Abstract
One of the daunting challenges facing modern medicine lies in the understanding and treatment of tumor heterogeneity. Most tumors show intra-tumor heterogeneity at both genomic and proteomic levels, with marked impacts on the responses of therapeutic targets. Therapeutic target-related gene expression pathways are affected by hypoxia and cellular stress. However, the finding that targets such as eukaryotic initiation factor (eIF) 4E (and its phosphorylated form, p-eIF4E) are generally homogenously expressed throughout tumors, regardless of the presence of hypoxia or other cellular stress conditions, opens the exciting possibility that malignancies could be treated with therapies that combine targeting of eIF4E phosphorylation with immune checkpoint inhibitors or chemotherapy.
Introduction
Owing to increased incidence, cancer is now the second most common cause of death in developed countries and the leading cause of death in individuals above 40 years of age [1]. The number of cancer-related deaths is expected to grow due to increases in life expectancy and lifestyle risk factors. Although current treatments have improved patient survival, the results remain dismal for advanced disease. For example, the 5-year survival rate is only 2% for stage IV lung cancer and 25% for breast cancer patients with metastatic disease [2]. In contrast, the 5-year survival of patients with in situ breast cancer exceeds 90%. Indeed, despite the development of a new arsenal of molecular targeted therapies over the last decade, patient survival with advanced cancer has improved by only 15% [1, 2]. One of the main reasons for these disappointing outcomes is the pervasive heterogeneous expression of drug targets within human tumors.
Cancer can be viewed as a group of heterogeneous diseases that arise from a small number of initiation events, but phenotypically diverge during progression due to environmental context (site of origin), the acquisition of different mutations required for survival, and individual patient responses to the tumor. Heterogeneity is observed at the genetic, proteomic, morphological, and environmental levels.
Tumor cell adaptability often leads to the use of redundant signaling pathways in response to stress, such as hypoxia and reduced nutrient availability. Within a tumor bed, variations in the “strength” of these stressor events and the corresponding responses can result in a significant degree of heterogeneity in gene expression, with some cells needing to respond more acutely than others. In this review, we underscore the nature of eukaryotic initiation factor (eIF) 4E in malignant tumors as a critical effector of cell signaling networks. We summarize the findings that the phosphorylated forms of eIF4E and 4E-BP1, termed p-eIF4E and p-4E-BP1, show a predominantly homogenous expression pattern within tumor beds, a feature that we predict to be actionable and to hold significant consequences for cancer therapy.
Clonal evolution during cancer progression
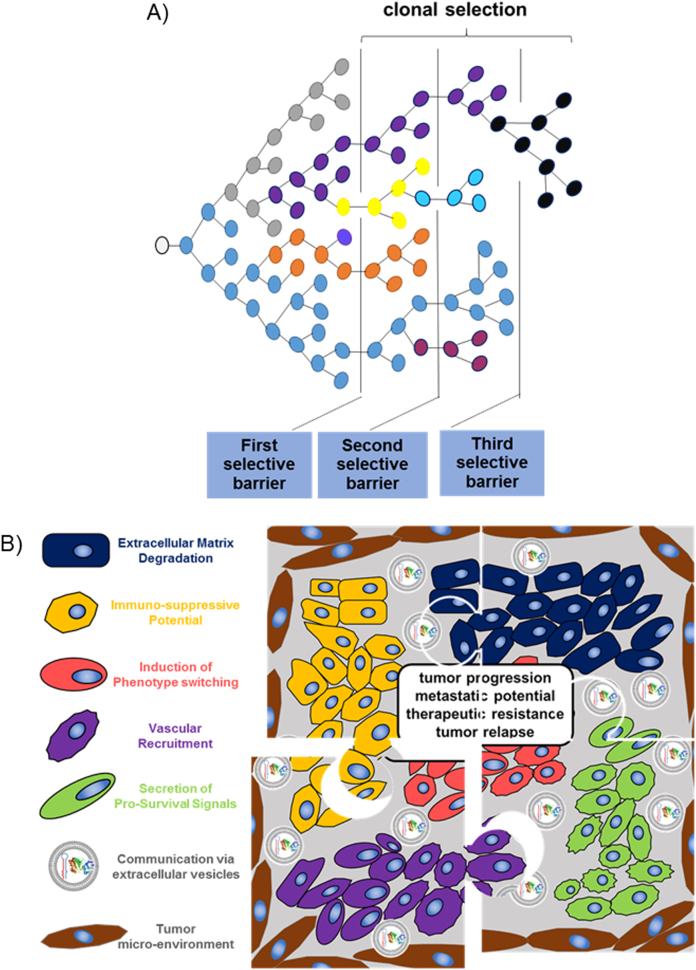
A complex molecular scenario is responsible for tumor initiation and tumor progression. It is well established that, in a single tumor, cell clones with novel genetic alterations arise constantly and are selected according to a Darwinian model [3]. Concomitantly, there is also cross talk between the tumor clones and the microenvironment that affect the ability of tumors to survive and proliferate. In fact, the cooperation between clones and the microenvironment is similar to that of a tumor consortium (Fig. 1) [4]. The new genetic alterations are driven by genetic instability, one of the hallmarks of tumor cells [5]. Only a small proportion of the total mutational burden is related to the process of clonal evolution because most are passenger mutations with no biological relevance [5]. In addition, treatments can alter clonal heterogeneity by selecting for more resistant cells or perturbing the microenvironmental conditions [6].
Fig. 1.
a Diagram representing clonal selection according to a Darwinian model. The best-adapted clones due to genetic or epigenetic advantages or with better interplay with neighboring cells will survive and proliferate, becoming the dominant clone until a new “selective barrier” appears. The tumor clonal composition varies over time, although, microscopically, these changes can be subtle or not evident. b Clonal cooperation and feature complementation. Puzzle diagram illustrating the contribution of individual cell clones with different tumor-promoting features to the formation of a tumor. The main feature of each clonal population within a tumor is shown as legend on the left side. The cooperation between different clones results in different functional consequences for the tumor, which are summarized in the middle of the figure
Epigenetic differences between clones are critical to tumor heterogeneity. Many of them are associated with an aberrant DNA methylation pattern, histone modification, or microRNA transcriptome and can be related to microenvironment factors [7]. Thus, both genome and epigenome diversity enables malignant tumor clones to acquire all of the capabilities to survive, proliferate, and invade neighboring or distant tissues [8–12].
