Abstract
In sub-Saharan Africa, maize is the key determinant of food security for smallholder farmers. The sudden outbreak of maize lethal necrosis (MLN) disease is seriously threatening the maize production in the region. Understanding the genetic basis of MLN resistance is crucial. In this study, we used four biparental populations applied linkage mapping and joint linkage mapping approaches to identify and validate the MLN resistance-associated genomic regions. All populations were genotyped with low to high density markers and phenotyped in multiple environments against MLN under artificial inoculation. Phenotypic variation for MLN resistance was significant and heritability was moderate to high in all four populations for both early and late stages of disease infection. Linkage mapping revealed three major quantitative trait loci (QTL) on chromosomes 3, 6, and 9 that were consistently detected in at least two of the four populations. Phenotypic variance explained by a single QTL in each population ranged from 3.9% in population 1 to 43.8% in population 2. Joint linkage association mapping across three populations with three biometric models together revealed 16 and 10 main effect QTL for MLN-early and MLN-late, respectively. The QTL identified on chromosomes 3, 5, 6, and 9 were consistent with the QTL identified by linkage mapping. Ridge regression best linear unbiased prediction with five-fold cross-validation revealed high accuracy for prediction across populations for both MLN-early and MLN-late. Overall, the study discovered and validated the presence of major effect QTL on chromosomes 3, 6, and 9 which can be potential candidates for marker-assisted breeding to improve the MLN resistance.
Electronic supplementary material
The online version of this article (10.1007/s11032-018-0829-7) contains supplementary material, which is available to authorized users.
Keywords: MLN, MCMV, QTL mapping, Joint linkage association mapping, Maize, GBS
Introduction
Maize is sub-Saharan Africa’s (SSA) most important staple food crop and is cultivated on more than 35 million hectares predominantly under rain-fed conditions and subject to the vagaries of weather (Shiferaw et al. 2011). The maize lethal necrosis (MLN) disease emerged as one of the major threats to the maize-based food security in SSA since 2011 (http://mln.cimmyt.org/). This devastating disease was first reported in September 2011 in the South Rift Valley of Kenya and by 2014, MLN was extensively reported in Kenya, Uganda, Tanzania, Rwanda, D.R. Congo, and Ethiopia (Wangai et al. 2012; Adams et al. 2014; Lukanda et al. 2014; Mahuku et al. 2015a, b). The MLN disease is caused by co-infection by two viruses—Maize Chlorotic Mottle Virus (MCMV) and Sugarcane Mosaic Virus (SCMV). Field observations indicated that MLN has affected almost all the commercially grown maize varieties in Kenya (De Groote et al. 2016), with yield losses ranging from 30 to 100% depending on the stage of disease infection and the environment.
Multi-mode transmission of both MLN-causing viruses is a significant challenge for effective management of the disease. SCMV is known to be transmitted by aphids in a non-persistent mode (Louie 1980; Tao et al. 2013) whereas MCMV is known to be transmitted by insect-vectors (including varied species of chrysomelid beetles and thrips) as well as contaminated seed (Jensen et al. 1991; Zhang et al. 2011). Application of chemical pesticides is resource-intensive for smallholders in eastern Africa and is not environment-friendly over a long term. Therefore, breeding for MLN resistance is a sustainable management option. The severity of MLN is widely influenced by favorable environments (Mahuku et al. 2015b). Screening germplasm under artificial inoculation against MLN is reliable, but is labor-intensive. Better understanding of the genetic basis of resistance to MLN can pave the way to accelerate the development of MLN-resistant germplasm.
