in a potent inhibition of tumor growth and angiogenesis [23] and the anti-angiogenic activity was associated with the N-terminal fragment of TSP2, which contains the procollagen homology domain and the type I repeats, but lacks the type 2 and 3 repeats and the C-terminal lectin domain [12]. It is of interest that TSP5 (also known as COMP, cartilage oligomeric matrix protein) contains type 2 and type 3 repeats and the C-terminal lectin domain, but not the procollagen homology or type 1 repeats domains, and fails to inhibit endothelial cell migration or corneal vascularization in comparison to full-length TSP2 [22]. Thus, the anti-angiogenic activity of TSP2 resides within the type 1 repeat domain [10–13, 46] but not the N-terminal globular domain or the procollagen homology domain [12]. TSP2 has a trimeric structure having three modules containing type 1 repeat domains and also three C-terminal lectin domains [47]. In contrast, sTSP2 is produced as a monomer, with a structure of almost the complete C-terminal lectin domain (lacking four first amino acids).
In line with these findings, our overexpression studies in vivo revealed that the newly identified sTSP2 variant that comprises the C-terminal lectin domain failed to inhibit tumor growth and angiogenesis. This is in agreement with our in vitro studies where TSP2, but not sTSP2, inhibited the migration of BECs. Recombinant sTSP2 also failed to inhibit the proliferation of growth factor-stimulated BECs and—when combined with TSP2—was able to block the inhibitory effect of TSP2. As the molecular mechanisms of action of TSP2 are currently not fully known [40, 48], and the published TSP2 concentrations used for inhibition of migration and proliferation of ECs in vitro are quite high (in μg range [20, 22]), it is difficult to determine the exact mechanism of action of sTSP2. Using intracellular Ca2+ live cell imaging, we found that treatment with VEGF-A activated Ca2+ signaling in 89% of the cells, whereas pre-treatment with TSP2 reduced the activation to 75% the cells. In contrast, sTSP2 pre-treatment had no effect (91% activated cells, data not shown). We also evaluated phospho-AKT (p-AKT) and total-AKT (t-AKT) by Western blot. We found a moderate decrease in phosphorylation of AKT by VEGF-A after pre-treatment with TSP2 (−32%), but not after pre-treatment with sTSP2 (−16%, data not shown). Quantification was performed in comparison to the control (PBS) treated ECs, and p-AKT expression was normalized to t-AKT. These rather moderate inhibitory effects of TSP2 are in accordance with previous studies of TSP1 and TSP1 repeats [49, 50] in vitro.
We propose that the sTSP2 expressed by tumor-associated vessels might compete with stromal TSP2 for binding to anti-angiogenic receptors and thus diminish the anti-angiogenic effects of TSP2. Overall, our study suggests a novel mechanism by which a potent endogenous inhibitor of angiogenesis loses its anti-angiogenic properties upon usage of an alternative transcription start site. The alternative transcription of TSP2 in the tumor microenvironment also represents a novel mechanism by which angiogenesis can be promoted in tumors. Further experiments are needed to understand the exact mechanism of competition between sTSP2 and TSP2. It will be of interest to investigate if sTSP2 is also expressed in tumor-associated vessels of other cancer types and in other conditions associated with angiogenesis.
Materials and methods
Tissue samples
Clinically annotated frozen cancer and normal tissue samples were obtained after radical cystectomy from the University Hospital Zurich (USZ). Tissue collection was approved by the SPUK-GGU-USZ Ethics Committee (KEK-StV-Nr 02/09) and written informed consent was obtained from each patient.
