Abstract Abstract
The genus Arthrinium includes important plant pathogens, endophytes and saprobes with a wide host range and geographic distribution. In this paper, 74 Arthrinium strains isolated from various substrates such as bamboo leaves, tea plants, soil and air from karst caves in China were examined using a multi-locus phylogeny based on a combined dataset of ITS rDNA, TEF1 and TUB2, in conjunction with morphological characters, host association and ecological distribution. Eight new species were described based on their distinct phylogenetic relationships and morphological characters. Our results indicated a high species diversity of Arthrinium with wide host ranges, amongst which, Poaceae and Cyperaceae were the major host plant families of Arthrinium species.
Keywords: Ascomycota, Morphology, Phylogeny, Systematics, Taxonomy
Introduction
Arthrinium Kunze is an anamorph-typified genus, which has been traditionally linked to the teleomorph-typified genus Apiospora Sacc. (Ellis 1971, Seifert et al. 2011). It is strikingly different from other anamorphic genera for the presence of basauxic conidiophores (Hughes 1953, Minter 1985). The traditional generic circumscription of Arthrinium was primarily based on morphological characters (e.g. conidial shape, conidiophores, sterile cells and the presence of setae) but has been regarded as too narrow (Ellis 1971, Minter 1985, Crous et al. 2013). It is now recognised that, at the generic level, conidial shape and the presence of setae are not reliable characters to infer phylogenetic relationships (Crous et al. 2013). For example, Arthrinium was regarded as being different from Cordella Speg. (1886) by the absence of setae amongst the clusters of specialised hyphae and different from Pteroconium Sacc. (1892) by the absence of sporodochia and pseudoparenchyma (Minter 1985). However, both genera have been reduced to the generic synonyms of Arthrinium, based on molecular phylogenetic data (Crous et al. 2013).
Arthrinium species are geographically widely distributed in various hosts. Many species of Arthrinium are associated with plants as endophytes or saprobes, as well as plant pathogens on some important ornamentals, e.g. A. phaeospermum causing culm rot on Phyllostachys viridis (Li et al. 2016); A. arundinis causing brown culm streak of Phyllostachys praecox (Chen et al. 2014). Moreover, A. phaeospermum has been reported for causing cutaneous infections of humans (Rai 1989, Zhao et al. 1990, de Hoog et al. 2000, Crous et al. 2013). Many Arthrinium species are also known to produce bioactive compounds with pharmacological and medicinal applications, such as A. arundinis and A. saccharicola isolated from a brown alga Sargassum sp., with good antifungal activities against some plant pathogenic fungi (Hong et al. 2015). Arthrinium saccharicola, A. sacchari and A. phaeospermum isolated from Miscanthus sp. are known to produce industrially important enzymes (Shrestha et al. 2015).
In this paper, eight new Arthrinium species are described and characterised based on morphological characters and phylogeny inferred from the combined ITS rDNA, TEF1 and TUB2 sequences dataset. Comparisons were made with morphologically similar and phylogenetically related species. Fungus-host distribution of Arthrinium species are summarised based on data from literature and this study.
Materials and method
Isolates
Diseased and healthy tissues of bamboo leaves and other plant hosts were collected from six provinces or municipalities in China (Chongqing, Guangxi, Guangdong, Guizhou, Jiangxi, Hunan). Tissue pieces (5 mm × 5 mm) were taken from the margin of leaf lesions and the surface sterilised with 75% ethanol for 1 min, 5% NaClO for 30 s, followed by rinsing in sterile distilled water for 1 min. The pieces were dried with sterilised paper towels and then placed on 1/4 PDA (potato dextrose agar) (Cai et al. 2009).
All cultures were preserved in the LC culture collection (personal culture collection of Lei Cai housed in the Institute of Microbiology, Chinese Academy of Sciences). Type specimens were deposited in Mycological Herbarium of the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), with ex-type living cultures deposited in China General Microbiological Culture Collection Center (CGMCC). Taxonomic information of the new taxa was deposited in MycoBank (www.MycoBank.org; Crous et al. 2004).
Morphology
Cultures were incubated on PDA for 7 d at 25 °C to measure the growth rates and on 2% malt agar with bamboo leaves to enhance sporulation. Morphological descriptions were based on cultures sporulating on MEA (malt extract agar) medium at room temperature (ca. 25 °C). Shape and size of microscopic structures were observed using a light microscope and colonies were assessed according to the colour charts of Rayner (1970). At least 50 conidiogenous cells and conidia were measured to calculate the mean size.
DNA extraction, PCR amplification and sequencing
Fresh fungal mycelia were taken from 7-d-old cultures growing on PDA and ground with the organisation disruptor FastPrep-48. Genomic DNA was extracted following the modified CTAB protocol as described in Guo et al. (2000).
Phylogenetic analyses were conducted using partial sequences of three loci, 5.8S nuclear ribosomal gene with the two flanking transcribed spacers (ITS), part of the translation elongation factor 1-alpha (TEF1) and beta-tubulin (TUB2). The ITS locus was amplified using the primer pair ITS1/ITS4 (Vilgalys and Hester 1990, White et al. 1990); TEF1 using EF1-728F/ EF-2 (O’Donnell et al. 1998, Carbone and Kohn 1999); and TUB2 using T1 (O’Donnell and Cigelnik 1997) and Bt-2b (Glass and Donaldson 1995).
PCR was performed in a 25 ml reaction containing 18.95 µl double distilled water, 2.5 µl 10 × PCR buffer, 0.3 µl dNTP mix (2.5 mM), 1 µl of each primer (10 mM), 1 µl DNA template and 0.25 µl Taq DNA polymerase (Genstar). The annealing temperatures were adjusted to 52 °C for ITS and TUB2, and 56 °C for TEF1. Purification and sequencing of the PCR amplicons were done by SinoGenoMax, Beijing.
Phylogenetic analysis
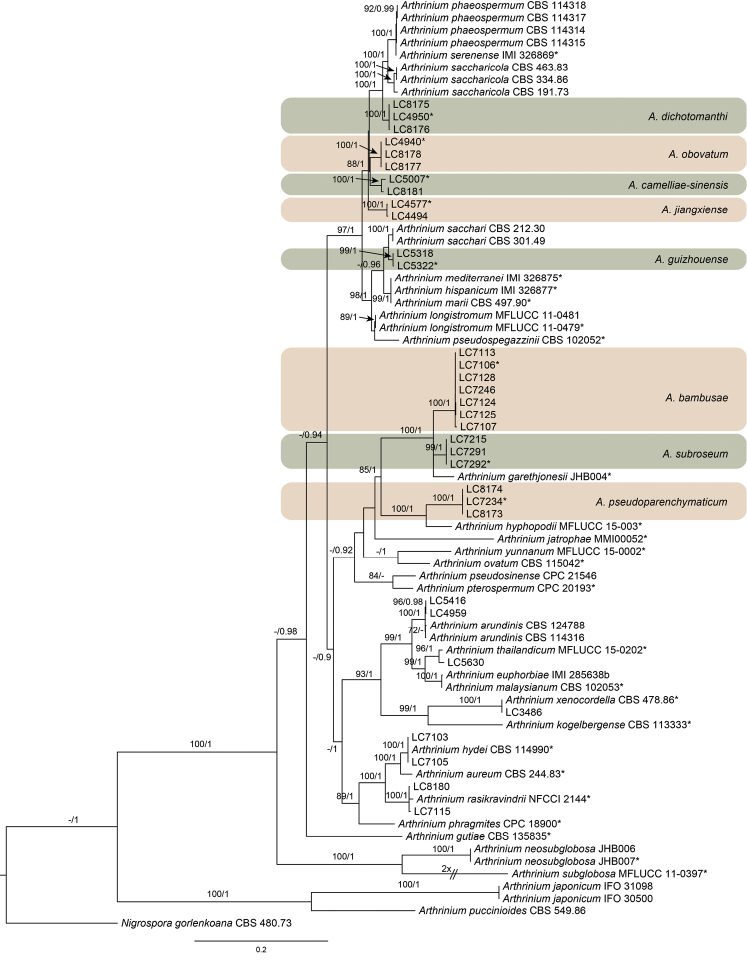
Sequences generated from the forward and reverse primers were used to obtain consensus sequences using MEGA v. 6.0 (Tamura et al. 2013). The concatenated tree was inferred based ITS, TUB2 and TEF1 sequences (Figure 1) using Bayesian and Maximum-likelihood analyses. Sequences were aligned using an online version of MAFFT v. 7 (available at http://mafft.cbrc.jp/alignment/server/). Ambiguous regions were excluded from the analyses and gaps were treated as missing data. Maximum-likelihood (ML) analysis was performed in RAxML v. 7.2.6 (Stamatakis and Alachiotis 2010), employing GTR models of evolution settings of the programme and bootstrap support obtained by running 1000 pseudo replicates. Maximum Likelihood bootstrap values (ML) equal to or greater than 70% are given above each node.