The microenvironment appears to be important for the selection of the best-adapted clones. Contributors include neighboring cells (e.g., fibroblasts and immune cells), growth factors, cytokines, hypoxia, and nutrient availability [13–16]. Hence, variations among tumor microenvironments may be responsible for some of the phenotypic heterogeneity observed within tumor beds. Consequently, the microenvironment may also have an impact on the selection of specific clones with different driver/maintenance mutations in topologically segregated areas of a tumor, which together affect the evolutionary trajectory of the disease (Fig. 1a).
Clonal accumulation and response to anticancer agents
The proteomic complexity of tumors must be fully understood to develop more effective therapeutic strategies. Pathologists have long recognized that not all cells within a given tumor express the same amounts of a large number of proteins. For example, the expression of cyclin D1 in mantle cell lymphoma is not homogeneous even when all of the tumor cells carry the signature CCND1-IGH translocation [17]. Likewise, hormone receptor expression in breast carcinoma is often non-homogenous and irregular within a given tumor bed. The protein intra-tumor heterogeneity is mirrored in some cases at the transcriptional level and has been well documented by single-cell RNA-sequencing in glioblastomas [18]. Moreover, the latter study noted variations in the expression level and differences in cell signaling receptor and cell proliferation markers. In addition, differences in post-translational modifications of a given protein among cancer cells in a single tumor have been documented [19, 20].
The consequences of this pervasive lack of uniformity between cancer cells are grim for patients. It is a major cause of treatment failure in many patients, particularly in those treated with molecular targeted therapies [21]. If a fraction of cancer cells in the tumor do not express a particular drug target or have evolved to no longer be dependent on its presence/activity, then it stands to reason that these cells will fail to be eliminated by targeted therapies. A case in point is HER2+ (human epidermal growth factor receptor 2) breast cancers, with the classification requiring that only 30% of the cells have to stain positive for HER2 by immunocytochemistry. Clearly, treatment with anti-HER2 therapies cannot be expected to be curative in such a context. Similarly, therapies based on rapamycin fail because of the uneven and heterogeneous expression of p-mTOR [22–24].
It is noteworthy that this same target expression issue may also affect the outcomes of immunotherapeutic approaches. Most patients with B cell malignancies who are treated with chimeric antigen receptor (CAR) T cells targeting CD19 will initially respond to therapy, but about 30% will relapse. The relapse appears to be because the tumor cells express a novel CD19 isoform arising from alternative splicing and lacking the exon encoding the antigenic epitope [25]. One way around this problem is to combine multiple therapies after tumor mutational profiling [26–28], but appropriate clinical trials are required to ensure that the resulting combination is not antagonistic.
Identifying the status of the protein synthesis machinery and the key regulators of translational control
Choosing targets whose expression levels do not vary significantly among cancer cells and whose expression cannot be extinguished (i.e., essential targets) is a logical step to deal with the issue of heterogeneity. Such potential targets include components of the translation apparatus, an essential biochemical process, with recent experience showing that an optimal therapeutic index can be obtained when this process is targeted in cancer cells [20, 21, 26, 27]
Two of the most important regulatory signal transduction pathways that modulate cellular translation rates are the RAS-RAF-ERK1/2 and PI3K-AKT/mTOR [29–31] pathways. These pathways are crucial to the development of targeted therapies because many of their components are changed in the vast majority of human cancers (e.g., HER2, PI3K, RAS, and RAF) [32]. What is generally underappreciated is that components of the translation regulatory machinery (namely, 4E-BP1, eIF4E, and eIF4A) involved in the ribosome recruitment phase of translation initiation fall under the control of these pathways (Fig. 2) [33, 34]. The eIF4F complex, consisting of eIF4E (the cap-binding protein), eIF4A (a DEAD-box RNA helicase), and eIF4G (a large scaffolding protein), regulates ribosome recruitment to mRNA templates [35]. This step in translation initiation is thought to be rate-limiting for protein synthesis. The assembly of eIF4F is regulated by mTOR via phosphorylation of 4E-BPs (of which there are three, with 4E-BP1 being the best studied), as well as of PDCD4 [36–39]. Binding of 4E-BP1 to eIF4E prevents eIF4F complex formation [40]. mTOR activation (as occurs in a broad range of human cancers) causes direct phosphorylation of 4E-BP1 and its dissociation from eIF4E to consequently stimulate eIF4F formation [41–43]. eIF4F discriminates between different mRNAs and therefore an increase in eIF4F levels or activity causes a selective change in the translatome. Although the features responsible for mRNA discrimination by eIF4F are not completely understood, cap accessibility and 5′ leader secondary structure are important contributors [44–46]. PDCD4 forms an inhibitory complex with eIF4A and phosphorylation of the former by S6K1/2 leads to its degradation and allows eIF4A to enter the eIF4F complex [38, 39]. In addition, eIF4E can be directly phosphorylated upon activation of the RAS-RAF-ERK1/2-MNK pathway or through p38 and this is also associated with a selective increase in translation, the mechanistic basis of which remains to be elucidated [47–51].
Fig. 2.
Schematic representing the signaling cascade regulating translation initiation. The key event is the dissociation of eIF4E from 4E-BP1 by different signaling pathways under normal growth conditions and in response to stress
4E-BP1 harbors seven phosphorylation sites and, although mTOR is the most prominent kinase targeting these sites, other kinases, such as CDK1, ATM, PI3K-AKT, ERK1/2, and PIM1, also phosphorylate 4E-BP1 [52, 53]. Therefore, 4E-BP1 phosphorylation can be the consequence of many different oncogenic events that modulate disparate signaling pathways or that occur via several mechanisms, including amplification or mutation of growth factor receptors or mutations in critical oncogenes (e.g., PTEN, ATM, p53, PI3K, or RAS). Our current understanding of perturbed translation initiation in cancer cells is that the eIF4E/4E-BP1 ratio is critical to sustain the oncogenic features of a transformed cell. Ultimately, this essential node may act as a “bottleneck” or “funnel factor” to sustain transformation, regardless of the upstream oncogenic alterations [54] (Fig. 2).
Expression of signaling factors in human tumors
In the past decade, by analyzing more than 2500 human tumor samples [55–60], we have assessed the expression of membrane receptors such as EGFR and HER2, components of the RAS/RAF/ERK and PI3K/AKT/mTOR pathways, and their effectors such as p70S6K, 4E-BP1, eIF4E, and p-eIF4E. We have found that increased amounts of total or p-eIF4E, as well as p-4E-BP1, are associated with malignant progression and adverse prognosis in several tumors, including breast, lung, ovary, endometrium, glioma, and prostate cancers, regardless of the upstream oncogenic alterations (Figs. 3 and 4) [22]. Other groups have confirmed the prognostic importance of these factors in additional tumor types (Table 1), including colon cancer [61, 62], nasopharyngeal carcinoma [63], hepatocellular carcinoma [64], astrocytomas [65], lung cancer [66, 67], and melanoma [68]. Importantly, eIF4E is a central regulator of metastatic progression [69–71] (Fig. 4).