LCM and transcriptional profiling of BECs from bladder tissue
We performed LCM on frozen sections of IBCs (6 patients; pT1–pT4)) and matched normal bladder tissue samples (tumor adjacent, lack of malignancy confirmed by a board-certified pathologist). Bladder tissue sections of 8 μm were fixed in acetone for 1 min at 4 °C. Blood vessels were stained using a biotinylated rabbit antibody directed against human von Willebrand factor (vWF; 0.25 mg/mL; A0082; Dako Cytomation) and Cy3-streptavidin (1:100) in a mixture of buffers A and B (all Life Technologies) containing Protector RNase inhibitor (2 U/mL; Roche). Slides were dehydrated in 75%, 95% and 100% ethanol, and xylene. Immediately afterward, LCM was performed using near infrared laser-based Arcturus Veritas LCM (Life Technologies). Up to 1 mm2 of vWF-immunostained blood vessels were isolated per patient sample, followed by RNA isolation, cDNA generation, and microarray hybridization as described [2]. The microarray raw data can be accessed at GEO under accession number GSE41614.
Analysis of human exon 1.0 ST array
In the human exon 1.0 ST array (Affymetrix), each exon is represented by a probeset comprising at least four oligonucleotides. Probesets with the highest level of supportive evidence (core probesets (n = 284,000)) that target all exons of transcripts annotated by Refseq were used for exon analysis. Raw fluorescence intensity value summarization is described extensively in Supplementary Material and Methods under: Human Exon 1.0 ST probe summarization.
Alternative splicing was investigated using three parallel analyses. To detect differential splicing/transcription, the effect of differential gene expression needs to be removed. A commonly used strategy to achieve this is to calculate a splicing index (SI, [17]) for each exon, which is denoted as the log 2 value of the ratio of expression levels of individual exons to overall gene level expression. The SI was used as a measure for differential splicing between two groups. An SI value of 0 then indicates equal expression of a particular exon in a transcript. SI values of 1 and −1 were used as arbitrary thresholds. We used this SI in an analysis of variance (ANOVA) model named MIDAS [15], using Genespring Gx 12 (Agilent) and Altanalyze [51] to test the null hypothesis that no alternative splicing occurs for a particular exon. For the sake of clarity, SI when used in the MIDAS ANOVA model is called MIDAS score. Exons with a MIDAS p value <0.05 were considered significant for exon inclusion/exclusion. Second, ANOSVA [16] as performed on raw exon values using the Partek Genomics Suite (Partek Incorporated). This method directly uses the probeset intensity signals of individual exons to detect differences in the expression of exons using an ANOVA model. Exons with an ANOSVA value of p < 0.01 were considered significant for exon inclusion/exclusion. A third analysis was performed using the FIRMA [18] using the statistical language R/Bioconductor [52]. FIRMA predicts alternative splicing of an exon based on the distance from the transcript expression estimate produced by the RMA model. The FIRMA score is a measure of this distance. A FIRMA score of 1 indicates that no splicing has occurred. FIRMA scores were compared using Bayesian statistics. FIRMA was performed using the default implementation of the aroma.affymetrix package in R together with logarithmic transformation, as described in the human exon array analysis vignette [53, 54]. A custom chip description file covering the exon array core probesets was used (HuEx-1_0-st-v2,coreR3,A20071112,EP.cdf, created by E Purdom). After obtaining the FIRMA scores per probeset per sample, the mean FIRMA score in both experimental groups and the difference between the groups was calculated. Finally, the probesets were mapped to transcript clusters and the transcripts were sorted by the FIRMA score. The FIRMA scores were also subjected to statistical testing using Bayesian statistics as implemented in the limma [55] statistical package to identify consistent splicing differences across samples. Exons whose FIRMA scores were lower than p < 0.001 were considered significant for exon inclusion/exclusion. A detailed description of the analysis is provided in Supplementary Material and Methods under: Comparison of different exon array methods of analysis.
SMARTer RACE cDNA amplification
Total RNA was isolated from bladder cancer-associated blood vasculature. 5′-end and 3′-end cDNA fragments were generated from total RNA using SMARTerTM RACE cDNA amplification technology according to the manufacturer’s instructions (Clontech). The 5′- and 3′-RACE PCR reaction was carried out following the manufacturer’s protocol using Universal Primer mix (UPM) and gene-specific primers (5′-GSP1 or 3′-GSP1). Nested 5′-RACE PCR was carried out using UPM as a forward primer and one of three nested gene-specific reverse primers (NGSP1, NGSP2, NGSP3). PCR products were separated on a 1% agarose gel. DNA was isolated from the gels using the QIAquick Gel Extraction kit (Qiagen) and sequenced by Sanger sequencing. Primer sequences are provided in Supplementary Table 2.