Figure 1.
Phylogenetic tree based on the combined ITS, TEF1 and TUB2 sequences alignment generated from a Maximum likelihood phylogenetic analysis. Bootstrap support values (>70%) and posterior probabilities (>0.9) are given at the nodes (ML/PP). The tree is rooted with Nigrospora gorlenkoana CBS 480.73. The novel species were highlighted (* indicates the ex-type cultures).
Bayesian analysis was conducted using MrBayes v. 3.2.1 (Ronquist et al. 2012) and the best nucleotide substitution model for each locus was calculated with jModelTest v. 2.1.4 (Posada 2008). Posterior probabilities (PP) (Zhaxybayeva and Gogarten 2002) were determined by Markov Chain Monte Carlo sampling (MCMC) under the estimated model of evolution. Four simultaneous Markov chains were run for 10 million generations and trees were sampled every 1000 generations. The run was stopped automatically when the average standard deviation of split frequencies fell below 0.01. The first 25% trees, which represented the burn-in phase of the analyses, were discarded and the remaining trees were used for calculating PP in the majority rule consensus tree. Sequences generated in this study were deposited in GenBank (Table 1) and the final matrices used for the phylogenetic analyses in TreeBASE (www.treebase.org; accession number: 21341).
Table 1.
Strains included in the phylogenetic analyses.
| Speices | Strain numbers1 | Hosts | Countries | GenBank accessions | ||
|---|---|---|---|---|---|---|
| ITS | TUB | TEF | ||||
| Arthirinium arundinis | CBS 114316 | Leaf of Hordeum vulgare | Iran | KF144884 | KF144974 | KF145016 |
| CBS 124788 | Living leaves of Fagus sylvatica | Switzerland | KF144885 | KF144975 | KF145017 | |
| LC4477 | Unknow host | China | KY494688 | KY705159 | KY705087 | |
| LC4493 | Phyllostachys sp. | China | KY494689 | KY806202 | KY705088 | |
| LC4650 | Osmanthus sp. | China | KY494695 | KY705165 | KY705094 | |
| LC4951 | Dichotomanthus tristaniaecarpa | China | KY494698 | KY705168 | KY705097 | |
| LC4959 | Bothrocaryum controversum | China | KY494699 | KY705169 | KY705098 | |
| LC5311 | Air in karst cave | China | KY494706 | KY705175 | KY705105 | |
| LC5312 | Air in karst cave | China | KY494707 | KY705176 | KY705106 | |
| LC5332 | Air in karst cave | China | KY494710 | KY705179 | KY705109 | |
| LC5394 | Soil in karst cave | China | KY494711 | KY705180 | KY705110 | |
| LC5416 | Water in karst cave | China | KY494712 | KY705181 | KY705111 | |
| LC7118 | Leaf of bamboo | China | KY494723 | KY705191 | KY705120 | |
| LC7122 | Leaf of bamboo | China | KY494726 | KY705194 | KY705123 | |
| LC7160 | Leaf of bamboo | China | KY494738 | KY705206 | KY705134 | |
| LC7211 | Leaf of bamboo | China | KY494739 | KY705207 | KY705135 | |
| LC7216 | Leaf of bamboo | China | KY494741 | KY705209 | KY705137 | |
| LC7218 | Leaf of bamboo | China | KY494742 | KY705210 | KY705138 | |
| LC7243 | Leaf of bamboo | China | KY494744 | KY705212 | KY705140 | |
| LC7252 | Leaf of bamboo | China | KY494747 | KY705215 | KY705143 | |
| LC7277 | Leaf of bamboo | China | KY494750 | KY705218 | KY705146 | |
| A. aureum | CBS 244.83* | Air | Spain | AB220251 | KF144981 | KF145023 |
| A. bambusae | LC7106* = CGMCC 3.18335 | Leaf of bamboo | China | KY494718 | KY705186 | KY806204 |
| LC7107 | Leaf of bamboo | China | KY494719 | KY705187 | KY705117 | |
| LC7113 | Leaf of bamboo | China | KY494720 | KY705188 | KY806205 | |
| LC7124 | Leaf of bamboo | China | KY494727 | KY705195 | KY806206 | |
| LC7125 | Leaf of bamboo | China | KY494728 | KY705196 | KY705124 | |
| LC7128 | Leaf of bamboo | China | KY494730 | KY705198 | KY705126 | |
| LC7246 | Leaf of bamboo | China | KY494745 | KY705213 | KY705141 | |
| A. camelliae-sinensis | LC5007* = CGMCC 3.18333 | Camellia sinensis | China | KY494704 | KY705173 | KY705103 |
| LC8181 | Brassica capestris | China | KY494761 | KY705229 | KY705157 | |
| A. dichotomanthi | LC4950* = CGMCC 3.18332 | Dichotomanthus tristaniaecarpa | China | KY494697 | KY705167 | KY705096 |
| LC8175 | Dichotomanthus tristaniaecarpa | China | KY494755 | KY705223 | KY705151 | |
| LC8176 | Dichotomanthus tristaniaecarpa | China | KY494756 | KY705224 | KY705152 | |
| A. euphorbiae | IMI 285638b | Bambusa sp. | Bangladesh | AB220241 | AB220288 | – |
| A. guizhouense | LC5318 | Air in karst cave | China | KY494708 | KY705177 | KY705107 |
| LC5322* =CGMCC3.18334 | Air in karst cave | China | KY494709 | KY705178 | KY705108 | |
| A. gutiae | CBS 135835 | Gut of a grasshopper | India | KR011352 | KR011350 | KR011351 |
| A. hispanicum | IMI 326877* | Maritime sand | Spain | AB220242 | AB220289 | – |
| A. hydei | CBS 114990* | Culms of Bambusa tuldoides | Hong Kong | KF144890 | KF144982 | KF145024 |
| LC7103 | Leaf of bamboo | China | KY494715 | KY705183 | KY705114 | |
| LC7105 | Leaf of bamboo | China | KY494717 | KY705185 | KY705116 | |
| A. hyphopodii | MFLUCC 15-0003* | Culms of Bambusa tuldoides | Thailand | KR069110 | – | – |
| A. japonicum | IFO 30500 | Carex despalata (dead leaf) | Japan | AB220262 | AB220309 | – |
| IFO 31098 | Carex despalata (leaf) | Japan | AB220264 | AB220311 | – | |
| A. garethjonesii | KUMCC 16-0202 | Dead culms of bamboo | China | KY356086 | – | – |
| A. jatrophae | MMI 00052* = MCC 1014 | Healthy petiole of Jatropha podagrica | India | JQ246355 | – | – |
| A. jiangxiense | LC2831 | Leaf of bamboo | China | KY494686 | KY80620106201 | KY705085 |
| LC4494 | Phyllostachys sp. | China | KY494690 | KY705160 | KY705089 | |
| LC4541 | Maesa sp. | China | KY494691 | KY705161 | KY705090 | |
| LC4547 | Machilus sp. | China | KY494692 | KY705162 | KY705091 | |
| LC4577* = CGMCC 3.18381 | Maesa sp. | China | KY494693 | KY705163 | KY705092 | |
| LC4578 | Camellia sinensis | China | KY494694 | KY705164 | KY705093 | |
| LC4993 | Phyllostachys sp. | China | KY494700 | KY806203 | KY705099 | |
| LC4997 | Phyllostachys sp. | China | KY494701 | KY705170 | KY705100 | |
| LC5001 | Phyllostachys sp. | China | KY494702 | KY705171 | KY705101 | |
| LC5004 | Phyllostachys sp. | China | KY494703 | KY705172 | KY705102 | |
| LC5015 | Imperata cylindrica | China | KY494705 | KY705174 | KY705104 | |
| A. jiangxiense | LC7104 | Leaf of bamboo | China | KY494716 | KY705184 | KY705115 |
| LC7154 | Leaf of bamboo | China | KY494736 | KY705204 | KY705132 | |
| LC7156 | Leaf of bamboo | China | KY494737 | KY705205 | KY705133 | |
| LC7275 | Leaf of bamboo | China | KY494749 | KY705217 | KY705145 | |