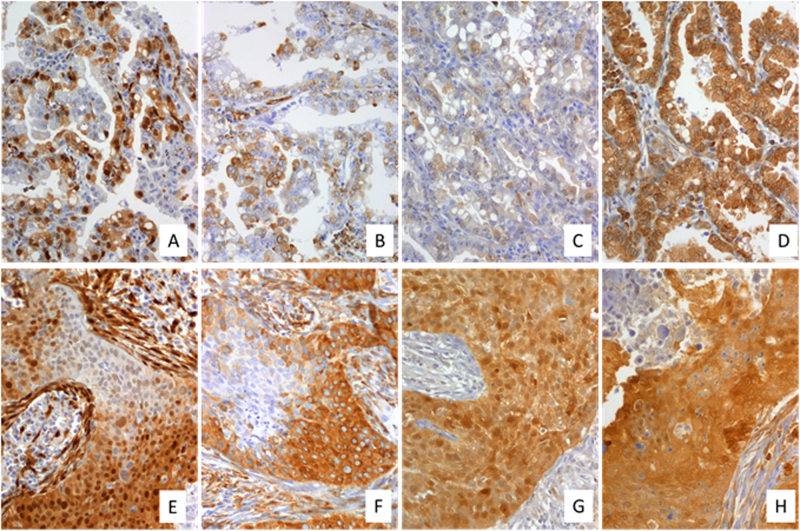
Fig. 3.
a–h Invasive ductal carcinoma a–g (×200), h (×400). Immunohistochemistry for a p-ERK1/2, b p-S6, c p-4E-BP1, d p-eIF4E, e p-AKT, and f p-mTOR. g, h GLUT-1 Immunohistochemistry was performed as described previously [58] using the following primary antibodies: p-eIF4E (Abcam, Ab76256), p-4E-BP1 (T37/46) (Cell Signaling, #2855), pS6 (S235/236) (Cell Signaling, #2211), p-ERK1/2 (T202/Y204) (Cell Signaling, #9101), p-mTOR (Ser2448) (Cell Signaling #2971); p-Akt (Ser473) (Cell Signaling #3787), GLUT-1 (Abcam, Ab652)
Fig. 4.
a–d Low-grade lung adenocarcinoma (×400). e–h High-grade lung squamous carcinoma (×200). Immunohistochemistry for p-ERK1/2 (a, e), p-S6 (b, f), p-4E-BP1 (c, g), and p-eIF4E (d, h). Immunohistochemistry was performed as described previously [58]
Table 1.
Prognostic significance of total and phosphorylated eIF4E and 4E‐BP1 in malignant tumors
| Primary tumor | Clinical significance | References |
|---|---|---|
| Bladder | p-4E-BP1 correlates with prognosis in patients with muscle-invasive bladder cancer | Nishikawa et al. [96] |
| Breast | Increased expression of eIF4E in invasive ductal carcinoma correlate with presence of lymph node metastasis | Hu et al. [97] |
| 4E-BP1 is an independent prognostic factor and is associated a poor response to endocrine therapy | Karlsson et al. [98] | |
| eIF4E predicts survival after anthracycline chemotherapy in breast cancer patients | Heikkinen et al. [99] | |
| eIF4E expression is related to breast cancer survival and it is modulated by 4E-BP1 | Coleman et al. [100] | |
| p-4E-BP1 correlates with grade and prognosis in breast cancer | Rojo et al. [60] | |
| High eIF4E is an independent predictor of recurrence in breast cancer. | Li et al. [101] | |
| Cervix | Overexpression of p-4E-BP1 predicts recurrence and reduced survival in cervical carcinoma | Benavente et al. [55] |
| CNS | p-eIF4E is an independent prognostic factor in astrocytoma | Martínez-Saez et al. [58] |
| p-4E-BP1 expression increase with tumor grade and predicts survival in astrocytomas | Korkolopoulou et al. [102] | |
| Colon | High 4E-BP1 expression is associated with poor prognosis | Chen et al. [103] |
| High expression of eIF4E is associated with advanced stage and poor prognosis | Chao et al. [61] | |
| Endometrium | p-4E-BP1 is associated with high-grade endometrial carcinomas and worse prognosis | Castellvi et al. [57] |
| p-4E-BP1 is associated with stage and high-grade tumors | Darb-Esfahani et al. [104] | |
| Esophagus | p-4E-BP1 expression after chemoradiotherapy is a predictor for recurrence and worse survival in esophageal carcinoma | Chao et al. [105] |
| p-4E-BP1 is associated with poor prognosis in early stage esophageal carcinoma | Yeh et al. [106] | |
| Head and neck | eIF4E expression is associated with tumor stage, lymph node metastasis and grade of differentiation | Han et al. [107] |
| p-eIF4E and p-MNK1 are independent prognostic factors in nasopharyngeal carcinoma | Zheng et al. [63] | |
| Kidney | p-4E-BP1 is associated with poor prognosis in Xp11.2 translocated renal cell carcinoma | Qu et al. [108] |
| p-4E-BP1 and eIF4E are independent prognostic factors in clear cell renal cell carcinoma | Campbell et al. [109] | |
| p-4E-BP1 is a prognostic predictor in patients with metastatic renal cell carcinoma | Nishikawa et al. [110] | |
| Liver | eIF4E overexpression is an independent indicator for overall survival in hepatocarcinoma | Jiang et al. [64] |
| p-4E-BP1 is overexpressed in cholangiocarcinomas with poor differentiation and lymph node metastasis, and is an independent prognostic factor | Fang et al. [111] | |
| Lung | p-4E-BP1 expression is associated with poor prognosis in small-cell lung cancer | Roh et al. [66] |
| p-4E-BP1 Thr70 predicts poor prognosis in non-small-cell lung cancer | Lee et al. [112] | |
| p-4E-BP1 and eIF4E are prognostic factors in stage I lung adenocarcinoma | Seki et al. [113] | |
| High eIF4E expression correlates with poor prognosis in lung adenocarcinomas | Wang et al. [114] | |
| Melanoma | eIF4E is associated with melanoma thickness and overall survival | Khosravi et al. [68] |
| p-4E-BP1 is associated with poor survival in melanoma | O’Reilly et al. [115] | |
| Ovary | p-4E-BP1 is a prognostic factor in ovarian cancer | Castellvi et al. [56] |
| Stomach | p-eIF4E is overexpressed in tumors with lymph node metastasis | Tapia et al. [116] |
