Abstract
In rare disease (RD) research, there is a huge need to systematically collect biomaterials, phenotypic, and genomic data in a standardized way and to make them findable, accessible, interoperable and reusable (FAIR). RD-Connect is a 6 years global infrastructure project initiated in November 2012 that links genomic data with patient registries, biobanks, and clinical bioinformatics tools to create a central research resource for RDs. Here, we present RD-Connect Registry & Biobank Finder, a tool that helps RD researchers to find RD biobanks and registries and provide information on the availability and accessibility of content in each database. The finder concentrates information that is currently sparse on different repositories (inventories, websites, scientific journals, technical reports, etc.), including aggregated data and metadata from participating databases. Aggregated data provided by the finder, if appropriately checked, can be used by researchers who are trying to estimate the prevalence of a RD, to organize a clinical trial on a RD, or to estimate the volume of patients seen by different clinical centers. The finder is also a portal to other RD-Connect tools, providing a link to the RD-Connect Sample Catalogue, a large inventory of RD biological samples available in participating biobanks for RD research. There are several kinds of users and potential uses for the RD-Connect Registry & Biobank Finder, including researchers collaborating with academia and the industry, dealing with the questions of basic, translational, and/or clinical research. As of November 2017, the finder is populated with aggregated data for 222 registries and 21 biobanks.
Introduction
Rare diseases (RDs) are usually life-threatening or chronically debilitating conditions with a very low prevalence and a high level of complexity. In the EU, a disease is considered rare when it affects no more than 1 per 2000 persons [1], in the United States, when it affects fewer than 200,000 people or about 1 in 1500 people [2]. Though individually rare, taken together RDs are common. There are around 6000–7000 rare genetic diseases [3], the most rare of which are estimated to affect about 1/2,000,000 patients [4].
Most RDs are untreatable, and research remains the only option for patients who are striving to get a right diagnosis and a treatment for their condition.
For RDs, supporting collaboration and optimizing the use of limited resources by data sharing is particularly needed, due to the low individual prevalence and the scarcity of information related to each disease [5]. We note here that by data sharing, we do not mean making data public, but rather controlled access to data under well-defined conditions [6].
In particular, RD registries, databases, and biobanks constitute key instruments for increasing knowledge, especially when they allow the pooling of -omics, clinical, and phenotypic data [7].
In the past decades, the advent of methods for large-scale data analyses, and the creation of comprehensive databases for the hosting and exchange of genetic and clinical data has allowed researchers to learn and discover more about the pathogenetic mechanisms underlying the diseases and response to drugs [8]. In the context of next-generation sequencing analysis, as well as clinical outcome measures, the availability of phenotypic data has become key to increase the chances of providing a molecular diagnosis to diseases correct gene identification [9].
The availability of biomaterials and of clinical and -omics data has thus become essential to study disease etiology and pathogenesis paving the way to highly effective targeted therapies in oncology [10] and in several RDs (e.g., Ivacaftor and lumacaftor as potentiator and corrector of CFTR gene for class III CFTR pathogenic variants [11]; Nusinersen for the treatment of spinal muscular atrophy (SMA), functionally converting splicing of the SMN2 into SMN1 gene, which codes for survival motor neuron (SMN) protein [12]).
Funded by the European Union’s Seventh Framework Programme under the International Rare Diseases Research Consortium (IRDiRC), a consortium of national and international funders investing in RD research projects/programs [13], RD-Connect (http://rd-connect.eu/) is a 6 years global infrastructure project initiated in November 2012. It aims to create a unified system and platform for reprocessing, sharing, and analyzing -omics data integrated with phenotypic data and biological samples [14]. The RD-Connect Genome-Phenome Analysis Platform (https://platform.rd-connect.eu/) is designed as key resource for stakeholders of RD research; including registries, biobanks, researchers, patients, etc. The platform is created with a view to preserve patient integrity by involving patient organizations through active two-way dialog between researchers and patients [14], as well as support from experts in ethical, legal, and social issues. RD-Connect is an important partner for European research projects and infrastructures such as the Biobanking and BioMolecular Resources Research Infrastructure-European Research Infrastructure Consortium (BBMRI-ERIC) [15] and ELIXIR, the Distributed Infrastructure for Life Science Information [16]. Close development with partner projects NeurOmics [17] and EURenOmics [18], implementing -omics technologies in the study of rare neurological and renal diseases, is ensuring its usability and relevance of RD-Connect tools for RD research.
The integration of phenotype data, biosamples, and -omics data requires coordinated efforts among different initiatives in order to ensure standardization, comparability, and reproducibility of research results.
In order to be suitable for integrated data analysis, sample and phenotype data need to be collected or mapped into standardized and “computable” form according to selected ontologies. Ontologies are a way to express concepts in standardized terminology that is ordered in a hierarchical/tree structure, often with annotations and links out to other information. Several existing ontologies are suitable for RD research. RD-Connect, in line with the IRDiRC, supports the adoption of the Human Phenotype Ontology (HPO) [19] and the Orphanet Rare Disease Ontology (ORDO) [20] in RD research.
Currently, the phenotype data that are processed in the RD-Connect genomics platform are those collected by partner projects via the software PhenoTips [21], an open source tool for collecting and analyzing phenotypic information for patients with genetic disorders using terms and annotations from the HPO.
With a view to extending the use of the platform to all RD researchers, and to promote intense data sharing among the RD community, including prospective as well as retrospective collections, RD-Connect is providing important advances for accessing RD patient registries and biobanks at an international level by developing dedicated catalogues or resources for RD research [22, 23].
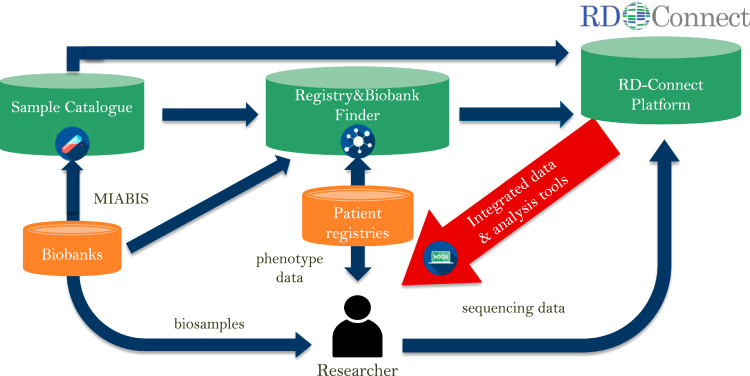
Here, we present the RD-Connect Registry & Biobank Finder (http://catalogue.rd-connect.eu/), a tool that helps to find RD biobanks and registries and provides information on the availability and accessibility of content in each database. RD-Connect Registry & Biobank Finder is also a portal to other RD-Connect tools, providing a direct link to the RD-Connect Sample Catalogue, a large inventory of RD biological samples available in participating biobanks for RD research (Fig. 1). RD-Connect Sample Catalogue is designed as a centralized sample-level catalogue, where researchers can select and request specific biosamples via the platform without needing to contact multiple biobanks separately for samples of interest. The RD-Connect Sample Catalogue is currently available as a beta version (https://samples.rd-connect.eu/); the production version is planned for release in the third quarter of 2017.
