Abstract
Mitochondrial genes and whole mitochondrial genome sequences are widely used as molecular markers in studying population genetics and resolving both deep and shallow nodes in phylogenetics. In animals the mitochondrial genome is generally composed of a single chromosome, but mystifying exceptions sometimes occur. We determined the complete mitochondrial genome of the millipede-parasitic nematode Ruizia karukerae and found its mitochondrial genome consists of two circular chromosomes, which is highly unusual in bilateral animals. Chromosome I is 7,659 bp and includes six protein-coding genes, two rRNA genes and nine tRNA genes. Chromosome II comprises 7,647 bp, with seven protein-coding genes and 16 tRNA genes. Interestingly, both chromosomes share a 1,010 bp sequence containing duplicate copies of cox2 and three tRNA genes (trnD, trnG and trnH), and the nucleotide sequences between the duplicated homologous gene copies are nearly identical, suggesting a possible recent genesis for this bipartite mitochondrial genome. Given that little is known about the formation, maintenance or evolution of abnormal mitochondrial genome structures, R. karukerae mtDNA may provide an important early glimpse into this process.
Introduction
The majority of metazoan mitochondrial genomes have a well-conserved structure and consist of a single circular chromosome, ranging from 14 to 20 kb and containing 37 genes: 13 protein-coding genes (PCGs) (atp6, atp8, cob, cox1–3, nad1–6 and nad4l), two ribosomal RNA (rRNA) genes (rrnL and rrnS) and 22 transfer RNA (tRNA) genes1,2. In nematodes, mitochondrial genomes are also fairly conserved in structure and gene content, although they differ from other metazoans in some features. For example, most nematode species lack an atp8 gene (except Trichinella spp. and Trichuris spp.3–8), and their tRNAs have unique secondary structures (no DHU arm in 20 tRNAs and no TΨC arm in two tRNAs, trnS1 and trnS2). Complete mitochondrial genomes have been reported from more than 176 nematode species since Caenorhabditis elegans and Ascaris suum were first published in 19929. Interestingly, in four nematode species the mitochondrial genome has been found to be divided into multiple chromosomes10–13. The reasons underlying these structural abnormalities are unclear, and the sequencing of additional mitogenomes is needed in order to better understand common features of this unusual phenomenon.
The mitochondrial genome has been used in many phylogenetic studies as a powerful molecular marker for resolving both deep and shallow nodes in various groups, including nematodes14–17. In recent decades, nematode mitochondrial genomes have provided independent confirmation of some phylogenetic hypotheses based on nuclear genes, and yielded insights into various evolutionary patterns such as convergent morphological evolution, and independent origins of plant parasitism18,19. In this study we report the complete mitochondrial genome sequence of Ruizia karukerae, a member of the infraorder Rhigonematomorpha, a group of about 150 named nematode species that have a direct parasitic life cycle and use millipedes as their final host20. The mitochondrial genome of R. karukerae is made up of two circular chromosomes of similar size, each encoding mostly different genes.
Results
The two circular mitochondrial chromosomes of R. karukerae
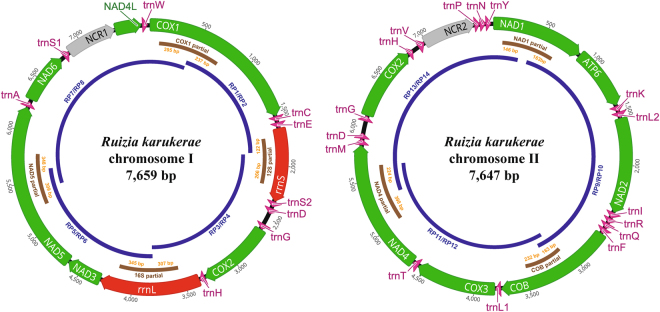
Initially, the PCR, sequencing, and assembly of four long PCR fragments [cox1-rrnS (1.5 kb), rrnS-rrnL (1.8 kb), rrnL-nad5 (1.7 kb) and nad5-cox1 (2.7 kb)] and partial fragment sequences (cox1, rrnS, rrnL and nad5) produced an unexpectedly small, circular molecule of mtDNA (termed chromosome I) consisting of 7,659 nucleotides and containing only six PCGs, two ribosomal genes and nine tRNA genes (Fig. 1, Table 1). Repeated attempts produced the same results. This suggested the presence of multiple chromosomes, since several key mitochondrial genes (including cob, nad1 and nad4) were missing from the sequence of chromosome I. To locate the remaining genes not found on our assembly of chromosome I, we determined the sequences of partial fragments of cob, nad1 and nad4 individually and designed three species-specific primer sets (RP9/RP10, RP11/RP12 and RP13/RP14) (Table 2) for long PCR. These were used to amplify and sequence three overlapping fragments [nad1-cob (2.8 kb), cob-nad4 (2.3 kb) and nad4-nad1 (2.5 kb)], which ultimately formed a second circular mtDNA molecule (termed chromosome II), which consisted of 7,647 nucleotides and contained seven PCGs and 16 tRNA genes (Fig. 1, Table 1). The entire circular mtDNA sequences of chromosomes I and II were assembled by confirming the sequence identity in the overlapping regions (3,655 bp of overlap including 7 independently amplified fragments with overlaps ranging from 122~398 bp) between the long PCR fragments and the partial gene fragments (see Fig. 1 and Table 2 for details). Confirmation of the sequence identity in the overlapping regions, and the primer walking strategy used for the long PCR fragments employed in this study, support the presence of a bipartite circular mitochondrial genome (chromosomes I and II), rather than an artefact resulting from nuclear copies of mitochondrial DNA (“numts”). The sequences from chromosome I and chromosome II were deposited in GenBank (accession numbers MF509850 and MF509851, respectively).
