Abstract
Molecularly targeted therapies and immune checkpoint inhibitors have markedly improved the therapeutic management of advanced lung cancer. However, it still remains the leading cause of cancer-related mortality worldwide, with disease stage at diagnosis representing the main prognostic factor. Detection of lung cancer at an earlier stage of disease, potentially susceptible of curative resection, can be critical to improve patients survival. Low-dose computed tomography (LDCT) screening of high-risk patients has been demonstrated to reduce mortality from lung cancer, but can be also associated with high false-positive rate, thus often resulting in unnecessary interventions for patients. Novel sensitive and specific biomarkers for identification of high-risk subjects and early detection that can be used alternatively and/or complement current routine diagnostic procedures are needed. Liquid biopsy has recently demonstrated its clinical usefulness in advanced NSCLC as a surrogate of tissue biopsy for noninvasive assessment of specific genomic alterations, thereby providing prognostic and predictive information. Different biosources from liquid biopsy, including cell free circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), exosomes and tumor-educated platelets (TEPs), have also been widely investigated for their potential role in lung cancer diagnosis. This review will provide an overview on the circulating biomarkers being evaluated for lung cancer detection, mainly focusing on results from most recent studies, the techniques developed to perform their assessment in blood and other biologic fluids and challenges in their clinical applications.
Keywords: Early-stage non-small cell lung cancer (Early-stage NSCLC), liquid biopsy, circulating cell-free tumor DNA
Introduction
Lung cancer is the most common cause of cancer-related deaths for both sexes (1,2). About 85% of lung cancers are non-small cell lung cancer (NSCLC), which comprises, mainly, adenocarcinoma, squamous cell carcinoma, and large-cell lung cancer. The high mortality is mainly attributable to its aggressiveness and to the fact that most lung tumors are generally detected at advanced, inoperable stages of disease. Only a minority of NSCLC patients are diagnosed with localized, early-stage disease and, for these patients, the optimal treatment remains surgical resection with curative intent (3). However, despite optimal surgical management, the probability of survival at 5 years remains unsatisfactory and is closely related to the stage of disease, ranging from about 90%, for patients with stage IA, to 60%, for those in stage II (4,5). At least a third of patients who undergo complete surgical resection will experience recurrent disease within 5 years, mainly systemic relapse, suggesting that early-stage NSCLC is frequently metastatic at diagnosis. Still, early detection of lung cancer emerges as the only valuable approach to diagnose the disease at an asymptomatic and potentially curable stage, in order to improve patient outcome. Early detection could be pursued with different approaches, such as the use of diagnostic imaging techniques, including low-dose computed tomography (LDCT), in high-risk individuals.
Among subjects enrolled in the National Lung Screening Trial (NLST), LDCT detected more than twice the number of early-stage lung cancers and was associated with a significant reduction in lung cancer-specific mortality compared to chest radiography (6). Also, all-cause mortality was reduced by 6.7%, indicating that screening resulted in no deleterious downstream effects that contributed to death. Despite its high sensitivity, LDCT screening has a relatively low specificity, which resulted in a high false-positive rate of 96.4%, with an overall 18.5% probability of overdiagnosis (7). Other European trials showed less significant results compared to the NLST trial in terms of reduced mortality, thus raising some concerns regarding LDCT implementation as a general screening approach in high-risk populations (8-10). Other screening trials and pooled analyses of similar European randomized trials could probably provide more data about the role of LDCT in reducing mortality (11). However, one major concern about the use of this radiological screening tool, beyond the high-false positive rate and the potential for overdiagnosis, is radiation exposure (12). There is a need for more specific, less invasive biomarkers that could be used alternatively or complementary to radiological approaches to better define the optimal risk cohort and improve cost-effectiveness of current radiological screening. In this context, the assessment of circulating biomarkers can represent an attractive strategy for lung cancer screening and early diagnosis.
In this review we will discuss the role of liquid biopsy to assess potential biomarkers for lung cancer screening and early detection.
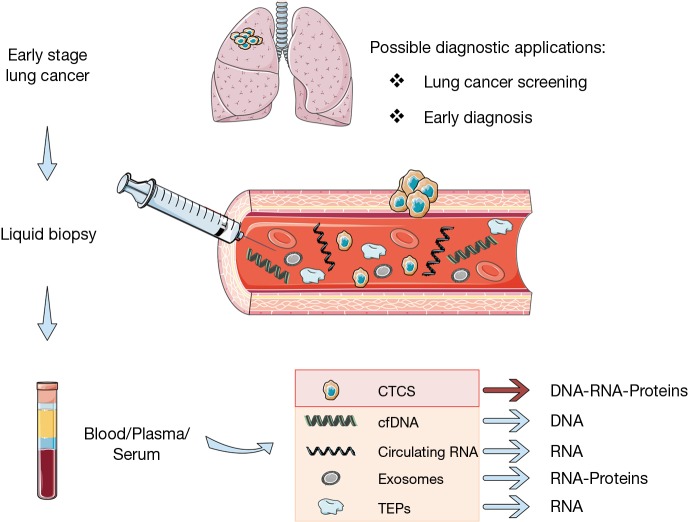
Clinical applications of liquid biopsy
Molecular profiling of tumor specimens at diagnosis currently represents the standard of care to guide treatment decisions in advanced NSCLC patients (13,14). However, tumor tissue biopsy is an invasive procedure with several limitations in clinical practice: it often provides insufficient materials, it represents a single snapshot of the tumor and is subject to selection bias resulting from tumor heterogeneity. Moreover, tumor tissue specimens cannot be easily obtained repeatedly to track tumor dynamics over time, because most diagnostic procedures are invasive and not acceptable for the patients. Liquid biopsy has widely demonstrated to be a viable surrogate for tumor tissue for noninvasive assessment of tumor specific biomarkers and can be potentially used for a variety of clinical and investigational applications (15). Indeed, different tumor-derived components can be isolated from blood, including circulating cell-free tumor DNA (cfDNA), cell-free tumor RNA (cfRNA), exosomes, tumor-educated platelets (TEP), and circulating tumor cells (CTCs) (Figure 1), and can be leveraged to uncover the molecular landscape of the tumor and capture the heterogeneity of metastatic cancers (15,16). Different body fluids (other than blood) can be a source of tumor-derived molecular information, such as urine, cerebrospinal fluid (CSF), saliva, and pleural effusions (17). Many studies have illustrated the potential of liquid biopsy approaches to predict and monitor treatment responses, to quantify minimal residual disease and assess the emergence of therapy resistance, mainly due to the feasibility to collect and analyze liquid biopsy specimens at various points before and during treatment, thus offering a dynamic portrait of tumor genomic changes over the course of disease. The best-known example of clinical application of liquid biopsy is the use of cfDNA genotyping for EGFR sensitizing- or T790M-mutations as surrogate of tumor tissue profiling to predict response and monitor development of resistance to EGFR TKI therapy (15,16,18). Beyond predictive and prognostic information, liquid biopsy can be exploited for diagnostic purposes, such as screening and early detection of lung cancer (Figure 1).
Figure 1.
Tumor-derived components that can be used as liquid biopsy for genetic testing.
Circulating cell-free tumor DNA
Circulating cell-free DNA derived from tumors is likely to represent the entire genomic landscape of the tumor and can serve as a liquid biopsy to analyze tumor-specific genetic and epigenetic alterations. In a seminal study, it was discovered that cancer patients had much higher levels of cell-free DNA in their blood compared to healthy individuals (19). In addition, higher cfDNA levels were found in patients with metastatic disease compared to non-metastatic cancer patients (19). Several years later, it was proven that plasma DNA found in patients with malignant diseases originates from tumor (also known as ctDNA) (20) and discrimination of ctDNA from cfDNA deriving from normal cells is possible because of the presence in ctDNA of specific genomic alterations, including mutations in oncogenes or tumor suppressor genes, gene amplifications or epigenetic changes, which are typically found in the cancer cell genome (15,16). For consistency in our review we will refer to cfDNA, considering that this is the pool from which ctDNA is derived. Although the mechanisms by which tumor DNA is released into the bloodstream has not been fully elucidated, it is thought to originate mainly from apoptotic and necrotic cells and, as the tumor grows and increases in volume, so do the number of necrotic and apoptotic cells as a consequence of cellular turnover (15,16,21-24).