Quantitative real-time PCR
The transcript variants of TSP2 were quantified in samples of bladder cancer-associated vessels and of vessels of healthy bladder tissue and in A431 cells using qRT-PCR with the AB 7900 HT Fast Real-Time PCR System and the 2−ΔΔCt method [56]. SYBR probes were designed spanning the exon–exon junction. Primer sequences are provided in Supplementary Table 2. Every reaction was normalized to the housekeeping gene RPLP0.
Expression vectors
For expressing sTSP2 in CHO-S cells and A431 cells, an expression construct was synthesized using GeneArt gene synthesis (Thermo Fisher) and cloned into the pcDNA3.1 (+) expression vector. The sequence contained a leader peptide sequence for efficient protein secretion (IgK; METDTLLLWVLLLWVPGSTG), a Kozak sequence and a terminal 6xHis-Tag (Supplementary Fig. S6). The full-length TSP2 plasmid has been described before [23].
Production of recombinant sTSP2
Twenty-four hours prior to transfection, CHO-S cells (Thermo Fisher, R800-07) were cultured (1 × 106 cells per mL) in ProCHO4 medium (without l-glutamine, nucleosides (HT), with HEPES, Pluronic F-68, Lonza) supplemented with 1% Pro-HT and 4% l-glutamine. For transient transfection of 1 × 108 cells, 500 μg polyethylenimine (PEI) transfection solution (1 μg/μL) was diluted in sterile 2.5 mL 150 mM NaCl and mixed with 125 μg plasmid DNA. The mixture was incubated for 10 min at room temperature. Cells were centrifuged for 10 min at 1,000 RPM and then resuspended in 50 mL of ProCHO4 medium. The PEI-DNA mixture was added carefully to the cells with subsequent incubation at 37 °C with shaking. After 4 h, 50 mL PowerCHO-2CD medium (without l-glutamine, nucleosides (HT), with HEPES, Pluronic F-68) supplemented with 1% Pro-HT and 4% l-glutamine was added to the cells, and cells were incubated at 32 °C with shaking. After 5–6 days, supernatant was harvested, and a low yield of protein was obtained. Supernatant was purified using immobilized metal-affinity chromatography, and purity was validated by western blot and Coomassie staining under reducing and non-reducing conditions. Recombinant protein concentration was determined using a NanoDrop (ND-1000 spectrophotometer), bicinchoninic acid assay (ThermoScientific), and Western blot.
Production of polyclonal antibody against the C-terminal end of TSP2
Recombinantly produced sTSP2 with a his-tag was used to immunize rabbits for antibody production (GenScript). The resulting antiserum was purified by column affinity purification (GenScript).
Western blot analyses
Proteins were isolated from 4 to 8,100 μm sections of bladder tissue from optimal-cutting temperature blocks containing tissue of invasive cancer bladder cancers (from some patients, several blocks from different parts of the tumor were available), using 350 μL of modified RIPA buffer lysis solution (pH 7.4, 50 mM Tris-HCl, 150 mM NaCl, 0.25% sodium dodecyl sulfate (SDS), 0.25% sodium deoxycholate, 1 mM EDTA, 0.1% Triton X-100, 1 tablet protease inhibitor cocktail: complete, Mini, Roche). After RIPA buffer was added to the sections, samples were frozen in liquid nitrogen for further processing. Tissue lysis was performed using the TissueLyser (Qiagen) twice for 2 min at 30 Hz. Tubes were then centrifuged at 16,000 × g and 4 °C for 20 min in a microcentrifuge. The supernatant were transferred to fresh tubes.