Figure 1.
A representation of the two circular mitochondrial chromosomes of Ruizia karukerae. All genes are encoded in the same direction, and the 22 tRNA genes are indicated by a single-letter abbreviation. The leucine and serine tRNA genes are marked according to their anticodon sequence as L1 (trnL1-uag), L2 (trnL2-uaa), S1 (trnS1-ucu) and S2 (trnS2-uga). Primer names (see Table 2 for details) are in blue lettering, and blue curved bars indicate long PCR fragments. Brown curved bars indicate partial gene fragments obtained by 7 independent PCR reactions. Orange numbers indicate length of sequence overlap between the partial sequences and ends of long PCR fragments.
Table 1.
Mitochondrial genome organization of Ruizia karukerae.
| Gene | Positions of nucleotide sequences | No. of nt (bp) | Initiation/termination codons | Intergenic sequence |
|---|---|---|---|---|
| Chromosome I | ||||
| cox1 | 1–1536 | 1536 | ATT/TAA | −2 |
| trnC | 1535–1590 | 56 | 2 | |
| trnE | 1593–1648 | 56 | 0 | |
| rrnS | 1649–2338 | 690 | 0 | |
| trnS2 | 2339–2393 | 55 | 12 | |
| trnD | 2406–2461 | 56 | 106 | |
| trnG | 2568–2623 | 56 | −1 | |
| cox2 | 2623−3312 | 690 | ATA/TAA | −1 |
| trnH | 3312–3365 | 54 | 0 | |
| rrnL | 3366–4306 | 941 | 0 | |
| nad3 | 4307–4642 | 336 | ATT/TAA | −1 |
| nad5 | 4642–6223 | 1582 | ATA/T | 0 |
| trnA | 6224–6278 | 55 | 19 | |
| nad6 | 6298–6771 | 474 | ATA/TAA | 29 |
| trnS1 | 6801–6865 | 65 | 0 | |
| NCR | 6866–7336 | 471 | 0 | |
| nad4l | 7337–7588 | 252 | ATT/TAA | 5 |
| trnW | 7594–7646 | 53 | 13 | |
| Chromosome II | ||||
| nad1 | 1–861 | 861 | ATT/TAA | 2 |
| atp6 | 864–1442 | 579 | ATG/TAG | 1 |
| trnK | 1444–1504 | 61 | 0 | |
| trnL2 | 1505–1559 | 55 | 1 | |
| nad2 | 1561–2449 | 889 | ATA/T | 0 |
| trnI | 2450–2509 | 60 | 0 | |
| trnR | 2510–2563 | 54 | −1 | |
| trnQ | 2563–2617 | 55 | −1 | |
| trnF | 2617–2670 | 54 | 1 | |
| cob | 2672–3760 | 1089 | ATG/TAG | 0 |
| trnL1 | 3761–3815 | 55 | −3 | |
| cox3 | 3813–4560 | 748 | ATA/T | 0 |
| trnT | 4561–4617 | 57 | 0 | |
| nad4 | 4618–5844 | 1227 | ATA/TAA | −2 |
| trnM | 5843–5901 | 59 | −1 | |
| trnD | 5901–5956 | 56 | 106 | |
| trnG | 6063–6118 | 56 | −1 | |
| cox2 | 6118–6807 | 690 | ATA/TAA | −1 |
| trnH | 6807–6860 | 54 | 57 | |
| trnV | 6918–6987 | 70 | 0 | |
| NCR | 6988–7476 | 489 | 0 | |
| trnP | 7477–7529 | 53 | −1 | |
| trnN | 7529–7583 | 55 | −2 | |
| trnY | 7582–7635 | 54 | 12 | |
Table 2.
Primers used to sequence the complete mitochondrial genomes of Ruizia karukerae.
| Primers | Sequence (5′-3′) | Source | Size of PCR fragment | Size of overlapping region (with long PCR fragment) |
|---|---|---|---|---|
| Chromosome I | ||||
| LCO1490 | GGTCAACAAATCATAAAGATATTGG | 48 | 655 bp | 395 bp (RP7/RP8) |
| HCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | 237 bp (RP1/RP2) | ||
| Nema_12S_F | GTTCCAGAATAATCGGCTA | This study | 465 bp | 122 bp (RP1/RP2) |
| Nema_12S_R | GCKATTGARGGATGYTTTGTACC | 266 bp (RP3/RP4) | ||
| Nema_16S_F_2 | TTAGTGTTGAAAAATCGTTC | This study | 678 bp | 307 bp (RP3/RP4) |
| Nema_16S_R | TCTYMCRAYGAAYTAAACTAATATC | 345 bp (RP5/RP6) | ||
| Nema_ND5_F | GTTCATAGAAGTACTTTGGTKACTGCTG | This study | 485 bp | 309 bp (RP5/RP6) |
| Nema_ND5_R | AAGACGMWAACWATAAMHAAAAGT | 348 bp (RP7/RP8) | ||
| RP1 | AGTCTGCATATGGCAGGTGTTAGC | This study | 1.5 kb | |
| RP2 | GGCTACCCGGGTACTAATCCG | |||
| RP3 | CAAACTGAAGTAAATTGGCAGGTGC | This study | 1.8 kb | |
| RP4 | CAATGGATTATGCTACTTTAATGTCC | |||
| RP5 | GGACATTAAAGTAGCATAATCCATTG | This study | 1.7 kb | |
| RP6 | GATTAAATAAGGTAACTTCCCTAAACCAC | |||
| RP7 | GATAGAGGAGGATATGAAGAAGGTAGTG | This study | 2.7 kb | |
| RP8 | GAGCTAACACCTGCCATATGCAGAC | |||
| Chromosome II | ||||
| Chroma_ND1_F_4 | GGCTTTTGTAACTCTTTATGAGCG | This study | 511 bp | 146 bp (RP13/RP14) |
| Chroma_ND1_R_2 | CCDCTNACYARYTCDCTYTC | 163 bp (RP9/RP10) | ||
| Chroma_Cob_F_2 | CARATRWSNTWTTGRGC | This study | 358 bp | 163 bp (RP9/RP10) |
| Chroma_Cob_R_2 | TAYCAYTCNGGNACAAYATG | 232 bp (RP11/RP12) | ||
| Chroma_ND4_F_1 | CATGTHGARGCDCCNAC | This study | 398 bp | 398 bp (RP11/RP12) |
| Chroma_ND4_R_3 | GTCCCAGCGTTAGTTAAAAATGTCA | 224 bp (RP13/RP14) | ||
| RP9 | GTAGAAGCCCCGACTACTGCTAG | This study | 2.8 kb | |
