Abstract
Deep learning techniques have recently emerged as promising decision supporting approaches to automatically analyze medical images for different clinical diagnosing purposes. Diagnosing of pulmonary nodules by using computer-assisted diagnosing has received considerable theoretical, computational, and empirical research work, and considerable methods have been developed for detection and classification of pulmonary nodules on different formats of images including chest radiographs, computed tomography (CT), and positron emission tomography in the past five decades. The recent remarkable and significant progress in deep learning for pulmonary nodules achieved in both academia and the industry has demonstrated that deep learning techniques seem to be promising alternative decision support schemes to effectively tackle the central issues in pulmonary nodules diagnosing, including feature extraction, nodule detection, false-positive reduction, and benign-malignant classification for the huge volume of chest scan data. The main goal of this investigation is to provide a comprehensive state-of-the-art review of the deep learning aided decision support for pulmonary nodules diagnosing. As far as the authors know, this is the first time that a review is devoted exclusively to deep learning techniques for pulmonary nodules diagnosing.
Keywords: Computer-aided diagnosis, convolutional neural network (CNN), deep learning, pulmonary nodules
Introduction
Deep learning techniques have recently emerged as promising decision supporting approaches to automatically analyze medical images for different clinical diagnosing purposes (1-10). The recent remarkable and significant progress in deep learning for pulmonary nodules achieved in both academia and the industry has demonstrated that deep learning techniques seem to be promising alternative decision support schemes to effectively tackle the central issues in pulmonary nodules diagnosing, including feature extraction, nodule detection, false-positive reduction (3,5,6,11-13).
Figure 1 depicts a rapid surge of interest in deep learning for detection of pulmonary nodules as well as for classification between benign and malignant in terms of the number of publications with respect to the time. Figure 1 demonstrates that the first paper dedicated to deep learning for classification of pulmonary nodules appeared in 2014 and the first paper for detection of pulmonary nodules appeared in 2015. Then deep learning for both tasks were emerging and booming especially in 2017. It is the motivation of this investigation to provide a comprehensive state-of-the-art review of the deep learning aided decision support for pulmonary nodules diagnosing.
Figure 1.
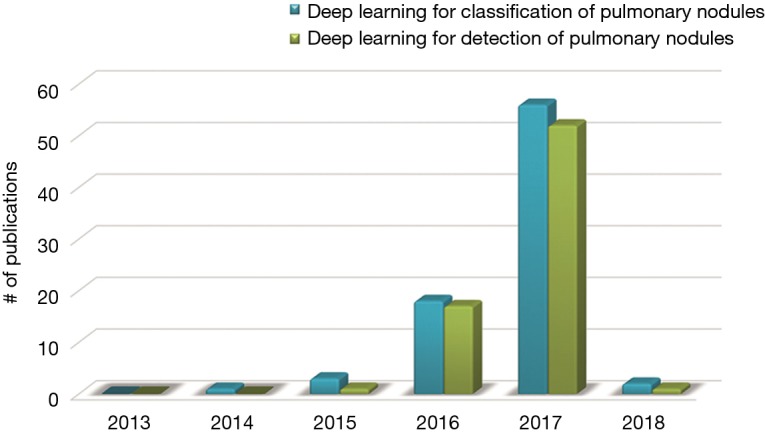
Rapid surge of interest in deep learning for detection of pulmonary nodules as well as for classification between benign and malignant in terms of the number of publications with respect to the time.
The paper is organized as follows. We first present the computer-assisted diagnostic techniques of pulmonary nodules developed in the deep learning era, including the two/three-dimensional, multiview, multi-stream, multi-scale convolutional neural networks (CNN), deep belief deep network, massive training network, and convolutional autoencoder network. Finally, we conclude this paper with future work.