An important role for macrophages in the release of tumor DNA fragments in the circulation via phagocytosis of necrotic neoplastic cells has been proposed (22,25). The fraction of ctDNA can vary, with some studies demonstrating very low concentrations of approximately 0.01–1% of cfDNA for early-stage disease, thus making quantification and analysis in blood very challenging. Novel, highly sensitive blood-based assays have been developed to test cfDNA at very low concentrations for most genomic abnormalities. These include polymerase chain reaction (PCR)-based techniques, including digital PCR (dPCR), droplet digital PCR (ddPCR), peptide nucleid acid (PNA) clamp-based PCR assay (Taqman assay), beads, emulsions, amplification and magnetics (BEAMing), pyrophosphorolysis-activated polymerization and next-generation sequencing (NGS) technologies (26). Different platforms have already been approved for clinical use (18,26). However, several difficulties remain, limiting the clinical implementation for most of these methodologies, particularly the lack of standardization.
After the initial report (19), other studies have shown higher cfDNA levels in cancer patients and also a correlation of these levels with disease burden or tumor stage, suggesting cfDNA may potentially have diagnostic and prognostic value. The concentration of plasma DNA, determined by real time quantitative PCR (qPCR) of the human telomerase reverse transcription gene (hTERT), was almost eight-times higher in patients with lung cancer than in matched controls from smoker patients and emerged as a strong risk factor for lung cancer development. This study suggested that plasma DNA assessment, by using this highly sensitive and specific real time qPCR assay, could represent a novel, noninvasive approach for identifying higher-risk individuals for lung cancer screening (17). In 35 cancer patients, a second plasma sample was collected, 3 to 15 months after surgery, and analyzed for plasma DNA concentration. Interestingly, median DNA concentration at follow-up was significantly lower in disease-free individuals as compared with the five patients with proven cancer relapse, suggesting a potential role of this liquid biopsy also in predicting recurrences, thus as a prognostic marker. The diagnostic and prognostic role of plasma DNA was assessed in another study from the same group in the setting of a lung cancer screening trial. Plasma DNA levels were assessed in a cohort of 1,035 heavy smokers at baseline and at time of lung cancer diagnosis. Although measurement of plasma DNA levels was not shown to improve the accuracy of lung cancer screening by spiral CT, a higher amount of plasma DNA at surgery was significantly associated with a poorer prognosis at 5-year survival, thus indicating the presence of a more aggressive disease (27).
Some subsequent studies demonstrated the reproducibility of the quantitative PCR assay used by Sozzi et al. and its ability in discriminating between healthy subjects and NSCLC patients (28,29). In another study by Gautschi et al., serum and plasma DNA concentrations, as assessed by real-time PCR with a standard protocol set up and validated at two oncology units, were significantly higher in NSCLC patients than in healthy controls, and increased plasma DNA concentrations were correlated with advanced tumor stage (P<0.003). Elevated serum and plasma DNA concentrations emerged as independent prognostic factors for survival.
The above data underline that quantitative analysis of circulating DNA by PCR-based assays could represent a potential tool to identify high-risk individuals; however, baseline assessment of plasma DNA level alone did not seem to improve the accuracy of lung cancer screening by spiral CT in heavy smokers included in a large study (27), suggesting cfDNA quantification could have a role in supplementing current screening and diagnostic tests, including chest CT scans and cytological/histological examination. Beyond quantification of circulating free DNA in patients, detection of genomic abnormalities, including specific mutations, in cfDNA could offer a more promising, noninvasive approach for early lung cancer detection, considering the high specificity of these tumor-related alterations. Due to the low frequency of some mutations and the fact that mutation-harboring cfDNA can be obscured by a relative excess of background wild-type DNA from normal cells, highly sensitive approaches, beyond mutation specific PCR assays, have been developed to analyze low-level mutant cfDNA, including NGS-based options.
In a recent study, by using the Cancer Personalized Profiling by deep Sequencing (CAPP-Seq), which is a highly sensitive method able to quantify cfDNA and detect multiple classes of somatic mutations, Newman and colleagues detected cfDNA in 100% of NSCLC patients with stages II–IV and in 50% of patients with stage I disease, thus suggesting its application also for early detection (30). In this study, cfDNA levels significantly correlated with tumor volume, were able to monitor treatment response and residual disease and detect disease progression earlier than radiological approaches. However, the role of cfDNA genotyping for somatic mutations in a screening setting to detect pre-clinical cancers, without prior knowledge from tumor tissue of the expected mutation, remains unknown. Moreover, biomarkers used for screening purposes in asymptomatic populations should be selected for their high specificity in distinguishing between individuals, with and without cancer. Recently, results from a study assessing the presence of TP53 mutations in the plasma cfDNA from SCLC cases and non-cancer controls, TP53 mutations were detected in 11% of the 225 non-cancer controls, thus suggesting they are not specific of cancer patients and pose some difficulties for the development of cfDNA screening tests (31).
Beyond somatic mutations, other biomarkers, including epigenetic changes, can be assessed in cfDNA for diagnostic purposes.
Gene methylation in cell-free tumor DNA
Epigenetic modifications, including methylation of the CpG islands in gene promoter regions, closely regulate expression of a large number of genes involved in malignant transformation, mainly tumor suppressor genes, and are commonly observed in tumors, including lung cancers. Hypermethylation has been shown to contribute to carcinogenesis and one of the main advantages of DNA methylation alterations, compared to other potential diagnostic biomarkers, is that they are remarkably stable, generally occur early during carcinogenesis and can be found in circulating DNA fragments from different body fluids, including blood and sputum (32-36). Methylation of circulating DNA can be assessed using different techniques, such as methylation-specific PCR (MSP), real-time quantitative MSP, multiplex-nested methylation-specific PCR and methyl-BEAMing. Although epigenetic alterations are not unique for any one tumor, some tumor suppressor genes are frequently methylated and downregulated in specific tumors, suggesting that noninvasive assessment of these candidate genes could be potentially useful for diagnostic and prognostic purposes (15,36).
There are multiple reports evaluating methylation of single or panels of tumor suppressor genes in plasma or serum of lung cancer patients as potential biomarkers. Methylation in specific tumor suppressor genes, including MGMT, p16, RASSF1A, DAPK and RAR-β, was more frequently observed in patients diagnosed with lung cancer than those with nonmalignant diseases, such as individuals undergoing bronchoscopy for abnormal findings in chest radiographs. Patients with methylation in at least one gene, and in ≥2 genes showed 5.28 (2.39–11.7; P<0.001) and 5.89 (1.53–22.7; P=0.010) times higher probability of having lung cancer, respectively, compared with those without any methylated genes. Notably, 16 (89%) of 18 tissue samples from patients with serum DNA methylation also had methylated genes (37).
Another study by Chinese investigators assessed the methylation profiles of NSCLC by MSP in tissues and plasma samples. Nine genes (APC, CDH13, KLK10, DLEC1, RASSF1A, EFEMP1, SFRP1, RAR-β and p16INK4A) demonstrated a significantly higher methylation frequency in NSCLC compared with the normal tissues (P≤0.001). A high concordance of the methylation status in matched tumor tissues and plasma samples was observed. A five-gene set (APC, RASSF1A, CDH13, KLK10 and DLEC1) achieved a sensitivity of 83.64% and a specificity of 74.0% for cancer diagnosis (38). The potential role of the methylation status in blood of retinoic acid receptor B2 (RARB2) and RAS association domain family 1 isoform A (RASSF1A) tumor suppressor genes for lung cancer diagnosis has been further demonstrated (39). These genes have been found frequently hypermethylated in lung tumors and correlated with poor prognosis.
The presence in plasma of methylated SHOX2 gene, a member of the homeobox family of genes that encode DNA binding transcription factors, has emerged as a sensitive and specific biomarker for lung cancer in different reports (40,41). Notably, SHOX2 gene hypermethylation was specifically associated with SCLC and squamous cell carcinoma and was shown to be frequently accompanied by gene amplification (40). Other genes have been found to be differentially methylated in cfDNA between patients with lung cancer and controls, including doublecortin like kinase 1 (DCLK1) (42) and septin9 (SEPT9) (43).
Most of the above studies were performed by selecting tumor-specific candidate genes to assess methylation in blood by different sensitive techniques, based on the recognition that specificity of methylated DNA as a biomarker can be lower compared with the presence of genomic alterations. This is because methylation changes can also occur in DNA of surrounding normal tissue and can overlap with methylated DNA from tumors (15). Indeed, in recent years, different epigenetic candidates in blood have been proposed, but none has been introduced in clinic yet, mainly due to the lack of standardized methods and large validation studies. Nowadays, high-throughput epigenomic studies could probably offer a deeper characterization of epigenetic variations in cancer with the potential of identifying more robust biomarkers to be assessed in circulating cell-free tumor DNA in blood or other biological samples from lung cancer patients, including sputum or bronchial fluid aspirates (44,45).