| RP10 | ACAAGCTTCTCCTTCCAGTCTCATG | |||
| RP11 | TGTTACATTTCTTGTTACCTTGGGC | This study | 2.3 kb | |
| RP12 | GTCCCAGCGTTAGTTAAAAATGTCA | |||
| RP13 | GTCTGTTCCAGAGGGATGGTAAAGCTCTAGC | This study | 2.8 kb | |
| RP14 | ATAACCACAAAGGCTACTGCGGGAG | |||
IUPAC nucleotide ambiguity codes used are W (A or T), R (A or G), K (G or T), S (C or G), Y (C or T), M (A or C), D (A, C or T), H (A, G or T) and N (A, C, G or T).
Gene content of R. karukerae mtDNA
The mitochondrial genome of R. karukerae contains 12 PCGs (atp6, cob, cox1-3, nad 1-6 and nad4L), two rRNA genes and 22 tRNA genes, and lacks an atp8 gene, a common feature in nematode mitochondrial genomes with the exception of Trichinella spp. and Trichuris spp.3–8. In the mitochondrial genome of R. karukerae, chromosome I contains six PCGs, two rRNA genes and nine tRNA genes, and chromosome II includes seven PCGs and 16 tRNA genes (Fig. 1, Table 1). All genes are encoded in the same direction, as is common in other known chromadorean nematode mitochondrial genomes (except for nad2 in Plectus acuminatus and P. aquatilis21). Interestingly, cox2, trnD, trnG and trnH were identified on both chromosomes (Fig. 1, Table 1). The trnG sequences on chromosomes I and II are identical, while the sequences of cox2, trnD, and trnH differ by one nucleotide between chromosomes. The nucleotide substitution between the two copies of the cox2 gene is a nonsynonymous mutation: a TAT codon encoding tyrosine in chromosome I is substituted by a TCT codon encoding serine in chromosome II. Tyrosine and serine are polar, non-charged hydrophilic amino acids, and therefore both duplicate copies of cox2 gene are assumed to be functional. In addition, both of the trnD and trnH genes on chromosomes I and II are also presumed to be functional because the nucleotide substitution had no effect on the tRNA secondary structure (Supplementary Fig. S1). Low levels of sequence difference between homologous gene copies on chromosome I and II suggests that a bipartite mitochondrial genome has evolved in this species relatively recently, although more work is needed to confirm this. The cox2 genes on the two chromosomes of R. karukerae are 99.9% similar, whereas between R. karukerae and its putative closest three sequenced relatives (Rhigonema thysanophora [Rhigonematomorpha], Ascaridia columbae and Cucullanus robustus22), cox2 sequence similarity ranges from 63% to 69%.
Protein-coding genes and codon usage
Among the 12 PCGs, six genes (cox2, nad5, nad6, nad2, cox3 and nad4) use ATA as the start codon, four (cox1, nad3, nad4l and nad1) start with ATT, and two (atp6 and cob) start with ATG (Table 1). As a termination codon, seven genes (cox1, cox2, nad3, nad6, nad4l, nad1 and nad4) were inferred to use TAA, two genes (atp6 and cob) to use TAG, and three genes (nad5, nad2 and cox3) to use the incomplete termination codon ‘T’.
The PCGs were biased towards T-rich codons (more than 2 Ts per triplet), similar to other chromadorean nematodes16,19,21–23. The three most commonly used codons from each chromosome were all T-rich: TTT (11.3%), TTA (8.4%) and ATT (6.2%) for chromosome I and TTT (11.3%), TTA (9.9%) and ATT (7.2%) for chromosome II (Supplementary Table S1). In contrast, the frequency of C-rich codons (two or more Cs per triplet) was only 3.3% of the total PCGs from chromosome I and 3.5% of the total PCGs from chromosome II.
Transfer RNA gene and the non-coding region
Twenty-two tRNA gene sequences, ranging in size from 53 bp (trnW) to 70 bp (trnV), are inferred to fold into secondary structures of tRNAs (Supplementary Fig. S1, Table 1). Twenty of the tRNAs contain an amino-acyl stem of 7 bp and a DHU arm and anticodon stem, but lack a TΨC arm structure. In contrast, trnS1 and trnS2 have a TΨC arm and lack a DHU arm. These tRNA structures are commonly found in other nematode species21. The trnD, trnG and trnH genes were found on both chromosome I and chromosome II (Fig. 1, Table 1). The trnD and trnH sequences on chromosome I differed from their corresponding sequences (homologous genes) on chromosome II by a single nucleotide, but their putative secondary structure forms were identical (Supplementary Fig. S1).