Deep learning aided decision support for pulmonary nodules diagnosing
It is well established that the deep learning techniques seamlessly integrate exploitation feature and tuning of performance so as to simplify the ad hoc analysis pipeline of conventional computer aided diagnosis. In this regard, several studies were dedicated to pulmonary nodule diagnosis with the assistance of deep learning techniques. Table 1 summarizes the deep learning aided decision support for pulmonary nodules diagnosing.
Table 1. Summary of the deep learning aided decision support techniques for pulmonary nodules diagnosing.
| Authors | Year | Methods | Purpose | Image format |
|---|---|---|---|---|
| Tu et al. (14) | 2017 | Two-dimensional CNN | Classification | Chest CT |
| Teramoto et al. (15) | 2016 | The shape and metabolic feature analysis + the CNN based feature extraction + two-step SVM classifiers | Features extraction | PET and CT |
| Anthimopoulos et al. (16) | 2016 | Two-dimensional CNN | Classification | Chest CT |
| Xie et al. (17) | 2018 | Co-occurrence matrix + Fourier shape descriptor + deep CNN + AdaBoosted back propagation neural network | Features extraction | Chest CT |
| Shen et al. (18) | 2017 | Multi-crop CNN | Classification | Chest CT |
| Setio et al. (19) | 2016 | Two-dimensional multi-view CNN | Detection | Chest CT |
| Sun et al. (20) | 2017 | LeCun model based CNN, deep belief network, and stacked denoising autoencoder (SDAE) | Classification | Chest CT |
| Ciompi et al. (21) | 2017 | Multi-stream multi-scale CNN | Classification | Chest CT |
| Yuan et al. (22) | 2017 | Multi-view multi-scale CNN + Fisher vector encodings + support vector machine | Features extraction | Chest CT |
| Dou et al. (23) | 2017 | Three-dimensional CNN | Detection | Volumetric CT |
| Liu et al. (24) | 2017 | Three-dimensional CNN | Classification | Chest CT |
| Hua et al. (25) | 2015 | Deep belief network (DBN) and CNN | Classification | Chest CT |
| Tajbakhsh and Suzuki (26) | 2017 | Massive-training artificial neural networks, AlexNet, rd-CNN, LeNet architecture, and sh-CNN | Detection | Chest CT |
| Chen et al. (27) | 2017 | Convolutional autoencoder neural network | Features extraction | Chest CT |
CNN, convolutional neural networks.
Two-dimensional convolutional neural networks
Tu et al. (14) trained a two-dimensional CNN for categorization of solid, part-solid and non-solid pulmonary nodules in computed tomography (CT) images. The CNN was structured with two convolutional layers (5×5 kernel size with stride 2), each followed by a max-pooling layer (2×2 kernel size with stride 2). The number of feature maps in the first and the second convolutional layer were 6 and 16, respectively. A fully-connected layer was then connected to the pooling-2 layer, and further followed by a Softmax activation layer for classification. For the regression task, the features computed at the final fully-connected layer of the trained CNN were used and input into a random forest regression for rating the nodules. Based on LIDC (Lung Image Database Consortium) dataset (28,29), experimental results show significant performance improvement by the CNN model over histogram analysis in both classification and regression.
Teramoto et al. (15) proposed an ensemble method to reduce false positives of solitary pulmonary nodules using both positron emission tomography (PET) and CT images through which both metabolic and anatomical information could be provided. A two-step classifier was developed in which rule-based classifier worked in the first stage, and two support vector machines (SVM) in the second stage. One SVM took 22 features (all features by CT, three SUV features by PET, and features extracted by CNN) as the input, while the second SVM took all features as input. Evaluation based on 104 PET/CT images showed that the ensemble method achieved a sensitivity of 90.1% with 4.9 FPs/case.