Fig. 1.
Registry & Biobank Finder and the RD-Connect infrastructure. Registry & Biobank Finder is integrated with other RD-Connect elements, in particular the RD-Connect Sample Catalogue and the Genomics Platform
RD-Connect Registry & Biobank Finder: a tool for RD registries and biobanks
In the field of RD registries, the possibility to find, access, integrate, and reuse existing data is still limited and there are many barriers to data sharing. Registries only exist for a small fraction of RDs and, conversely, more than one registry exists for certain RDs [24]. Very few registries collect data in a standardized way; most restrict access to external users and are not linked to other databases [25]. Information on the number of RD patients included in a registry is rarely readily available to external users. Registries may provide information in periodic reports or in the scientific literature or, more rarely, via a public website.
Even though directories of biobanks exist, it is difficult to understand which biobank to choose from and whether such biobanks hold trustworthy biological samples for research. Having identified potential biobanks, researchers would then need to navigate individual websites for catalogues to identify actual biological samples or writing several e-mails to request information on sample collections.
Currently, several lists and directories of biobanks and registries are available to the research community on both common and rare diseases, but none of them is providing information on the number of patients or samples included in the databases (Table 1).
Table 1.
Directories including RD biobanks and registries
| Name | Focused on rare diseases? | Focus of directory | Coverage | Information available | URL | Reference |
|---|---|---|---|---|---|---|
| EuroBioBank | Yes | Biobank | Europe/International | Biospecimen collections, disease names, aggregated data (No. of samples), biobank contacts, country | http://www.eurobiobank.org/ | [28] |
| Telethon Network of Genetic Biobanks | Yes | Biobank | Italy | Biospecimen collections, disease names, number of patients, proband families, biobank contacts, metadata | http://biobanknetwork.telethon.it/ | [60] |
| BBMRI-ERIC | No | Biobank | Europe | Type of samples, metadata, biobank name, country, contact details | www.bbmri-eric.eu | [34] |
| NIH Rare Diseases Registry (RaDaR) Program (formerly Global Rare Disease Registry -GRDR) | Yes | Registry | USA | List of registries previously participating in the (GRDR) | https://ncats.nih.gov/radar | [24] |
| Parent | No | Registry | Europe | Registry name, country, primary purpose, affiliation (public/private), geographical coverage, primary observational unit, governing board, data sharing | http://patientregistries.eu/ror | [61] |
| NIH lists | No | Registry | USA | Name of the registry and URL with related information | https://www.nih.gov/health-information/nih-clinical-research-trials-you/list-registries | [62] |
| RoPR | No | Registry | USA | catalogue of registries:classification and purpose, contact and conditions of access, progress reports, common data elements | https://patientregistry.ahrq.gov/ | [63] |
| RD-Connect Registry & Biobank Finder | Yes | Biobank/Registry | International | catalogue of biobanks and registries: | http://catalogue.rd-connect.eu/ | [64] |
| biobank/registry name, country, affiliation, contact details, aggregated data (N. of cases/samples), metadata (study documents, CRF). | ||||||
| Orphanet | Yes | Biobank/Registry | International | biobank/registry name, country, affiliation (public/private), principal investigator, institution, contact details | http://www.orpha.net/consor/cgi-bin/ResearchTrials_RegistriesMaterials.php?lng=EN | [33] |
| P3G | No | Biobank/Registry | International | catalogue of biobanks including study description, networks, questionnaires, physical and cognitive measures, DNA processing and procedures to access. | http://www.p3gobservatory.org/catalogs.htm | [65] |
| ENGAGE | No | Biobank/Registry | International | Cohorts for which data and/or specimens can be made available | http://www.euengage.org/data%20access%20catalogue.html | [66] |
The RD-Connect Registry & Biobank Finder is meant to concentrate information that is currently sparse on different repositories (inventories, databases, websites, scientific journals, technical reports, etc.), starting from identifying registries and biobanks that are collecting data on patients affected by a RD, and on the number of cases or biological samples included in each database. Such basic information, if appropriately checked, is fundamental for any researcher who is trying to estimate the prevalence of one or more RDs, or for any public or private entity that is planning to organize a clinical trial on a certain RD and needs to know if there are enough patients to conduct the study. This information also provides a view on the volume of patients seen by the clinical centers that are coordinating or collaborating with a RD registry, and can be of interest for the coordination of the newly established European Reference Networks for care and research on RD [26, 27].
Establishing a directory of high-quality RD biobanks linked to a centralized sample catalogue helps researchers to quickly gain access to relevant biological samples for experiments [28].
The Finder consists of a standardized view of key elements of participating databases called ID-Cards (Supplementary Material), in order to provide users with the same type of information in the same way for all databases, and increase databases comparability. Key elements include unique identifiers for diseases (ORPHAcodes, OMIM, ICD-10, etc) that in a linked data environment [29] can be linked to larger knowledge networks, for instance via mapping between diseases and phenotypes as provided by the HPO Project [30].
A human readable ID is displayed in the top left-hand corner of the ID-Card for each individual biobank and registry entry. The identifier is mapped to Orphanet identifiers of registries and biobanks.
Technically, the Registry & Biobank Finder is built on an OpenSource Portal Framework, Liferay [31], and based on additional OpenSource portlets built in the framework of RD-Connect. Liferay is one of the leading solutions for horizontal portals according to Gartner analysis [32]. It is a framework based on Java, and enables developers to create enterprise-grade applications for the web. The system has a strong built-in security system and a flexible data management capability. The platform developed is complementary to other catalogues like orpha.net [33] and BBMRI catalogue [34] and it is possible to share the information between these different catalogues.
All users can browse the directory for the registries and/or biobanks for the RD of interest. Researchers can search the directory for biobanks or registries jointly or separately (filter by type) by biobank/registry name, disease name, ORPHAcode, OMIM and ICD-10 codes, synonyms, and other keywords related to the database.