Linkage mapping is commonly used to detect the quantitative trait loci (QTL) in biparental populations (Mackay et al. 2009). The QTL conferring resistance to SCMV and other major virus diseases in maize has been investigated in several studies particularly in temperate germplasm (Wisser et al. 2006; Redinbaugh and Pratt 2009; Ding et al. 2012; Zambrano et al. 2014; Li et al. 2016). Genome-wide association study (GWAS) on tropical maize germplasm showed that MLN is controlled by few loci with major effects and many loci with minor effects (Gowda et al. 2015). Joint linkage association mapping (JLAM) with multiple biparental populations offers an additional advantage over other mapping approaches by combining the high power of QTL detection from linkage analyses with the fine resolution of association mapping (Yu et al. 2008; Liu et al. 2011). However, the benefits of linkage mapping and JLAM have not been explored yet to understand the genetic architecture of MLN resistance.
In SSA, development and deployment of improved maize germplasm with enhanced yield and yield stability in disease-prone environments are the topmost priority (Cairns et al. 2013). Successful deployment of climate-resilient improved maize germplasm depends largely on improvement of relevant adaptive traits, including resistance to MLN, maize streak virus (MSV), Northern corn leaf blight (NCLB), gray leaf spot (GLS), and ear rots. Identifying and validating genomic regions conferring resistance to MLN and developing production markers can significantly accelerate the efforts on rapid development and deployment of elite, multiple stress-tolerant maize germplasm in SSA.
Genomic selection (GS) is rapidly gaining importance in plant breeding to accelerate genetic gain (Crossa et al. 2010, 2013, 2017; Vivek et al. 2017; Zhao et al. 2012; Zhang et al. 2017). Predicting and identifying the best resistant or best performing lines before phenotyping from the selected biparental populations is one of the most important applications of GS in maize breeding. Moderate to high accuracy has been reported in biparental populations of maize (Zhao et al. 2012; Zhang et al. 2015, 2017). In this study, our aim was to improve the understanding of the genetic architecture of MLN resistance in tropical maize germplasm, including identification/validation of genomic regions associated with MLN resistance. We applied linkage mapping, JLAM, and GS on four different biparental populations genotyped with low to high density markers and phenotyped in multiple environments in Kenya, under artificial inoculation with optimum combinations of MLN-causing viruses. The specific objectives were to (i) investigate the phenotypic variation for MLN resistance; (ii) identify/validate the genomic regions associated with MLN resistance by linkage mapping and JLAM; and (iii) evaluate the potential of GS for improving MLN resistance.
Materials and methods
Plant materials and field trials
Four biparental maize populations from the Global Maize Program of International Maize and Wheat Improvement Center (CIMMYT) were evaluated in MLN screening facility under artificial inoculation. Population 1 comprised of 229 F3 families from the cross, CML543 × LaPostaSeqC7-F71-1-2-1-2-B-B-B-B, population 2 comprised of 200 F3 families from the cross, CML543 × CML444, and population 3 comprised of 260 F3 families from the cross, CML444 × CML539. In addition, population 4 comprised of 124 F3 families obtained from the cross, Mo37 × CML144 were also used in this study. All 689 F3 families from the first three populations were crossed with a common single-cross tester (CML312 × CML442) from the opposite heterotic pool, whereas population 4 was used as per se for phenotypic evaluation. The testcross progenies and F3 families of population 4 were evaluated in one row (3 m) plots with two replications in three seasons in two locations during 2012 to 2014 in Kenya. The locations were Narok (latitude 01° 05′ S, longitude 35° 52′ E, 1827 m above sea level (masl), clay loam texture) and Naivasha (latitude 0° 43′ S, longitude 36° 26′ E, 2086 masl, clay sandy loam soil texture). All standard agronomic management practices were followed. All lines were evaluated in replicated trials with α-lattice design.