| p-4E-BP1 is a prognostic factor in gastric cancer patients and correlates with advanced stage | Jiao et al. [117] |
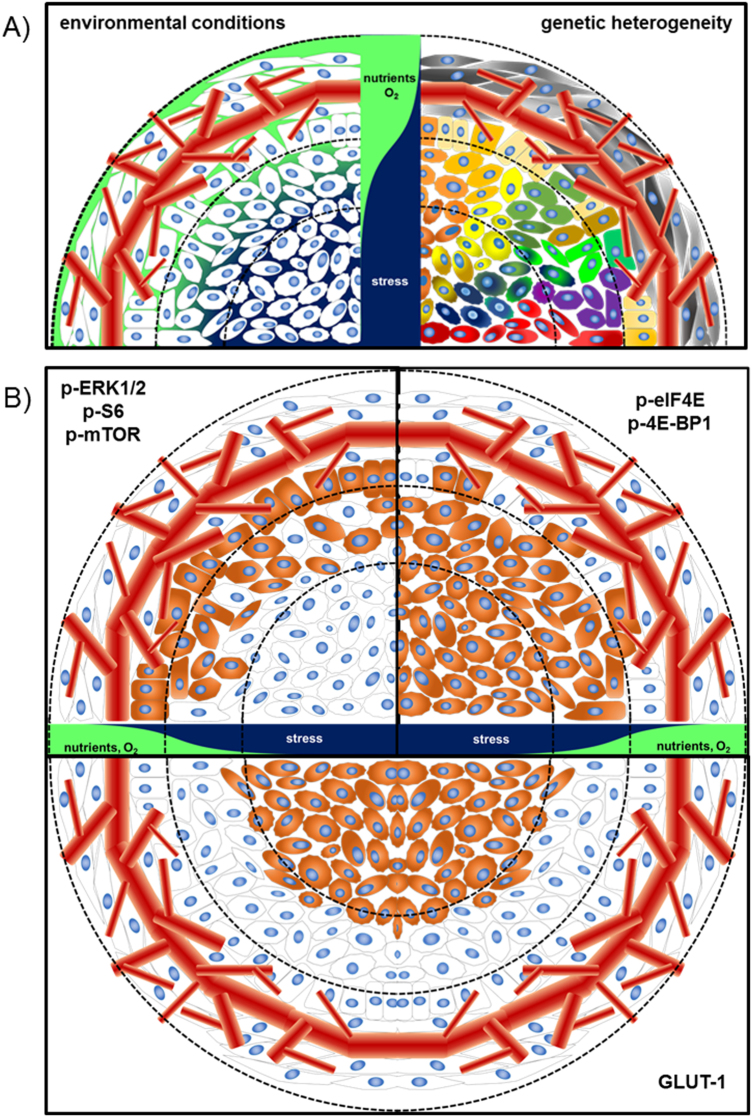
Thus, the eIF4E/4E-BP1 node appears to act as a restriction point for essential oncogenic features such as self-sufficiency in growth signals and should serve as a highly relevant molecular marker of malignant potential. Interestingly, the expression of eIF4E and 4E-BP1 and their phosphorylated forms is apparent even in the presence of upstream receptor or kinase overexpression (e.g., AKT, mTOR, or ERK), suggesting that other mechanisms are involved in their regulation. The expression of p-AKT or p-mTOR is highly heterogeneous within a tumor, whereas the expression of 4E-BP1 and eIF4E is more homogeneous (Fig. 3) [22]. This may be due to the activation status of the global growth signaling and proliferative network in being able to maintain a certain flux threshold rather than the necessity of maintaining activity of a specific player. Even in tumors showing constitutive expression of EGFR and HER2, the global gene expression program is not necessarily permanently fixed or homogeneous in all cells. Interestingly, the geographic context of the tumor cell may impinge on the expression levels of these pathways [72]. For example, some markers are more highly expressed at the invasive front or around necrotic areas, suggesting that ischemia or other microenvironmental factors impinge on their expression or activity (Figs. 3 and 5).
Fig. 5.
a Schematic drawing of a tumor and the tumor microenvironment. Left: Cells within a tumor exposed to different microenvironmental cues. Nutrient and O2 supply decreases from the periphery to the center of the tumor while stress conditions are elevated. Right: Genetic alterations of cells within a tumor are depicted by different colors. Importantly, genetic alterations are not strictly limited to the different environmental conditions. b Scheme depicting the staining pattern of different proteins involved in the signaling cascade regulating translation initiation (top), as shown in Figs. 3 and 4. GLUT1 serves as a marker for hypoxic conditions within the tumor (bottom)
Perspectives in tumor heterogeneity beyond genetics
The microenvironment has a key role in selecting the best-adapted cancer clone and can alter communication networks between different cancer cell types. The aberrant information flow in cancer cells leads to alterations in gene regulatory networks that support the cancer hallmarks [73] and can be influenced by features such as cytokines, exosomes, hypoxia, starvation, and oxidative stress (Fig. 5). In this respect, modulating the translational program is thought to ensure the expression of factors, which confer resistance to cellular stress [74] (Fig. 3). In most malignant cells, the cap-dependent pathway is highly upregulated and interference with this translational program has been shown to be an attractive venue for novel therapeutics that ultimately prevent the adaptation of tumor cells to stress conditions. The main therapeutic approaches targeting the 5′ cap-dependent translational machinery (summarized in ref. [33]) are directed against, the expression of eIF4E [75, 76], the interaction between eIF4E-4G [77, 78], the binding of eIF4F complex to the 5′-cap structure [79, 80], the eIF4A helicase activity [81–84], the phosphorylation status of eIF2α [85, 86], and the kinase activity of MNK1/2 [87–91]. Among the different strategies that prevent 5′ cap-dependent translation under stress conditions, it is believed that inhibition of MNKs may be a powerful way to increase the efficacy of other anti-tumor agents, as phosphorylation of its downstream target eIF4E has been shown to confer resistance to cellular stress, genomic damage, lack of nutrients, and oxidative stress (Fig. 5) [92, 93]. In fact, several companies are developing inhibitors of MNK1/2 activity [94], and at least one of them (eFT508) is already being studied in a clinical phase II trial. Impressive data have been obtained from preclinical models of diffuse large B cell lymphoma, non-small-cell lung carcinoma, and breast adenocarcinoma. Moreover, the inhibitor eFT508 enhances the efficiency of anti-PDL1 checkpoint blockade inhibitors [95]. Similarly, blockage of 4E-BP1 phosphorylation by inhibition of upstream signaling activity (e.g., mTOR) will decrease eIF4F levels and dampen cap-dependent translation and tumor cell growth [40].