| A. kogelbergense | CBS 113333* | Dead culms of Restionaceae | South Africa | KF144892 | KF144984 | KF145026 |
| A. longistromum | MFLUCC 11-0481* | Decaying bamboo culms | Thailand | KU940141 | – | – |
| MFLUCC 11-0479 | Decaying bamboo culms | Thailand | KU940142 | – | – | |
| A. malaysianum | CBS 102053* | Macaranga hullettii stem colonised by ants | Malaysia | KF144896 | KF144988 | KF145030 |
| A. marii | CBS 497.90* | Air | Spain | AB220252 | KF144993 | KF145035 |
| A. mediterranei | IMI 326875* | Air | Spain | AB220243 | AB220290 | – |
| A. mytilomorphum | DAOM 214595* | Dead blades of Andropogon sp. | India | KY494685 | – | – |
| A. neosubglobosa | JHB006 | Dead culms of bamboo | China | KY356089 | – | – |
| KUMCC 16-0203 | Dead culms of bamboo | China | KY356090 | – | – | |
| A. obovatum | LC4940* = CGMCC 3.18331 | Lithocarpus sp. | China | KY494696 | KY705166 | KY705095 |
| LC8177 | Lithocarpus sp. | China | KY494757 | KY705225 | KY705153 | |
| LC8178 | Lithocarpus sp. | China | KY494758 | KY705226 | KY705154 | |
| A. ovatum | CBS 115042* | Arundinaria hindsii | Hong Kong | KF144903 | KF144995 | KF145037 |
| A. paraphaeospermum | MFLU 16-1974 | Dead clumps of Bambusa sp. | Thailand | KX822128 | – | – |
| A. phaeospermum | CBS 114314 | Leaf of Hordeum vulgare | Iran | KF144904 | KF144996 | KF145038 |
| CBS 114315 | Leaf of Hordeum vulgare | Iran | KF144905 | KF144997 | KF145039 | |
| CBS 114317 | Leaf of Hordeum vulgare | Iran | KF144906 | KF144998 | KF145040 | |
| CBS 114318 | Leaf of Hordeum vulgare | Iran | KF144907 | KF144999 | KF145041 | |
| A. phragmites | CPC18900* | Culms of Phragmites australis | Italy | KF144909 | KF145001 | KF145043 |
| A. pseudoparenchymaticum | LC7234* = CGMCC 3.18336 | Leaf of bamboo | China | KY494743 | KY705211 | KY705139 |
| LC8173 | Leaf of bamboo | China | KY494753 | KY705221 | KY705149 | |
| LC8174 | Leaf of bamboo | China | KY494754 | KY705222 | KY705150 | |
| A. pseudosinense | CPC 21546* | Leaf of bamboo | The Netherlands | KF144910 | – | KF145044 |
| A. pseudospegazzinii | CBS 102052* | Macaranga hullettii stem colonised by ants | Malaysia | KF144911 | KF145002 | KF145045 |
| A. pterospermum | CPC 20193* | Leaf lesion of Machaerina sinclairii | Australia | KF144913 | KF145004 | KF145046 |
| A. puccinioides | CBS 549.86 | Leaf of Lepidosperma gladiatum | Germany | AB220253 | AB220300 | – |
| A. rasikravindrii | CBS 337.61 | Cissus sp. | The Netherlands | KF144914 | – | – |
| CPC 21602 | Rice | Thailand | KF144915 | – | – | |
| MFLUCC 15-0203 | Dead bamboo culms | Thailand | KU940143 | – | – | |
| MFLUCC 11-0616 | Dead bamboo culms | Thailand | KU940144 | – | – | |
| NFCCI 2144* | Soil | Svalbard | JF326454 | – | – | |
| LC5449 | Soil in karst cave | China | KY494713 | KY705182 | KY705112 | |
| LC7115 | Leaf of bamboo | China | KY494721 | KY705189 | KY705118 | |
| LC7117 | Leaf of bamboo | China | KY494722 | KY705190 | KY705119 | |
| LC7119 | Leaf of bamboo | China | KY494724 | KY705192 | KY705121 | |
| LC7120 | Leaf of bamboo | China | KY494725 | KY705193 | KY705122 | |
| LC7126 | Leaf of bamboo | China | KY494729 | KY705197 | KY705125 | |
| LC7129 | Leaf of bamboo | China | KY494731 | KY705199 | KY705127 | |
| LC7135 | Leaf of bamboo | China | KY494732 | KY705200 | KY705128 | |
| LC7139 | Leaf of bamboo | China | KY494733 | KY705201 | KY705129 | |
| LC7141 | Leaf of bamboo | China | KY494734 | KY705202 | KY705130 | |
| LC7142 | Leaf of bamboo | China | KY494735 | KY705203 | KY705131 | |
| LC7251 | Leaf of bamboo | China | KY494746 | KY705214 | KY705142 | |
| LC7254 | Leaf of bamboo | China | KY494748 | KY705216 | KY705144 | |
| LC8179 | Brassica capestris | China | KY494759 | KY705227 | KY705155 | |
| LC8180 | Brassica capestris | China | KY494760 | KY705228 | KY705156 | |
| A. sacchari | CBS 212.30 | Phragmites australis | United Kingdom | KF144916 | KF145005 | KF145047 |
| CBS 301.49 | Bamboo | Indonesia | KF144917 | KF145006 | KF145048 | |
| A. saccharicola | CBS 191.73 | Air | The Netherlands | KF144920 | KF145009 | KF145051 |
| CBS 334.86 | Dead culms of Phragmites australis | France | AB220257 | KF145010 | KF145052 | |
| CBS 463.83 | Dead culms of Phragmites australis | The Netherlands | KF144921 | KF145011 | KF145053 | |
| A. serenense | IMI 326869* | Food, pharmaceutical excipients, atmosphere and home dust | Spain | AB220250 | AB220297 | – |
| A. subglobosum | MFLUCC 11-0397* | Dead bamboo culms | Thailand | KR069112 | – | – |
| A. subroseum | LC7215 | Leaf of bamboo | China | KY494740 | KY705208 | KY705136 |
| LC7291 | Leaf of bamboo | China | KY494751 | KY705219 | KY705147 | |
| LC7292* =CGMCC3.18337 | Leaf of bamboo | China | KY494752 | KY705220 | KY705148 | |
| A. thailandicum | MFLUCC 15-0202* | Dead bamboo culms | Thailand | KU940145 | – | – |
| LC5630 | Rotten wood | China | KY494714 | KY806200 | KY705113 | |
| A. xenocordella | CBS 478.86* | Soil from roadway | Zimbabwe | KF144925 | KF145013 | KF145055 |
| LC3486 | Camellia sinensis | China | KY494687 | KY705158 | KY705086 | |
| A. yunnanum | MFLUCC 15-0002* | Decaying bamboo culms | China | KU940147 | – | – |
| N. gorlenkoana | CBS 480.73 | Vitis vinifera | Kazakhstan | KX986048 | KY019456 | KY019420 |
*= type strains, strains and sequences generated in this study are shown in bold.
1 CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CGMCC: China General Microbiological Culture Collection; CPC: Culture collection of Pedro Crous, housed at the Westerdijk Fungal Biodiversity Institute; DAOM: Canadian Collection of Fungal Cultures, Ottawa, Canada; DSM: Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany; IMI: Culture collection of CABI Europe UK Centre, Egham, UK; IFO: Institute for Fermentation, Osaka; LC: Working collection of Lei Cai, housed at CAS, China; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; MCC: Microbial Culture Collection of India; NFCCI: National Fungal Culture Collection of India.
Fungus-host distribution of Arthrinium species
To determine the distribution of Arthrinium species on host/substrate, the number of species occurred on each host (based on family level) was counted based on data from this study, relevant literature and the USDA fungal database (https://nt.ars-grin.gov/fungaldatabases/). The proportion account for the known 66 species in Arthrinium (Index Fungorum) was illustrated in a histogram. Four species with an unknown host range were not included in this analysis.