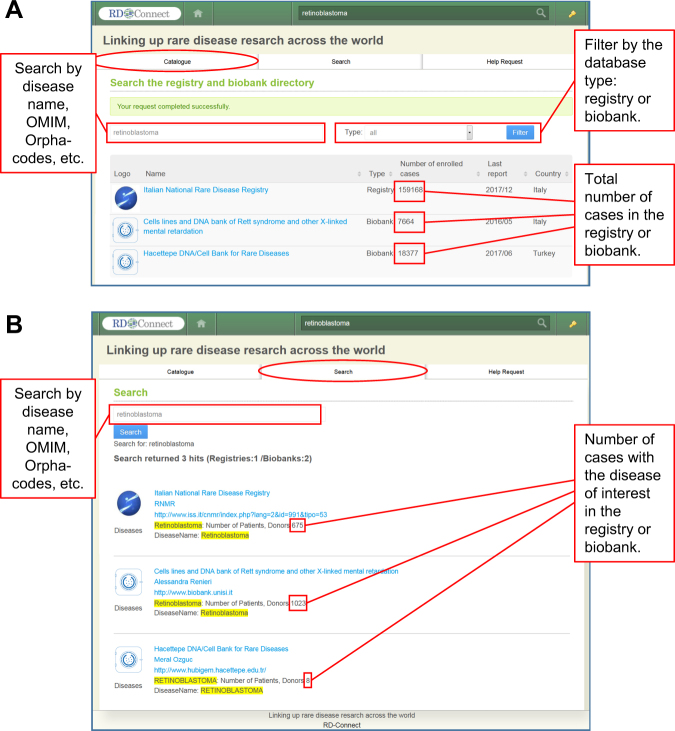
Currently, users can make their search at two different levels (Fig. 2). The “Catalogue” level (general user reporting system) displays the list of relevant databases and the total number of cases registered for all diseases in each database (Fig. 2a). The “Search” level filters the results by specific RD and displays only the cases of interest. (Fig. 2b). This filtering function is especially helpful when a researcher needs to identify specific RD cases in multi-disease biobanks and registries.
Fig. 2.
Search strategies in the Registry & Biobank Finder. To help researchers find registries and biobanks, the Registry & Biobank Finder offers two types of search engines: “Catalogue” and “Search”. a In the “Catalogue” tab, users can search the directory against different keywords, such as the disease name and its synonyms, OMIM- and Orphacodes (upper left). The results can be filtered by the database type and display only registries or only biobanks (upper right). The search results display the list of found databases with the information on their type, the total number of cases (patients or biosamples) for all diseases collected in the database, date of last data update, and the country (lower right). b The “Search” tab is particularly useful for searching the numbers of cases with the disease of interest in each found database (marked in red)
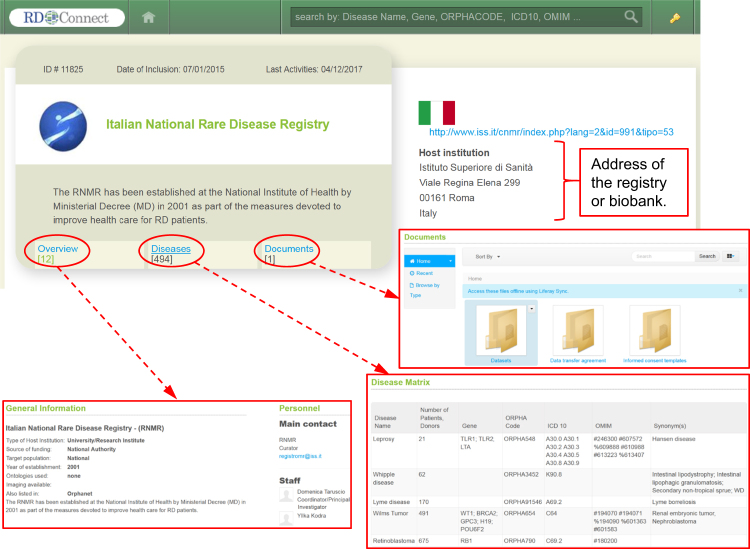
More information on a given registry/biobank, is displayed in its individual ID-Card, consisting of the “Overview”, “Diseases”, and “Documents” sections (Fig. 3). The Overview provides general information such as address and contact data, sources of funding, ontologies used, associated data, and imaging availability, associated biobanks which are storing biosamples for RD patients included in the registry with a link to the biobank ID-Cards, and participation to RD registry networks.
Fig. 3.
An ID-Card of an individual database. The detailed description of each registry or biobank in the directory is presented as an ID-Card with the address and three tabs: “Overview”, “Diseases” and “Documents”. “Overview” provides general information about the database, such as the contact data, source of funding, target population and membership in networks. “Diseases” opens a Disease Matrix, which lists all the diseases for which the registry/biobank has collected data, with the numbers of cases (patients or donors) and the mutations covered. The “Documents” tab provides access to various documents, such as database’s data transfer agreement, templates for informed consent and publications
The “Diseases” section contains a “Disease Matrix” (DM), which is a list indicating the number of RD patients or biological samples included in a registry or biobank according to their RD or RD group. Each RD in the DM is described by the disease name; gene; ORPHAcode; ICD-10; OMIM number, and synonym(s).
The “Search” strategy of the Registry & Biobank Finder is based on the information reported in the DM. The level of detail in the DM may vary according to the level of granularity used to describe cases or samples through different coding systems. For example, the cases included in a database may be described by ORPHAcodes associated to groups of diseases (e.g., rare genetic neurological disorder), or by single diseases (e.g., Duchenne and Becker muscular dystrophy).
For the scope of data sharing, the use of a more detailed description is encouraged
The “Documents” section contains relevant documents, such as study protocols, case report forms, standard operating procedures, informed consent templates, approval by the research ethics committee, data transfer agreements, and publications. When available, registries provide templates to request access (i.e., Material and Data transfer Agreement – MTA, DTA) that can be used to contact the database.
To help users navigate the Registry & Biobank Finder and other systems, RD-Connect provides video tutorials on its YouTube channel (https://www.youtube.com/channel/UCwwcUPJZfyWGaW13Lvao7Ag). This includes a tutorial for end users on how to browse the Registry & Biobank Finder as well as a tutorial addressed to registry and biobank managers, demonstrating how to add their database to the Finder.