DNA methylation of the SHOX2 gene locus was reported to be a sensitive and specific biomarker based on the analysis of bronchial fluids (46). In this case-control study, the relative abundance of methylated SHOX2 gene copies, assessed by using the real-time PCR based HeavyMethyl technology, in bronchial aspirates obtained during bronchoscopy were found to significantly correlate with the presence of lung cancer. Indeed, SHOX2 methylation allowed to distinguish between malignant and benign lung diseases with a sensitivity of 68% and a high specificity of 95% (46). The CE-marked in vitro diagnostic (CE IVD) test ‘Epi proLung BL Reflex Assay’ (Epigenomics) detects the relative amount of methylated SHOX2 gene fragments in a background of normal DNA in a real-time PCR assay based on TaqMan technology from bronchial fluid specimens. This test has been successfully validated as a sensitive and reliable lung cancer diagnostic tool (46,47). The test may be used as an aid in the diagnosis of a malignant lung disease alongside other diagnostic procedures to allow confirmation of lung cancer based on analysis of bronchial lavage samples, even when cytology results are negative or suspect. In a recent, appealing study, two genome-wide DNA methylation datasets including stage I NSCLC were used to select a specific epigenetic signature whose diagnostic value was then validated by pyrosequencing into 4 independent cohorts, including formalin-fixed paraffin-embedded (FFPE) tissues, bronchial aspirates/lavages and sputum samples from lung cancer patients and cancer-free individuals (44). Pyrosequencing, a targeted-region validation technique, was chosen because it represents a robust and quantitative method able to detect multiple CpGs not only in FFPE tissues but also in minimally and noninvasive samples as biologic fluids with potential use in daily basis clinical settings. The signature, consisting of top four selected genes: CDO1 (cysteine dioxygenase type 1), BCAT1 (branched chain amino-acid transaminase 1), TRIM58 (tripartite motif containing 58) and ZNF177 (zinc finger transcription factor 177), yielded a high specificity and sensitivity for early lung cancer detection in minimally and noninvasive samples (44), thus suggesting it as a potentially useful tool to refine the risk categorization within current diagnostic protocols, in order to improve lung cancer diagnosis and, in turn, patient outcomes.
CTCs
CTCs originate by cell detachment from the primary tumor mass into circulation. Indeed, CTCs are considered to contribute to cancer progression and development of metastases. CTCs have been isolated in blood of patients with different solid tumors, including lung cancer, at variable concentrations depending on tumor types and stage of diseases. Various studies have demonstrated a potential role of CTCs as prognostic biomarkers or in predicting and monitoring response to different treatments, including targeted therapies (16,48-51). Compared to ctDNA, CTCs allow comprehensive studies at the DNA, RNA, and protein level as well as functional studies including the establishment of cell lines and xenografts (16,52). However, tumor cells in circulation show substantial apoptosis and fragility, thus requiring more complex laboratory procedures for isolation from other blood cells, enumeration and characterization (53). Multiple techniques have been developed and present great variability in CTC detection rates, sensitivity, and specificity (16,49,54).
To date, the only U.S. Food and Drug Administration (FDA) approved technology for CTCs detection and quantification with prognostic purposes in metastatic breast, colorectal, or prostate cancer is the CellSearchTM (Veridex LLC), that utilizes magnetic beads coated with anti-epithelial cell-adhesion molecule (EpCAM) antibodies to isolate CTCs (55-58). Another antibody-mediated capture technology has been developed, the “CTC-chip”, which has shown superior detection rates of CTCs of approximately 100% in metastatic tumors, including lung cancer. CTC-chip is a microfluidic-based platform capable of separating viable CTCs from peripheral blood samples on the basis of the interaction of target CTCs with EpCAM-coated microposts under laminar flow conditions, and without requisite pre-labeling or processing of samples. The CTC-chip successfully identified CTCs in the peripheral blood of patients with solid tumors with approximately 50% purity (59). Despite its usefulness, EpCAM-based methods can have some drawbacks for CTCs detection and capture in epithelial tumors, and can be thus limited in their performance by the intrinsic variability of these cells. Indeed, isolation of CTCs in NSCLC was lower compared to other tumors and a proportion of NSCLC CTCs can be missed because undergo epithelial to mesenchymal transition (EMT) and down-regulate their epithelial markers during progression (55,60-62). In addition to CellSearch, other label-free approaches to isolate CTCs which do not rely on the expression of specific cell surface markers but instead on other inherent properties, such as size (i.e., ISET; RareCell Diagnostics, Paris, France), deformability, or dielectric susceptibility, and/or negative selection of WBCs have been developed and tested in solid tumors (61-64). In lung cancers, different techniques have been used to detect and characterize CTCs and evaluate their role as predictive and/or prognostic biomarkers (51,65-76). In addition, different subtypes of CTCs with distinct molecular profiles have been detected in lung cancer (60), thus offering a useful source of material for genetic testing, including evaluation of EGFR or KRAS mutations, or MET expression, thereby potentially being valuable, noninvasive biomarkers to predict response and track the emergence of resistance to targeted therapies (60,77-80). As demonstrated by recent data, the identification of CTCs may provide not only important prognostic, but also diagnostic information. Ilie et al. showed CTCs could be detected in patients with chronic obstructive pulmonary disease (COPD) without clinically detectable lung cancer (73). The study included 168 (68.6%) patients with COPD and 77 subjects without COPD (31.4%), including 42 control smokers and 35 nonsmoking healthy individuals. Patients with COPD were monitored annually by low-dose spiral CT. CTCs were detected by ISET in 3% of patients with COPD (5 of 168 patients). The annual surveillance of the CTC-positive COPD patients by CT-scan screening detected lung nodules 1 to 4 years after CTC detection, leading to prompt surgical resection and histopathologic diagnosis of early-stage lung cancer, whereas, no CTCs were detected in the control smoking and nonsmoking healthy individuals (73). The study suggests that CTCs can be predictive of lung cancer onset several months before objective detection.
Another study showed that Folate Receptor (FR)-positive CTCs were feasible diagnostic biomarkers in patients with NSCLC, including early-stage tumors. In fact, with a threshold of 8.64 CTC units, the method showed a sensitivity of 73.2% and a specificity of 84.1% for the diagnosis of NSCLC, with a sensitivity of 67.2% in stage I disease. Compared with the existing clinical biomarkers, such as neuron-specific enolase (NSE), carcinoembryonic antigen (CEA), cancer antigen 125 (CA125), cyfra21-1, and squamous cell carcinoma antigen (SCC Ag), the method showed the highest diagnostic efficiency (area under the curve, 0.823; 95% confidence interval, 0.773–0.874) (81).
In another work, the ISET technique was used to detect CTCs in asymptomatic patients with stage I NSCLC before surgery. Fiorelli and coll., demonstrated that malignant circulating cells isolated by ISET from the peripheral blood of 77 patients with malignant and benign lung lesions (60 and 17 patients, respectively), may be a valid biomarker in the diagnostic workup of lung cancer (74).
Although there is some positive evidence, the value of CTCs as reliable diagnostic biomarkers in the clinic in NSCLC has not yet been established due to the lack of standardization of current methods, and variability in pre-analytical and analytical processes. The use of novel, more sensitive assays can further improve detection of lung cancer cells in early stages (76).
microRNAs
miRNAs are short non-coding RNA sequences that regulate gene expression by binding to specific target RNA messenger sequences (mRNAs) and are frequently dysregulated in cancer. miRNAs are the most abundant cfRNA molecules in the blood, and can be carried in exosomes, apoptotic bodies, protein–miRNA complexes, and tumour-educated platelets (TEP) (16). A single miRNA can target hundreds of mRNAs, thereby regulating the expression of a large number of genes involved in crucial cancer processes, including proliferation, cell migration, and apoptosis. The landscape of miRNAs in blood seems to correlate with that of the solid tumours from which they originate (16,82,83), therefore different cancer types have distinct miRNA expression patterns. The stability and substantial tissue-specificity of miRNAs implicate they serve as valuable, potential non-invasive biomarkers for the detection of various cancers, including lung cancer.
Various studies have investigated the role of differential circulating miRNA expression when discriminating between NSCLC patients and healthy controls. Compared to normal subjects, the expression profile of miRNAs in lung cancer serum was significantly different, with identification of 63 new miRNAs, including miR-122, miR-221, by Solexa technology (83).