Results
Phylogeny
The combined ITS, TUB2 and TEF1 dataset contained 75 strains, with Nigrospora gorlenkoana CBS 480.73 as the out group. For the Bayesian analyses, the best-fit models TrN+I+G, GTR+I+G, HKY+I+G were selected for ITS, TUB2 and TEF1 loci, respectively. The ML analysis showed the same tree topology as that obtained in the Bayesian analysis. All the Arthrinium strains in this study separated into 13 clades, representing five known (A. arundinis, A. hydei, A. rasikravindrii, A. thailandicum, A. xenocordella) and eight new species (Figure 1). The eight new species clustered in distinct clades with high bootstrap supports (Figure 1). Phylogenetic analyses based on an individual locus were also conducted (not shown) and the generated trees are similar to the one generated from the combined multi-locus dataset (Figure 1).
Host associated with Arthrinium species
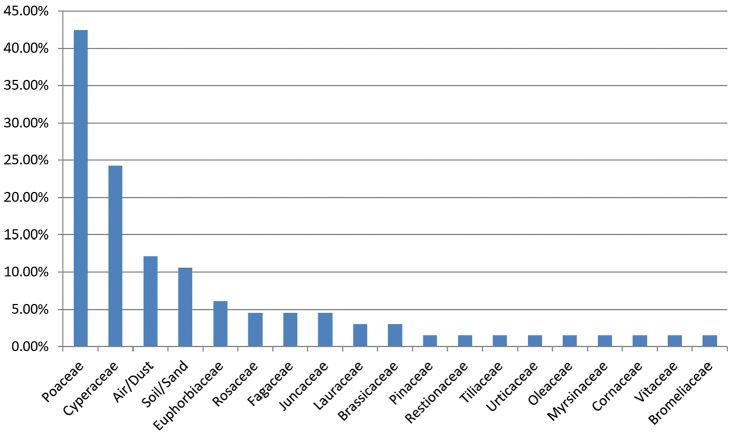
The histogram in Figure 2 shows that Arthrinium species were widely distributed amongst 17 plant families, including Brassicaceae, Bromeliaceae, Cornaceae, Cyperaceae, Euphorbiaceae, Fagaceae, Juncaceae, Lauraceae, Myrsinaceae, Oleaceae, Pinaceae, Poaceae, Restionaceae, Rosaceae, Tiliaceae, Urticaceae and Vitaceae. Arthrinium species were also isolated from air, dust, soil and sand. The proportion of species occurring on each host family was assessed (Figure 2). Poaceae and Cyperaceae were the major host families for Arthrinium, which accounted for 42.42% and 24.24% of species in Arthrinium respectively.
Figure 2.
A Histogram to show fungus-host distribution of Arthrinium species.
Taxonomy
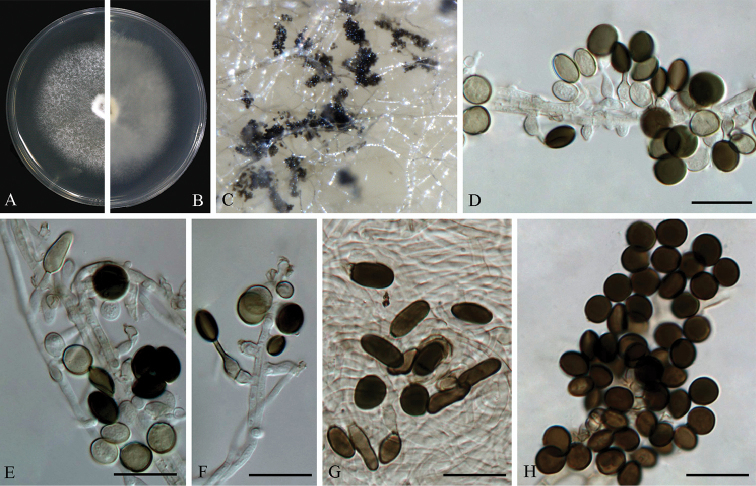
Arthrinium bambusae
M. Wang & L. Cai sp. nov.
824906
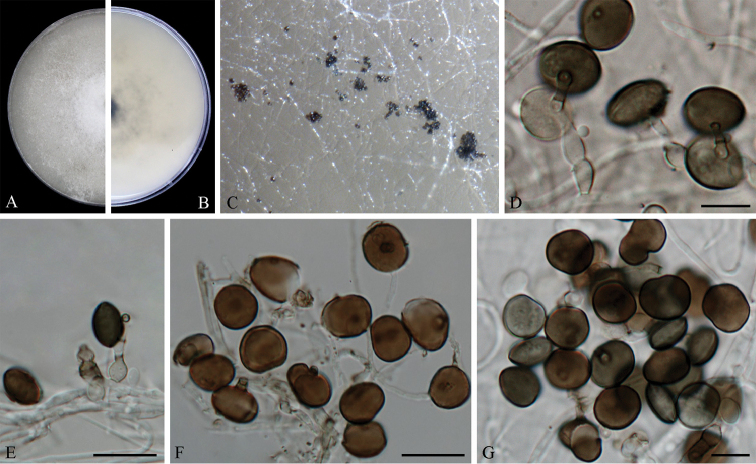
Figure 3.
Arthrinium bambusae (from ex-holotype strain CGMCC 3.18335) A–B 7 d old cultures on PDA C Colony on MEA producing conidia masses D–F Conidiogenous cells giving rise to conidia G Conidia. Scale bars = 10 μm.
Type.
CHINA, Guangdong Province, on bamboo leaves, 10 Jul. 2016, D.W. Xiao, (holotype: HMAS 247187; culture ex-type: CGMCC 3.18335 = LC7106).
Etymology.
Named after the host of the holotype.
Description.
Hyphae hyaline, branched, septate, 1.5–5.0 μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells erect, aggregated in clusters on hyphae, hyaline to pale brown, smooth, doliiform to ampulliform, or lageniform, 4.0–12.0 × 3.0–7.0 μm (x̄ = 6.6 ± 1.8 × 4.8 ± 0.9, n = 30). Conidia olivaceous to brown, smooth to finely roughened, subglobose to ellipsoid, 11.5–15.5 × 7.0–14.0 μm (x̄ = 13.2 ± 0.8 × 11.4 ± 1.2, n = 50).
Culture characteristics.
On PDA, colonies flat, spreading, margin circular, with abundant aerial mycelia, surface and reverse white to grey. On MEA, colonies flat, spreading, surface and reverse brown to black.
Additional specimens examined.
CHINA, Jiangxi Province, on bamboo leaves, 10 Jul. 2016, Q. Xiong, living culture LC7246; Guangdong Province, on bamboo leaves, 10 Jul. 2016, D.W. Xiao, living culture LC7107; ibid. living culture LC7113; ibid. living culture LC7124; ibid. living culture LC7125; ibid. living culture LC7128.
Notes.
Seven strains representing A. bambusae clustered in a well-supported clade closely related to A. subroseum (98% sequence similarity in ITS; 92% in TUB2; 96% in TEF1). Arthrinium bambusae differs from A. subroseum in the morphology of conidiophore (reduced to conidiogenous cells in A. bambusae vs. erect or ascending, clustered in groups in A. subroseum). Moreover, A. bambusae does not produce pigment on the PDA.
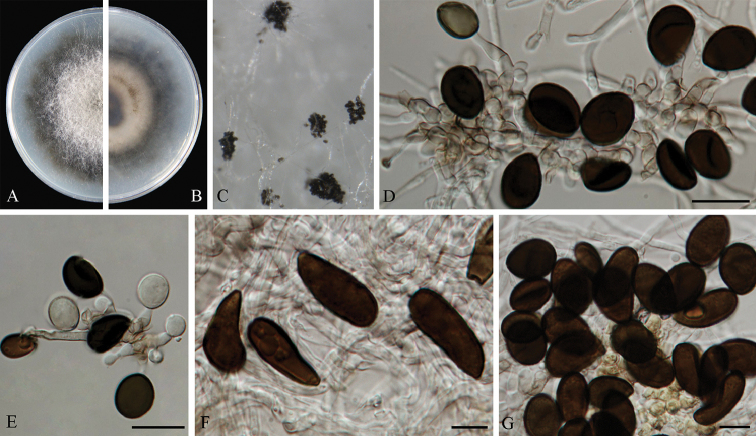
Arthrinium camelliae-sinensis
M. Wang, F. Liu & L. Cai sp. nov.
824907
Figure 4.
Arthrinium camelliae-sinensis (from ex-holotype strain CGMCC 3.18333) A–B 7 d old cultures on PDA C Colony on MEA with bamboo leaves producing conidia masses D–F Conidiogenous cells giving rise to conidia G Conidia. Scale bars = 10 μm.
Type.
CHINA, Jiangxi Province, on Camellia sinensis, 22 Apr. 2013, Q. Chen, (holotype: HMAS 247186; culture ex-type: CGMCC 3.18333 = LC5007).