Population of the RD-Connect Registry & Biobank Finder
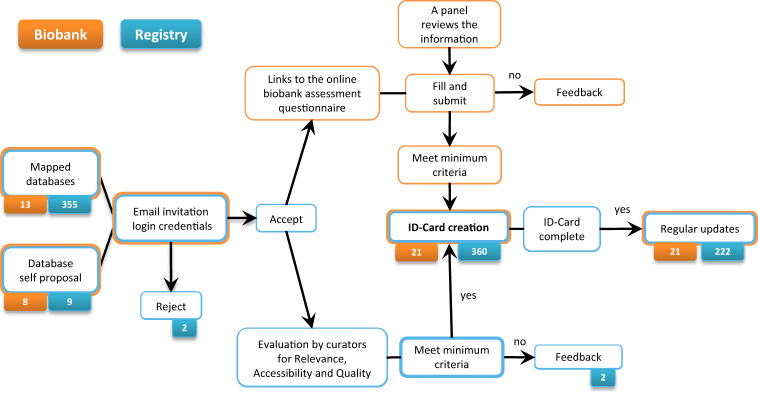
A backend database was first created with mapping of existing RD registries and biobanks starting from existing inventories (see Table 1). An invitation strategy was planned in order to increase the probability of acceptation and the success of the RD-Connect ID-Cards (Fig. 4)
Fig. 4.
Inclusion of biobanks and registries in the Registry & Biobank Finder. The process of inclusion and evaluation of biobanks and registries in the Registry & Biobank Finder (mapped and self-proposed)
Inclusion of registries
Based on the mapped registries, RD-Connect curators invited as early adopters the databases that were familiar with the program, in the endeavor to get a “snowball effect”.
In particular, the invitation was directed to registries in the RD-Connect Core Implementation Group (CIG) [35], some of which also tested the system and provided feedback, registries collaborating with the US National Center for Advancing Translational Sciences (NCATS) Global Rare Disease Patient Registry Data Repository (NIH/NCATS/GRDR) program [36], registries collaborating with the RD-Connect project partner Invitae (formerly, Patientcrossroads) [35], and registries participating to the RD-Connect partner projects NeurOmics [17] and EURenOmics [18].
The registries that accepted the invitation were evaluated according to three main criteria:
Relevance: The registry should be relevant for the study of RDs and include clinical data; registries dealing with common diseases and registries that were not focused on clinical outcomes (i.e., on the experience of illness) were excluded.
Accessibility: Information about the registry should be easily available, ideally on a project website, or on the website of an organization (university, hospital, etc.) or patient association. Information could be also displayed in clinicaltrial.gov, Orphanet, or in works published by controlled and updated sources (peer reviewed journals, periodic reports, etc.). The registries for which no information or poor or old information was available were excluded.
Quality: High-quality data are considered to be one of the most important elements in the establishment and maintenance of a registry. Data quality control is possible through a systematic examination of data (internal or external audit process) against a number of dimensions: completeness; validity; coherence and comparability; accessibility; usefulness; timeliness and prevention of duplicate records. Since it is impossible for curators to evaluate directly the quality of a registry, two elements were considered: availability of information on standards and procedures for quality management and availability of peer reviewed publications. The last point was considered as particularly relevant as most databases that are disseminating their results have undergone a quality check through the peer review process and they may be more keen to share other data in the future [36]. However, since registries are only asked to share aggregated data in the Finder, quality evaluation was rather permissive in this phase of the project, with the plan to intensify quality checks for those registries that will decide to share de-identified data in the future.
Since the end of 2015, the Finder has been open to receive online submissions through the “Propose” section, as described above, and 8 registries have been added to the directory after curators’ evaluation and approval, and self-completed by the proponents.
As of November 2017, the RD-Connect Registry & Biobank Finder included aggregated data of more than 222 RD registries from around the world with a strong participation of US registries (24.8%), followed by International registries (18%), Italian (12.6%), French (7.7%), German (5.9%), United Kingdom (5.4%), and Spanish registries (4.5%) (Table 2).
Table 2.
RD registries by country and dimension
| RD registries by country and dimension | No | (%) |
|---|---|---|
| Country | ||
| Australia | 5 | (2.3) |
| Austria | 3 | (1.4) |
| Belgium | 2 | (1) |
| Bulgaria | 10 | (4.5) |
| Canada | 4 | (1.8) |
| Czech Republic | 5 | (2.3) |
| France | 17 | (7.7) |
| Germany | 13 | (5.9) |
| International | 40 | (18) |
| Ireland | 3 | (1.4) |
| Italy | 28 | (12.6) |
| Japan | 2 | (1) |
| Spain | 10 | (4.5) |
| United Kingdom | 12 | (5.4) |
| United States | 55 | (24.8) |
| Others | 13 | (5.9) |
| Dimension (no. of cases included) | ||
| >5000 | 13 | (5.9) |
| 1000–5000 | 61 | (27.5) |
| 500–1000 | 41 | (18.5) |
| 100–500 | 82 | (36.9) |
| <100 | 25 | (11.3) |
Included registries are categorized into five main sizes or dimensions, according to the number of cases included: <100, 100–500, 500–1000, 1000–5000, and >5000. The most frequent dimension in participating registries was the one counting 100–500 registered cases (Table 2). The most represented disease categories in RD registries are rare neurological diseases (17.1%), rare neuromuscular diseases (14%), rare hematological diseases (11.3%), rare pulmonary diseases (9.5%), rare renal diseases (8.1%), and rare hereditary metabolic disorders (7.2%) (Table 3).
Table 3.
Distribution of RD registries according to disease category
| RD registries by disease categorya | No | (%) |
|---|---|---|
| Rare bone diseases | 5 | (2.3) |
| Rare cancers and tumors | 11 | (5) |
| Rare connective tissue and musculoskeletal diseases | 2 | (0.9) |
| Rare craniofacial anomalies and ENT (ear, nose and throat) disorders | 1 | (0.5) |
| Rare endocrine diseases | 4 | (1.8) |
| Rare eye diseases | 6 | (2.7) |
| Rare gastrointestinal diseases | 3 | (1.4) |
| Rare hematological diseases | 25 | (11.3) |
| Rare hepatic diseases | 5 | (2.3) |
| Rare hereditary metabolic disorders | 16 | (7.2) |
| Rare immunological and auto-inflammatory diseases | 10 | (4.5) |
| Rare malformations and developmental anomalies and rare intellectual disabilities | 17 | (7.7) |
| Rare multi-systemic vascular diseases | 3 | (1.4) |
| Rare neurological diseases | 38 | (17.1) |
| Rare neuromuscular diseases | 31 | (14) |
| Rare pulmonary diseases | 21 | (9.5) |
| Rare renal diseases | 18 | (8.1) |
| Rare skin disorders | 1 | (0.5) |
| Rare urogenital diseases | 1 | (0.5) |
| Others | 4 | (1.8) |
aAddendum to EUCERD Recommendations of January 2013
Two registries have opted out indicating their wish not to be involved.