On chromosome I, a non-coding region (designated NCR1) with a total length of 471 bp was found between trnS1 and nad4l. On chromosome II, a non-coding region (designated NCR2) 489 bp in length was located between trnV and trnP (Fig. 1, Table 1). The A + T contents of the non-coding regions on chromosome I and chromosome II were 73% and 70.6%, respectively (Supplementary Table S2).
Mitochondrial gene arrangement of R. karukerae
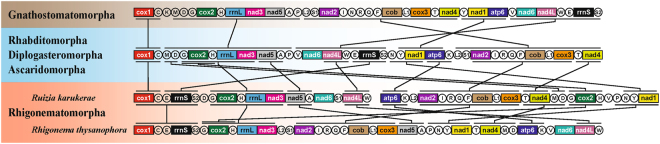
The two species of Rhigonematomorpha for which mitochondrial genomes are now available (i.e., R. karukerae and R. thysanophora) share many gene clusters even though their gene order is not identical. Specifically, nad4l-trnW-cox1-trnC-trnE-rrnS-trnS2, trnG-cox2-trnH-rrnL-nad3, and nad5-trnA are shared between R. thysanophora and chromosome I of R. karukerae, and atp6-trnK, nad2-trnI-trnR-trnQ-trnF-cob-trnL1-cox3, trnT-nad4-trnM-trnD, trnG-cox2-trnH, and trnP-trnN-trnY-nad1 are shared between R. thysanophora and chromosome II of R. karukerae (Fig. 2). Although mitochondrial phylogenies support a sister relationship among Rhigonematomorpha, Ascaridomorpha and Gnathostomatomorpha22,24, gene order in rhigonematomorphs is more similar to the most common gene order pattern among Ascaridomorpha, Diplogasteromorpha and Rhabditomorpha: they have many shared gene clusters: cox1-trnC, trnM-trnD, trnG-cox2-trnH-rrnL-nad3, nad5-trnA, nad4l-trnW, trnE-rrnS-trnS2, trnN-trnY-nad1, atp6-trnK, nad2-trnI-trnR-trnQ-trnF-cob-trnL1-cox3 and trnT-nad4.
Figure 2.
Linearized comparison of the mitochondrial gene arrangement of Ruizia karukerae and Rhigonema thysanophora (Rhigonematomorpha) and one species representative of Ascaridomorpha, Diplogasteromorpha, Gnathostomatomorpha and Rhabditomorpha, respectively. 22 tRNA genes are indicated by a single-letter abbreviation. The two leucine and two serine tRNA genes are marked according to their separate anticodon sequence, as L1 (trnL1-uag), L2 (trnL2-uaa), S1 (trnS1-ucu), and S2 (trnS2-uga). The non-coding regions are not shown.
Discussion
As presented in this study, the mitochondrial genome of R. karukerae consists of two circular chromosomes. There are a few other metazoan species in which the mitochondrial genome is known to be divided into multiple chromosomes. In such multipartite genomes, the size and the number of chromosomes vary widely. For example, Brachionus plicatilis and B. koreanus (rotifers), Liposcelis bostrychophila (booklouse), Rhabditophanes sp. KR3021 (nematode) and Globodera ellingtonae (nematode) all have two circular, evenly sized mitochondrial chromosomes12,13,25–27; however, the insect Scirtothrips dorsalis (thrip) has two chromosomes of very different sizes (14.3 kb and 0.9 kb)28. In contrast, the potato cyst nematodes (Globodera pallida and G. rostochiensis) have a least six circles10,11,29, and several lice species have mitochondrial genomes with 9 to 20 circles ranging from 1.5 to 5 kb30–37. In a few animals even the circular structure of the mitochondrial genome is not conserved: linearized mitochondrial genomes have been reported from an isopod (Armadillidium vulgare) and some cnidarians38,39.
The number of mitochondrial chromosomes in R. karukerae is the same as G. ellingtonae and Rhabditophanes sp.12,13, but the size of its two chromosomes (less than 8 kb each) is more similar to those of G. pallida, a nematode with at least six mitochondrial chromosomes (each 6.4–9.5 kb in size) or Rhabditophanes sp. (9.3 kb and 9.2 kb) rather than those of G. ellingtonae (17.8 kb and 14.4 kb)10,12,13,29. A common feature of G. ellingtonae, Rhabditophanes sp. and G. pallida is that they have abnormally long NCSs (non-coding sequences). The two mitochondrial chromosomes of G. ellingtonae have lengthy, intergenic NCSs between almost every gene (7 bp-8.1 kb for chromosome I and 1 bp-7.2 kb chromosome II)13. The longest NCSs in both chromosomes of Rhabditophanes sp. are each 2.6 kb12. In G. pallida, all six of its mitochondrial chromosomes contain many NCSs (3 to 7.4 kb in length)10,29. Abnormally lengthy NCSs in multipartite mitochondrial genomes are also found in some other metazoan groups: for example, two mitochondrial chromosomes of B. plicatilis (rotifer) have long NCSs (4.9 kb each)25 and L. bostrychophila (booklouse) has a relatively large number of intergenic NCSs (1–485 bp)26. In contrast, the NCSs of R. karukerae are more or less similar in size to those found in typical nematode species with a single mitochondrial chromosome (i.e., genes are abutting or the intergenic spaces are relatively short) (Fig. 1, Table 1).