Anthimopoulos et al. (16) firstly applied CNN for classification of patterns of interstitial lung diseases (ILDs) (30,31) including micronodules, ground glass opacity, reticulation, honeycombing, and consolidation. The CNN was trained based on dataset of 14,696 image patches by the first-order gradient-based Adam optimizer. Comparative analysis demonstrated the effectiveness of the proposed CNN over the state-of-the-art methods, including SVM classifiers with intensity texton based features (32), k nearest neighbor classifier with local binary patterns and intensity histograms features (33), random forest classifier with local discrete cosine transform and gray level histogram features (34), LeNet (35), AlexNet (36) and VGG-Net (37).
Xie et al. (17) proposed a hybrid feature extraction approach in which the gray level co-occurrence matrix was employed for texture description, Fourier shape descriptor for characterizing the heterogeneous nodules, and deep CNN for learning the feature representation of nodules. Then, the AdaBoosted back propagation neural network was trained for classification based on the aforementioned extracted features.
Shen et al. (18) investigated the likelihood of nodule malignancy in CT images by multi-crop CNN in which a novel multi-crop pooling strategy was proposed to crop different regions from convolutional feature maps and apply max-pooling different times. Experimental results showed that the proposed method achieves state-of-the-art nodule classification performance.
Setio et al. (19) proposed a two-dimensional multi-view CNN for false positive reduction of pulmonary nodules, for which the inputs were fed with two-dimensional patches extracted from differently oriented planes. Their approach reached sensitivities of 85.4% and 90.1% at 1 and 4 false positives per scan, respectively on 888 scans of LIDC-IDRI dataset (28,29).
Sun et al. (20) compared the performances of three multichannel ROI based deep structured algorithms [i.e., LeCun model based CNN (35), deep belief network (DBN) (38), and stacked denoising autoencoder] which were based on extracting automatically generated features with traditional computer aided diagnosis (CADx) systems using hand-crafted features including density (i.e., average intensity, standard deviation, and entropy), texture (i.e., GLCM, wavelet, LBP and SIFT) and morphological features (area, circularity, and ratio of semi-axis) for lung nodule CT image diagnosis in all of the LIDC 1,018 cases (28,29). Their experiments showed that the CNN achieved AUC of 0.899±0.018, which was higher than traditional CADx with the AUC of 0.848±0.026.
To provide fair comparisons of performances from different nodule detection and false positive reduction algorithms, Setio et al. (39) set up the LUng Nodule Analysis 2016 (LUNA16) challenge based on the LIDC-IDRI data set of 888 chest CT scans, which is the largest publicly available reference database so far (28,29). Seven systems have been applied to the complete nodule detection track and five systems have been applied to the false positive reduction track. They investigated the impact of combining individual systems on the detection performances. It was observed that the leading solutions employed CNN and used the provided set of nodule candidates. The combination of these solutions achieved an excellent sensitivity of over 95% at less than 1.0 false-positive per scan. Moreover, the LIDC-IDRI reference standard has been updated by identifying nodules that were missed in the original LIDC-IDRI annotation process.