Inclusion of biobanks
All RD biobanks can apply to participate in the RD-Connect Platform and be included in the Registry & Biobank Finder. The potential and benefits of the RD-Connect Platform for biobanks are explained in the promotional video (https://youtu.be/tLbn0748shk). To ensure that RD researchers have access to high-quality biological samples, participating RD biobanks undergo an assessment to ensure a set of minimum criteria is met (Fig. 4). The biobank assessment process was adapted from the EuroBioBank, a recognized European high-quality RD biobank network, from its membership application model [28]. RD-Connect Registry & Biobank Finder serves as the portal to manage this assessment process. Self-proposed and invited RD biobanks receive individual access to the online questionnaire, which includes questions on the general information of the biobank (Overview), disease focus, sample collections, use of biobanking SOPs, implementation of quality standards, collection of informed consent, management, and availability of their sample catalogue. Some answers from the questionnaire then form a base of the information on the biobanks provided in the Finder. A sample of the questionnaire is available on the RD-Connect website (www.rd-connect.eu) with a video tutorial explaining the application process (https://youtu.be/x5b70fLY5L8). The RD biobanks are required to accept the terms laid out by the RD-Connect Code of Practice [37], a document that contains definitions and governance responsibilities of participants on data usage across the RD-Connect Platform. A declaration by the biobanks to share their sample data sets is considered compulsory as a part of participation to RD-Connect.
Once the biobank submits the completed questionnaire, the curator forwards the application to the RD-Connect Biobank Assessment Panel. The panel is composed of experts in the field of RD biobanking, biobank IT infrastructure, and a patient representative, who provides independent review on their own personal capacities. The panel reviews the application and the outcome of whether the minimum criteria (RD focus, sample quality assurance plans, use of informed consent, evidence of biobanking activity, and the curriculum of the biobank director) for participation are met is fed back to the biobank within 4 weeks. Once the acceptance notification has been sent, the curator publishes the biobank’s ID-Card, rendering it visible to users. Successful biobanks subsequently receive information to assist them in uploading data related to their sample collections to the RD-Connect Sample Catalogue.
The first biobank ID-Card was published in February 2015. Before moving on to invite RD biobanks mapped by RD-Connect, two associated partner biobanks of the project were asked to test the portal and provide feedback on usability and clarity of the questions for biobank assessment. Amendments were made to the portal, and similarly to the RD registries, priorities were given to include project collaborators and associate partner biobanks. The first invitations were extended to EuroBioBank members [28]. Since the opening of the portal, it has also received online self-proposals from seven biobanks via the “Propose” section. These expressions of interest from RD biobanks to participate were prioritized for processing and assessment. By 21 November 2017, biobank ID-Cards have been published, including two biobanks who self-proposed and were positively evaluated.
Discussion
RD-Connect aims to promote the culture of data sharing among RD researchers. RD-Connect Registry & Biobank Finder enables more intensive sharing of patient disease information and biosamples. Researchers can find registries and biobanks storing disease data or biosamples for their disease of interest. Database managers working on the same or related RDs can increase mutual visibility, paving the way to new scientific collaborations. It also allows researchers to find registries and biobanks storing data or biosamples for their disease of interest.
In its first phase of activity, the Registry & Biobank Finder has made an increasing amount of aggregated data and metadata from participating databases available, thus contributing to making RD registry and biobank data more findable, accessible, interoperable and reusable (FAIR) [38]. By displaying the number of cases included in participating databases, as well as the connections between participating biobanks and registries, the Registry & Biobank Finder is making RD data more “findable”. It is making biobank data more “accessible” by providing a link to the RD-Connect Sample Catalogue. Furthermore, propagating access information as it is provided by the registered sources fits a federated model, letting registries maintain the prerogative to decide on providing access (or not) upon request. The finder can contribute to “interoperability” by standardizing the minimum information that it collects, but to fully exploit its capabilities also the sources must be “interoperable”. Therefore, RD-Connect and ELIXIR support contributors with tools and training activities such as “Bring Your Own Data” workshops (BYOD) and the International Summer School on RD registries [39]. The RD-Connect platform and ELIXIR website provide more information. Finally, we are working on a FAIR Data Point API for the Registry & Biobank Finder to enable retrieval of data in linkable format, particularly via the resource description framework and richly annotated with ontologies. This addresses the machine-readability criterion of FAIR data and renders the data from the finder more “reusable” for future analyses across multiple sources.
However, some barriers to participation have become apparent especially for registries, as not all invited registries are replying to the invitation and among participants not all are completing their ID-Cards and in particular the DM. Indeed, of the 222 registries with a complete DM, less than half were provided upon personal communication (44%), while in most cases data were retrieved by finder curators in the registry’s website (21%) and in published papers (33%).
We speculate that the most probable reason for this barrier is the lack of available time researchers have to complete and update the profiles.
Other barriers to participation reported by PIs and registry curators include the need to ask permission to the registry steering committee or governing board before revealing aggregated data and metadata to the finder and no foreseen advantage in making aggregated data publicly available.
While the general advantages of data sharing for the scientific community are easily understood, the advantages of data sharing for researchers may not be immediately profitable even though funding agencies [40–43], journals [44], and scientific societies [45] are promoting data sharing among scientists at all levels.
Several tentative strategies were put in place to increase participation:
(1) make more targeted invitations, i.e., disease or event related; (2) send periodic newsletters; (3) organize training courses for participants; (4) reduce registry owners’ efforts by pre-filling the DMs of registries; however, this implies an increased effort for curators.
Stronger incentives to participation would consist in creating opportunities for more citations in high-impact journals but also outside conventional metrics [46]. For example, in the biobank setting, the sharing of bioresources is incentivized by the BRIF initiative (Bioresource Research Impact Factor project), which consists in keeping a link between the initiators/implementers of a bioresource and the impact of the biological resource in scientific/academic publications [47].
The same tools and principles could be easily applied to registry data, to maximize their accessibility and reuse while recognizing efforts involved in their maintenance.
Another possibility to promote participation, already discussed in appropriate forums, would be to provide economic incentives to databases, in the form of fees for single data access requests or more extensive subscriptions with third parties, including access by third parties to part of the data or to the entire database for a determined period of time, and related to a specific research project. Pay-for-access services would be offered to different stakeholders, such as “data hunting” companies and Centres for Research Organizations, pharmaceutical companies, foundations, researchers working in academia, and in other public or non-profit organizations.
The Registry & Biobank Finder may support registries in agreeing a “rate table” (price list) according to the rarity of the disease and according to the kind of data released: i.e., aggregate data or statistics; unit record data; linked data; access to a patient recruitment service for clinical trials, etc.
As foreseen in important health data repositories like some National Cancer Services [48, 49], additional costs may be foreseen for the more complex and time-consuming requests (e.g., for rich annotation with ontologies) to cover platform or registry’s costs of retrieving and processing data. Otherwise, to support the most accessible but least accessed databases, the Registry & Biobank Finder might consider linking financial support for registries according to predefined accessibility profiles starting from databases sharing very general aggregated data to those sharing more detailed aggregated data (i.e., by patient’s age, sex, age at onset, etc.), to those also sharing patient de-identified data. However, this approach would require that the finder centralizes and mediates access requests and permissions for registries, which is beyond the current scope of the project.