Among nematodes with multipartite mitogenomes, there is considerable variation both in the characteristics of duplicated genes and the sequence divergence between duplicated copies. The duplicated genes found on both circular chromosomes (chromosome I and II) of R. karukerae comprise cox2 and three tRNA genes (trnD, trnG and trnH), accounting for 856 bp (Fig. 1, Table 1), and their sequences are identical or nearly so: the two copies of trnG are the same, whereas the other genes (cox2, trnD and trnH) are nearly identical to their homologous copies, each differing by only a single base pair. In comparison, the duplicated region (2.6 kb) in each of the mitochondrial chromosomes of Rhabditophanes sp. is identical12, while in G. ellingtonae the two mitochondrial chromosomes share both an NCS and several copies of nonfunctional pseudogenes (highly degraded or truncated), with no homologous, functional genes shared between the two circles13. In G. pallida, several protein-coding genes are found on more than one circle, but many of the duplicated genes are nonfunctional due to frame-shift or nonsense mutations29. Features of the duplicated regions of mitochondrial chromosomes in other non-nematode groups are somewhat similar: the rotifer B. plicatilis has a duplicated region 4.9 kb in size containing the tRNA(L) genes and a non-coding region25, while in the booklouse L. bostrychophila, each 0.9 kb duplicated region is identical and includes three tRNA genes (trnA, trnE and trnM) and a putative control region26.
To explain how multipartite mitogenomes arise, various models have been suggested, such as intramolecular homologous recombination40, or genome replication13 followed by gene deletion41; however, a broad consensus has not yet been reached. Multipartite mitochondrial genomes with circular or linear forms have evolved independently in disparate, unrelated taxa (insects, rotifers, and nematodes [circular form]; sponges, hydras, box jellyfish [linear form])42, but it is still unclear how multiple chromosomes can arise from a single circular chromosome.
The two main arguments for why multipartite mitochondrial genomes exist are genetic drift and/or an unknown evolutionary advantage to having multiple mitochondrial chromosomes26,43,44 (but see45). There is no evidence favoring one of these arguments over another for R. karukerae, but there is some evidence that natural selection may favor shorter mitochondrial genomes. For example, in cultured human cells with both 11 kb and 16 kb mitochondrial circles, smaller circles became more numerous over time while the larger circles decreased in number46; and in heteroplasmic crickets, smaller mitochondrial genomes are transmitted to offspring more frequently than larger genomes43. Alternatively, multiple mitochondrial chromosomes may exist because a specific replisome gene (the mitochondrial single-strand binding protein, mtSSB that aids in DNA replication) was previously lost or mutated. The abnormality in this gene would prevent a full-size mitochondrial chromosome from being replicated, but would still allow smaller chromosomes to exist. In lice, it has been argued that the absence of this gene is responsible for multipartite mitochondrial genomes33. Although it is unknown exactly how or why multipartite genomes arise, previous work has noted a correlation with blood-feeding30 or a parasitic life-style47. All nematode species thus far recorded as having multipartite mitochondrial genomes, including the present study, are parasitic species (G. ellingtonae, G. pallida, G. rostochiensis [plant parasitic]; Rhabditophanes sp. KR3021, R. karukerae [animal parasitic]). However, comparatively few free-living nematode mitogenomes have been sequenced (compared to parasitic forms), and there is no clear evidence that parasitic life styles are correlated with multipartite mitochondrial genomes, nor has any work conclusively demonstrated that multipartite mitogenomes would be an advantage for parasites. Much more research is needed to better elucidate the evolutionary mechanisms leading to unusual mitochondrial genome structures.
Mitochondrial DNA genes have a relatively long history of use in phylogenetics2 and phylogeography48,49. More recently, phylogenetic comparisons of invertebrates based on complete mitochondrial genomes have been used for assessing deep relationships, and also for comparing species sharing more recent common ancestry6,50–54. This range of resolution is possible because mitochondrial genomes are composed of genes with very different rates of substitution. Faster evolving genes track more recent evolutionary events, whereas more conserved genes (and protein sequences) are informative for some deeper divergences. Disadvantages of mitochondrial DNA include that its multiple genes are inherited as a single locus, and certain groups of organisms show substantial nucleotide bias across genes. For nematodes, mtDNA genomes represent one of the main loci that have been used to infer phylogenetic relationships spanning the phylum. NCBI contains complete mitochondrial genomes for 176 nematode species. In contrast, nuclear ribosomal genes are a much more extensively sampled locus, with thousands of nematode species sequenced for 18S (SSU) rDNA55,56. This difference in taxon sampling between SSU and mitogenomes precludes detailed comparisons of phylogenetic results, but the main phylogenetic framework resulting from analysis of these separate loci is concordant21,57 despite some notable specific differences16,19 that will need to be tested through sampling of additional loci from nuclear genomes.