In summary, tumor heterogeneity must first and foremost be considered by a treating oncologist after a cancer diagnosis and be a key factor in the determination of a therapeutic target following mutation profiling. We know that intra-tumor heterogeneity is dynamic, occurs at multiple levels, and follows a Darwinian model. Still unresolved is the number of biopsies or sections required from the primary specimen to determine the extent of molecular target heterogeneity. Moreover, relapses and metastases need to be analyzed to understand how they differ from the primary tumor. Given the complexities of these issues, collaboration among oncologists, radiologists, pathologists, bioinformaticians, and molecular biologists is required to offer the best care to patients. Finally, it is clearly of paramount importance to explore intervention strategies that target critical factors involved in regulating translation, such as eIF4E. With the rigorous evaluation of combinations of small-molecule eIF4E or MNK1/2 inhibitors with other therapeutics (e.g., cytotoxics, targeted therapies, immunotherapy), the issue of proteomic heterogeneity can start to be therapeutically addressed.
Funding
SRYC acknowledges support from Fondo de Investigaciones Sanitarias (P1170185 and PI 14/01320), Redes Temáticas de Investigación Cooperativa en Salud (RTICC, RD 12/0036/0057), Generalitat de Catalunya (AGAUR, 2014, 1131), and CIBERONC 2017.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interests.
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 Cancer Groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2016;3:524–48. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yong M, Jensen AO, Jacobsen JB, Norgaard M, Fryzek JP, Sorensen HT. Survival in breast cancer patients with bone metastases and skeletal-related events: a population-based cohort study in Denmark (1999-2007) Breast Cancer Res Treat. 2011;129:495–503. doi: 10.1007/s10549-011-1475-5. [DOI] [PubMed] [Google Scholar]
- 3.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramon y Cajal S, Capdevila C, Hernandez-Losa J, De Mattos-Arruda L, Ghosh A, Lorent J, et al. Cancer as an ecomolecular disease and a neoplastic consortium. Biochim Biophys Acta. 2017;1868:484–99. doi: 10.1016/j.bbcan.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–60. [PubMed] [Google Scholar]
- 6.McGranahan N, Swanton C. Biological and therapeutic impact of intra-tumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerji CR, Severini S, Caldas C, Teschendorff AE. Intra-tumour signalling entropy determines clinical outcome in breast and lung cancer. PLoS Comput Biol. 2015;11:e1004115. doi: 10.1371/journal.pcbi.1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai X, Chen A, Bai Z. Integrative investigation on breast cancer in ER, PR and HER2-defined subgroups using mRNA and miRNA expression profiling. Sci Rep. 2014;4:6566. doi: 10.1038/srep06566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–10. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolch W, Halasz M, Granovskaya M, Kholodenko BN. The dynamic control of signal transduction networks in cancer cells. Nat Rev Cancer. 2015;15:515–27. doi: 10.1038/nrc3983. [DOI] [PubMed] [Google Scholar]
- 12.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Gregorio A, Bowling S, Rodriguez TA. Cell competition and its role in the regulation of cell fitness from development to cancer. Dev Cell. 2016;38:621–34. doi: 10.1016/j.devcel.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia. 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends Genet. 2009;25:30–8. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–37. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jares P, Colomer D, Campo E. Molecular pathogenesis of mantle cell lymphoma. J Clin Invest. 2012;122:3416–23. doi: 10.1172/JCI61272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–28. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 20.Singh DK, Ku CJ, Wichaidit C, Steininger RJ, 3rd, Wu LF, Altschuler SJ. Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities. Mol Syst Biol. 2010;6:369. doi: 10.1038/msb.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rybinski B, Yun K. Addressing intra-tumoral heterogeneity and therapy resistance. Oncotarget. 2016;7:72322–42. doi: 10.18632/oncotarget.11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramon y Cajal S, De Mattos-Arruda L, Sonenberg N, Cortes J, Peg V. The intra-tumor heterogeneity of cell signaling factors in breast cancer: p4E-BP1 and peIF4E are diffusely expressed and are real potential targets. Clin Transl Oncol. 2014;16:937–41. doi: 10.1007/s12094-014-1203-9. [DOI] [PubMed] [Google Scholar]
- 23.Faes S, Demartines N, Dormond O. Resistance to mTORC1 inhibitors in cancer therapy: from kinase mutations to intratumoral heterogeneity of kinase activity. Oxid Med Cell Longev. 2017;2017:1726078. doi: 10.1155/2017/1726078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabernero J, Rojo F, Calvo E, Burris H, Judson I, Hazell K, et al. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: a phase I tumor pharmacodynamic study in patients with advanced solid tumors. J Clin Oncol. 2008;26:1603–10. doi: 10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Sotillo E, Harrington C, Wertheim G, Paessler M, Maude SL, et al. Repeated loss of target surface antigen after immunotherapy in primary mediastinal large B cell lymphoma. Am J Hematol. 2017;92:E11–E3. doi: 10.1002/ajh.24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodon J, Saura C, Dienstmann R, Vivancos A, Ramon y Cajal S, Baselga J, et al. Molecular prescreening to select patient population in early clinical trials. Nat Rev Clin Oncol. 2012;9:359–66. doi: 10.1038/nrclinonc.2012.48. [DOI] [PubMed] [Google Scholar]
- 27.Doroshow JH, Kummar S. Translational research in oncology–10 years of progress and future prospects. Nat Rev Clin Oncol. 2014;11:649–62. doi: 10.1038/nrclinonc.2014.158. [DOI] [PubMed] [Google Scholar]
- 28.Webster RM. Combination therapies in oncology. Nat Rev Drug Discov. 2016;15:81–2. doi: 10.1038/nrd.2016.3. [DOI] [PubMed] [Google Scholar]