Participation response from RD biobanks was more positive in comparison to registries. This may have resulted from the limited number and heterogeneity of participating biobanks, as well as from the intrinsic nature of biobanks to distribute biological samples for research, where a culture of sharing and the need to be visible already exist [49]. Biobanks are also becoming aware of the paradigm shift in the genomic era, where research has become more data intensive and huge amounts of NGS and -omics data derived from biological samples are generated as a part of personalized medicine research [50]. Importantly, biobanks are poised to respond to new opportunities and demands by developing new business plans, partnerships, and cost recovery models, to ensure that sustainability and relevance of the biobank operation is maintained [51–53]. The RD-Connect Platform provides an opportunity for RD biobanks to enrich their data sets by linking RD biological samples to phenotypic and -omics data. As RD biobanks tend to be small units with limited resources, participating biobanks can easily recognize the benefit from being in a supported community created by the RD-Connect project, for a shared effort in data harmonization and learning from experiences [28].
RD-Connect and the Registry & Biobank Finder represent a driving case for developing life science data infrastructure [16]. The low frequency of rare disease cases and the scattering of rare disease data resources across institutes and countries, combined with the consequential need to combine data within legal and ethical constraints, provide an excellent target for developing a federated data infrastructure.
As the number of data sharing initiatives increases, researchers, pharmaceutical companies, and regulators also see value in supporting these initiatives and we may agree that “data sharing, in one form or another, is becoming inevitable” [54].
There are several kinds of users and potential uses for the RD-Connect Registry & Biobank Finder, including academic researchers and researchers collaborating with the industry, dealing with the questions of basic, translational, and/or clinical research.
Every effort must be done in order to keep the dialog open to internal and external users’ requests and solicitations in order to reply to their research questions in a satisfactory way.
The Registry & Biobank Finder provides an essential building block in the global infrastructure for the RD domain. It makes biobanks and registries across the globe findable in a controlled manner, and gives stakeholders, including clinicians, researchers, and funders, access to key information about these resources. This can foster collaboration and allow valorization of the registered sources. RD-Connect and the Registry & Biobank Finder must also keep patients and families involved in their development since they are the main actors of RD research.
The increased sharing and moving of patients’ data and samples across national borders through dedicated platforms like RD-Connect is to be acknowledged by patients and their families, who must be informed on all foreseen benefits and risks of international collaboration among RD researchers. This can be done via the informed consent process to participate in registries and biobanks, including a description and an explanation of data sharing platforms like RD-Connect, but also through the active involvement of patient representatives and patient organizations in the governance and advisory activities [55].
The possibility to share more aggregated data and match patient data with data on the same patient coming from other databases via the use of unique identifiers [56], which is foreseen in the development of the RD-Connect platform, requires dynamic solutions for the informed consent and return of secondary findings to patients [57]. We are therefore investigating how machine-readable consent that is directly attached to data elements can be used to check consent efficiently [58].
The design and development of the RD-Connect Registry & Biobank Finder is related to the activity of the RD-Connect project, which warrant its sustainability until the end of 2018. Solutions are under study for the long-term maintenance, growth, and stability of the Registry & Biobank Finder, including the involvement and support of Orphanet [33], of the European platform for rare diseases registries that is under study at the Joint Research Centre [59], the involvement of ERNs, which are required to promote collaborative research within the Network and reinforce research and epidemiological surveillance, through setting up of shared registries [26, 27] and the inclusion of its activities in the framework of ELIXIR [16] and an existing European Research Infrastructures Consortium (ERIC) such as BBMRI-ERIC [15].
Electronic supplementary material
Acknowledgements
This work has been supported by the European Union Seventh Framework Programme (FP7/20072013) under grant agreements no. 305444 (RD-Connect). RD-Connect has a main role in funding authors contributing to the study design, data collection and analysis, decision to publish, or preparation of the manuscrip. NeurOmics (no. 305121, http://rd-neuromics.eu/) and EURenOmics (no. 305608, http://www.eurenomics.eu/) have had mainly a role of data providers, since several registries participating to the Registry & Biobank Finder collaborate with the two projects. We thank ODEX4all (NWO 650.002.002), ELIXIR funded through participating member states, and ELIXIR-EXCELERATE funded through the European Commission within the Research Infrastructures Programme of Horizon 2020 (grant agreement number 676559) for collaborating to the study design and software development especially with a view to interoperability with other systems (MR, MT). MF and MM are funded by Fondazione Telethon (Project GTB12001; Telethon Network of Genetic Biobanks).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-017-0085-z) contains supplementary material, which is available to authorized users.
References
- 1.Council of the European Union. Council Recommendation of 8 June 2009 on an action in the field of rare diseases (2009/c 151/02); Official Journal of the European Union 3/7/2009:C 151/7-10. http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:C:2009:151:0007:0010:EN:PDF. Accessed 20 Nov 2017.
- 2.U.S. Government. 107th Congress Public Law 280. Rare Diseases Act of 2002. https://www.gpo.gov/fdsys/pkg/PLAW-107publ280/html/PLAW-107publ280.htm. Accessed 20 Nov 2017.
- 3.Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14:681–91. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- 4.Hennekam RCM. Care for patients with ultra-rare disorders. Eur J Med Gen. 2011;54:220–4. doi: 10.1016/j.ejmg.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Boycott K, Rath A, Chong JX, et al. International cooperation to enable the diagnosis of all rare genetic diseases. Am J Hum Gen. 2017;100:695–705. doi: 10.1016/j.ajhg.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mascalzoni D, Dove ES, Rubinstein Y, et al. International charter of principles for sharing bio-specimens and data. Eur J Hum Genet. 2015;23:721–8. doi: 10.1038/ejhg.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brookes AJ, Robinson PN. Human genotype–phenotype databases: aims, challenges and opportunities. Nat Rev Genet. 2015;16:702–15. doi: 10.1038/nrg3932. [DOI] [PubMed] [Google Scholar]
- 8.Johnston L, Thompson R, Turner C, Bushby K, Lochmüller H, Straub V. The impact of integrated omics technologies for patients with rare diseases. Expert opinion on orphan. Drugs. 2014;2:1–9. doi: 10.1517/21678707.2014.974554. [DOI] [Google Scholar]
- 9.Baynam G, Walters M, Claes P, et al. Phenotyping: targeting genotype’s rich cousin for diagnosis. J Paediatr Child Health. 2015;51:381–6. doi: 10.1111/jpc.12705. [DOI] [PubMed] [Google Scholar]
- 10.Verma M. Personalized medicine and cancer. J Pers Med. 2012;2:1–14. doi: 10.3390/jpm2010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quon BS, Rowe SM. New and emerging targeted therapies for cystic fibrosis. BMJ. 2016;352:i859. doi: 10.1136/bmj.i859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanetta C, Nizzardo M, Simone C, et al. Molecular therapeutic strategies for spinal muscular atrophies: current and future clinical trials. Clin Ther. 2014;36:128–40. doi: 10.1016/j.clinthera.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 13.International Rare Diseases Research Consortium. Policies and guidelines to maximize impact. http://www.irdirc.org. Accessed 20 Nov 2017. [DOI] [PMC free article] [PubMed]
- 14.Thompson R, Johnston L, Taruscio D, et al. RD-Connect: an integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. J Gen Intern Med. 2014;29(Suppl 3):780–7. doi: 10.1007/s11606-014-2908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biobanking and Biomolecular Resources Infrastructure –European Research Infrastructure Consortium. http://www.bbmri-eric.eu/. Accessed 20 Nov 2017.