Tissue lysates containing 60–260 μg of protein, 2 μg recombinant human TSP2 (R&D System), and 0.2 μg sTSP2 of protein (assessed using the BCA protein assay) were subjected to SDS-polyacrylamide gel electrophoresis using Bolt 4–12% gradient Bis-Tris gels (Invitrogen). Proteins were transferred to a polyvinylidene fluoride membrane (EMD Millipore) and the membranes were incubated at 4 °C with a rabbit anti-sTSP2 (3.225 μg/mL, GenScript) overnight. Membranes were then probed with an anti-rabbit immunoglobulin G horseradish peroxidase conjugate (1:5000, GenScript) and the ECL System (ThermoScientific) was used for detection.
Cell culture
Primary human dermal BECs [57] were cultured as described [2] and tested negatively for mycoplasma contamination. Human A431 squamous cell carcinoma cells were purchased from the American Type Culture Collection and cultured in DMEM + Glutamax (Thermo Fisher) with 10% fetal bovine serum (FBS).
Cell proliferation and migration assays
A431 cells (103/well) or BECs (4 × 103/well) were seeded onto black clear-bottom 96-well plates (Costar) and left to attach for 24 h at 37 °C and 5% CO2. Cells were starved overnight in media containing only 1% FBS. Afterwards, cells were cultured for 72 h and viable cells were quantified using the MUH fluorimetric method (6 wells per condition) [58]. The monolayer wound healing assay was performed as previously described [59]. In brief, two cross-shaped scratches were made into a confluent BEC monolayer (5 wells per condition). After washing, cells were pre-treated with TSP2 (100 ng/mL) or sTSP2 (100 ng/mL) or a combination of TSP and sTSP2 for 4 h, followed by treatment with VEGF-A for 16 h. Plain medium was used as a control. Images of the scratches were taken immediately after scratching and 16 h after VEGF-A addition. The surface areas of the cell-free zones were measured and the % scratch closure was determined using TScratch software [59]. The assays were repeated at least three times. Results from one representative experiment are shown.
In vivo tumorigenesis assay
Tumor studies were performed in accordance with animal protocols approved by the Kantonales Veterinaramt Zurich. NOD-SCID strain NOD.CB17-Prkdcscid/J (Tyrc/Tyrc: Haplotype H2g7) albino mice were used (Charles River). A431 cells (2 × 106), stably transfected with full-length TSP2 or sTSP2 or the empty expression vector pcDNA3.1 (+), were injected intradermally into both flanks of 10–12-week-old mice (10 mice per group, 18–25 g, males and females). Mice were ear marked and randomly assigned codes for the grouping into experimental groups. This was done before the start of the study and the codes were revealed only after analysis of the data to ensure blinded observation. Animal welfare was monitored by physical appearance, behavior, and body weight. The size of the tumors was determined by external measurements of the tumor diameter every 2 days post-injection using a digital caliper. Tumor volume (V) was estimated as V = (4/3 × π × (1/2 × smaller diameter)2 × 1/2 × larger diameter). Animals were sacrificed under anesthesia (64 mg/kg ketamine, 10 mg/kg xylazine, 2 mg/kg acetopromazine) after 2 weeks post-injection when the total tumor load per mouse reached approximately 1 cm3. Tumors were harvested for mRNA and protein expression analyses, and tissue sections were stained as described [60], using a monoclonal rat MECA-32 antibody (2.5 μg/mL, 553849, BD Pharmingen), an Alexa Fluor 488-coupled secondary antibodies (1:200, Invitrogen), and the nuclear-dye Hoechst 33342 (10 μg/mL, Invitrogen). The animal study was repeated twice. Data from one representative study is shown.
Image acquisition after immunohistochemistry of mouse tumors
Images were captured on an Axioskop 2 mot plus microscope, equipped with an AxioCam MRc camera and Plan-APOCHROMAT 10×/0.45 MA and Plan-NEOPLAN 20×/0.5 NA objectives (all from Zeiss). Image acquisition in the individual fluorescence channels was accomplished using Axio Vision 4.8 software (Zeiss). The stained areas were quantified using ImageJ.