Etymology.
Named with the host plant of the type.
Description.
Hyphae hyaline, branched, septate, 2.0–4.5 μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells erect, aggregated in clusters, hyaline to pale brown, smooth, doliiform to ampulliform, 4.0–9.5 × 3.0–6.0 μm (x̄ = 6.1 ± 1.4 × 4.4 ± 0.9, n = 30). Conidia brown to dark brown, smooth, globose to subglobose, 9.0–13.5 × 7.0–12.0 μm (x̄ = 11.1 ± 0.9 × 10.1 ± 1.0, n = 50).
Culture characteristics.
On PDA, colonies flat, margin circular, initially white, becoming greyish on surface, reaching 9 cm in 7 days at 25 °C. On MEA, with sparse aerial mycelia, surface dirty white, reverse pale luteous.
Other specimens.
CHINA, Hubei Province, on Brassica campestris, 31 Mar. 2016, Y.Z. Zhao, living culture LC8181 = LF1498.
Notes.
Two strains representing A. camelliae-sinensis clustered in a well-supported clade and appeared closely related to A. jiangxiense (97% sequence similarity in ITS; 94% in TUB2; 94% in TEF1) and A. obovatum (98% sequence similarity in ITS; 95% in TUB2; 93% in TEF1). While A. camelliae-sinensis is distinct from A. jiangxiense in its larger conidia (globose or subglobose, 9.0–13.5 × 7.0–12.0 μm in A. camelliae-sinensis vs. surface view 7.5–10.0 μm diam, side view 4.5–7.0 μm diam in A. jiangxiense) and conidiogenous cell arrangement (aggregated irregularly on hyphae vs. scattered on hyphae in A. jiangxiense) and distinct from A. obovatum in the lack of obovoid conidia (see the note under A. obovatum).
Arthrinium dichotomanthi
M. Wang & L. Cai sp. nov.
824908
Figure 5.
Arthrinium dichotomanthi (from ex-holotype strain CGMCC 3.18332) A–B 7 d old cultures on PDA C Colony on MEA producing conidia masses D–F Conidiogenous cells giving rise to conidia G Conidia. Scale bars = 10 μm.
Type.
CHINA, Chongqing, on Dichotomanthus tristaniaecarpa, 20 Dec. 2012, L. Cai, (holotype: HMAS 247185; culture ex-type: CGMCC 3.18332 = LC4950).
Etymology.
Named after the host from which it was isolated.
Description.
Hyphae hyaline, branched, septate, 1.5–5.0 μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells erect, aggregated in clusters on hyphae, hyaline to pale brown, smooth, doliiform to clavate or lageniform, 5.5–11.0 × 3.0–5.0 μm (x̄ = 7.9 ± 1.4 × 4.0 ± 0.5, n = 30). Conidia brown to dark brown, smooth to finely roughened, globose, subglobose to lenticular, with a longitudinal germ slit, 9.0–15.0 × 6.0–12.0 μm (x̄ = 12.0 ± 1.4 × 8.5 ± 1.1, n = 50).
Culture characteristics.
On PDA, colonies umbonate, margin irregular, with sparse aerial mycelia. Colonies creamy-white to greyish without patches reverse, reaching 9 cm in 7 days at 25 °C. On MEA, colonies flat, spreading, surface and reverse pale luteous.
Other specimens.
CHINA, Chongqing, on Dichotomanthus tristaniaecarpa, 20 Dec. 2012, L. Cai, living culture LC8175 = WM529; ibid. living culture LC8176 = WM 530.
Notes.
Three strains representing A. dichotomanthi formed a distinct clade closely related to A. phaeospermum (Corda) M.B. Ellis (99% sequence similarity in ITS; 96% in TUB2; 96% in TEF1), A. serenense Larrondo & Calvo (99% sequence similarity in ITS; 95% in TUB2) and A. saccharicola F. Stevens (99% sequence similarity in ITS; 95% in TUB2; 97% in TEF1). Arthrinium dichotomanthi differs from A. phaeospermum and A. saccharicola in its larger conidia (globose or subglobose, 9.0–15.0 × 6.0–12.0 μm in A. dichotomanthi vs. surface view (9–)10(–12) μm diam, side view 6–7 μm diam in A. phaeospermum, surface view (7–)8–9(–10) μm diam, side view (4–)5(–6) μm diam in A. saccharicola) and from A. serenense by the absence of odour on the MEA colony (Larrondo 1990).
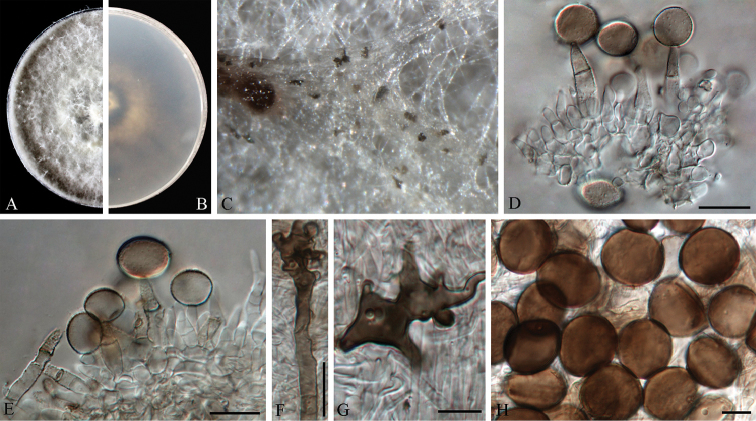
Arthrinium guizhouense
M. Wang & L. Cai sp. nov.
824909
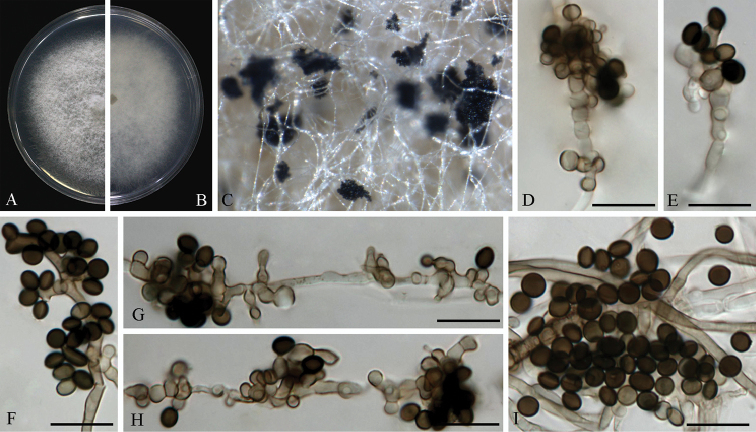
Figure 6.
Arthrinium guizhouense (from ex-holotype strain CGMCC 3.18334) A–B 6 d old cultures on PDA C Colony on MEA producing conidia masses D–H Conidiogenous cells giving rise to conidia I Conidia. Scale bars = 10 μm.
Type.
CHINA, Guizhou Province, from the air in karst cave, 23 Jul. 2014, Z.F. Zhang, (holotype: HMAS 247188; culture ex-type: CGMCC 3.18334 = LC5322).
Etymology.
Named after the province where type was collected, Guizhou province.
Description.
Hyphae hyaline, branched, septate, 1.5–5.0 μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells erect, aggregated in clusters on hyphae, pale brown, smooth, subglobose, ampulliform or doliiform, 3.5–8.0 × 3.0 – 4.5 μm (x̄ =5.1 ± 1.08 × 3.7 ± 0.49, n = 30). Conidia dark brown to black, smooth to finely roughened, globose or subglobose, occasionally elongated to ellipsoidal, with a longitudinal, hyaline, thin, germ slit, 5.0–7.5 × 4.0–7.0 μm (x̄ = 6.1 ± 0.5 × 5.5 ± 0.6, n = 50).
Culture characteristics.
On PDA, colonies flat, woolly, margin circular, with moderate aerial mycelia, surface initially white, becoming greyish and reverse with black patches, reaching 9 cm in 9 days at 25 °C. On MEA, surface dirty white with patches of olivaceous-grey and reverse greyish.
Other specimens examined.
CHINA, Guizhou Province, from the air in karst cave, 23 Jul. 2014, Z.F. Zhang, living culture LC5318.
Notes.