- 16.ELIXIR: A distributed infrastructure for life-science information. https://www.elixir-europe.org/. Accessed 22 June 2017.
- 17.NEurOmics: Integrated European Project on Omics Research of Rare Neuromuscular and Neurodegenerative Diseases. http://rd-neuromics.eu/. Accessed 20 Nov 2017.
- 18.EURenOmics: Cutting edge technologies for rare kidney diseases. http://www.eurenomics.eu/. Accessed 20 Nov 2017.
- 19.Köhler S, Vasilevsky N, Engelstad M, et al. The human phenotype ontology in 2017. Nucl Acids Res. 2017;45(D1):D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasant D, Chanas L, Malone J, et al. ORDO: an ontology connecting rare disease, epidemiology and genetic data. 2014. Phenotype day @ ISMB2014.
- 21.Girdea M, Dumitriu S, Fiume M, et al. PhenoTips: patient phenotyping software for clinical and research use. Hum Mutat. 2013;34:1057–65. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 22.López E, Thompson R, Gainotti S, et al. Overview of existing initiatives to develop and improve access and data sharing in rare disease registries and biobanks worldwide. Expert Opin. Orphan Drugs. 2016. 10.1080/21678707.2016.1188002.
- 23.Monaco L, Crimi M, Wang CM. The challenge for a European network of biobanks for rare diseases taken up by RD-Connect. Pathobiology. 2014;81:231–6. doi: 10.1159/000358492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubinstein YR, Groft SC, Bartek R, et al. Creating a global rare disease patient registry linked to a rare diseases biorepository database: Rare Disease-HUB (RD-HUB) Contemp Clin Trials. 2010;31:394–404. doi: 10.1016/j.cct.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taruscio D, Gainotti S, Mollo E, et al. The current situation and needs of rare disease registries in Europe. Public Health. Genomics. 2013;16:288–98. doi: 10.1159/000355934. [DOI] [PubMed] [Google Scholar]
- 26.Directive 2011/24/EU of the European Parliament and of the Council of 9 March 2011 on the application of patients’ rights in cross-border healthcare. Official Journal of the European Union L 88/62. Strasbourg, 4.4.2011.
- 27.European Commission. Commission Delegated Decision of 10 March 2014. Setting out criteria and conditions that European Reference Networks and healthcare providers wishing to join a European Reference Network must fulfil. Annex I point 5. (2014/286/EU) Official Journal of the European Union L 147/71.
- 28.Mora M, Angelini C, Bignami F, et al. The EuroBioBank Network: 10 years of hands-on experience of collaborative, transnational biobanking for rare diseases. Eur J Hum Genet. 2015;23:1116–23. doi: 10.1038/ejhg.2014.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bizer C, Heath T, Berners-Lee T. Linked data—the story so far. IJSWIS. 2009;5:1–22. doi: 10.4018/jswis.2009081901. [DOI] [Google Scholar]
- 30.Köhler S, Doelken SC, Mungall CJ, et al. The human phenotype ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42(D1):D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liferay Digital Experience Platform. https://www.liferay.com/it (Accessed 20 Nov 2017).
- 32.Murphy J, Phifer G, Tay G, Revang M. Gartner: Critical Capabilities for Horizontal Portals. November 15, 2016. Available at: https://www.liferay.com/company/gartner-critical-capabilities?utm_source=blog%20post&utm_medium=content&utm_content=gartner%20mq%20blog%20post&utm_term=post%20cta&utm_campaign=70170000000vO6L. (Accessed 20 Nov 2017.).
- 33.Rare Disease Registries in Europe, Orphanet Report Series, Rare Diseases collection, January 2016. http://www.orpha.net/orphacom/cahiers/docs/GB/Registries.pdf. Accessed 20 Nov 2017.
- 34.Holub P, Swertz M, Reihs R, van Enckevort D, Müller H, Litton JE. BBMRI-ERIC directory: 515 biobanks with over 60 million biological samples. Biopreserv Biobank. 2016;14:559–62. doi: 10.1089/bio.2016.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.RD-Connect Core Implementation Group. http://rd-connect.eu/phenotypic-data/core-implementation-group/. (last access November 20, 2017).
- 36.Piwowar HA. Who shares? who doesn’t? factors associated with openly archiving raw research data. PLoS ONE. 2011;6:e18657. doi: 10.1371/journal.pone.0018657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.RD-Connect Code of Practice. http://rd-connect.eu/rdcon/files/RD-Connect-Code-of-Practice20151109.pdf. Accessed 20 Nov 2017.
- 38.Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carta C, Roos M, Gainotti S, et al. Linked data approach: some results and considerations from the first RD-Connect Bring Your Own Data meeting in Rome. The European Society of Human Genetics 2015, Glasgow, Poster PS16.33. http://www.abstractsonline.com/Plan/ViewAbstract.aspx?sKey=07d9c7ae-8512-4d13-a2c9-834b7e89a648&cKey=8f546af4-114d-456b-8b4c-c383c8b827b4&mKey=cabdedda-497c-457e-8481-34a866ab3681. Accessed 20 Nov 2017.
- 40.National Science Foundation. Award & Administration Guide (AAG) Chapter VI.D.4. Dissemination and Sharing of Research Results. http://www.nsf.gov/bfa/dias/policy/dmp.jsp. Accessed 20 Nov 2017.
- 41.IRDIRC. Policies & Guidelines. April 2013. http://www.irdirc.org/policies-guidelines/. Accessed 20 Nov 2017.