Multi-stream multi-scale convolutional neural network
Ciompi et al. (21) proposed a deep learning system based on multi-stream multi-scale CNN, which classified nodule type referring to the PanCan model as solid, non-solid, part-solid, calcified, perifissural and spiculated type. The proposed system consisted of nine streams of CNN, grouped into three sets of three streams. Each set of streams was fed with axial, coronal and sagittal view of nodule patches respectively at the same scale and with the same resolution. Different sets of streams processed the same triplets of patches at different scales, namely 10, 20 and 40 mm. The input 2D data was then processed by a series of convolutional and pooling layers. Each multi-stream network had the same architecture and shared the parameters across the three streams, but the parameters were optimized independently at each scale. The multi-stream networks at different scales were merged in a final fully-connected layer. At last, a soft-max layer with six neurons computed the probability for the six classes. It is worth mentioning that the multi-stream multi-scale CNN processed raw CT data without any pretreatment such as nodule segmentation or judgement of nodule size. The data used to train deep learning system were derived from Multicentric Italian Lung Detection (MILD) trail. After the initial screening, the data sets consisted of 1,805 nodules from 943 subjects, and then were divided into two sets: a training set (1,352 nodules) and a validation set (453 nodules), while the latter played a supervisory role during training. In order to expand the training set, several different samples were extracted from same nodule by rotating, and the nodule center was shifted randomly within a sphere of 1 mm radius. Finally, about 500,000 training samples were generated, of which about 80,000 each type. The test data were obtained from the Danish Lung Cancer Screening Trial (DLCST), independent with training data, containing 639 nodules from 468 subjects. In order to compare the performance of deep learning system with human, Ciompi et al. selected nodules randomly (162 in total, 27 in each category) from the testing data and asked three experienced human observers to label nodule type. The comparison results showed human observers had a moderate to substantial agreement, with Cohen’s kappa coefficient k between 0.59 and 0.75. The level of computer observer agreement was increased with the number of scales. Especially, the k value between computer with 3-scale architecture and each human observer was between 0.58 and 0.67. Furthermore, nodule classification performances in terms of accuracy and F-measure per nodule type were computed for each pair of human observer and for the computer (3-scale) versus observers. The results showed that the average performance between human observers were comparable to the average performance of computer and observers, with an average accuracy of 72.9% and 69.6%. A similar trend could be observed by the F measure. In addition, they trained two linear SVM classifiers based on intensity features and unsupervised features. The DLCST test data were used to compare the performance of the proposed CNN with SVM classifiers and annotations from DLCST were considered as the reference standard. The results showed that three deep learning systems were much better than two classical machine learning approaches, with an average accuracy of 78.9% versus 33.5%. In order to get insights of nodule features learned by CNN, t-Distributed Stochastic Neighbor Embedding algorithm was applied to multidimensional scaling of nodule in the last fully-connected layer of the network. It could be observed that clearly defined clusters of nodules with similar features could be identified. Besides, the values of precision and recall per nodule (classified with 3-scale network) were computed, which showed classification of solid, calcified and non-solid nodules has good performance. The low value of precision and recall for part-solid, perifissural and spiculated nodules can be compensated in the future by adding more training samples.
Yuan et al. (22) extracted statistical features by multi-view multi-scale CNN and geometrical features by Fisher vector encodings based on scale-invariant feature transform. Then they utilized multiple kernel learning to unify the statistical and geometrical features by weight adjustment of different kernels, and classified nodule types via multi-class support vector machine.
Three-dimensional convolutional neural network
To take full advantage of three-dimensional spatial contextual information of pulmonary nodules, Dou et al. (23) proposed three-dimensional CNNs for false positive reduction of pulmonary nodule in volumetric CT scans, where the input of each of the three different three-dimensional CNN was fed with image with different receptive field so that multiple levels of contextual information surrounding the suspicious nodule could be encoded. Then the prediction probabilities from the three three-dimensional CNNs for given candidates were fused with weighted linear combination.
Liu et al. (24) developed three-dimensional CNN to classify pulmonary nodule malignancy from low-dose chest CT scans. The dataset contained 326 nodules from two large lung cancer screening studies, the National Lung Cancer Screening Trial (NLST) and Early Lung Cancer Action Program (ELCAP). What’s more, the size and type of nodules in the dataset were evenly distributed. Two three-dimensional CNN architectures were utilized. The former consisted of two three-dimensional convolutional and two fully connected layers with input image of size 16×16×16, while the later contained one more convolutional layer than the former with input image of size 32×32×32. The three-dimensional CNN was trained and tested using fivefold cross-validation and was evaluated based on the ROC curves. In addition, several ensembles of the three-dimensional CNN and traditional machine learning classifiers based on handcraft three-dimensional image features were also explored. The comparisons between the single and ensemble models demonstrated that complementary information could be learned by the three-dimensional CNN and the conventional models. Besides, both the three-dimensional CNN single model and the ensemble models with three-dimensional CNN achieved better performance than using only traditional models. In this study, although the best performance model with AUC of 0.780 was not sufficient for diagnosis, the three-dimensional CNN has great potential for improving the lung cancer screening.