In the study by Foss et al., miRNA profiling was performed on total RNA extracted from serum of early-stage NSCLC and controls by using the GenoExplorer microRNA Expression System (GenoSensor Corporation, Tempe, AZ, USA) (84). Quantitative polymerase chain reaction was used to validate the profiling results in the discovery set and in an additional validation set of NSCLC patients and controls (individuals with a history of smoking tobacco). Two miRNAs, miR-1254 and miR-574-5p, were significantly increased in early-stage NSCLC samples compared to the controls, with 82% and 77% of sensitivity and specificity, respectively, in the discovery cohort and 73% and 71% of sensitivity and specificity, in the validation cohort. Despite the small sample size of patients and controls analyzed, this study suggests a potential role of these two miRNAs for noninvasive lung cancer screening (84).
Heegaard et al. analyzed and compared the expression levels of 30 selected miRNAs in a larger sample size of early-stage NSCLC patients and controls in serum and plasma by using qRT-PCR. The study reported an increased serum expression of miR-29c in NSCLC patients, while a set of 7 miRNAs (miR-146b, miR-221, let-7a, miR-155, miR-17-5p, miR-27a and miR-106a) was significantly reduced compared with case-controls. Notably, overall expression levels in serum did not correlate well with levels in plasma in which no significant differences of miRNAs levels were observed between patients compared to controls (85), suggesting these two sources may not be comparable regarding miRNA expression.
Shen et al. demonstrated the diagnostic value of a set of plasma miRNAs, miRNA-21, miRNA-126, miRNA-210, and miRNA-486-5p, evaluated by qRT-PCR assay and whose altered expression had been previously validated in lung tumor tissues, in distinguishing NSCLC from healthy controls, with 86.2% sensitivity and 96.55% specificity (86). Notably, these miRNAs have higher sensitivity (91.67%) in diagnosis of lung adenocarcinomas compared with squamous cell carcinomas (82.35%) (P<0.05), even if the number of sample size to perform this analysis was small (87). In another study from the same group, plasma expression levels of miR-21, miR-210 and miR-486-5p demonstrated a 76% of sensitivity and 85% specificity to discriminate patients with CT-detected malignant solitary pulmonary nodules from benign lesions (88).
A high rate of sensitivity (81.33%) and specificity (86.76%) was demonstrated for the combination of a panel consisting of plasma miR-155, miR-197 and miR-182, assessed by real-time RT-PCR, which was able to discriminate lung cancer from controls in a cohort of 74 lung cancer patients, including those with stage I of disease, and 68 cancer-free subjects (89).
Boeri and colleagues reported the identification and validation of plasma-based miRNA signatures from patients in two independent LDCT screening studies (INT/IEO and MILD trials) who provided plasma samples before and at the time of disease detection (90). High-throughput miRNA expression profiles of plasma samples were first assessed by using TaqMan microfluidic cards and then selected miRNAs analyzed by quantitative measurement by RT-PCR. Interestingly, miRNAs that were found deregulated in tissue specimens were rarely detected in plasma samples, underlying the high tissue-specificity of miRNAs and suggesting a potential predictive role of plasma miRNAs independent from tissue specimens. Different altered circulating miRNAs involved in crucial cellular processes such as aging (mir-19b, mir-17, mir-106), bronchioalveolar and hematopoietic stem cells renewal (mir-486, mir-106a, 142-3p), tumor recurrence in stage I NSCLC (mir-27b; mir-106a; mir-19b; mir-15b mir-16, mi-21) and lung cancer aggressiveness (mir-221) defined specific signatures which were able to categorize patients into different risk groups and had strong diagnostic and prognostic value. The authors stated that identification of miRNA signatures in plasma samples, collected 1–2 years before disease, predict cancer development and prognosis, suggesting their potential usefulness in the selection of high-risk individuals who need to undergo spiral-CT surveillance (90). In a subsequent large validation study from the same group, including participants of the MILD study of LDCT versus observation, this prespecified miRNA signature classifier (MSC) formed by 24 miRNAs assessed by a quantitative RT-PCR-based assay, demonstrated significant diagnostic performance for detecting the presence of malignant disease, with 87% sensitivity and 81% specificity across both arms. The signature was able to distinguish lung cancers from the large majority of benign LDCT-detected pulmonary nodules. Moreover, combination of MSC with LDCT reduced consistently the false-positive rate of the LDCT alone, thus suggesting a synergistic approach in which MSCs could complement LDCT screening and improve its effectiveness for lung cancer screening by avoiding further exams in a large proportion of individuals and unnecessary invasive diagnostic follow-up (91). The MSC had also a significant prognostic value, with risk groups significantly associated with survival (91). In 2013 the BioMILD trial started. The investigators propose a large prospective study in heavy smokers volunteers based on plasma miRNA profiling to assess its efficacy as a first line screening test for lung cancer detection. This study will enroll 4,000 volunteers older than 50 years. Volunteers will undergo LDCT and blood withdrawal, in order to include them in a program of active surveillance on the basis of their miRNA risk profile. Assessment of miRNA expression profile will use a custom-made microfluidic card containing the 24 miRNAs previously identified in the diagnostic signatures. On the basis of results of the MILD trial, the screening algorithm is decided according to the combination of LDCT and MSC (92).
In a multicenter study, Wang et al., recruited 221 NSCLC patients, 161 healthy controls and 56 subjects with benign nodules from China and America (93). Initial miRNA screening was performed using the TaqMan Low Density Array (TLDA) followed by confirmation with RT-qPCR. Results from this study showed that a panel of 5 miRNAs, including miR-483-5p, miR-193a-3p, miR-25, miR-214 and miR-7, were significantly elevated in NSCLC patients compared to controls included in the Chinese cohorts. In these cohorts, the panel showed a remarkable accuracy of diagnosis. In a blind trial, the diagnostic value of the panel was tested and results demonstrated it was able to correctly classify 95% NSCLC cases and 84% controls. Most importantly, the panel was capable of distinguishing lung cancer from benign nodules and allowed correct prediction of 86% and 95% stage I–II tumors in the Chinese and American cohorts, respectively, suggesting its function as a reliable diagnostic indicator of NSCLC in patients of different ethnicities (93). In another study, miRNA profiles were determined by using a real-time RT-qPCR (Taqman) and it was found that three miRNAs (miR-125a-5p, miR-25 and miR-126) were significantly decreased in the serum of early stage lung cancer patients compared to healthy control subjects. The combination of these 3 miRNAs could discriminate patients with lung cancer with 87.5% sensitivity and 87.5% specificity. Furthermore, the 3 miRNAs could distinguish different stages of lung cancer patients from healthy controls, thus supporting the panel had the potential for the early detection of lung cancer (94).
Montani et al., designed a multi-tiered study (95) to validate a blood test based on serum miRNAs (96) in high-risk individuals (heavy smokers, older than 50 years) enrolled in the Continuous Observation of Smoking Subjects (COSMOS) lung cancer screening trial (86) and lung cancer patients diagnosed outside of the screening. They initially refined a previous serum 34-miRNA signature using a “Calibration set” of 24 subjects, leading to reduction of the signature to 13 miRNAs (the “miRTest”: miR-92a-3p, miR-30b-5p, miR-191-5p, miR-484, miR-328-3p, miR-30c-5p, miR-374a-5p, let-7d-5p, miR-331-3p, miR-29a-3p, miR-148a-3p, miR-223-3p, miR-140-5p), which maintained the same performance as the original signature (Corr. ≥0.96) and could be more easily translated into clinical practice. The miR-Test was then validated in the “Validation Set” of 1008 subjects in the COSMOS trial. Risk scores between this set of lung cancer patients and healthy subjects were, statistically, significantly different. The overall accuracy, sensitivity, and specificity of the miR-Test were 74.9%, 77.8%, and 74.8%, respectively. The miR-Test was also able to distinguish between nonmalignant lung diseases (NMD) and lung cancer in an independent cohort and maintained comparable sensitivity when tested in more advanced lung cancer (stage II and III) patients diagnosed outside of the COSMOS trial. Notably, most individuals with NMD (126 out of 164, 76.8%) were miR-Test negative and the negative predictive value (NPV) was greater than 99%, meaning that negative-risk individuals could safely avoid subsequent LDCT scan. This is relevant because many LDCT screening trials were characterized by high rate of false-positive findings. The sensitivity and NPV of miR-Test are comparable with LDCT alone, indicating that the miR-Test could be used as a first-line screening tool. Overall, these data suggest that this blood-based test has sufficient accuracy and robustness to be used as noninvasive tool for lung cancer screening in high-risk individuals for subsequent LDCT, in order to reduce radiological screening costs and unnecessary LDCT for individuals without lung cancer (95).