Artificial inoculation of MLN viruses
Stock isolates of MCMV and SCMV, collected from MLN hotspot areas in Kenya, were further confirmed by enzyme-linked immunosorbent assay (ELISA). In an MLN quarantine facility established in Naivasha, Kenya, both viruses were propagated on a susceptible hybrid, H614, in isolated greenhouses. Infected leaf samples collected from the field were cut into small pieces and ground in a mortar and pestle in extraction buffer (10 mM potassium–phosphate, pH 7.0). The resulting sap extract was centrifuged for 2 min at 12,000 rpm. Carborundum was added to decanted sap extract at the rate of 0.02 g/ml. The susceptible hybrid H614 at two leaves stage was inoculated by rubbing sap extract onto the leaves. These infected plants served as a source of inoculum for large-scale field trials. Two separate, sealed greenhouses were maintained for SCMV and MCMV inoculum production. Three weeks before harvesting the plants for field inoculation, random samples from the inoculated plants were tested with ELISA from the SCMV and MCMV greenhouses to confirm the inoculum purity.
Keeping the uniform disease pressure across field trials is crucial to get high-quality data. After several experiments, we optimized the optimum combination of SCMV and MCMV to have maximum MLN infection on maize plants. The ratio of 4 parts of SCMV and 1 part of MCMV (weight/weight) combination was more ideal (Gowda et al. 2015; Mahuku et al. 2015b). We used this optimized combination of SCMV and MCMV viruses (ratio of 4:1) and inoculated twice at the fourth and fifth week after planting. Plants were inoculated using a motorized, backpack mist blower (Solo 423 MistBlower, 12 ltr capacity). An open nozzle (2-in. diameter) was used to deliver inoculum spray at a pressure of 10 kg/cm2. Presence of both viruses in the field trials was confirmed by ELISA once disease symptoms were apparent (approximately 2 weeks post inoculation). MLN disease severity was visually scored on each plot in an ordinal scale of 1 (highly resistant, with no disease symptoms) to 5 (highly susceptible, leading to necrosis and death). Data were recorded twice as “early stage of infection” (21 days 1st post inoculation; hereafter referred to as “MLN-early”) and “late stage of infection” (42 days 1st post inoculation; hereafter referred to as “MLN-late”).
Phenotypic evaluation
The testcross progenies from the first three populations (Pop1, Pop2, and Pop3) were evaluated in two seasons in Naivasha and one season in Narok, in separate but adjacent field trials connected with four common checks, whereas the fourth population (Pop4) was evaluated separately at two seasons in Naivasha. For the analyses, each season was treated as different locations. Observed outliers were excluded from analysis. Since MLN data were based on ordinal scales, it was evaluated to know whether the data meets the assumptions of the applied statistical model (independent, normally distributed, and constant variance; Rawlings et al. 1998). For each population, residuals plot and histogram across locations revealed that the MLN data meets all the model assumptions, and consequently, data was not transformed.
Analyses of variance for each of the location and across locations for each population were carried out using the PROC MIXED procedure with restricted maximum likelihood (REML) option in SAS 9.2 (SAS Institute 2010). Variance components were determined by following linear mixed model: Yijko = μ + gi + lj + rkj + bojk + eijko, where Yijko was the disease severity of the ith genotype at the jth environment in the kth replication of the oth incomplete block, μ was an intercept term, gi was the genetic effect of the ith genotype, lj was the effect of the jth environment, rkj was the effect of the kth replication at the jth environment, bojk was the effect of the oth incomplete block in the kth replication at the jth environment, and eijko was the residual. Locations and replications were treated as fixed effects, and genotype and incomplete blocks as random effects. For JLAM, combined analyses of the first three populations were carried out to calculate best linear unbiased predictions (BLUPs) and total variance components by using MEATA-R software (http://hdl.handle.net/11529/10201). Heritability (H2) on an entry-mean basis was calculated as the ratio of genotypic to phenotypic variance.
Molecular analyses
Six parental lines and their F3 progenies were genotyped with preselected, polymorphic, low-density SNPs by Monsanto Company, using a TaqMan assay (http://www.appliedbiosystems.com website), under the Water Efficient Maize for Africa (WEMA) project. In addition, the first three populations were also genotyped with high-density markers by genotyping-by-sequencing (GBS) at the Institute for Genomic Diversity, Cornell University, Ithaca, USA, as per the procedure described in earlier studies (Elshire et al. 2011; Glaubitz et al. 2014; Gowda et al. 2015). The detailed information on low-density markers were described in previous study (Semagn et al. 2013).