Materials and Methods
Specimen sampling and molecular methods
Nematode specimens were obtained from Anadenobolus monilicornis (millipedes) collected from the John Pennecamp Coral Reef State Park, Key Largo, Florida, USA by R. Carreno. The specimens were identified based on morphological features and measurements58. Total genomic DNA was extracted using a commercial kit (Epicentre MasterPure DNA Purification Kit; Epicentre Co.) following the manufacturer’s protocol. Four partial DNA fragments from four different genes (cox1, rrnS, rrnL and nad5) were amplified by polymerase chain reaction (PCR) using a universal primer set (LCO1490/HCO219859) for cox1 and three nematode-specific primer sets (Nema_12S_F/Nema_12S_R for rrnS, Nema_16S_F_2/Nema_16S_R for rrnL and Nema_ND5_F/Nema_ND5_R for nad5), designed directly from conserved regions of nematode mitochondrial genes (Table 2). PCR amplifications were carried out using TaKaRa Ex Taq (Takara) in a total volume of 50 ul containing 2 ul template DNA, 10 pmol of each primer, 1.25 u of Ex Taq polymerase, 1X Ex Taq buffer and 0.2 mM dNTP mixture, with the following amplification conditions: one initial denaturing step at 95 °C for 1 min followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 47 °C for 30 s, extension at 72 °C for 1 min, and final elongation at 72 °C for 10 min. Four primer pairs (RP1/RP2, RP3/RP4, RP5/RP6 and RP7/RP8) were designed from the sequences of the partial fragments (cox1, rrnS, rrnL and nad5) (Table 2) and used to obtain four overlapping long PCR fragments ranging from 1.5 kb to 2.7 kb: cox1-rrnS (1.5 kb), rrnS-rrnL (1.8 kb), rrnL-nad5 (1.7 kb) and nad5-cox1 (2.7 kb). Long PCR reactions consisted of 2 ul template DNA, 10 pmol of each primer, 2.5 unit LA Taq polymerase (TaKaRa), 1X LA Taq buffer, 0.4 mM dNTP mixture, 2.5 mM MgCl2 and 29.5 ul distilled water with the following amplification conditions: one cycle of initial denaturing at 95 °C for 1 min followed by 40 cycles of denaturation at 95 °C for 30 s, annealing and extension at 55 °C to 65 °C for 3 min to 10 min, followed by a final extension at 68 °C for 10 min. The amplified PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN Co.) following standard protocols. The sequences of the PCR-amplified fragments were determined for both strands using Big Dye Terminator Cycle-Sequencing (Applied Biosystems) and a primer walking strategy. The sequence of a complete strand of mtDNA was assembled by checking the sequences of the overlapping regions of the long PCR fragments and partial fragments obtained from the four different genes (cox1, rrnS, rrnL and nad5). Initially only a 7,659 bp contig (chromosome I) was obtained, containing six PCGs, two rDNAs, and nine tRNAs (Fig. 1). To locate the other genes, three partial fragments of three protein-coding genes missing from chromosome I (cob, nad1 and nad4) were amplified using three nematode specific primer sets (Chroma_ND1_F_4/Chroma_ND1_R_2 for nad1, Chroma_ND4_F_1/Chroma_ND4_R_3 for nad4 and Chroma_Cob_F_2/Chroma_Cob_R_2 for cob) (Table 2), and then sequenced. Using three species-specific primer sets (RP9/RP10, RP11/RP12 and RP13/RP14) designed from cob, nad1 and nad4 partial sequences (Table 2), three overlapping fragments were amplified by long-PCR and sequenced. The sequence of the complete strand of the second contig (chromosome II) was assembled by checking the sequences of the overlapping regions of the three long PCR fragments and the partial fragments obtained from the cob, nad1 and nad4 genes (Fig. 1).
Gene annotation
The 12 mitochondrial protein-coding genes and two ribosomal RNA genes of R. karukerae were identified using the annotation program DOGMA60 and ORF finder (NCBI), and were confirmed by comparing nucleotide sequences with those from closely related nematodes. Putative secondary structures of 22 tRNA genes were inferred using the program tRNAscan-SE61 and verified by examining potential tRNA secondary structures and anticodon sequences.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (NRF-2013R1A1A2005898), the Basic Science Research Program through the National Research Foundation of Korea (NRF) by the Ministry of Science, ICT & Future Planning (NRF-2015R1A4A1041997), a grant from the Collaborative Genome Program (20140428) funded by the Ministry of Oceans and Fisheries, Korea, and a grant from the Animal and Plant Quarantine Agency (No. Z-1541785-2013-15-03). We thank Dr. Ramon Carreno for providing R. karukerae specimens.
Author Contributions
T.K., E.K., C.P. and Y.J.B. performed molecular experiments and analyzed the data. T.K., E.K., S.A.N. and J.K.P. wrote the paper. J.K.P. designed and coordinated the study. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25759-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wolstenholme DR. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992;141:173–216. doi: 10.1016/S0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 2.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavrov DV, Brown WM. Trichinella spiralis mtDNA: a nematode mitochondrial genome that encodes a putative ATP8 and normally structured tRNAs and has a gene arrangement relatable to those of coelomate metazoans. Genetics. 2001;157:621–637. doi: 10.1093/genetics/157.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu GH, et al. Characterization of the complete mitochondrial genomes of two whipworms Trichuris ovis and Trichuris discolor (Nematoda: Trichuridae) Infect. Genet. Evol. 2012;12:1635–1641. doi: 10.1016/j.meegid.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Liu GH, et al. Clear genetic distinctiveness between human-and pig-derived Trichuris based on analyses of mitochondrial datasets. PLoS Neglect. Trop. Dis. 2012;6:e1539. doi: 10.1371/journal.pntd.0001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu GH, et al. Mitochondrial and nuclear ribosomal DNA evidence supports the existence of a new Trichuris species in the endangered françois’ leaf-monkey. PLoS One. 2013;8:e66249. doi: 10.1371/journal.pone.0066249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohandas N, et al. Mitochondrial genomes of Trichinella species and genotypes - a basis for diagnosis, and systematic and epidemiological explorations. Int. J. Parasitol. 