In another study, the evaluation of miRNA expression in serum samples of NSCLC patients and healthy subjects by using a fluorescence quantum dots liquid bead array, revealed that five miRNAs (miR-16-5p, miR-17b-5p, miR-19-3p, miR-20a-5p, and miR-92-3p) were significantly downregulated in patients with cancer, while miR-15b-5p was upregulated. Notably, miR-15b-5p, miR-16-5p, and miR-20a-5p emerged as independent diagnostic factors for the identification of patients with NSCLC after adjustment for patient’s age and sex (97).
Due to the number of studies testing distinct circulating miRNAs as potential diagnostic biomarkers in lung cancer and leading to inconsistent results, a very recent systematic review and metanalysis was carried out. A total of 134 studies, with 6,919 patients with lung cancer and 7,064 controls, were included in the analysis. Overall analysis showed that circulating miRNAs had a good diagnostic performance in lung cancers, also in early-stage, especially the use of combined miRNAs and in Caucasian populations. Moreover, some miRNAs, such as miR-21-5p, miR-223-3p, miR-155-5p and miR-126-3p, emerged as most promising as potential biomarkers (98). Another noninvasive source of circulating miRNA is sputum (99).
Mir-21 expression in sputum specimens, assessed by RT-PCR, was significantly higher in cancer patients and was able to distinguish them from cancer-free subjects, showing 69.66% sensitivity and 100% specificity in diagnosis of lung cancer (100). In another study, miR-21, together with other 3 miRNAs (miR-486, miR-375 and miR-200b) previously selected in lung tumor tissues and then assessed on sputum samples of a case-control cohort, were able to distinguish lung adenocarcinoma patients from normal subjects with 80.6% sensitivity and 91.7% specificity (101). The same group demonstrated that miR-31 and miR-210, evaluated by RT-PCR in the sputum of lung cancer patients and cancer-free smokers, significantly improved specificity of CT alone for lung cancer diagnosis (102). The previous panel, added with also miR-21, was validated in different testing sets including patients with either malignant or benign solitary pulmonary nodules (SNPs), so confirming the performance of these miRNAs for diagnosis of malignant SPNs. The higher PPV (84%) of the biomarkers as compared with only 2% PPV of LDCT indicates that the biomarkers could result in much less overdiagnosis (103). In a meta-analysis of 28 published studies on the performance of miRNAs in body fluids including sputum as potential diagnostic biomarkers in a total of 2,121 NSCLC patients and 1,582 healthy control participants, the overall sensitivity and specificity for NSCLC detection was 75% and 79%, respectively. However, the diagnostic performance of miRNAs was superior in Caucasian compared with Asian patients and in blood compared with sputum samples, and multiple miRNA assays were more accurate compared with single miRNA assays (104).
Overall, the above data suggest a role for some miRNAs, especially when combined, as biomarkers for early cancer diagnosis, to be used alone or complementary to current standard screening procedures, including LDCT, for selection of high-risk patients to address further diagnostic exams. However, the great inter-variability among studies, most of which were single-center including small samples size, in terms of patient populations and stages of diseases included, deregulated miRNAs analyzed and methodologies used to assess circulating miRNAs, cannot allow a precise definition of the exact clinical role these biomarkers could have for screening and lung cancer diagnosis and management, suggesting that further, larger validation studies are still needed. Moreover, the importance of altered miRNAs relies on their target genes, thus suggesting the need for more in depth functional analysis to assess the role of deregulated genes, including their potential as prognostic markers or as therapeutic targets.
Exosomes
Exosomes are extracellular vesicles (EVs) of 40–100 nm in size that are released by several cell types, including cancer cells, into the extracellular space and a variety of body fluids. Several studies demonstrated that exosomes have a key role in the cell-cell communication: they have been shown to contain proteins as well as a range of nucleic acids, including DNA, mRNAs, and miRNAs, which can be transferred to target cells, thereby modulating the activities of these recipient cells. They have been shown a key role in tumor biology, including tumor growth, progression and drug resistance, and were recently indicated as important actors in metastatic niche preparation (16,105-107). Exosomes can be extracted from body fluids by normal density-gradient centrifugation or can be isolated through ultracentrifugation, visualized by transmission microscopy, or selected based on the presence of specific protein markers, such as the tetraspanin proteins CD63, CD9, and CD81. Over the past decade, impressive advances have been made in the development of novel exosome isolation methods, including extracellular vesicle array or immunobeads precipitation (16,108). Once harvested from biological fluid, they can be analyzed for miRNA, and thus for somatic mutations, splice variants, gene fusions and gene or protein expression profiling by using different techniques including RT-PCR, sequencing techniques, Western Blot or ELISA. Unlike circulating miRNAs, exosomal miRNAs are enriched in the circulatory system and protected from RNase degradation. Indeed, exosomal miRNAs have been extensively investigated in lung cancer for several clinical applications, including as potential diagnostic biomarkers (109).
A group of 12 exosomal miRNAs (miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212, and miR-214), selected based on a previous report demonstrating their potential diagnostic utility on tumor biopsies, was reported to be specifically expressed in patients with lung adenocarcinoma compared with a control group. Intriguingly, in four lung adenocarcinoma cases in which paired tumor and plasma samples were examined, there was a close correlation between circulating miRNAs of tumor-derived exosomes and tumor miRNAs, indicating the potential to analyze plasma as surrogate of tumor tissue (110).
A wide range microRNAs analysis (742 microRNAs), performed by quantitative RT-PCR, led to identification of 4 exosomal miRNAs (miR-378a, miR-379, miR-139-5p, and miR-200b-5p) able to distinguish patients with carcinomas from a control group of healthy former smokers (with 97.5% sensitivity and 72.0% specificity). A further group of 6 miRNAs (miR-151a-5p, miR-30a-3p, miR-200b-5p, miR-629, miR-100 and miR-154-3p) emerged as potential diagnostic biomarkers to discriminate between lung adenocarcinoma and granuloma (111). In a recent study, an extensive tumor-derived exosomal miRNA profiling by using miRNA-seq revealed a unique pattern of expression between stage I adenocarcinoma and SCC patients paired with healthy individuals. The miRNA profiles showed a sensitivity of 80.65% and of 83.33% and a specificity of 91.67%, and of 90.32% for adenocarcinoma and SCC diagnosis, respectively. These data suggest that these miRNAs may be suitable as highly sensitive, noninvasive biomarkers for early NSCLC diagnosis (112).
Despite promising results, different non-neoplastic cell types, including blood or endothelial cells, are known to release exosomes, thus making the clinical application of tumor cell–derived exosomal miRNAs for early NSCLC diagnosis challenging. Exosomes can be isolated in cancer patients from other body fluids, including pleural effusions (113), and could be investigated for other biomarkers, including oncogenic alterations, potentially useful for a wider range of clinical applications (16,109).
Tumor educated platelets (TEPs)
Blood platelet is the second most abundant cell type in peripheral blood. Platelets are circulating enucleated cell fragments that originate from megakaryocytes in bone marrow, known for their role in hemostasis. Platelets interact with tumor cells and affect tumor growth, invasion and establishment of distant metastasis (114,115). The interaction with tumor cells relies on the sequestration of tumor-associated biomolecules by platelets. Indeed, platelets can directly ingest circulating mRNA (116). Moreover, external signals, including activation of platelet surface receptors and lipopolysaccharide-mediated platelets activation induce specific splice variants of pre-mRNAs in circulating platelets, giving rise to unique mRNA profiles with potential applicability to cancer diagnostics (117-121).
A seminal study by Best and colleagues, characterized the platelet mRNA profiles of 283 samples, including 228 from early or advanced metastatic cancer patients (six tumor types, including NSCLC, colorectal cancer, glioblastoma, pancreatic cancer, hepatobiliary cancer and breast cancer) and 55 healthy individuals, by using SMARTer mRNA amplification and sequencing (122). Among the 5003 RNAs detected in all the samples, a total of 1,453 of mRNAs showed to be increased and 793 were decreased in TEPs as compared to platelet samples from healthy donors. Increased TEP mRNAs were enriched for biological processes such as vesicle-mediated transport and the cytoskeletal protein binding while decreased mRNAs were strongly involved in RNA processing and splicing. The mRNA profiles were able to distinguish patients with localized and metastasized tumors from healthy individuals with 96% accuracy. In addition to the pan-cancer diagnosis, the TEP mRNA profiles also distinguished healthy donors and patients with specific types of cancer, as demonstrated by the unsupervised hierarchical clustering of differential platelet mRNA levels of healthy donors and all six individual tumor types. Across six different tumor types, the location of the primary tumor was correctly identified with 71% accuracy (122). Moreover, targetable oncogenic drivers, such as MET or HER2-positive, and mutant KRAS, EGFR, or PIK3CA tumors were accurately distinguished using surrogate TEP mRNA profiles, suggesting platelets may provide diagnostic information and a strong indication on tumor type and molecular subclass. However, they were not able to measure significant differences between non-metastasized and metastasized tumor, thus suggesting these TEP mRNA profiles do not have the power to discriminate between certain stages of cancer (122). This was surprising, because experimental work has shown an influence of blood platelets on tumor cell dissemination and establishment of metastasis (121-123), but this could possibly be explained by the low number of patients analyzed for each tumor type and the great heterogeneity among the tumors analyzed.