Statistical analyses
Based on previous experience with the cellular assays used here, we performed more than four replicates (for proliferation assay: 6 and for migration assay: 5). For the animal studies, we used 10 mice per group. Statistical analyses were performed using the version 5.00 of Prism (GraphPad Software Inc.). Data distribution was checked for normality by visual observation of the variation of data points within experimental groups. For most comparisons, ANOVA or a two-tailed unpaired Student’s t test were employed as indicated. Differences were considered statistically significant at p < 0.05.
Electronic supplementary material
Supplementary Figures legend and Supplementary Material and Methods
Acknowledgements
We thank Peter Camenzind, Chloé Chong, Daniel Bezar, Luca Ducoli, and Jeannette Scholl for excellent technical assistance.
Financial support
This work was supported by Swiss National Science Foundation grant 310030B_147087, Advanced European Research Council Grant LYVICAM, Krebsliga Schweiz and Krebsliga Zurich (to MD).
Author contributions
FR, VIO, and MD designed the experiments and wrote the manuscript. FR. developed the methodology. FR, SYY, SP, SS, SBB, and GV performed the majority of experiments and data analyses. CP acquired human tissue samples. SK and SS provided research assistance for animal experiments. JWS provided assistance with the bioinformatics analysis of splicing. All authors have read the manuscript and have approved the submitted manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
The online version of this article (10.1038/s41388-018-0129-z) contains supplementary material, which is available to authorized users.
References
- 1.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–6. doi: 10.1056/NEJM197108122850711. [DOI] [PubMed] [Google Scholar]
- 2.Roudnicky F, Poyet C, Wild P, Krampitz S, Negrini F, Huggenberger R, et al. Endocan is upregulated on tumor vessels in invasive bladder cancer where it mediates VEGF-A-induced angiogenesis. Cancer Res. 2013;73:1097–106. doi: 10.1158/0008-5472.CAN-12-1855. [DOI] [PubMed] [Google Scholar]
- 3.Roudnicky F, Dieterich LC, Poyet C, Buser L, Wild P, Tang D, et al. High expression of insulin receptor on tumour-associated blood vessels in invasive bladder cancer predicts poor overall and progression-free survival. J Pathol. 2017;242:193–205. doi: 10.1002/path.4892. [DOI] [PubMed] [Google Scholar]
- 4.Balar AV, Apolo AB, Ostrovnaya I, Mironov S, Iasonos A, Trout A, et al. Phase II study of gemcitabine, carboplatin, and bevacizumab in patients with advanced unresectable or metastatic urothelial cancer. J Clin Oncol. 2013;31:724–30. doi: 10.1200/JCO.2012.42.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Lassmann T, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–70. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 8.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell. 2016;164:805–17. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson DW, Volpert OV, Pearce SF, Schneider AJ, Silverstein RL, Henkin J, et al. Mol Pharmacol. 1999;55:332–8. doi: 10.1124/mol.55.2.332. [DOI] [PubMed] [Google Scholar]
- 11.Huwiler KG, Vestling MM, Annis DS, Mosher DF. Biophysical characterization, including disulfide bond assignments, of the anti-angiogenic type 1 domains of human thrombospondin-1. Biochemistry. 2002;41:14329–39. doi: 10.1021/bi026463u. [DOI] [PubMed] [Google Scholar]
- 12.Noh YH, Matsuda K, Hong YK, Kunstfeld R, Riccardi L, Koch M, et al. An N-terminal 80 kDa recombinant fragment of human thrombospondin-2 inhibits vascular endothelial growth factor induced endothelial cell migration in vitro and tumor growth and angiogenesis in vivo. J Invest Dermatol. 2003;121:1536–43. doi: 10.1046/j.1523-1747.2003.12643.x. [DOI] [PubMed] [Google Scholar]