Linkage mapping
For the first three populations (Pop1, Pop2, and Pop3), the GBS data was filtered with a minor allele frequency (MAF) of 0.05 and a minimum count of 95% of the sample size. Then, only marker loci homozygous for both parents and polymorphic between the two parents were retained in all populations. After quality screening, set of uniformly distributed, polymorphic SNPs was selected. For each marker locus, observed genotype frequencies were checked for deviations from Mendelian segregation ratios and allele frequency of 0.5 using a χ2 test. High-quality molecular data were used to construct genetic linkage maps. For population 4, a linkage map was constructed using low-density markers. Individual linkage maps for each population were constructed by using QTL IciMapping software ver 4.0 (Meng et al. 2015; http://www.isbreeding.net). Linkage analyses of SNPs were conducted using the Kosambi (1944) mapping function with a minimum logarithm of odds (LOD) of 3.0 and a maximum distance of 30 cM between two loci.
For each population, BLUPs across locations for MLN-early and MLN-late disease scores were used to detect QTL based on inclusive composite interval mapping (ICIM) implemented in the software QTL IciMapping V.4 (Meng et al. 2015). With the ICIM method, the walking step in QTL scanning of 1 cM and a relaxed LOD threshold of 3.0 were chosen for declaring putative QTL. The origin of the favorable alleles for MLN resistance was identified based on sign of the additive effects of each QTL.
Joint linkage association mapping
For JLAM, GBS-based SNPs from the first three populations were used. For quality screening, in each population, SNPs which were either monomorphic between the parents, or had missing value of > 5%, or had a minor allele frequency of < 0.05 were discarded from analysis. After these quality checks, 15,000 high-quality GBS SNPs were retained for JLAM analyses across populations. BLUPs calculated across populations and environments were used in JLAM studies.
For the JLAM approaches, an additive genetic model was chosen for the testcross progenies (Utz et al. 2000). We used three multiple regression approaches for JLAM and each of these models was explained in detail by Liu et al. (2011) and Würschum et al. (2012). In brief, we applied a two-step procedure for QTL detection. In a first step, stepwise multiple linear regression was used to select a cofactors based on the Schwarz Bayesian Criterion (SBC, Schwarz 1978). Cofactors were selected by using Proc GLM SELECT implemented in the statistical software SAS 9.2 (SAS Institute 2010). In the second step, we calculated a P value for the F test by using a full model (including SNP effect) versus a reduced model (without SNP effect) (Reif et al. 2010). Genome-wide scans for QTL were implemented in R version 3.2.5 (R Development Core Team 2013).
The first model, model A, for QTL detection is as follows: Trait = Cofactors + Marker, it includes both cofactors and marker effects across populations (for details, see Reif et al. 2010; Liu et al. 2011). In model B, a population effect is included as additional effect to correct the population stratification: Trait = Pop + Cofactors + Marker. In the third model (model C), both cofactors and marker effects were modeled as nested within populations: Trait = Pop + Cofactors (Pop) + Marker (Pop). Bonferroni–Holm procedure (Holm 1979) was used to declare markers significantly (P < 0.05) associated with MLN disease resistance. The total proportion of phenotypic variance explained by the detected QTL was calculated by fitting all significant SNPs simultaneously in a linear model to obtain adjusted R2 (Utz et al. 2000). Principal components (PC) were calculated using TASSEL ver 5.0 (Bradbury et al. 2007).