- 29.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–45. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 30.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 31.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–8. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Luca A, Maiello MR, D’Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets. 2012;16:S17–27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 33.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015;14:261–78. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 34.Siddiqui N, Sonenberg N. Signalling to eIF4E in cancer. Biochem Soc Trans. 2015;43:763–72. doi: 10.1042/BST20150126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2:277–98. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 36.von Manteuffel SR, Gingras AC, Ming XF, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1996;93:4076–80. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–7. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–71. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 40.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr., et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–7. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 41.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–64. [PMC free article] [PubMed] [Google Scholar]
- 42.Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 43.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–37. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–8. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–66. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 46.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, et al. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–94. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buxade M, Parra-Palau JL, Proud CG. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases) Front Biosci. 2008;13:5359–73. doi: 10.2741/3086. [DOI] [PubMed] [Google Scholar]
- 48.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–20. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–33. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheper GC, van Kollenburg B, Hu J, Luo Y, Goss DJ, Proud CG. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem. 2002;277:3303–9. doi: 10.1074/jbc.M103607200. [DOI] [PubMed] [Google Scholar]
- 51.Topisirovic I, Ruiz-Gutierrez M, Borden KL. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 2004;64:8639–42. doi: 10.1158/0008-5472.CAN-04-2677. [DOI] [PubMed] [Google Scholar]
- 52.Yanagiya A, Suyama E, Adachi H, Svitkin YV, Aza-Blanc P, Imataka H, et al. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol Cell. 2012;46:847–58. doi: 10.1016/j.molcel.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velasquez C, Cheng E, Shuda M, Lee-Oesterreich PJ, Pogge von Strandmann L, Gritsenko MA, et al. Mitotic protein kinase CDK1 phosphorylation of mRNA translation regulator 4E-BP1 Ser83 may contribute to cell transformation. Proc Natl Acad Sci USA. 2016;113:8466–71. doi: 10.1073/pnas.1607768113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Armengol G, Rojo F, Castellvi J, Iglesias C, Cuatrecasas M, Pons B, et al. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67:7551–5. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 55.Benavente S, Verges R, Hermosilla E, Fumanal V, Casanova N, Garcia A, et al. Overexpression of phosphorylated 4E-BP1 predicts for tumor recurrence and reduced survival in cervical carcinoma treated with postoperative radiotherapy. Int J Radiat Oncol Biol Phys. 2009;75:1316–22. doi: 10.1016/j.ijrobp.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 56.Castellvi J, Garcia A, Rojo F, Ruiz-Marcellan C, Gil A, Baselga J, et al. Phosphorylated 4E binding protein 1: a hallmark of cell signaling that correlates with survival in ovarian cancer. Cancer. 2006;107:1801–11. doi: 10.1002/cncr.22195. [DOI] [PubMed] [Google Scholar]
- 57.Castellvi J, Garcia A, Ruiz-Marcellan C, Hernandez-Losa J, Peg V, Salcedo M, et al. Cell signaling in endometrial carcinoma: phosphorylated 4E-binding protein-1 expression in endometrial cancer correlates with aggressive tumors and prognosis. Hum Pathol. 2009;40:1418–26. doi: 10.1016/j.humpath.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 58.Martinez-Saez E, Peg V, Ortega-Aznar A, Martinez-Ricarte F, Camacho J, Hernandez-Losa J, et al. peIF4E as an independent prognostic factor and a potential therapeutic target in diffuse infiltrating astrocytomas. Cancer Med. 2016;5:2501–12. doi: 10.1002/cam4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pons B, Peg V, Vazquez-Sanchez MA, Lopez-Vicente L, Argelaguet E, Coch L, et al. The effect of p-4E-BP1 and p-eIF4E on cell proliferation in a breast cancer model. Int J Oncol. 2011;39:1337–45. doi: 10.3892/ijo.2011.1118. [DOI] [PubMed] [Google Scholar]
- 60.Rojo F, Najera L, Lirola J, Jimenez J, Guzman M, Sabadell DM, et al. 4E-binding protein 1, A cell signaling hallmark in breast cancer that correlates with pathologic grade and prognosis. Clin Cancer Res. 2007;13:81–9. doi: 10.1158/1078-0432.CCR-06-1560. [DOI] [PubMed] [Google Scholar]
- 61.Chao MW, Wang LT, Lai CY, Yang XM, Cheng YW, Lee KH, et al. eIF4E binding protein 1 expression is associated with clinical survival outcomes in colorectal cancer. Oncotarget. 2015;6:24092–104. doi: 10.18632/oncotarget.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen YT, Tsai HP, Wu CC, Wang JY, Chai CY. Eukaryotic translation initiation factor 4E (eIF-4E) expressions are associated with poor prognosis in colorectal adenocarcinoma. Pathol Res Pract. 2017;213:490–495. doi: 10.1016/j.prp.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 63.Zheng J, Li J, Xu L, Xie G, Wen Q, Luo J, et al. Phosphorylated Mnk1 and eIF4E are associated with lymph node metastasis and poor prognosis of nasopharyngeal carcinoma. PLoS ONE. 2014;9:e89220. doi: 10.1371/journal.pone.0089220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang XM, Yu XN, Huang RZ, Zhu HR, Chen XP, Xiong J, et al. Prognostic significance of eukaryotic initiation factor 4E in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2016;142:2309–17. doi: 10.1007/s00432-016-2232-2. [DOI] [PubMed] [Google Scholar]
- 65.Fan W, Wang W, Mao X, Chu S, Feng J, Xiao D, et al. Elevated levels of p-Mnk1, p-eIF4E and p-p70S6K proteins are associated with tumor recurrence and poor prognosis in astrocytomas. J Neurooncol. 2016;131:485–93. doi: 10.1007/s11060-016-2327-2. [DOI] [PubMed] [Google Scholar]
- 66.Roh MS, Lee JH, Kang KW, Nam HY, Jung SB, Kim K, et al. Phosphorylated 4E-binding protein 1 expression is associated with poor prognosis in small-cell lung cancer. Virchows Arch. 2015;467:667–73. doi: 10.1007/s00428-015-1860-2. [DOI] [PubMed] [Google Scholar]