2014;44:1073–1080. doi: 10.1016/j.ijpara.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Hawash MBF, Andersen LO, Gasser RB, Stensvold CR, Nejsum P. Mitochondrial genome analyses suggest multiple Trichuris species in humans, baboons, and pigs from different geographical regions. PLoS Neglect. Trop. Dis. 2015;9:e0004059. doi: 10.1371/journal.pntd.0004059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okimoto R, Macfarlane JL, Clary DO, Wolstenholme DR. The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics. 1992;130:471–498. doi: 10.1093/genetics/130.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armstrong MR, Block VC, Phillips MS. A multipartite mitochondrial genome in the potato cyst nematode Globodera pallida. Genetics. 2000;154:181–192. doi: 10.1093/genetics/154.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson T, Block VC, Dowton M. Sequence and characterization of six mitochondrial subgenomes from Globodera rostochiensis: Multipartite structure is conserved among close nematode relatives. J. Mol. Evol. 2007;65:308–315. doi: 10.1007/s00239-007-9007-y. [DOI] [PubMed] [Google Scholar]
- 12.Hunt VL, et al. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat. Genet. 2016;48:299–307. doi: 10.1038/ng.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips WS, et al. The mitochondrial genome of Globodera ellingtonae is composed of two circles with segregated gene content and differential copy numbers. BMC Genomics. 2016;17:706. doi: 10.1186/s12864-016-3047-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu L, Li YW, Ryder OA, Zhang YP. Analysis of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol. Biol. 2007;7:198. doi: 10.1186/1471-2148-7-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morin PA, et al. Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res. 2010;20:908–916. doi: 10.1101/gr.102954.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JK, et al. Monophyly of clade III nematodes is not supported by phylogenetic analysis of complete mitochondrial genome sequences. BMC Genomics. 2011;12:392. doi: 10.1186/1471-2164-12-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cameron SL. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 18.Sultana T, et al. Comparative analysis of complete mitochondrial genome sequences confirms independent origins of plant-parasitic nematodes. BMC Evol. Biol. 2013;13:12. doi: 10.1186/1471-2148-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, et al. Mitochondrial genomes advance phylogenetic hypotheses for Tylenchina (Nematoda: Chromadorea) Zool. Scr. 2015;44:446–462. doi: 10.1111/zsc.12112. [DOI] [Google Scholar]
- 20.Hunt DJ. A synopsis of the Rhigonematidae (Nematoda), with an outline classification of the Rhigonematida. Afro-Asian J. Nematol. 1996;6:137–150. [Google Scholar]
- 21.Kim J, et al. Phylogenetic analysis of two Plectus mitochondrial genomes (Nematoda: Plactida) supports a sister group relationship between Plectida and Rhabditida within Chromadorea. Mol. Phylogenet. Evol. 2017;107:90–102. doi: 10.1016/j.ympev.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Kim T, et al. Phylogeny of Rhigonematomorpha based on the complete mitochondrial genome of Rhigonema thysanophora (Nematoda: Chromadorea) Zool. Scr. 2014;43:289–303. doi: 10.1111/zsc.12047. [DOI] [Google Scholar]
- 23.Kim T, Kim J, Nadler SA, Park JK. The complete mitochondrial genome of Koerneria sudhausi (Diplogasteromorpha: Nematoda) supports monophyly of Diplogasteromorpha within Rhabditomorpha. Curr. Genet. 2016;62:391–403. doi: 10.1007/s00294-015-0536-4. [DOI] [PubMed] [Google Scholar]
- 24.Liu GH, Shao R, Cai XQ, Li WW, Zhu XQ. Gnathostoma spinigerum mitochondrial genome sequence: a novel gene arrangement and its phylogenetic position within the class Chromadorea. Sci. Rep. 2015;5:12691. doi: 10.1038/srep12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suga K, Welch DBM, Tanaka Y, Sakakura Y, Hagiwarak A. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol. Biol. Evol. 2008;25:1129–37. doi: 10.1093/molbev/msn058. [DOI] [PubMed] [Google Scholar]
- 26.Wei DD, et al. The multipartite mitochondrial genome of Liposcelis bostrychophila: insights into the evolution of mitochondrial genomes in bilateral animals. PloS One. 2012;7:e33973. doi: 10.1371/journal.pone.0033973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang DS, et al. Complete mitochondrial genome of the monogonont rotifer, Brachionus koreanus (Rotifera, Brachionidae). Mitochondr. DNA. 2014;25:29–30. doi: 10.3109/19401736.2013.775274. [DOI] [PubMed] [Google Scholar]
- 28.Dickey AM, et al. A novel mitochondrial genome architecture in thrips (Insecta: Thysanoptera): extreme size asymmetry among chromosomes and possible recent control region duplication. BMC Genomics. 2015;16:439. doi: 10.1186/s12864-015-1672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibson T, et al. The mitochondrial subgenomes of the nematode Globodera pallida are mosaics: Evidence of recombination in an animal mitochondrial genome. J. Mol. Evol. 2007;64:463–471. doi: 10.1007/s00239-006-0187-7. [DOI] [PubMed] [Google Scholar]