Arthrinium guizhouense is closely related to A. sacchari (Speg.) M.B. Ellis (99% sequence similarity in ITS; 99% in TUB2; 94% in TEF1). Morphologically, A. guizhouense and A. sacchari are very similar in conidial size, but A. guizhouensis produces relatively shorter conidiogenous cells (3.5–8.0 μm in A. guizhouense vs. 5–12 μm in A. sacchari).
Arthrinium jiangxiense
M. Wang & L. Cai sp. nov.
824910
Figure 7.
Arthrinium jiangxiense (from ex-holotype strain CGMCC 3.18381) A–B 5 d old cultures on PDA C Colony on MEA producing conidia masses D–F Conidiogenous cells giving rise to conidia G Elongated conidia H Conidia. Scale bars = 10 μm.
Type.
CHINA, Jiangxi Province, on Maesa sp., 05 Sept. 2013, Y.H. Gao, (holotype: HMAS 247183; culture ex-type: CGMCC3.18381 = LC4577).
Etymology.
Named after the province where the most strains of this species were collected, Jiangxi.
Description.
Hyphae hyaline, branched, septate, 1.5–5.0 μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells erect, scattered or aggregated in clusters on hyphae, hyaline to pale brown, smooth, ampulliform, 6.0–15.0 × 2.5–5.0 μm (x̄ = 9.7 ± 2.6 × 3.7 ± 0.6, n = 30), apical neck 2.5–6.0 μm long, basal part 3.0–9.0 μm long. Conidia brown, smooth to finely roughened, granular, globose to ellipsoid in surface view, 7.5–10.0 μm diam (x̄ = 8.7 ± 0.6, n = 50), lenticular in side view, with longitudinal, pale germ slit, 4.5–7.0 μm diam (x̄ = 5.8 ± 0.6, n = 50). Sterile cells forming on solitary loci on hyphae, brown, finely roughened, subcylindrical to clavate.
Culture characteristics.
On PDA, colonies flat, woolly, margin circular, with sparse aerial mycelia, initially white, becoming greyish due to sporulation, reaching 9 cm in 10 days at 25 °C, on MEA, sienna with patches of luteous, reverse luteous to sienna.
Other specimens examined.
CHINA, Hunan Province, on bamboo, 22 Sept. 2010, L. Cai, living culture LC2831; Jiangxi Province, on Phyllostachys sp., 05 Sept. 2013, Y.H. Gao, living culture LC4494; on Phyllostachys sp., 22 Apr. 2013, Q. Chen, living culture LC4993; ibid. living culture LC4497; ibid. living culture LC5001; ibid. living culture LC5004; on Imperata cylindrical, 22 Apr. 2013, Q. Chen, living culture LC5015; on Maesa sp., 05 Sept. 2013, Y.H. Gao, living culture LC4541; on Machilus sp., 05 Sept. 2013, Y.H. Gao, living culture LC4547; on Camellia sinensis, 05 Sept. 2013, Y.H. Gao, living culture LC4578; on bamboo, 01 Jul. 2016, J.E. Huang, living culture LC7104; ibid. living culture LC7154; ibid. living culture LC7156; ibid. living culture LC7275.
Notes.
Two strains representing Arthrinium jiangxiense clustered in a well-supported clade and appeared closely related to A. camelliae-sinensis (97% sequence similarity in ITS; 94% in TUB2; 94% in TEF1). While A. jiangxiensis is distinct from A. camelliae-sinensis in its smaller conidia (surface view 7.5–10.0 μm diam, side view 4.5–7.0 μm diam in A. jiangxiensis vs. globose or subglobose, 9.0–13.5 × 7.0–12.0 μm in A. camelliae-sinensis) and conidiogenous cell arrangements (conidiogenous cells scattered on hyphae vs. aggregated irregularly on hyphae in A. jiangxiense).
Arthrinium obovatum
M. Wang & L. Cai sp. nov.
824911
Figure 8.
Arthrinium obovatum (from ex-holotype strain CGMCC 3.18331) A–B 7 d old cultures on PDA C Colony on MEA producing conidia masses D–E Conidiogenous cells giving rise to conidia F Obovoid conidia G Globose to subglobose conidia. Scale bars = 10 μm.
Type.
CHINA, Chongqing, on Lithocarpus sp., 20 Dec. 2012, L. Cai, (holotype: HMAS 247184; culture ex-type: CGMCC 3.18331 = LC4940).
Etymology.
Referring to the production of the large obovoid conidia.
Description.
Hyphae hyaline to pale brown, branched, septate, 1.5–5.0 μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells erect, aggregated in clusters on hyphae, pale brown, smooth, subcylindrical or clavate, 5.5–13.5 × 2.5–5.0 μm (x̄ = 8.7 ± 2.4 × 3.6 ± 0.6, n = 30). Conidia dark brown, roughened, globose to subglobose, 11.0–16.5 μm (x̄ = 13.8 ± 1.5, n = 50) in diam.; obovoid, 16.0–31.0 × 9.0–16.0 μm (x̄ = 23.0 ± 2.7 × 12.7 ± 1.4, n = 50), occasionally elongated to ellipsoidal.
Culture characteristics.
On PDA, colonies flat, spreading, margin circular, initially white, becoming olivaceous-grey on surface, reverse smoke-grey with patches of olivaceous grey, reaching 9 cm in 7 days at 25 °C. On MEA, surface olivaceous grey in the central and luteous around, reverse with patches of olivaceous grey.
Other specimens examined.
CHINA, Chongqing, on Lithocarpus sp., 20 Dec. 2012, L. Cai, living culture LC8177; ibid. living culture LC8178.
Notes.
Arthrinium obovatum is the only species that produces obovoid conidia (Figure 8F) in this genus, a character distinctly different from other species (Ellis 1965, 1976, Gjaerum 1967, Pollack and Benjamin 1969, Hudson et al. 1976, Calvo and Guarro 1980, Khan and Sullia 1980, Samuels et al. 1981, von Arx 1981, Koskela 1983, Kirk 1986, Larrando and Calvo 1990, 1992, Müller 1992, Bhat and Kendrick 1993, Hyde et al. 1998, Jones et al. 2009, Singh et al. 2012, Crous et al. 2013, 2015, Sharma et al. 2014, Senanayake et al. 2015, Senanayake et al. 2015, Hyde et al. 2016, Dai et al. 2016a, b).
Arthrinium pseudoparenchymaticum
M. Wang & L. Cai sp. nov.
824912
Figure 9.
Arthrinium pseudoparenchymaticum (from ex-holotype strain CGMCC 3.18336) A–B 8 d old cultures on PDA C Colony on MEA producing conidia masses D–E Conidiogenous cells giving rise to conidia F–G Dentate conidia H Globose conidia. Scale bars = 10 μm.
Type.
CHINA, Guangdong Province, on bamboo, Jul. 2016, D.W. Xiao, (holotype: HMAS 247189; culture ex-type: CGMCC 3.18336 = LC7234).
Etymology.
Referring to the pseudoparenchymatous hyphae.
Description.
Hyphae hyaline to pale brown, branched, septate, 1.5–5.0 μm diam., pseudoparenchymatous. Conidiophores aggregated in hyaline to light brown sporodochia, smooth, usually unbranched, up to 40 μm long, 3–6 μm width. Conidiogenous cells hyaline to pale yellow, smooth to finely roughened, subcylindrical to doliiform, 8.0–18.5 × 3.0–8.5μm (x̄ = 13.7 ± 3.2 × 5.4 ± 1.2, n = 30). Conidia pale to dark brown, smooth, finely guttulate, globose to subglobose, 13.5–27.0 × 12.0–23.5 μm (x̄ = 20.2 ± 2.5 × 17.1 ± 2.4, n = 50). Sometimes lobed or dentate, polygonal or irregular in surface view.
Culture characteristics.
On PDA, colonies flat, spreading, margin circular, with moderate aerial mycelia, initially white, becoming grey on surface, reverse smoke-grey without patches, reaching 9 cm in 8 days at 25 °C. On MEA, surface pale luteous to grey with abundant mycelia, reverse greyish without patches.
Other specimens examined.
CHINA, Guangdong Province, on bamboo, Jul. 2016, D.W. Xiao, living culture LC8173; ibid. living culture LC8174.
Notes.
Arthrinium pseudoparenchymaticum is closely related to A. hyphopodii (94% sequence similarity in ITS), but differs in its much larger conidia (13.5–27.0 × 12.0–23.5 μm vs. 5–10 × 4–8 μm), the absence of hyphopodia and the presence of dentate conidia.
Arthrinium subroseum
M. Wang & L. Cai sp. nov.
824913
Figure 10.