- 67.Lee HW, Lee EH, Lee JH, Kim JE, Kim SH, Kim TG, et al. Prognostic significance of phosphorylated 4E-binding protein 1 in non-small-cell lung cancer. Int J Clin Exp Pathol. 2015;8:3955–62. [PMC free article] [PubMed] [Google Scholar]
- 68.Khosravi S, Tam KJ, Ardekani GS, Martinka M, McElwee KJ, Ong CJ. eIF4E is an adverse prognostic marker of melanoma patient survival by increasing melanoma cell invasion. J Invest Dermatol. 2015;135:1358–67. doi: 10.1038/jid.2014.552. [DOI] [PubMed] [Google Scholar]
- 69.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–4. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 70.Nasr Z, Robert F, Porco JA, Jr., Muller WJ, Pelletier J. eIF4F suppression in breast cancer affects maintenance and progression. Oncogene. 2013;32:861–71. doi: 10.1038/onc.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015;75:250–63. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buckley NE, Forde C, McArt DG, Boyle DP, Mullan PB, James JA, et al. Quantification of HER2 heterogeneity in breast cancer-implications for identification of sub-dominant clones for personalised treatment. Sci Rep. 2016;6:23383. doi: 10.1038/srep23383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 74.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–37. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 75.De Benedetti A, Joshi-Barve S, Rinker-Schaeffer C, Rhoads RE. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol Cell Biol. 1991;11:5435–45. doi: 10.1128/mcb.11.11.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hong DS, Kurzrock R, Oh Y, Wheler J, Naing A, Brail L, et al. A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin Cancer Res. 2011;17:6582–91. doi: 10.1158/1078-0432.CCR-11-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghosh B, Benyumov AO, Ghosh P, Jia Y, Avdulov S, Dahlberg PS, et al. Nontoxic chemical interdiction of the epithelial-to-mesenchymal transition by targeting cap-dependent translation. ACS Chem Biol. 2009;4:367–77. doi: 10.1021/cb9000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zochowska M, Piguet AC, Jemielity J, Kowalska J, Szolajska E, Dufour JF, et al. Virus-like particle-mediated intracellular delivery of mRNA cap analog with in vivo activity against hepatocellular carcinoma. Nanomedicine. 2015;11:67–76. doi: 10.1016/j.nano.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 79.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–67. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 80.Cencic R, Desforges M, Hall DR, Kozakov D, Du Y, Min J, et al. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J Virol. 2011;85:6381–9. doi: 10.1128/JVI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS ONE. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsumuraya T, Ishikawa C, Machijima Y, Nakachi S, Senba M, Tanaka J, et al. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem Pharmacol. 2011;81:713–22. doi: 10.1016/j.bcp.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 83.Kuznetsov G, Xu Q, Rudolph-Owen L, Tendyke K, Liu J, Towle M, et al. Potent in vitro and in vivo anticancer activities of des-methyl, des-amino pateamine A, a synthetic analogue of marine natural product pateamine A. Mol Cancer Ther. 2009;8:1250–60. doi: 10.1158/1535-7163.MCT-08-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest. 2008;118:2651–60. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen L, Aktas BH, Wang Y, He X, Sahoo R, Zhang N, et al. Tumor suppression by small-molecule inhibitors of translation initiation. Oncotarget. 2012;3:869–81. doi: 10.18632/oncotarget.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schewe DM, Aguirre-Ghiso JA. Inhibition of eIF2alpha dephosphorylation maximizes bortezomib efficiency and eliminates quiescent multiple myeloma cells surviving proteasome inhibitor therapy. Cancer Res. 2009;69:1545–52. doi: 10.1158/0008-5472.CAN-08-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Konicek BW, Stephens JR, McNulty AM, Robichaud N, Peery RB, Dumstorf CA, et al. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res. 2011;71:1849–57. doi: 10.1158/0008-5472.CAN-10-3298. [DOI] [PubMed] [Google Scholar]
- 88.Knauf U, Tschopp C, Gram H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol Cell Biol. 2001;21:5500–11. doi: 10.1128/MCB.21.16.5500-5511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu J, Chen A, Joy J, Xavier VJ, Ong EH, Hill J, et al. Rational design of resorcylic acid lactone analogues as covalent MNK1/2 kinase inhibitors by tuning the reactivity of an enamide Michael acceptor. ChemMedChem. 2013;8:1483–94. doi: 10.1002/cmdc.201300231. [DOI] [PubMed] [Google Scholar]
- 90.Ramalingam S, Gediya L, Kwegyir-Afful AK, Ramamurthy VP, Purushottamachar P, Mbatia H, et al. First MNKs degrading agents block phosphorylation of eIF4E, induce apoptosis, inhibit cell growth, migration and invasion in triple negative and Her2-overexpressing breast cancer cell lines. Oncotarget. 2014;5:530–43. doi: 10.18632/oncotarget.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Diab S, Teo T, Kumarasiri M, Li P, Yu M, Lam F, et al. Discovery of 5-(2-(phenylamino)pyrimidin-4-yl)thiazol-2(3H)-one derivatives as potent Mnk2 inhibitors: synthesis, SAR analysis and biological evaluation. ChemMedChem. 2014;9:962–72. doi: 10.1002/cmdc.201300552. [DOI] [PubMed] [Google Scholar]
- 92.Martínez A, Sesé M, Losa HJ, Robichaud N, Sonenberg N, Aasen T, et al. Phosphorylation of eIF4E confers resistance to cellular stress and DNA-damaging agents through an interaction with 4E-T: a rationale for novel therapeutic approaches. PLoS ONE. 2015;10:e0123352. doi: 10.1371/journal.pone.0123352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Z, Sun Y, Qu M, Wan H, Cai F, Zhang P. Inhibiting the MNK-eIF4E-beta-catenin axis increases the responsiveness of aggressive breast cancer cells to chemotherapy. Oncotarget. 2017;8:2906–15. doi: 10.18632/oncotarget.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dreas A, Mikulski M, Milik M, Fabritius CH, Brzozka K, Rzymski T. Mitogen-activated protein kinase (MAPK) interacting kinases 1 and 2 (MNK1 and MNK2) as targets for cancer therapy: recent progress in the development of MNK inhibitors. Curr Med Chem. 2017;24:3025–53. doi: 10.2174/0929867324666170203123427. [DOI] [PubMed] [Google Scholar]
- 95.Webster KR, Goel VK, Staunton J, Hung INJ, Parker GS, Stumpf CR, et al. eFT508: an oral, potent and highly selective inhibitor of MNK1 and MNK2, promotes anti-tumor immunity as a monotherapy and in combination with immune checkpoint blockade. Cancer Res. 2017;77:596.