- 13.Vogel T, Guo NH, Krutzsch HC, Blake DA, Hartman J, Mendelovitz S, et al. Modulation of endothelial cell proliferation, adhesion, and motility by recombinant heparin-binding domain and synthetic peptides from the type I repeats of thrombospondin. J Cell Biochem. 1993;53:74–84. doi: 10.1002/jcb.240530109. [DOI] [PubMed] [Google Scholar]
- 14.Laajala E, Aittokallio T, Lahesmaa R, Elo LL. Probe-level estimation improves the detection of differential splicing in Affymetrix exon array studies. Genome Biol. 2009;10:R77. doi: 10.1186/gb-2009-10-7-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genom. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cline MS, Blume J, Cawley S, Clark TA, Hu JS, Lu G, et al. ANOSVA: a statistical method for detecting splice variation from expression data. Bioinformatics. 2005;21(Suppl 1):i107–115. doi: 10.1093/bioinformatics/bti1010. [DOI] [PubMed] [Google Scholar]
- 17.Srinivasan K, Shiue L, Hayes JD, Centers R, Fitzwater S, Loewen R, et al. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37:345–59. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Purdom E, Simpson KM, Robinson MD, Conboy JG, Lapuk AV, Speed TP. FIRMA: a method for detection of alternative splicing from exon array data. Bioinformatics. 2008;24:1707–14. doi: 10.1093/bioinformatics/btn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freeman LA. Cloning full-length transcripts and transcript variants using 5’ and 3’ RACE. Methods Mol Biol. 2013;1027:3–17. doi: 10.1007/978-1-60327-369-5_1. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong LC, Bjorkblom B, Hankenson KD, Siadak AW, Stiles CE, Bornstein P. Thrombospondin 2 inhibits microvascular endothelial cell proliferation by a caspase-independent mechanism. Mol Biol Cell. 2002;13:1893–905. doi: 10.1091/mbc.e01-09-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panetti TS, Chen H, Misenheimer TM, Getzler SB, Mosher DF. Endothelial cell mitogenesis induced by LPA: inhibition by thrombospondin-1 and thrombospondin-2. J Lab Clin Med. 1997;129:208–16. doi: 10.1016/S0022-2143(97)90141-4. [DOI] [PubMed] [Google Scholar]
- 22.Volpert OV, Tolsma SS, Pellerin S, Feige JJ, Chen H, Mosher DF, et al. Inhibition of angiogenesis by thrombospondin-2. Biochem Biophys Res Commun. 1995;217:326–32. doi: 10.1006/bbrc.1995.2780. [DOI] [PubMed] [Google Scholar]
- 23.Streit M, Riccardi L, Velasco P, Brown LF, Hawighorst T, Bornstein P, et al. Thrombospondin-2: a potent endogenous inhibitor of tumor growth and angiogenesis. Proc Natl Acad Sci USA. 1999;96:14888–93. doi: 10.1073/pnas.96.26.14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Streit M, Velasco P, Brown LF, Skobe M, Richard L, Riccardi L, et al. Overexpression of thrombospondin-1 decreases angiogenesis and inhibits the growth of human cutaneous squamous cell carcinomas. Am J Pathol. 1999;155:441–52. doi: 10.1016/S0002-9440(10)65140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Streit M, Stephen AE, Hawighorst T, Matsuda K, Lange-Asschenfeldt B, Brown LF, et al. Systemic inhibition of tumor growth and angiogenesis by thrombospondin-2 using cell-based anti-angiogenic gene therapy. Cancer Res. 2002;62:2004–12. [PubMed] [Google Scholar]
- 26.Yee KO, Streit M, Hawighorst T, Detmar M, Lawler J. Expression of the type-1 repeats of thrombospondin-1 inhibits tumor growth through activation of transforming growth factor-beta. Am J Pathol. 2004;165:541–52. doi: 10.1016/S0002-9440(10)63319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckanovich RJ, Sasaroli D, O’Brien-Jenkins A, Botbyl J, Hammond R, Katsaros D, et al. Tumor vascular proteins as biomarkers in ovarian cancer. J Clin Oncol. 2007;25:852–61. doi: 10.1200/JCO.2006.08.8583. [DOI] [PubMed] [Google Scholar]