- 30.Shao R, Kirkness EF, Barker SC. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse. Pediculus humanus. Genome Res. 2009;19:904–912. doi: 10.1101/gr.083188.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao R, Zhu XQ, Barker SC, Herd K. Evolution of extensively fragmented mitochondrial genomes in the lice of humans. Genome Biol. Evol. 2012;4:1088–1101. doi: 10.1093/gbe/evs088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao R, et al. Fragmented mitochondrial genomes in two suborders of parasitic lice of eutherian mammals (Anoplura and Rhynchophthirina, Insecta) Sci. Rep. 2015;5:17389. doi: 10.1038/srep17389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cameron SL, Yoshizawa K, Mizukoshi A, Whiting MF, Johnson KP. Mitochondrial genome deletions and minicircles are common in lice (Insecta: Phthiraptera) BMC Genomics. 2011;12:394. doi: 10.1186/1471-2164-12-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang H, Barker SC, Shao R. Substantial variation in the extent of mitochondrial genome fragmentation among blood-sucking lice of mammals. Genome Biol. Evol. 2013;5:1298–1308. doi: 10.1093/gbe/evt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong WG, et al. Fragmented mitochondrial genomes are present in both major clades of the blood-sucking lice (suborder Anoplura): evidence from two Hoplopleura rodent lice (family Hoplopleuridae) BMC Genomics. 2014;15:751. doi: 10.1186/1471-2164-15-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong WG, Song S, Jin DC, Guo XG, Shao R. Fragmented mitochondrial genomes of the rat lice, Polyplax asiatica and Polyplax spinulosa: intra-genus variation in fragmentation pattern and a possible link between the extent of fragmentation and the length of life cycle. BMC Genomics. 2014;15:44. doi: 10.1186/1471-2164-15-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song SD, Barker SC, Shao R. Variation in mitochondrial minichromosome composition between blood-sucking lice of the genus Haematopinus that infest horses and pigs. Parasite. Vector. 2014;7:144. doi: 10.1186/1756-3305-7-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcadé I, et al. Structure and evolution of the atypical mitochondrial genome of Armadillidium vulgare (Isopoda, Crustacea) J. Mol. Evol. 2007;65:651–659. doi: 10.1007/s00239-007-9037-5. [DOI] [PubMed] [Google Scholar]
- 39.Kayal E, et al. Evolution of linear mitochondrial genomes in medusozoan cnidarians. Genome Biol. Evol. 2011;4:1–12. doi: 10.1093/gbe/evr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fauron C, Casper M, Gao Y, Moore B. The maize mitochondrial genome: dynamic, yet functional. Trends Genet. 1995;11:288–235. doi: 10.1016/S0168-9525(00)89056-3. [DOI] [PubMed] [Google Scholar]
- 41.Melov S, Lithgow GJ, Fischer DR, Tedesco PM, Johnson TE. Increased frequency of deletions in the mitochondrial genome with age of Caenorhabditis elegans. Nucleic Acids Res. 1995;23:1419–1425. doi: 10.1093/nar/23.8.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavrov DV, Pett W. Animal mitochondrial DNA as we do not know it: mt-genome organization and evolution in nonbilaterian lineages. Genome Biol. Evol. 2016;8:2896–2913. doi: 10.1093/gbe/evw195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rand DM, Harrison RG. Mitochondrial DNA transmission genetics in crickets. Genetics. 1986;114:955–970. doi: 10.1093/genetics/114.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rand DM. ‘Why genomes in pieces?’ revisited: Sucking lice do their own thing in mtDNA circle game. Genome Res. 2009;19:700–702. doi: 10.1101/gr.091132.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyce TM, Zwick ME, Aquadro CF. Mitochondrial DNA in the bark weevils: size, structure and heteroplasmy. Genetics. 1989;123:825–836. doi: 10.1093/genetics/123.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayashi JL, Ohta S, Kikuchi A, Takemitsu M, Goto Y. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA. 1991;88:10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burger G, Gary MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Moritz C, Dowling TE, Brown WM. Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 1987;18:269–292. doi: 10.1146/annurev.es.18.110187.001413. [DOI] [Google Scholar]
- 49.Avise JC. Phylogeography: retrospect and prospect. J. Biogeogr. 2009;36:3–15. doi: 10.1111/j.1365-2699.2008.02032.x. [DOI] [Google Scholar]
- 50.Hu M, Gasser RB. Mitochondrial genomes of parasitic nematodes-progress and perspectives. Trends Parasitol. 2006;22:78–84. doi: 10.1016/j.pt.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 51.Waeschebach A, Telford MJ, Porter JS, Littlewood DTJ. The complete mitochondrial genome of Flustrellidra hispida and the phylogenetic position of Bryozoa among the Metazoa. Mol. Phylogenet. Evol. 2006;40:195–207. doi: 10.1016/j.ympev.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 52.Littlewood DTJ, Lockyer AE, Webster BL, Johnston DA, Le TH. The complete mitochondrial genomes of Schistosoma haematobium and Schistosoma spindale and the evolutionary history of mitochondrial genome changes among parasitic flatworms. Mol. Phylogenet. Evol. 2006;39:452–467. doi: 10.1016/j.ympev.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 53.Park JK, et al. A common origin of complex life cycles in parasitic flatworms: evidence from the complete mitochondrial genome of Microcotyle sebastis (Monogenea: Platyhelminthes) BMC Evol. Biol. 2007;11:1–13. doi: 10.1186/1471-2148-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sultana T, Han H, Park JK. Comparison of complete mitochondrial genomes of pine wilt nematode Bursaphelenchus xylophilus and Bursaphelenchus mucronatus (Nematoda: Aphelenchoidea) and development of a molecular tool for species identification. Gene. 2013;520:39–46. doi: 10.1016/j.gene.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Holterman M, et al. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- 56.van Megen H, et al. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology. 2009;11:927–950. doi: 10.1163/156854109X456862. [DOI] [Google Scholar]
- 57.Liu GH, et al. Mitochondrial phylogenomics yields strongly supported hypotheses for ascaridomorph nematodes. Sci. Rep. 2016;6:39248. doi: 10.1038/srep39248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carreno RA, Ordosch D, Koltek JK, Hamill DR, Tuhela L. First United States records of the Rhigonematid genera Heth and Ruizia (Nematoda: Rhigonematida) from the introduced millipede, Anadenobolus monilicornis (Diplopoda: Rhinocricidae) in Key Largo, Florida, USA. Comp. Parasitol. 2013;80:225–232. doi: 10.1654/4614.1. [DOI] [Google Scholar]
- 59.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994;3:294–299. [PubMed] [Google Scholar]
- 60.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with Dogma. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 61.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genome sequences. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.