The physical positions of all the markers mapped in all four populations are known. We developed integrated physical map by including all the markers from four populations. All the QTL detected for MLN-early and MLN-late in each populations and JLAM were mapped on this integrated physical map. The 60-bp source sequences of the significant SNP were used to perform BLAST searches against the ‘B73’ RefGen_v2 (http://blast.maizegdb.org/home.php?a=BLAST_UI). Within the local LD block including associated SNPs, the filtered genes in MaizeGDB (http://www.maizegdb.org) containing directly or adjacent to each significant SNP were considered as possible candidate genes for MLN resistance.
Genome-wide prediction
For genome-wide prediction, 2000 common SNPs for each of the three populations which had no missing values and distributed uniformly across the genome were selected. For GS, the ridge regression best linear unbiased prediction (RR-BLUP; Whittaker et al. 2000) method was used (Zhao et al. 2012). Prediction accuracy of the GS approach was evaluated using five-fold cross-validation with 100 times repetitions. The accuracy of GS was calculated as rGS = rMP/h, where h refers to the square root of heritability and rMP is the correlation between observed and predicted phenotypes (Dekkers 2007). The prediction accuracies for MLN resistance were compared based on random markers and combination of random markers with significant markers detected through JLAM.
Further, to understand the effect of different training populations on prediction accuracy, GS was applied to predict within and across biparental populations. We estimated the marker effect and predicted the genomic breeding values in two different scenarios as follows: scenario 1a: Estimation of marker effects was performed across populations, and prediction accuracy was assessed across populations; scenario 1b: Estimation of marker effects was performed across populations, and prediction set was drawn from within each population. In scenario 2, estimation of marker effects and prediction of genomic breeding values were performed within each segregating population. For scenario 1a and b, estimation of marker effects was based on the genotypic variance of the total populations. In contrast, scenario 2 was based on the estimates of the average genotypic variance and heritability within segregating populations.
Results
Among the six parental lines used in this study CML543, CML539, and CML144 are moderately resistant or tolerant with mean score of 2.1, 2.2, and 2.1 for MLN-early and 2.3, 2.5, and 2.4, for MLN-late, respectively. Whereas, CML444, LaPostaSeqC7-F7,1 and Mo37 are susceptible to MLN with the mean score of 2.8, 3.1, and 3.3 for MLN-early, and 3.4, 3.6, and 4.1, for MLN-late, respectively. We observed a wide variation for MLN disease severity at both early and late stages of infection (Fig. S1). The analyses across environments revealed significant (P < 0.01) variances for genotypes, and environments, for both MLN-early and MLN-late in all four populations (Table 1). Genotype × environment interaction variances were significant for first two populations for both MLN-early and MLN-late. Genotypic variances among populations (σ2G-Among = 0.021 and 0.042 for MLN-early and MLN-late, respectively) were of the same magnitude as those of within populations (σ2G-Within = 0.020 and 0.023 for MLN-early and MLN-late, respectively). The estimates of broad-sense heritability were moderate to high ranging from 0.34 to 0.83 for MLN-early and 0.44 to 0.89 for MLN-late scores. Consequently, phenotyping in multiple locations under artificial inoculations resulted in high-quality data representing an excellent resource to study the genetic architecture of MLN disease resistance.
Table 1.