Platelets have also been assessed to identify predictive biomarkers to targeted agents. Indeed, Nilsson et al. have analyzed EML4-ALK rearrangements by RT-PCR in platelets and plasma isolated from blood obtained from 77 patients with non-small cell lung cancer, 38 of whom have EML4-ALK rearranged tumors (65% sensitivity and 100% specificity for detection) (124). In the subset of 29 patients treated with crizotinib, progression-free survival was 3.7 months for patients with EML4-ALK+ platelets and 16 months for those with EML4-ALK− platelets (P=0.02). TEPs may prove useful for predicting and monitoring outcome to crizotinib, thereby improving clinical decisions based on radiographic imaging alone. Finally, authors reported that monitoring of EML4-ALK rearrangements in the platelets of a patient over a period of 30 months revealed crizotinib resistance two months prior to radiographic disease progression (124).
More robust data, possibly deriving from prospective, case-control studies, with larger sample size and homogeneous populations, are needed to further define the diagnostic value of platelet RNA, alone or complementary, to other circulating biomarker or current standard diagnostic procedures.
Conclusions
The ability to study genomic profile of cancer cells through noninvasive sampling of blood or other body fluids represents one of the most exciting and rapidly improving fields in lung cancer. Recently, liquid biopsy has largely demonstrated to be a surrogate of tumor tissue for noninvasive assessment of tumor-linked genetic and epigenetic alterations and has recently entered into clinical use for predicting response and monitor resistance to targeted therapies in the setting of advanced disease. Because of its several clinical advantages compared to tissue biopsy and, thanks to development of newer, sensitive technologies to analyse a larger number of biomarkers, liquid biopsy has been suggested for a wide range of clinical applications, including early diagnosis of lung cancer as a potentially curable disease. Different blood-based biosources, such as ctDNA, CTCs, exosomes and TEPs have been evaluated with distinct technologies for biomarkers that could be used to supplement the clinic in lung cancer diagnosis. Some reports have shown positive results for certain biomarkers, such as specific miRNAs profile, that could be complementary and improve the performance of standard radiological screening tests. One of the mainstays of a screening test, beyond its sensitivity and specificity, is that it should be reliable and cost-effective. So far, the ideal biosource and molecular biomarker to be used in the clinic for diagnostic purposes in lung malignancies have not yet been defined. Indeed, there is a great heterogeneity among studies in terms of patient population, blood-based biomarker analyzed and, moreover, different techniques have been used for genetic and epigenetic analyses. Furthermore, most were single center studies or included small patient sample size. More robust data from large, multicenter validation studies are still needed before the implementation of each of these promising blood-based biomarkers in the clinical setting. Clinical implementation of highly specific, sensitive and reproducible tools to be used for lung cancer screening and early detection remains an achievement to be pursued to improve the management and prognosis of these patients.
Acknowledgements
None.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality wordwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 3.Santarpia M, Altavilla G, Pitini V, et al. Personalized treatment of early-stage non-small-cell lung cancer: the challenging role of EGFR inhibitors. Future Oncol 2015;11:1259-74. 10.2217/fon.14.320 [DOI] [PubMed] [Google Scholar]
- 4.Kris MG, Gaspar LE, Chaft JE, et al. Adjuvant Systemic Therapy and Adjuvant Radiation Therapy for Stage I to IIIA Completely Resected Non–Small-Cell Lung Cancers: American Society of Clinical Oncology/Cancer Care Ontario Clinical Practice Guideline Update. J Clin Oncol 2017;35:2960-74. 10.1200/JCO.2017.72.4401 [DOI] [PubMed] [Google Scholar]
- 5.Chansky K, Detterbeck FC, Nicholson AG, et al. The IASLC Lung Cancer Staging Project: external validation of the revision of the TNM stage groupings in the eighth edition of the TNM classification of lung cancer. J Thorac Oncol 2017;12:1109-21. [DOI] [PubMed] [Google Scholar]
- 6.National Lung Screening Trial Research Team , Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. 10.1056/NEJMoa1102873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patz EF, Jr, Pinsky P, Gatsonis C, et al. Overdiagnosis in low-dose computed tomography screening for lung cancer. JAMA Intern Med 2014;174:269-74. 10.1001/jamainternmed.2013.12738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paci E, Puliti D, Lopes Pegna A, et al. Mortality, survival and incidence rates in the ITALUNG randomised lung cancer screening trial. Thorax 2017;72:825-31. 10.1136/thoraxjnl-2016-209825 [DOI] [PubMed] [Google Scholar]
- 9.Wille MM, Dirksen A, Ashraf H, et al. Results of the randomized danish lung cancer screening trial with focus on high-risk profiling. Am J Respir Crit Care Med 2016;193:542-51. 10.1164/rccm.201505-1040OC [DOI] [PubMed] [Google Scholar]
- 10.Infante M, Sestini S, Galeone C, et al. Lung cancer screening with low-dose spiral computed tomography: evidence from a pooled analysis of two Italian randomized trials. Eur J Cancer Prev 2017;26:324-9. 10.1097/CEJ.0000000000000264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma D, Newman TG, Aronow WS. Lung cancer screening: history, current perspectives, and future directions. Arch Med Sci 2015;11:1033-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aberle DR, Abtin F, Brown K. Computed tomography screening for lung cancer: has it finally arrived? Implications of the national lung screening trial. J Clin Oncol 2013;31:1002-8. 10.1200/JCO.2012.43.3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanna N, Johnson D, Temin S, et al. Systemic Therapy for Stage IV non-small-cell lung cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol 2017;35:3484-515. 10.1200/JCO.2017.74.6065 [DOI] [PubMed] [Google Scholar]
- 14.Santarpia M, Altavilla G, Salazar MF, et al. Tyrosine kinase inhibitors for non-small-cell lung cancer: finding patients who will be responsive. Expert Rev Respir Med 2011;5:413-24. 10.1586/ers.11.27 [DOI] [PubMed] [Google Scholar]
- 15.Santarpia M, Karachaliou N, González-Cao M, et al. Feasibility of cell-free circulating tumor DNA testing for lung cancer. Biomark Med 2016;10:417-30. 10.2217/bmm.16.6 [DOI] [PubMed] [Google Scholar]
- 16.Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. 10.1038/nrclinonc.2017.14 [DOI] [PubMed] [Google Scholar]
- 17.Sozzi G, Conte D, Leon M, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol 2003;21:3902-8. 10.1200/JCO.2003.02.006 [DOI] [PubMed] [Google Scholar]
- 18.Karachaliou N, Sosa AE, Molina MA, et al. Possible application of circulating free tumor DNA in non-small cell lung cancer patients. J Thorac Dis 2017;9:S1364-72. 10.21037/jtd.2017.09.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed] [Google Scholar]
- 20.Stroun M, Anker P, Maurice P, et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989;46:318-22. 10.1159/000226740 [DOI] [PubMed] [Google Scholar]
- 21.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed] [Google Scholar]
- 22.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA 2005;102:16368-73. 10.1073/pnas.0507904102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 24.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi JJ, Reich CF, Pisetsky DS. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology 2005;115:55-62. 10.1111/j.1365-2567.2005.02130.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayo-de-Las-Casas C, Garzón Ibáñez M, Jordana-Ariza N, et al. An update on liquid biopsy analysis for diagnostic and monitoring applications in non-small cell lung cancer. Expert Rev Mol Diagn 2018;18:35-45. 10.1080/14737159.2018.1407243 [DOI] [PubMed] [Google Scholar]
- 27.Sozzi G, Roz L, Conte D, et al. Plasma DNA quantification in lung cancer computed tomography screening: five-year results of a prospective study. Am J Respir Crit Care Med 2009;179:69-74. 10.1164/rccm.200807-1068OC [DOI] [PubMed] [Google Scholar]
- 28.Paci M, Maramotti S, Bellesia E, et al. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer 2009;64:92-7. 10.1016/j.lungcan.2008.07.012 [DOI] [PubMed] [Google Scholar]