DBN
Hua et al. (25) proposed DBN and CNN for classification of the malignant or benign nature of pulmonary nodules in CT image without actually computing the morphology and texture features. The DBN was established by constructing stacked restrict Boltzmann machines (RBMs) iteratively with three hidden layers and a visible layer. Each RBM was trained based on the stochastic gradient descent method. The CNN was constructed from training data with the gradient back-propagation algorithm, consisted of two convolutional layers, two pooling layers and one last fully-connected layer. In order to evaluate the performance of DBN and CNN learning framework, geometric descriptors (scale invariant feature transform and local binary pattern) with K-nearest neighbor method and fractal analysis technique with support vector machine were implemented as baselines. A total of 2,545 annotated nodules were selected from the IIDC dataset. The results showed that both DBN and CNN outperformed the feature-based methods, which confirmed deep learning techniques was effective in identifying pulmonary nodules on CT image.
Massive-training artificial neural networks
Tajbakhsh and Suzuki (26) compared two massive-training artificial neural networks (MTANNs) and four state-of-the-art CNN (AlexNet, rd-CNN, LeNet architecture, and sh-CNN) in detection of lung nodules and distinction between benign and malignant lung nodules in CT. Based on the database of LDCT consisting of 38 scans with a total of 1,057 sections, their experiments demonstrated that the performance of MTANNs was higher than that of CNN when using only limited training data. For nodule detection, MTANNs achieved 100% sensitivity at 2.7 false positives per patient.
Convolutional autoencoder neural network (CANN)
To properly train the deep learning model, a large data set of labeled data is usually required. However, due to the issues arising from protection of privacy and labor-intensive labeling tasks, it is great challenge also pressing need to set up large labeled CT dataset for assistance of deep network functioning. In this regard, Chen et al. (27) proposed a CANN to perform unsupervised features learning for lung nodule on the unlabeled data. Experimental results showed that the proposed CANN could accurately automatically extract the pulmonary nodule features, and achieve faster labeling while avoiding the intensive laboring on labeling via the hand-crafted features.
Challenges and future developments
As observed from the aforementioned literature investigate and analysis, despite deep learning techniques have emerged as promising decision supporting approaches to automatically diagnosing pulmonary nodules, including feature extraction, nodule detection, false-positive reduction, and benign-malignant classification, there are still several challenges brought about to the deep learning for diagnosing pulmonary nodules. In this section, we identify the challenges and highlight opportunities for future work.
Scarce high-quality labeled dataset—manipulation of labeled dataset
One of the crucial issues which hinder the prevailing of the deep learning in medical image analysis is the shortage of the labeled dataset. To effectively train a deep learning model in which tons of parameters need to be estimated, a large volume of labeled data is required. There are thus pressing needs for setting up large-scale dataset for the purpose of detection of pulmonary nodules, just as the ImageNet which addresses the imposing challenges arising from the natural image analysis. However, differing significantly from the ImageNet for general purpose, it needs everlasting endeavors, tremendous efforts, and sophistication in evaluation of disease patterns by the professionals to build up the dataset for medical purpose. Several data pre-processing techniques mimicked from clinical practice can be adapted and adopted by deep learning to enhance and enlarge the labeled dataset.
First, multi-scale patches containing the regions of interest could be generated from a helicopter view (i.e., zoom in and zoom out) of the medical image in axial, coronal and sagittal views of Cartesian coordinate system. Usually the clinical radiologist recognizes the disease patterns following a routine that whole pixel information is explored to detect promising regions containing suspected nodule(s), then the zones are subject to much more scrutiny to identify the presence of real nodule(s) (40). The diagnosing process then starts all over again until all nodules are detected. By observing the clinical radiologist’s diagnosing procedure, it is found that the patches containing the suspected nodule(s) are examined in multiple different scales. Thus, in the learning phase of the networks, the multi-scale patches of the medical image should be used as the training examples. The by-product benefit along with the multi-scale patches is an enlarged volume of labeled data.