Analysis of variance components for MLN disease severity evaluated across two to three environments with four different F3 mapping populations
| Trait | σ 2 G | σ 2 GE | σ 2 e | h 2 |
|---|---|---|---|---|
| CML543 × LaPostaSeqC7-F71 | ||||
| MLN-early | 0.05* | 0.02* | 0.17 | 0.58 |
| MLN-late | 0.06* | 0.06* | 0.23 | 0.52 |
| CML543 × CML444 | ||||
| MLN-early | 0.04* | 0.01* | 0.18 | 0.53 |
| MLN-late | 0.07* | 0.02* | 0.24 | 0.60 |
| CML539 × CML444 | ||||
| MLN-early | 0.02* | 0.01 | 0.21 | 0.34 |
| MLN-late | 0.03* | 0.02* | 0.20 | 0.44 |
| M037 × CML144 | ||||
| MLN-early | 0.08* | 0.001 | 0.09 | 0.83 |
| MLN-late | 0.21* | 0.001 | 0.15 | 0.89 |
| Across three pops | ||||
| MLN-early | 0.03* | 0.002* | 0.19 | 0.25 |
| MLN-late | 0.04* | 0.03 | 0.15 | 0.35 |
*Significance at < 0.05 level of probability
Linkage maps were constructed for all four F3 populations. The number of progenies, SNPs, map length, and average genetic distance between SNPs for each population are presented in supplementary Table S1. The QTL mapping results revealed that the number of QTL associated with MLN-early varied from 2 to 8, with total phenotypic variance explained ranging from 38.8% in population 4 to 56.8% in population 2 (Table 2). In contrast, the number of QTL associated with resistance to MLN at late stage (MLN-late) varied from 3 to 6, with total phenotypic variance explained ranging from 37 to 58.6%. One QTL each in population 1 and population 4, three QTL in population 2, and two QTL on population 3 were consistently detected for both MLN-early and MLN-late data. The proportion of phenotypic variance explained by single QTL in each population ranged from 3.9 to 37.8% for population 1, 4.2 to 43.8% for population 2, 4.8 to 14.1% for population 3, and 9.6 to 16.6% for population 4 (Table 2). For MLN-early, three major QTL located on chromosomes 3, 6, and 9 were consistently detected in at least two populations. For MLN-late, one QTL each located on chromosomes 3 and 6 was consistently expressed in two populations (Table 2).
Table 2.
Detection of QTL associated with resistance to MLN at early and late stages of disease infection, and their physical positions and genetic effects of the QTL in four F3 populations
| Trait | QTL namea | Chr | Position (cM) | LOD | PVE (%) | Add | Dom | PVE (%) | QTL confidence interval | Physical position (bp) | Fav parent | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Left marker | Right marker | Left | Right | ||||||||||
| CML543 × LaPostaSeqC7-F71 | |||||||||||||
| MLN-early | qMLN_03-01 | 3 | 0 | 3.80 | 3.96 | − 0.09 | 0.17 | 48.22 | S3_1326446 | S3_1337663 | 1326446 | 1337663 | LPS_F71 |
| qMLN_03-130 | 3 | 157 | 27.48 | 37.80 | 0.27 | 0.00 | PZA00413_20 | PZA02299_16 | 125192432 | 130082791 | CML543 | ||
| qMLN_06-04 | 6 | 5 | 5.12 | 5.61 | 0.08 | 0.02 | S6_3869709 | S6_4015237 | 3869709 | 4015237 | CML543 | ||