Arthrinium subroseum (from ex-holotype strain CGMCC3.18337) A–B 10 d old cultures on PDA C Colony on MEA producing conidia masses D–E Conidiogenous cells giving rise to conidia F–G Conidia. Scale bars = 10 μm.
Type.
CHINA, Jiangxi Province, on bamboo, 1 Jul. 2016, J.E. Huang, (holotype: HMAS 247190; culture ex-type: CGMCC3.18337 = LC7292).
Etymology.
Named after the colour of colony on PDA, pinkish.
Description.
Hyphae hyaline to pale brown, branched, septate, 1.5–6.0 μm diam. Conidiophores hyaline to pale brown, smooth, erect or ascending, simple, flexuous, subcylindrical, clustered in groups. Conidiophores aggregated in brown sporodochia, smooth, hyaline to brown, up to 20 μm long, 2–4.5 μm width. Conidiogenous cells pale brown, smooth, doliiform to subcylindrical, 3.0–6.5 × 2.0–5.0 μm (x̄ = 4.7 ± 1.2 × 3.7 ± 0.9, n = 30). Conidia pale brown to dark brown, smooth, globose to subglobose or ellipsoidal, 12.0–17.5 × 9.0–16.0 μm (x̄ = 14.9 ± 1.4 × 11.8 ± 1.8, n = 50).
Culture characteristics.
On PDA, colonies flat, spreading, margin circular, with moderate aerial mycelia, initially white, becoming light pink on surface, reverse peach-puff without patches, reaching 10 cm in 8 days at 25 °C. On MEA, surface blackish-green with abundant mycelia, reverse with patches of greyish.
Other specimens.
CHINA, Jiangxi Province, on bamboo, 1 Jul. 2016, J.E. Huang, living culture LC7215; ibid. living culture LC7291.
Notes.
Three strains representing A. subroseum clustered in a well-supported clade, closely related to A. garethjonesii (94% sequence similarity in ITS) and A. bambusae (98% sequence similarity in ITS; 92% in TUB2; 96% in TEF1). However, A. subroseum differs from A. bambusae in the morphology of conidiophores (erect or ascending, clustered in groups in A. subroseum vs. reduced to conidiogenous cells in A. bambusae). Arthrinium subroseum is not morphologically comparable to A. garethjonesii, whose asexual morph is undetermined (Dai et al. 2016b).
Discussion
Arthrinium, Cordella and Pteroconium share similar morphological characters, e.g. basauxic conidiophores with terminal and intercalary polyblastic conidiogenous cells and brown, unicellular conidia with a pallid germ slit (Ellis 1971, Hyde et al. 1998). Crous et al. (2013) reduced both Cordella and Pteroconium as generic synonyms of Arthrinium based on molecular phylogenetic data and regarded traditionally applied morphological characters in distinguishing these genera as phylogenetically insignificant. This study added eight novel species and our data are in good accordance with that of Crous et al. (2013). For example, A. pseudoparenchymaticum is sporodochial and pseudoparenchymatous, which would be classified as Pteroconium in the traditional taxonomy. However, the multi-locus (ITS, TEF1 & TUB2) tree (Figure. 1) shows that A. pseudoparenchymaticum is phylogenetically distant from A. pterospermum (syn. P. pterospermum, the type of “Pteroconium”).
Currently there are 70 recognised species in Arthrinium (Index Fungorum), occurring on a wide variety of both living and decaying plant materials. It is noteworthy that Arthrinium species showed distinct preference for growing on two graminaceous families, Poaceae and Cyperaceae, amongst which, Bambusa (Poaceae) and Carex (Cyperaceae) are two of the most common host genera for Arthrinium species. For example, seven species have been recorded from Carex spp., i.e. A. austriacum Petr. (1959), A. caricicola Kunze (1817), A. globosum Koskela (1983), A. kamtschaticum Tranzschel & Woron (1914), A. morthieri Fuckel (1870), A. muelleri Ellis (1976) and A. naviculare Rostr. (1886). Bamboo has been widely known as a favourable host for Arthrinium, e.g. A. hyphopodii, A. longistromum, A. subglobosum, A. thailandicum and A. yunnanum (Senanayake et al. 2015, Dai et al. 2016). In this study, three new species (A. bambusae, A. subroseum and A. pseudoparenchymaticum) were also isolated from bamboo. In addition, three species (A. arundinis, A. guizhouense, and A. rasikravindrii) were isolated from air and soil from karst caves, where have been shown to encompass a high fungal diversity (Jiang et al. 2017, Zhang et al. 2017).
In addition to the Arthrinium species from China, we also tried to resolve the phylogenetic status of Arthrinium mytilomorphum Bhat & W.B. Kendr. (Bhat and Kendrick 1993) in the current study. DNA extraction from the type specimen of A. mytilomorphum (DAOM 214595) was prohibited but DAOM provided a DNA sample. Unfortunately, we only managed to obtain an ITS sequence from this DNA sample, while the amplifications of all other protein coding genes were unsuccessful. The ITS phylogenetic tree (not shown here) shows that A. mytilomorphum is closely related to A. subroseum (99 % sequence similarity in ITS), while the morphology of these two species are very different from each other. Conidia of A. mytilomorphum are dark brown, fusiform or navicular, measuring 20–30 × 6–8.5 μm, slightly bowed down and asymmetric (Figure 11), while those of A. subroseum are pale brown to dark brown, globose or subglobose, measuring 12–17.5 × 9–16 μm.
Figure 11.
Arthrinium mytilomorphum (from holotype DAOM 214595) A–B Overview of the type specimen C–F Conidiogenous cells giving rise to conidia G Conidia. Scale bars = 10 μm.
Teleomorph-typified genus Apiospora was treated as a synonym of anamorph-typified genus Arthrinium on the basis that Arthrinium is older and more commonly used in literature (Crous et al. 2013). However, only three of the 58 recorded Apiospora species have been properly linked to their known Arthrinium counterparts, i.e. Arthrinium hysterinum (syn. Ap. bambusae) (Sivanesan 1983, Kirk 1986); Arthrinium arundinis (syn. Ap. montagnei) (Hyde 1998); Arthrinium sinense (syn. Ap. sinensis) (Réblová et al. 2016). In addition, molecular data of only four Apiospora species (Ap. bambusae, Ap. montagnei, Ap. setosa and Ap. sinensis) are available, in which only A. bambusae and A. sinensis have type-derived sequences. A comprehensive taxonomic revision of this taxonomic group awaits fresh collection and epitypification of many Apiospora species and, based on which, phylogenetic links with Arthrinium species could be established.
Supplementary Material
Acknowledgments
We thank Peng Zhao, Qian Chen, Yahui Gao and Zhifeng Zhang for providing strains and technical assistance. We kindly appreciated the curator of Agriculture and Agri-Food Canada herbarium and Dr. Wen Chen in Ottawa Research and Development Centre AAFC for providing DNA samples and microscope slides of the type specimen of Arthrinium mytilomorphum. This work was financially supported by the National Science Fund for Distinguished Young Scholars of China (NSFC 31725001) and the Frontier Science Research Project of the Chinese Academy of Sciences (QYZDB-SSW-SMC044).
Citation
Wang M, Tan X-M, Liu F, Cai L (2018) Eight new Arthrinium species from China. MycoKeys 34: 1–24. https://doi.org/10.3897/mycokeys.34.24221
References
- von Arx JA. (1981) The genera of fungi sporulating in pure culture (3rd edn). Cramer Vaduz, 424 pp.
- Bhat DJ, Kendrick WB. (1993) Twenty-five new conidial fungi from the Western Ghats and the Andaman Islands (India). Mycotaxon 49: 19–90. [Google Scholar]
- Cai L, Hyde KD, Taylor PW, Weir B, Waller J, Abang MM, Zhang JZ, Yang YL, Phoulivong S, Liu ZY, Prihastuti H. (2009) A polyphasic approach for studying Colletotrichum. Fungal Diversity 39: 204.