Secondly, multi-angle patches containing the regions of interest could be generated in orthogonal coordinate systems rather than the widely used Cartesian coordinate system. For instance, under the spherical and cylindrical coordinate systems, rotation of the multi-scale patches by small angles could generate more labeled data. Vision of the multi-angle and multi-scale patches is in accordance with the vision diagnosing of the clinical radiologist. Meanwhile, the vision type neural network, such as deep CNN, can easily deal with such date due to its inherent properties of rotation and shift invariance on the image.
Overfitting and convergence issues—innovative deep network and learning strategy
Much progress achieved by deep learning for diagnosis of pulmonary nodules has been made using supervised learning schemes which are trained to replicate/imitate the decisions of human experts (2,41). Nevertheless, it is very expensive, time-consuming and unreliable to set up large labeled dataset through experienced radiologist. Thus, the small amount of training samples based supervised learning schemes leads to overfitting and convergence issues. We noticed that in the most challenging domains of artificial intelligence, such as in the game of Go, Alpha Zero was introduced which was solely based on reinforcement learning, without relying on human guidance or domain knowledge beyond game rules (42). And the Alpha Zero surpassed the previous AlphaGo (41). Such leaping advances in artificial intelligence and computer sciences also shed light on the artificial intelligence for medical diagnosing, particularly for pulmonary nodules diagnosis, and there are pressing needs to develop and design novel diagnosing models and algorithms which could achieve super-physician competences from an initial state of tabula rasa, through novel self-play, self-learning, self-feedback mechanisms beyond the currently prevailing supervised learning scheme.
Conclusions
This paper has given a comprehensive review of the deep learning aided decision support for pulmonary nodules diagnosing. The recent remarkable progresses in diagnosing of pulmonary nodules achieved by deep learning have been reported, which could be seen as promising and alternative decision support schemes to effectively address the feature extraction, detection, false-positive reduction, and benign-malignant classification. The deep learning techniques (e.g., two/three-dimensional, multiview, multi-stream, multi-scale, deep belief deep network, massive training network, and convolutional autoencoder network) were covered and analyzed in this survey. Although we are not quite sure that all landmark researches have been involved in this paper, we do hope that the goal has been met that we claimed to provide a research skeleton of success stories of deep learning for pulmonary nodules diagnosing. Our future work would provide a longer view about the evolving progress of the pulmonary nodules diagnosing from classical approaches to deep learning aided decision support.
Acknowledgements
The authors wish to thank the anonymous referees and Editors of this special issue for their constructive comments. The authors thank Royal Honored Prof. Michiel Keyzer (SOW-VU, Vrije Universiteit Amsterdam), Emeritus Prof. Yihui Jin, Prof. Ling Wang (Tsinghua University), Prof. Jikun Huang (Peking University) for their support.