- 96.Nishikawa M, Miyake H, Behnsawy HM, Fujisawa M. Significance of 4E-binding protein 1 as a therapeutic target for invasive urothelial carcinoma of the bladder. Urol Oncol. 2015;33:166.e9–15. doi: 10.1016/j.urolonc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 97.Hu A, Sun M, Yan D, Chen K. Clinical significance of mTOR and eIF4E expression in invasive ductal carcinoma. Tumori. 2014;100:541–6. doi: 10.1700/1660.18176. [DOI] [PubMed] [Google Scholar]
- 98.Karlsson E, Perez-Tenorio G, Amin R, Bostner J, Skoog L, Fornander T, et al. The mTOR effectors 4EBP1 and S6K2 are frequently coexpressed, and associated with a poor prognosis and endocrine resistance in breast cancer: a retrospective study including patients from the randomised Stockholm tamoxifen trials. Breast Cancer Res. 2013;15:R96. doi: 10.1186/bcr3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heikkinen T, Korpela T, Fagerholm R, Khan S, Aittomaki K, Heikkila P, et al. Eukaryotic translation initiation factor 4E (eIF4E) expression is associated with breast cancer tumor phenotype and predicts survival after anthracycline chemotherapy treatment. Breast Cancer Res Treat. 2013;141:79–88. doi: 10.1007/s10549-013-2671-2. [DOI] [PubMed] [Google Scholar]
- 100.Coleman LJ, Peter MB, Teall TJ, Brannan RA, Hanby AM, Honarpisheh H, et al. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br J Cancer. 2009;100:1393–9. doi: 10.1038/sj.bjc.6605044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li BD, Gruner JS, Abreo F, Johnson LW, Yu H, Nawas S, et al. Prospective study of eukaryotic initiation factor 4E protein elevation and breast cancer outcome. Ann Surg. 2002;235:732–8. doi: 10.1097/00000658-200205000-00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Korkolopoulou P, Levidou G, El-Habr EA, Piperi C, Adamopoulos C, Samaras V, et al. Phosphorylated 4E-binding protein 1 (p-4E-BP1): a novel prognostic marker in human astrocytomas. Histopathology. 2012;61:293–305. doi: 10.1111/j.1365-2559.2012.04236.x. [DOI] [PubMed] [Google Scholar]
- 103.Chen Y, Wang J, Fan H, Xie J, Xu L, Zhou B. Phosphorylated 4E-BP1 is associated with tumor progression and adverse prognosis in colorectal cancer. Neoplasma. 2017;64:787–94. doi: 10.4149/neo_2017_518. [DOI] [PubMed] [Google Scholar]
- 104.Darb-Esfahani S, Faggad A, Noske A, Weichert W, Buckendahl AC, Muller B, et al. Phospho-mTOR and phospho-4EBP1 in endometrial adenocarcinoma: association with stage and grade in vivo and link with response to rapamycin treatment in vitro. J Cancer Res Clin Oncol. 2009;135:933–41. doi: 10.1007/s00432-008-0529-5. [DOI] [PubMed] [Google Scholar]
- 105.Chao YK, Chuang WY, Yeh CJ, Chang YS, Wu YC, Kuo SY, et al. High phosphorylated 4E-binding protein 1 expression after chemoradiotherapy is a predictor for locoregional recurrence and worse survival in esophageal squamous cell carcinoma patients. J Surg Oncol. 2012;105:288–92. doi: 10.1002/jso.22097. [DOI] [PubMed] [Google Scholar]
- 106.Yeh CJ, Chuang WY, Chao YK, Liu YH, Chang YS, Kuo SY, et al. High expression of phosphorylated 4E-binding protein 1 is an adverse prognostic factor in esophageal squamous cell carcinoma. Virchows Arch. 2011;458:171–8. doi: 10.1007/s00428-010-0994-5. [DOI] [PubMed] [Google Scholar]
- 107.Han M, Wang W, Wang L, Jiang Y. Expression of eukaryotic initiation factor 4 E in hypopharyngeal carcinoma. J Int Med Res. 2014;42:976–83. doi: 10.1177/0300060514527912. [DOI] [PubMed] [Google Scholar]
- 108.Qu Y, Zhao R, Wang H, Chang K, Yang X, Zhou X, et al. Phosphorylated 4EBP1 is associated with tumor progression and poor prognosis in Xp11.2 translocation renal cell carcinoma. Sci Rep. 2016;6:23594. doi: 10.1038/srep23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Campbell L, Jasani B, Griffiths DF, Gumbleton M. Phospho-4e-BP1 and eIF4E overexpression synergistically drives disease progression in clinically confined clear cell renal cell carcinoma. Am J Cancer Res. 2015;5:2838–48. [PMC free article] [PubMed] [Google Scholar]
- 110.Nishikawa M, Miyake H, Harada K, Fujisawa M. Expression level of phosphorylated-4E-binding protein 1 in radical nephrectomy specimens as a prognostic predictor in patients with metastatic renal cell carcinoma treated with mammalian target of rapamycin inhibitors. Med Oncol. 2014;31:792. doi: 10.1007/s12032-013-0792-4. [DOI] [PubMed] [Google Scholar]
- 111.Fang Z, Lu L, Tian Z, Luo K. Overexpression of phosphorylated 4E-binding protein 1 predicts lymph node metastasis and poor prognosis of Chinese patients with hilar cholangiocarcinoma. Med Oncol. 2014;31:940. doi: 10.1007/s12032-014-0940-5. [DOI] [PubMed] [Google Scholar]
- 112.Lee HW, Park MI, Kim MS, Kim SH, Roh MS, Kim K, et al. Overexpression of phosphorylated 4E-binding protein 1 and its clinicopathological significances in gastric cancer. Pathol Res Pract. 2015;211:298–302. doi: 10.1016/j.prp.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 113.Seki N, Takasu T, Sawada S, Nakata M, Nishimura R, Segawa Y, et al. Prognostic significance of expression of eukaryotic initiation factor 4E and 4E binding protein 1 in patients with pathological stage I invasive lung adenocarcinoma. Lung Cancer. 2010;70:329–34. doi: 10.1016/j.lungcan.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 114.Wang R, Geng J, Wang JH, Chu XY, Geng HC, Chen LB. Overexpression of eukaryotic initiation factor 4E (eIF4E) and its clinical significance in lung adenocarcinoma. Lung Cancer. 2009;66:237–44. doi: 10.1016/j.lungcan.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 115.O’Reilly KE, Warycha M, Davies MA, Rodrik V, Zhou XK, Yee H, et al. Phosphorylated 4E-BP1 is associated with poor survival in melanoma. Clin Cancer Res. 2009;15:2872–8. doi: 10.1158/1078-0432.CCR-08-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tapia O, Riquelme I, Leal P, Sandoval A, Aedo S, Weber H, et al. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 2014;465:25–33. doi: 10.1007/s00428-014-1588-4. [DOI] [PubMed] [Google Scholar]
- 117.Jiao X, Pan J, Qian J, Luo T, Wang Z, Yu G, et al. Overexpression of p-4ebp1 in Chinese gastric cancer patients and its correlation with prognosis. Hepatogastroenterology. 2013;60:921–6. doi: 10.5754/hge12865. [DOI] [PubMed] [Google Scholar]