- 42.Wellcome Trust. Policy on data management and sharing. http://www.wellcome.ac.uk/About-us/Policy/Policy-and-position-statements/WTX035043.htm. Accessed 20 Nov 2017.
- 43.National Institutes of Health. Final NIH Statement on sharing research data. February 26, 2003. http://grants.nih.gov/grants/guide/notice-files/NOT-OD-03-032.html. Accessed 20 Nov 2017.
- 44.Taichman DB, Backus J, Baethge C. Sharing clinical trial data — a proposal from the International Committee of Medical. J Ed N Engl J Med. 2016;374:384–6. doi: 10.1056/NEJMe1515172. [DOI] [PubMed] [Google Scholar]
- 45.Nelson B. Data sharing: empty archives. Nature. 2009;461:160–3. doi: 10.1038/461160a. [DOI] [PubMed] [Google Scholar]
- 46.Gewin V. Data sharing: an open mind on open data. Nature. 2016;529:117–9. doi: 10.1038/nj7584-117a. [DOI] [PubMed] [Google Scholar]
- 47.Bravo E, Calzolari A, De Castro P, et al. Developing a guideline to standardize the citation of bioresources in journal articles (CoBRA) BMC Med. 2015;13:33. doi: 10.1186/s12916-015-0266-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapman N. National Cancer Intelligence Network. Access to the National Cancer Data Repository. UK. 21 Feb 2011. http://www.ncin.org.uk/view?rid=12. Accessed 20 Nov 2017.
- 49.Gaskell G, Gottweis H. Biobanks need publicity. Nature. 2011;471:159–60. doi: 10.1038/471159a. [DOI] [PubMed] [Google Scholar]
- 50.Vaught J. Biobanking comes of age: the transition to biospecimen science. Annu Rev Pharmacol Toxicol. 2016;56:211–28. doi: 10.1146/annurev-pharmtox-010715-103246. [DOI] [PubMed] [Google Scholar]
- 51.van Ommen GJ, Törnwall O, Bréchot C, et al. BBMRI-ERIC as a resource for pharmaceutical and life science industries: the development of biobank-based expert centres. Eur J Hum Genet. 2015;23:893–900. doi: 10.1038/ejhg.2014.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macheiner T, Huppertz B, Bayer M, Sargsyan K. Challenges and driving forces for business plans in biobanking. Biopreserv Biobank. 2017;15:121–5. doi: 10.1089/bio.2017.0018. [DOI] [PubMed] [Google Scholar]
- 53.Clément B, Yuille M, Zaltoukal K, Wichmann HE, Anton G, Parodi B. Public biobanks: calculation and recovery of costs. Sci Transl Med. 2014;6:261fs45. doi: 10.1126/scitranslmed.3010444. [DOI] [PubMed] [Google Scholar]
- 54.Merson L, Gaye O, Guerin PJ. Avoiding data dumpsters -toward equitable and useful data sharing. NEJM 2016. 10.1056/NEJMp1605148. [DOI] [PubMed]
- 55.McCormack P, Kole A. Setting up strategies: patient inclusion in biobank and genomics research in Europe. 7th European Conference on Rare Diseases and Orphan Products. Berlin, Germany: BioMed Central Ltd; 2014. .
- 56.Hansson MG, Hanns L, Olaf R, et al. The risk of re-identification versus the need to identify individuals in rare disease research. Eur J Hum Genet. 2016. 10.1038/ejhg.2016.52. [DOI] [PMC free article] [PubMed]
- 57.Gainotti S, Turner C, Woods S, et al. Improving the informed consent process in international collaborative rare disease research: effective consent for effective research. Eur J Hum Genet. 2016; 24:12485–54. 10.1038/ejhg.2016.2. [DOI] [PMC free article] [PubMed]
- 58.Dyke SOM, Philippakis AA, Rambla De Argila J, et al. Consent codes: upholding standard data use conditions. PLoS Genet. 2016;12:e1005772. doi: 10.1371/journal.pgen.1005772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicholl C. Towards a European platform for rare diseases registries. Orphanet J Rare Dis. 2014;9(Suppl 1):O6. doi: 10.1186/1750-1172-9-S1-O6. [DOI] [Google Scholar]
- 60.Filocamo M, Baldo C, Goldwurm S, et al. Telethon Network of Genetic Biobanks Staff. Telethon Network of Genetic Biobanks: a key service for diagnosis and research on rare diseases. Orphanet J Rare Dis. 2013;8:129. doi: 10.1186/1750-1172-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meglic M, Doupi P, Pristas I, Skalkidis Y, Zaletel M, Orel A. PARENT joint action: increasing the added value of patient registries in a cross-border setting. MEDINFO. 2013;192:1161. doi: 10.3233/978-1-61499-289-9-1161. [DOI] [PubMed] [Google Scholar]
- 62.U.S. Department of Health & Human Services, National Institutes of Health. NIH Clinical research trials and you - list of registries. https://www.nih.gov/health-information/nih-clinical-research-trials-you/list-registries. Accessed 20 Nov 2017.
- 63.U.S. Department of Health & Human Services, Agency for Healthcare Research and Quality. Registry of patient registries (RoPR). https://patientregistry.ahrq.gov/. Accessed 20 Nov 2017.
- 64.Torreri P, Gainotti S, De Paulis F, et al. RD-Connect ID-Cards of biobanks and registries: making RD data findable, accessible, interoperable and reusable. Poster presented at the 3rd International Rare Diseases Research Consortium (IRDiRC) Conference, Paris, February 8–9, 2016. 10.6084/m9.figshare.4674922.v1. Accessed 22 June 2017.
- 65.Public Population Project in Genomics and Society (P3g). http://www.p3g.org/. Accessed 20 Nov 2017.
- 66.European Network for Genetic and Genomic Epidemiology (Engage). http://www.euengage.org/index.html. Accessed 20 Nov 2017.
- 67.https://www.liferay.com/it. Accessed 20 Nov 2017.
- 68.Murphy J, Phifer G, Tay G, Revang M Gartner. Critical capabilities for horizontal portals. 15 November 2016. https://www.liferay.com/company/gartner-critical-capabilities?utm_source=blog%20post&utm_medium=content&utm_content=gartner%20mq%20blog%20post&utm_term=post%20cta&utm_campaign=70170000000vO6L. Accessed 20 Nov 2017.
- 69.Cancer Institute NSW. How to access cancer data managed by Cancer Institute NSW. Request linked data. http://www.cherel.org.au/pricing. Accessed 20 Nov 2017.
- 70.US Department of Health and Human Services, National Institutes of Health, National Center for Advancing Translational Sciences, Rare Diseases Registry (RaDaR) Program. https://rarediseases.info.nih.gov/radar (last access November 20, 2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.