Funding: This work was supported in part by Key Research Program of Frontier Sciences, Chinese Academy of Sciences (QYZDB-SSW-SYS020).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Schmidhuber J. Deep learning in neural networks: An overview. Neural Netw 2015;61:85-117. 10.1016/j.neunet.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 2.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 3.Greenspan H, van Ginneken B, Summers RM. Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Trans Med Imaging 2016;35:1153-9. 10.1109/TMI.2016.2553401 [DOI] [Google Scholar]
- 4.Ravì D, Wong C, Deligianni F, et al. Deep Learning for Health Informatics. IEEE J Biomed Health Inform 2017;21:4-21. 10.1109/JBHI.2016.2636665 [DOI] [PubMed] [Google Scholar]
- 5.Bibault JE, Giraud P, Burgun A. Big Data and machine learning in radiation oncology: State of the art and future prospects. Cancer Lett 2016;382:110-7. 10.1016/j.canlet.2016.05.033 [DOI] [PubMed] [Google Scholar]
- 6.Miotto R, Wang F, Wang S, et al. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform 2017. [Epub ahead of print]. 10.1093/bib/bbx044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou SK, Greenspan H, Shen D. Deep Learning for Medical Image Analysis. London: Elsevier/Academic Press, 2017. [Google Scholar]
- 8.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 9.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016;316:2402-10. 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 11.van Ginneken B. Fifty years of computer analysis in chest imaging: rule-based, machine learning, deep learning. Radiol Phys Technol 2017;10:23-32. 10.1007/s12194-017-0394-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris MA, Saboury B, Burkett B, et al. Reinventing Radiology: Big Data and the Future of Medical Imaging. J Thorac Imaging 2018;33:4-16. 10.1097/RTI.0000000000000311 [DOI] [PubMed] [Google Scholar]
- 13.Valente IRS, Cortez PC, Neto EC, et al. Automatic 3D pulmonary nodule detection in CT images: A survey. Comput Methods Programs Biomed 2016;124:91-107. 10.1016/j.cmpb.2015.10.006 [DOI] [PubMed] [Google Scholar]
- 14.Tu X, Xie M, Gao J, et al. Automatic Categorization and Scoring of Solid, Part-Solid and Non-Solid Pulmonary Nodules in CT Images with Convolutional Neural Network. Sci Rep 2017;7:8533. 10.1038/s41598-017-08040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teramoto A, Fujita H, Yamamuro O, et al. Automated detection of pulmonary nodules in PET/CT images: Ensemble false-positive reduction using a convolutional neural network technique. Med Phys 2016;43:2821-7. 10.1118/1.4948498 [DOI] [PubMed] [Google Scholar]
- 16.Anthimopoulos M, Christodoulidis S, Ebner L, et al. Lung Pattern Classification for Interstitial Lung Diseases Using a Deep Convolutional Neural Network. IEEE Trans Med Imaging 2016;35:1207-16. 10.1109/TMI.2016.2535865 [DOI] [PubMed] [Google Scholar]
- 17.Xie Y, Zhang J, Xia Y, et al. Fusing texture, shape and deep model-learned information at decision level for automated classification of lung nodules on chest CT. Information Fusion 2018:102-10. 10.1016/j.inffus.2017.10.005 [DOI] [Google Scholar]
- 18.Shen W, Zhou M, Yang F, et al. Multi-crop Convolutional Neural Networks for lung nodule malignancy suspiciousness classification. Pattern Recognition 2017;61:663-73. 10.1016/j.patcog.2016.05.029 [DOI] [Google Scholar]
- 19.Setio AA, Ciompi F, Litjens G, et al. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans Med Imaging 2016;35:1160-9. 10.1109/TMI.2016.2536809 [DOI] [PubMed] [Google Scholar]
- 20.Sun W, Zheng B, Qian W. Automatic feature learning using multichannel ROI based on deep structured algorithms for computerized lung cancer diagnosis. Comput Biol Med 2017;89:530-9. 10.1016/j.compbiomed.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 21.Ciompi F, Chung K, van Riel SJ, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep 2017;7:46479. 10.1038/srep46479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan J, Liu X, Hou F, et al. Hybrid-feature-guided lung nodule type classification on CT images. Computers & Graphics 2018;70:288-99.</jrn> 10.1016/j.cag.2017.07.020 [DOI] [Google Scholar]
- 23.Dou Q, Chen H, Yu LQ, et al. Multilevel Contextual 3-D CNNs for False Positive Reduction in Pulmonary Nodule Detection. IEEE Trans Biomed Eng 2017;64:1558-67. 10.1109/TBME.2016.2613502 [DOI] [PubMed] [Google Scholar]
- 24.Liu S, Xie Y, Jirapatnakul A, et al. Pulmonary nodule classification in lung cancer screening with three-dimensional convolutional neural networks. J Med Imaging (Bellingham) 2017;4:041308. 10.1117/1.JMI.4.4.041308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua KL, Hsu CH, Hidayati SC, et al. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Ther 2015;8:2015-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tajbakhsh N, Suzuki K. Comparing two classes of end-to-end machine-learning models in lung nodule detection and classification: MTANNs vs. CNNs. Pattern Recognition 2017;63:476-86. 10.1016/j.patcog.2016.09.029 [DOI] [Google Scholar]
- 27.Chen M, Shi X, Zhang Y, et al. Deep Features Learning for Medical Image Analysis with Convolutional Autoencoder Neural Network. IEEE Transactions on Big Data 2017;PP:1.