- Calvo A, Guarro J. (1980) Arthrinium aureum sp. nov. from Spain. Transactions of the British Mycological Society 75: 156–157. https://doi.org/10.1016/S0007-1536(80)80208-7 [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. https://doi.org/10.2307/3761358 [Google Scholar]
- Chen K, Wu XQ, Huang MX, Han YY. (2014) First report of brown culm streak of Phyllostachys praecox caused by Arthrinium arundinis in Nanjing, China. Plant Disease 98: 1274. https://doi.org/10.1094/PDIS-02-14-0165-PDN [DOI] [PubMed]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Groenewald JZ. (2013) A phylogenetic re-evaluation of Arthrinium. IMA fungus 4: 133–154. https://doi.org/10.5598/imafungus.2013.04.01.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Le Roux JJ. et al. (2015) Fungal planet description sheets: 371–399. Persoonia 35: 264–327. https://doi.org/10.3767/003158515X690269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DQ, Jiang HB, Tang LZ, Bhat DJ. (2016b) Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere 7: 1332–1345. https://doi.org/10.5943/mycosphere/7/9/7 [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2017a) Bambusicolous fungi. Fungal Diversity 82: 1–105. https://doi.org/10.1007/s13225-016-0367-8 [Google Scholar]
- Ellis MB. (1965) Dematiaceous Hyphomycetes. VI. Mycological Papers 103: 1–46. [Google Scholar]
- Ellis MB. (1971) Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew, 608 pp. [Google Scholar]
- Ellis MB. (1976) More Dematiaceous Hyphomycetes. CAB International Mycological Institute, Kew, 507 pp. [Google Scholar]
- Farr DF, Rossman AY. (2017) Fungal Databases, U.S. National Fungus Collections, ARS, USDA. https://nt.ars-grin.gov/fungaldatabases/
- Gjaerum HB. (1967) Arthrinium morthieri, A. fuckelii n. sp., and A. ushuvaiense. Nytt magasin for Botanikk 14: 1–6. [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LD, Hyde KD, Liew ECY. (2000) Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytologist 147: 617–630. https://doi.org/10.1046/j.1469-8137.2000.00716.x [DOI] [PubMed] [Google Scholar]
- de Hoog GS, Guarro J, Gene J. et al. (2000) Atlas of Clinical Fungi (2nd edn). Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands, 1–1160.
- Hong JH, Jang S, Heo YM, Min M, Lee H, Lee YM, Lee H, Kim JJ. (2015) Investigation of marine-derived fungal diversity and their exploitable biological activities. Marine Drugs 13: 4137–4155. https://doi.org/10.3390/md13074137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson HJ, McKenzie EHC, Tommerup IC. (1976) Conidial states of Apiospora Sacc. Transactions of the British Mycological Society 66: 359–362. https://doi.org/10.1016/S0007-1536(76)80075-7 [Google Scholar]
- Hughes SJ. (1953) Conidiophores, conidia, and classification. Canadian Journal of Botany 31: 577–659. https://doi.org/10.1139/b53-046 [Google Scholar]
- Hyde KD, Fröhlich J, Taylor JE. (1998) Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia 50: 21–80. [Google Scholar]
- Hyde KD, Hongsanan S, Jeewon R. et al. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. https://doi.org/10.1007/s13225-016-0373-x [Google Scholar]
- Jiang JR, Cai L, Liu F. (2017) Oligotrophic fungi from a carbonate cave, with three new species of Cephalotrichum. Mycology 8(3): 164–177. https://doi.org/10.1080/21501203.2017.1366370 [Google Scholar]
- Jones EBG, Sakayaroj J, Suetrong S. et al. (2009) Classification of marine Ascomycota, anamorphic taxa and Basidiomycota. Fungal Diversity 35: 1–187. [Google Scholar]
- Khan KR, Sullia SB. (1980) Arthrinium phaeospermum var. indicum var. nov., a new market pathogen of cowpea, garden pea and french bean. Acta Botanica Indica 8: 103–104. [Google Scholar]
- Kirk PM. (1986) New or interesting microfungi. XV. Miscellaneous hyphomycetes from the British Isles. Transactions of the British Mycological Society 86: 409–428. https://doi.org/10.1016/S0007-1536(86)80185-1 [Google Scholar]
- Koskela P. (1983) Arthrinium glahosum, a new hyphomycetous species. Karstenia 23: 13–14. https://doi.org/10.29203/ka.1983.218 [Google Scholar]
- Larrondo JV, Calvo MaA. (1990) Two new species of Arthrinium from Spain. Mycologia 82: 396–398. https://doi.org/10.2307/3759915 [Google Scholar]
- Larrondo JV, Calvo MaA. (1992) New contributions to the study of the genus Arthrinium. Mycologia 84: 475–478. https://doi.org/10.2307/3760203 [Google Scholar]
- Li BJ, Liu PQ, Jiang Y, Weng QY, Chen QH. (2016) First report of culm rot caused by Arthrinium phaeospermum on Phyllostachys viridis in China. Plant Disease 100: 1013. https://doi.org/10.1094/PDIS-08-15-0901-PDN
- Minter DW. (1985) A re-appraisal of the relationships between Arthrinium and other hyphomycetes. Plant Sciences 94: 281–308. [Google Scholar]
- Müller E. (1992) A new parasitic species of Apiospora. Boletin de la Sociedad Argentina de Botanica, La Plata 28: 201–203. [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. https://doi.org/10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences 95: 2044–2049. https://doi.org/10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack FG, Benjamin CR. (1969) https://doi.org/10.2307/3757360
- Posada D. (2008) jModelTest: phylogenetic model averaging. Molecular biology and evolution 25: 1253–1256. https://doi.org/10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Rai MK. (1989) Mycosis in man due to Arthrinium phaeospermum var. indicum. First case report: mykose durch Arthrinium phaeospermum var. indicum beim Menschen. Erstbericht. Mycoses 32: 472–475. https://doi.org/10.1111/j.1439-0507.1989.tb02285.x [DOI] [PubMed] [Google Scholar]
- Rayner RW. (1970) A Mycological Colour Chart. Commonwealth Mycological Institute, Kew, 34 pp. [Google Scholar]
- Réblová M, Miller AN. et al. (2016) Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7: 131–153. https://doi.org/10.5598/imafungus.2016.07.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels GJ, McKenzie EHC, Buchanan DE. (1981) Ascomycetes of New Zealand 3. Two new species of Apiospora and their Arthrinium anamorphs on bamboo. New Zealand Journal of Botany 19: 137–149. https://doi.org/10.1080/0028825X.1981.10425113 [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, Kendrick B. (2011) The genera of hyphomycetes. CBS Biodiversity Series 9, Utrecht, the Netherlands, 1–997.
- Senanayake IC, Maharachchikumbura SS, Hyde KD, Bhat JD, Jones EG, McKenzie EH, Dai DQ, Daranagama DA, Dayarathne MC, Goonasekara ID, Konta S. (2015) Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Diversity 73: 73–144. https://doi.org/10.1007/s13225-015-0340-y [Google Scholar]
- Sharma R, Kulkarni G, Sonawane MS, Shouche YS. (2014) A new endophytic species of Arthrinium (Apiosporaceae) from Jatropha podagrica. Mycoscience 55: 118–123. https://doi.org/10.1016/j.myc.2013.06.004 [Google Scholar]
- Shrestha P, Ibáñez AB, Bauer S, Glassman SI, Szaro TM, Bruns TD, Taylor JW. (2015) Fungi isolated from Miscanthus and sugarcane: biomass conversion, fungal enzymes, and hydrolysis of plant cell wall polymers. Biotechnology for Biofuels 8: 1. https://doi.org/10.1186/s13068-015-0221-3 [DOI] [PMC free article] [PubMed]
- Singh SM, Yadav LS, Singh PN, Hepat R, Sharma R, Singh SK. (2012) Arthrinium rasikravindrii sp. nov. from Svalbard, Norway. Mycotaxon 122: 449–460. https://doi.org/10.5248/122.449 [Google Scholar]
- Sivanesan A. (1983) Studies on ascomycetes. Transactions of the British Mycological Society 81: 313–332. https://doi.org/10.1016/S0007-1536(83)80084-9 [Google Scholar]
- Stamatakis A, Alachiotis N. (2010) Time and memory efficient likelihood-based tree searches on phylogenomic alignments with missing data. Bioinformatics 26: 132–139. https://doi.org/10.1093/bioinformatics/btq205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. https://doi.org/10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee SJ, Taylor JL. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18(1): 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1 [Google Scholar]
- Zhang ZF, Liu F, Zhou X, Liu XZ, Liu SJ, Cai L. (2017) Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia 39(1): 1–31. https://doi.org/10.3767/persoonia.2017.39.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YM, Deng CR, Chen X. (1990) Arthrinium phaeospermum causing dermatomycosis, a new record of China. Acta Mycologica Sinica 9: 232–235. [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3: 4. https://doi.org/10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.