- 29.Catarino R, Coelho A, Araújo A, et al. Circulating DNA: diagnostic tool and predictive marker for overall survival of NSCLC patients. PLoS One 2012;7:e38559. 10.1371/journal.pone.0038559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Cuesta L, Perdomo S, Avogbe PH, et al. Identification of Circulating Tumor DNA for the early detection of small-cell lung cancer. EBioMedicine 2016;10:117-23. 10.1016/j.ebiom.2016.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esteller M, Herman J.G. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol 2002;196:1-7. 10.1002/path.1024 [DOI] [PubMed] [Google Scholar]
- 33.Zöchbauer-Müller S, Minna JD, Gazdar AF. Aberrant DNA methylation inlung cancer: biological and clinical implications. Oncologist 2002;7:451-7. 10.1634/theoncologist.7-5-451 [DOI] [PubMed] [Google Scholar]
- 34.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 2012;13:679-92. 10.1038/nrg3270 [DOI] [PubMed] [Google Scholar]
- 35.Tomasetti M, Amati M, Neuzil J, et al. Circulating epigenetic biomarkers in lung malignancies: From early diagnosis to therapy. Lung Cancer 2017;107:65-72. 10.1016/j.lungcan.2016.05.023 [DOI] [PubMed] [Google Scholar]
- 36.Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in non-invasive cancer screening. Curr Mol Med 2010;10:123-32. 10.2174/156652410790963303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujiwara K, Fujimoto N, Tabata M, et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res 2005;11:1219-25. [PubMed] [Google Scholar]
- 38.Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 2011;303:21-8. 10.1016/j.canlet.2010.12.011 [DOI] [PubMed] [Google Scholar]
- 39.Ponomaryova AA, Rykova EY, Cherdyntseva NV, et al. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer 2013;81:397-403. 10.1016/j.lungcan.2013.05.016 [DOI] [PubMed] [Google Scholar]
- 40.Kneip C, Schmidt B, Seegebarth A, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol 2011;6:1632-8. 10.1097/JTO.0b013e318220ef9a [DOI] [PubMed] [Google Scholar]
- 41.Konecny M, Markus J, Waczulikova I, et al. The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma 2016;63:246-53. [DOI] [PubMed] [Google Scholar]
- 42.Powrózek T, Krawczyk P, Nicoś M, et al. Methylation of the DCLK1 promoter region in circulating free DNA and its prognostic value in lung cancer patients. Clin Transl Oncol 2016;18:398-404. 10.1007/s12094-015-1382-z [DOI] [PubMed] [Google Scholar]
- 43.Powrózek T, Krawczyk P, Kucharczyk T, et al. Septin 9 promoter region methylation in free circulating DNA-potential role in noninvasive diagnosis of lung cancer: preliminary report. Med Oncol 2014;31:917. 10.1007/s12032-014-0917-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diaz-Lagares A, Mendez-Gonzalez J, Hervas D, et al. A novel epigenetic signature for early diagnosis in lung cancer. Clin Cancer Res 2016;22:3361-71. 10.1158/1078-0432.CCR-15-2346 [DOI] [PubMed] [Google Scholar]
- 45.Nikolaidis G, Raji OY, Markopoulou S, et al. DNA methylation biomarkers offer improved diagnostic efficiency in lung cancer. Cancer Res 2012;72:5692-701. 10.1158/0008-5472.CAN-12-2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt B, Liebenberg V, Dietrich D, et al. SHOX2 DNA methylation for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer 2010;10:600. 10.1186/1471-2407-10-600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dietrich D, Kneip C, Raji O, et al. Performance evaluation of the DNA methylation biomarker SHOX2 for the aid in diagnosis of lung cancer based on the analysis of bronchial aspirates. Int J Oncol 2012;40:825-32. [DOI] [PubMed] [Google Scholar]
- 48.Krebs MG, Metcalf RL, Carter L, et al. Molecular analysis of circulating tumour cells—biology and biomarkers. Nat Rev Clin Oncol 2014;11:129-44. 10.1038/nrclinonc.2013.253 [DOI] [PubMed] [Google Scholar]
- 49.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 2014;4:650-61. 10.1158/2159-8290.CD-13-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013;59:110-8. 10.1373/clinchem.2012.194258 [DOI] [PubMed] [Google Scholar]
- 51.Hofman V, Ilie M, Long E, et al. Detection of circulating tumor cells from lung cancer patients in the era of targeted therapy: promises, drawbacks and pitfalls. Curr Mol Med 2014;14:440-56. 10.2174/1566524014666140414205455 [DOI] [PubMed] [Google Scholar]
- 52.Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, et al. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res 2016;5:466-82. 10.21037/tlcr.2016.10.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luke JJ, Oxnard GR, Paweletz CP, et al. Realizing the potential of plasma genotyping in an age of genotype-directed therapies. J Natl Cancer Inst 2014;106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623-31. 10.1038/nrc3820 [DOI] [PubMed] [Google Scholar]
- 55.Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897-904. 10.1158/1078-0432.CCR-04-0378 [DOI] [PubMed] [Google Scholar]
- 56.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- 57.Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. 10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- 58.de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- 59.Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450:1235-9. 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanssen A, Wagner J, Gorges TM, et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci Rep 2016;6:28010. 10.1038/srep28010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Millner LM, Linder MW, Valdes R., Jr Circulating tumor cells: a review of present methods and the need to identify heterogeneous phenotypes. Ann Clin Lab Sci 2013;43:295-304. [PMC free article] [PubMed] [Google Scholar]
- 62.Raimondi C, Nicolazzo C, Gradilone A. Circulating tumor cells isolation: the "post-EpCAM era". Chin J Cancer Res 2015;27:461-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vona G, Sabile A, Louha M, et al. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol 2000;156:57-63. 10.1016/S0002-9440(10)64706-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Desitter I, Guerrouahen BS, Benali-Furet N, et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res 2011;31:427-41. [PubMed] [Google Scholar]
- 65.Hofman V, Bonnetaud C, Ilie MI, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res 2011;17:827-35. 10.1158/1078-0432.CCR-10-0445 [DOI] [PubMed] [Google Scholar]
- 66.Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay™ and the isolation by size of epithelial tumor cell method. Int J Cancer 2011;129:1651-60. 10.1002/ijc.25819 [DOI] [PubMed] [Google Scholar]
- 67.Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. 10.1200/JCO.2010.28.7045 [DOI] [PubMed] [Google Scholar]
- 68.Hofman V, Long E, Ilie M, et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology 2012;23:30-8. 10.1111/j.1365-2303.2010.00835.x [DOI] [PubMed] [Google Scholar]
- 69.Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 2012;7:306-15. 10.1097/JTO.0b013e31823c5c16 [DOI] [PubMed] [Google Scholar]
- 70.Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. 10.1200/JCO.2010.33.3716 [DOI] [PubMed] [Google Scholar]
- 71.Hosokawa M, Yoshikawa T, Negishi R, et al. Microcavity array system for size-based enrich- ment of circulating tumor cells from the blood of patients with small-cell lung cancer. Anal Chem 2013;85:5692-8. 10.1021/ac400167x [DOI] [PubMed] [Google Scholar]
- 72.Wang J, Wang K, Xu J, et al. Prognostic significance of circulating tumor cells in non-small-cell lung cancer patients: a meta-analysis. PLoS One 2013;8:e78070. 10.1371/journal.pone.0078070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ilie M, Hofman V, Long-Mira E, et al. "Sentinel" circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS One 2014;9:e111597. 10.1371/journal.pone.0111597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fiorelli A, Accardo M, Carelli E. Circulating tumor cells in diagnosing lung cancer: clinical and morphologic analysis. Ann Thorac Surg 2015;99:1899-905. 10.1016/j.athoracsur.2014.11.049 [DOI] [PubMed] [Google Scholar]
- 75.Hanssen A, Loges S, Pantel K, et al. Detection of circulating tumor cells in non-small cell lung cancer. Front Oncol 2015;5:207. 10.3389/fonc.2015.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu Y, Liu B, Ding F, et al. Circulating tumor cell detection: A direct comparison between negative and unbiased enrichment in lung cancer. Oncol Lett 2017;13:4882-6. 10.3892/ol.2017.6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung cancer cells. N Engl J Med 2008;359:366-77. 10.1056/NEJMoa0800668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Punnoose EA, Atwal SK, Spoerke JM, et al. Molecular biomarker analyses using circulating tumor cells. PLoS One 2010;5:e12517. 10.1371/journal.pone.0012517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. 10.1158/1078-0432.CCR-11-3148 [DOI] [PubMed] [Google Scholar]