- 28.Armato S, McLennan G, McNitt-Gray M, et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A Completed Public Database of CT Scans for Lung Nodule Analysis. Med Phys 2010;37:3416-7. 10.1118/1.3469350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armato SG, McLennan G, Bidaut L, et al. The Lung Image Database Consortium , (LIDC) and Image Database Resource Initiative (IDRI): A Completed Reference Database of Lung Nodules on CT Scans. Med Phys 2011;38:915-31. 10.1118/1.3528204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King TE. Clinical advances in the diagnosis and therapy of the interstitial lung diseases. Am J Respir Crit Care Med 2005;172:268-79. 10.1164/rccm.200503-483OE [DOI] [PubMed] [Google Scholar]
- 31.Demedts M, Costabel U. ATS/ERS international multidisciplinary consensus classification of the idiopathic interstitial pneumonias. Eur Respir J 2002;19:794-6. 10.1183/09031936.02.00492002 [DOI] [PubMed] [Google Scholar]
- 32.Gangeh MJ, Sorensen L, Shaker SB, et al. A Texton-Based Approach for the Classification of Lung Parenchyma in CT Images. Med Image Comput Comput Assist Interv 2010;13:595-602. [DOI] [PubMed] [Google Scholar]
- 33.Sørensen L, Shaker SB, de Bruijne M. Quantitative Analysis of Pulmonary Emphysema Using Local Binary Patterns. IEEE Trans Med Imaging 2010;29:559-69. 10.1109/TMI.2009.2038575 [DOI] [PubMed] [Google Scholar]
- 34.Anthimopoulos M, Christodoulidis S, Christe A, et al. Classification of Interstitial Lung Disease Patterns Using Local DCT Features and Random Forest. Conf Proc IEEE Eng Med Biol Soc 2014:6040-3. [DOI] [PubMed] [Google Scholar]
- 35.Lecun Y, Bottou L, Bengio Y, et al. Gradient-based learning applied to document recognition. Proceedings of the IEEE 1998;86:2278-324. 10.1109/5.726791 [DOI] [Google Scholar]
- 36.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Advances in neural information processing systems, 2012. [Google Scholar]
- 37.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. International Conference on Learning Representations 2015. [Google Scholar]
- 38.Hinton GE, Osindero S, Teh YW. A Fast Learning Algorithm for Deep Belief Nets. Neural Comput 2006;18:1527-54. 10.1162/neco.2006.18.7.1527 [DOI] [PubMed] [Google Scholar]
- 39.Setio AA, Traverso A, de Bel T, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med Image Anal 2017;42:1-13. 10.1016/j.media.2017.06.015 [DOI] [PubMed] [Google Scholar]
- 40.Lo SC, Lin JS, Freedman MT, et al. Computer-assisted diagnosis of lung nodule detection using artificial convolution neural-network. Proc SPIE Med Imag, Image Process 1993:859-69. [Google Scholar]
- 41.Silver D, Huang A, Maddison CJ, et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016;529:484-9. 10.1038/nature16961 [DOI] [PubMed] [Google Scholar]
- 42.Silver D, Schrittwieser J, Simonyan K, et al. Mastering the game of Go without human knowledge. Nature 2017;550:354-9. 10.1038/nature24270 [DOI] [PubMed] [Google Scholar]