- 28.Dieterich LC, Mellberg S, Langenkamp E, Zhang L, Zieba A, Salomaki H, et al. Transcriptional profiling of human glioblastoma vessels indicates a key role of VEGF-A and TGFbeta2 in vascular abnormalization. J Pathol. 2012;228:378–90. doi: 10.1002/path.4072. [DOI] [PubMed] [Google Scholar]
- 29.Pepin F, Bertos N, Laferriere J, Sadekova S, Souleimanova M, Zhao H, et al. Gene expression profiling of microdissected breast cancer microvasculature identifies distinct tumor vascular subtypes. Breast Cancer Res. 2012;14:R120. doi: 10.1186/bcr3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borgia B, Roesli C, Fugmann T, Schliemann C, Cesca M, Neri D, et al. A proteomic approach for the identification of vascular markers of liver metastasis. Cancer Res. 2010;70:309–18. doi: 10.1158/0008-5472.CAN-09-2939. [DOI] [PubMed] [Google Scholar]
- 31.Castronovo V, Waltregny D, Kischel P, Roesli C, Elia G, Rybak JN, et al. A chemical proteomics approach for the identification of accessible antigens expressed in human kidney cancer. Mol Cell Proteom. 2006;5:2083–91. doi: 10.1074/mcp.M600164-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Rybak JN, Roesli C, Kaspar M, Villa A, Neri D. The extra-domain A of fibronectin is a vascular marker of solid tumors and metastases. Cancer Res. 2007;67:10948–57. doi: 10.1158/0008-5472.CAN-07-1436. [DOI] [PubMed] [Google Scholar]
- 33.Carnemolla B, Borsi L, Balza E, Castellani P, Meazza R, Berndt A, et al. Enhancement of the antitumor properties of interleukin-2 by its targeted delivery to the tumor blood vessel extracellular matrix. Blood. 2002;99:1659–65. doi: 10.1182/blood.V99.5.1659. [DOI] [PubMed] [Google Scholar]
- 34.Neri D, Bicknell R. Tumour vascular targeting. Nat Rev Cancer. 2005;5:436–46. doi: 10.1038/nrc1627. [DOI] [PubMed] [Google Scholar]
- 35.Hawighorst T, Velasco P, Streit M, Hong YK, Kyriakides TR, Brown LF, et al. Thrombospondin-2 plays a protective role in multistep carcinogenesis: a novel host anti-tumor defense mechanism. EMBO J. 2001;20:2631–40. doi: 10.1093/emboj/20.11.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunstfeld R, Hawighorst T, Streit M, Hong YK, Nguyen L, Brown LF, et al. Thrombospondin-2 overexpression in the skin of transgenic mice reduces the susceptibility to chemically induced multistep skin carcinogenesis. J Dermatol Sci. 2014;74:106–15. doi: 10.1016/j.jdermsci.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, et al. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 2001;11:422–35. doi: 10.1101/gr.GR1547R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Detmar M. The role of VEGF and thrombospondins in skin angiogenesis. J Dermatol Sci. 2000;24:S78–84. doi: 10.1016/S0923-1811(00)00145-6. [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D, Christofori G, Naik P, Arbeit J. Transgenic mouse models of tumour angiogenesis: the angiogenic switch, its molecular controls, and prospects for preclinical therapeutic models. Eur J Cancer. 1996;32A:2386–93. doi: 10.1016/S0959-8049(96)00401-7. [DOI] [PubMed] [Google Scholar]
- 40.Lawler J, Detmar M. Tumor progression: the effects of thrombospondin-1 and -2. Int J Biochem Cell Biol. 2004;36:1038–45. doi: 10.1016/j.biocel.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 41.O’Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, et al. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–85. doi: 10.1016/S0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 42.O’Reilly MS, Holmgren L, Shing Y, Chen C, Rosenthal RA, Moses M, et al. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell. 1994;79:315–28. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 43.Kyriakides TR, Zhu YH, Smith LT, Bain SD, Yang Z, Lin MT, et al. Mice that lack thrombospondin 2 display connective tissue abnormalities that are associated with disordered collagen fibrillogenesis, an increased vascular density, and a bleeding diathesis. J Cell Biol. 1998;140:419–30. doi: 10.1083/jcb.140.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyriakides TR, Leach KJ, Hoffman AS, Ratner BD, Bornstein P. Mice that lack the angiogenesis inhibitor, thrombospondin-2, mount an altered foreign body reaction characterized by increased vascularity. Proc Natl Acad Sci USA. 1999;96:4449–54. doi: 10.1073/pnas.96.8.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kyriakides TR, Tam JW, Bornstein P. Accelerated wound healing in mice with a disruption of the thrombospondin 2 gene. J Invest Dermatol. 1999;113:782–7. doi: 10.1046/j.1523-1747.1999.00755.x. [DOI] [PubMed] [Google Scholar]
- 46.Iruela-Arispe ML, Lombardo M, Krutzsch HC, Lawler J, Roberts DD. Inhibition of angiogenesis by thrombospondin-1 is mediated by 2 independent regions within the type 1 repeats. Circulation. 1999;100:1423–31. doi: 10.1161/01.CIR.100.13.1423. [DOI] [PubMed] [Google Scholar]
- 47.Carlson CB, Lawler J, Mosher DF. Structures of thrombospondins. Cell Mol Life Sci. 2008;65:672–86. doi: 10.1007/s00018-007-7484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawler PR, Lawler J. Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2. Cold Spring Harb Perspect Med. 2012;2:a006627. doi: 10.1101/cshperspect.a006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren B, Song K, Parangi S, Jin T, Ye M, Humphreys R, et al. A double hit to kill tumor and endothelial cells by TRAIL and antiangiogenic 3TSR. Cancer Res. 2009;69:3856–65. doi: 10.1158/0008-5472.CAN-08-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun J, Hopkins BD, Tsujikawa K, Perruzzi C, Adini I, Swerlick R, et al. Thrombospondin-1 modulates VEGF-A-mediated Akt signaling and capillary survival in the developing retina. Am J Physiol Heart Circ Physiol. 2009;296:H1344–1351. doi: 10.1152/ajpheart.01246.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38:W755–762. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bengtsson M, Simpson K, Bullard J, Hansen K. Aroma.affymetrix: a generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. Berkeley, CA: Department of Statistics, Univ. California; 2008.
- 54.Rodrigo-Domingo M, Waagepetersen R, Bodker JS, Falgreen S, Kjeldsen MK, Johnsen HE, et al. Reproducible probe-level analysis of the Affymetrix Exon 1.0 ST array with R/Bioconductor. Brief Bioinform. 2014;15:519–33. doi: 10.1093/bib/bbt011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Hirakawa S, Hong YK, Harvey N, Schacht V, Matsuda K, Libermann T, et al. Identification of vascular lineage-specific genes by transcriptional profiling of isolated blood vascular and lymphatic endothelial cells. Am J Pathol. 2003;162:575–86. doi: 10.1016/S0002-9440(10)63851-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Detmar M, Imcke E, Ruszczak Z, Orfanos CE. Effects of recombinant tumor necrosis factor-alpha on cultured microvascular endothelial cells derived from human dermis. J Invest Dermatol. 1990;95:219S–22S. doi: 10.1111/1523-1747.ep12875807. [DOI] [PubMed] [Google Scholar]
- 59.Geback T, Schulz MM, Koumoutsakos P, Detmar M. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques. 2009;46:265–74. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- 60.Huggenberger R, Ullmann S, Proulx ST, Pytowski B, Alitalo K, Detmar M. Stimulation of lymphangiogenesis via VEGFR-3 inhibits chronic skin inflammation. J Exp Med. 2010;207:2255–69. doi: 10.1084/jem.20100559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures legend and Supplementary Material and Methods