- 80.Mayo C, Ortega FG, Giménez-Capitán A, et al. CK-coated magnetic-based beads as a tool to isolate circulating tumor cells (CTCs) in human tumors. Transl Lung Cancer Res 2013;2:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu Y, Chen Z, Dong J, et al. Folate receptor–positive circulating tumor cells as a novel diagnostic biomarker in non-small cell lung cancer. Transl Oncol 2013;6:697-702. 10.1593/tlo.13535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mitchell PS, Parkin RK, Kroh RK, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008;105:10513-8. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- 84.Foss KM, Sima C, Ugolini D, et al. MiR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol 2011;6:482-8. 10.1097/JTO.0b013e318208c785 [DOI] [PubMed] [Google Scholar]
- 85.Heegaard NH, Schetter AJ, Welsh JA, et al. Circulating micro-RNA expression profiles in early stage non-small cell lung cancer. Int J Cancer 2012;130:1378-86. 10.1002/ijc.26153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Veronesi G, Bellomi M, Mulshine JL, et al. Lung cancer screening with lowdose computed tomography: a non-invasive diagnostic protocol for baseline lung nodules. Lung Cancer 2008;61:340-9. 10.1016/j.lungcan.2008.01.001 [DOI] [PubMed] [Google Scholar]
- 87.Shen J, Todd NW, Zhang H, et al. Plasma microRNAs as potential biomarkers for non-smal- cell lung cancer. Lab Invest 2011;91:579-87. 10.1038/labinvest.2010.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen J, Liu Z, Todd NW, et al. Diagnosis of lung cancer individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer 2011;11:374. 10.1186/1471-2407-11-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng D, Haddading S, Wang Y, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol 2011;4:575-86. [PMC free article] [PubMed] [Google Scholar]
- 90.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A 2011;108:3713-8. 10.1073/pnas.1100048108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sozzi G, Boeri M, Rossi M, et al. Clinical Utility of a Plasma-Based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD Trial Study. J Clin Oncol 2014;32:768-73. 10.1200/JCO.2013.50.4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Plasma microRNA profiling as first-line screening test for lung cancer detection: a prospective study (BIOMILD). ClinicalTrials.gov [NCT02247453]. Available online: https://clinicaltrials.gov/ct2/show/NCT02247453
- 93.Wang C, Ding M, Xia M, et al. A Five-miRNA panel identified from a multicentric case–control study serves as a novel diagnostic tool for ethnically diverse non-small-cell lung cancer patients. EBioMedicine 2015;2:1377-85. 10.1016/j.ebiom.2015.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang P, Yang D, Zhang H, et al. Early detection of lung cancer in serum by a panel of microRNA biomarkers. Clin Lung Cancer 2015;16:313-9.e1. 10.1016/j.cllc.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 95.Montani F, Marzi M., Dezi F, et al. miR-Test: a blood test for lung cancer early detection. J Natl Cancer Inst 2015;107:djv063. 10.1093/jnci/djv063 [DOI] [PubMed] [Google Scholar]
- 96.Bianchi F, Nicassio F, Marzi M, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med 2011;3:495-503. 10.1002/emmm.201100154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fan L, Qi H, Teng J, et al. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol 2016;37:7777-84. 10.1007/s13277-015-4608-3 [DOI] [PubMed] [Google Scholar]
- 98.Yang Y, Hu Z, Zhou Y, et al. The clinical use of circulating microRNAs as non-invasive diagnostic biomarkers for lung cancers. Oncotarget 2017;8:90197-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sheervalilou R, Ansarin K, Fekri Aval S, et al. An update on sputum MicroRNAs in lung cancer diagnosis. Diagn Cytopathol 2016;44:442-9. 10.1002/dc.23444 [DOI] [PubMed] [Google Scholar]
- 100.Xie Y, Todd NW, Liu Z, et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer 2010;67:170-6. 10.1016/j.lungcan.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu L, Todd NW, Xing L, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer 2010;127:2870-8. 10.1002/ijc.25289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shen J, Liao J, Guarnera MA, et al. Analysis of MicroRNAs in sputum to improve computed tomography for lung cancer diagnosis. J Thorac Oncol 2014;9:33-40. 10.1097/JTO.0000000000000025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xing L, Su J, Guarnera MA, et al. Sputum microRNA Biomarkers for identifying lung cancer in indeterminate solitary pulmonary nodules. Clin Cancer Res 2015;21:484-9. 10.1158/1078-0432.CCR-14-1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang H, Wu S, Zhao L, et al. Clinical use of microRNAs as potential non-invasive biomarkers for detecting non-small cell lung cancer: a meta-analysis. Respirology 2015;20:56-65. 10.1111/resp.12444 [DOI] [PubMed] [Google Scholar]
- 105.Mathivanan S, Ji H, Simpson R J. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010;73:1907-20. 10.1016/j.jprot.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 106.Mittelbrunn M, Sánchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol 2012;13:328-35. 10.1038/nrm3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weidle UH, Birzele F, Kollmorgen G, et al. The multiple roles of exosomes in metastasis. Cancer Genomics Proteomics 2017;14:1-15. 10.21873/cgp.20015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rani S, O'Brien K, Kelleher FC, et al. Isolation of exosomes for subsequent mRNA, microRNA, and protein profiling. Methods Mol Biol 2011;784:181-95. 10.1007/978-1-61779-289-2_13 [DOI] [PubMed] [Google Scholar]
- 109.Reclusa P, Taverna S, Pucci M, et al. Exosomes as diagnostic and predictive biomarkers in lung cancer. J Thorac Dis 2017;9:S1373-82. 10.21037/jtd.2017.10.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rabinowits G, Gerçel-Taylor C, Day JM, et al. Exosomal MicroRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009;10:42-6. 10.3816/CLC.2009.n.006 [DOI] [PubMed] [Google Scholar]
- 111.Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 2013;8:1156-62. 10.1097/JTO.0b013e318299ac32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jin X, Chen Y, Chen H, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res 2017;23:5311-9. 10.1158/1078-0432.CCR-17-0577 [DOI] [PubMed] [Google Scholar]
- 113.Han HS, Yun J, Lim SN, et al. Downregulation of cell-free miR-198 as a diagnostic biomarker for lung adenocarcinoma-associated malignant pleural effusion. Int J Cancer 2013;133:645-52. 10.1002/ijc.28054 [DOI] [PubMed] [Google Scholar]
- 114.Leslie M. Cell biology. Beyond clotting: the powers of platelets. Science 2010;328:562-4. 10.1126/science.328.5978.562 [DOI] [PubMed] [Google Scholar]
- 115.McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol 2014;16:717-27. 10.1038/ncb3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nilsson RJ, Balaj L, Hulleman E, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011;118:3680-3. 10.1182/blood-2011-03-344408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Denis MM, Tolley ND, Bunting M, et al. Escaping the nucleat confines: signal-dependent pre-mRNA splicing in anucleated platelets. Cell 2005;122:379-91. 10.1016/j.cell.2005.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Calverley DC, Phang TL, Choudhury QG, et al. Significant downregulation of platelet gene expression in metastatic lung cancer. Clin Transl Sci 2010;3:227-32. 10.1111/j.1752-8062.2010.00226.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brown GT, McIntyre TM. Lipopolysaccharide signaling without a nucleus:kinase cascades stimulate platelet sheding of proinflammtory IL 1-β-rich microparticles. J Immunol 2011;186:5489-96. 10.4049/jimmunol.1001623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schubert S, Weyrich AS, Rowley JW. A tour through the transcriptional landscape of platelets. Blood 2014;124:493-502. 10.1182/blood-2014-04-512756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Joosse SA, Pantel K. Tumor-educated platelets as liquid biopsy in cancer patients. Cancer Cell 2015;28:552-4. 10.1016/j.ccell.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 122.Best MG, Sol N, Kooi I, et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 2015;28:666-76. 10.1016/j.ccell.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kuznetsov HS, Marsh T, Markens BA, et al. Identification of luminal breast cancers that establish a tumor-supportive macroenvironment defined by proangiogenic platelets and bone marrow-derived cells. Cancer Discov 2012;2:1150-65. 10.1158/2159-8290.CD-12-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nilsson RJA, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. 10.18632/oncotarget.6279 [DOI] [PMC free article] [PubMed] [Google Scholar]