| MLN-late | qMLN_01-252 | 1 | 68 | 3.09 | 3.41 | − 0.08 | 0.00 | 48.23 | S1_251157170 | PZA02269_4 | 251157170 | 252721946 | LPS_F71 |
| qMLN_03-130 | 3 | 157 | 21.37 | 27.16 | 0.29 | − 0.03 | PZA00413_20 | PZA02299_16 | 125192432 | 130082791 | CML543 | ||
| qMLN_06-19 | 6 | 46 | 8.52 | 9.40 | 0.14 | − 0.06 | S6_18924381 | S6_21021616 | 18924381 | 21021616 | CML543 | ||
| qMLN_08-174 | 8 | 222 | 5.06 | 5.45 | − 0.10 | 0.03 | S8_174216280 | S8_174222902 | 174216280 | 174222902 | LPS_F71 | ||
| CML444 × CML543 | |||||||||||||
| MLN-early | qMLN_01-246 | 1 | 304 | 4.57 | 4.99 | 0.07 | − 0.02 | 56.75 | S1_246489667 | S1_246540548 | 246489667 | 246540548 | CML543 |
| qMLN_03-146 | 3 | 123 | 30.07 | 43.84 | 0.25 | 0.01 | S3_146251234 | S3_146250249 | 146251234 | 146250249 | CML543 | ||
| qMLN_06-85 | 6 | 197 | 5.73 | 6.12 | 0.08 | 0.01 | S6_85416016 | S6_96909472 | 85416016 | 96909472 | CML543 | ||
| qMLN_08-123 | 8 | 94 | 3.81 | 4.23 | 0.02 | − 0.11 | S8_123055373 | S8_123469828 | 123055373 | 123469828 | CML543 | ||
| qMLN_10-81 | 10 | 213 | 4.22 | 4.43 | − 0.07 | 0.00 | S10_81827920 | S10_81829237 | 81827920 | 81829237 | CML444 | ||
| MLN-late | qMLN_01-241 | 1 | 282 | 7.02 | 10.46 | 0.00 | − 0.24 | 58.61 | S1_237487786 | S1_241184216 | 237487786 | 241184216 | CML543 |
| qMLN_03-146 | 3 | 124 | 20.08 | 24.26 | 0.26 | − 0.03 | S3_146250249 | S3_146251923 | 146250249 | 146251923 | CML543 | ||
| qMLN_05-190 | 5 | 251 | 9.55 | 10.44 | − 0.16 | 0.01 | S5_190677275 | PZA00352_23 | 190677275 | 191075557 | CML444 | ||
| qMLN_05-199 | 5 | 349 | 15.36 | 17.74 | 0.19 | 0.00 | S5_199499548 | S5_199499538 | 199499548 | 199499538 | CML543 | ||
| qMLN_05-207 | 5 | 432 | 4.06 | 4.25 | − 0.04 | 0.13 | S5_206890892 | PZA02015_11 | 206890892 | 207464707 | CML444 | ||
| qMLN_06-85 | 6 | 176 | 10.32 | 10.90 | 0.15 | 0.06 | S6_85203511 | S6_85206463 | 85203511 | 85206463 | CML543 | ||
| CML539 × CML444 | |||||||||||||
| MLN-early | qMLN_01-148 | 1 | 72 | 3.24 | 4.84 | − 0.07 | 0.01 | 42.15 | ZM00148950 | ZM00145696 | 148500000 | 160300000 | CML539 |
| qMLN_03-27 | 3 | 69 | 4.17 | 6.91 | 0.02 | − 0.11 | S3_27365043 | S3_29611811 | 27365043 | 29611811 | CML444 | ||
| qMLN_03-126 | 3 | 350 | 6.32 | 10.55 | 0.00 | 0.14 | S3_122493752 | S3_126171099 | 122493752 | 126171099 | CML444 | ||
| qMLN_04-117 | 4 | 59 | 5.54 | 9.13 | 0.09 | − 0.04 | ZM00148564 | ZM00146885 | 106500000 | 117900000 | CML444 | ||
| qMLN_05-131 | 5 | 150 | 4.33 | 6.46 | 0.08 | 0.02 | ZM00147053 | ZM00148930 | 115900000 | 131900000 | CML444 | ||
| qMLN_06-03 | 6 | 185 | 5.42 | 8.96 | − 0.05 | − 0.11 | S6_2385933 | S6_3039514 | 2385933 | 3039514 | CML539 | ||
| qMLN_09-100 | 9 | 66 | 4.37 | 7.49 | − 0.08 | − 0.02 | S9_99440296 | S9_100608734 | 99440296 | 100608734 | CML539 | ||
| qMLN_10-71 | 10 | 32 | 5.42 | 8.21 | 0.09 | 0.01 | S10_71178058 | S10_71665925 | 71178058 | 71665925 | CML444 | ||
| MLN-late | qMLN_03-159 | 3 | 487 | 5.48 | 9.13 | − 0.10 | 0.03 | 36.95 | S3_159801859 | S3_158524074 | 159801859 | 158524074 | CML539 |
| qMLN_05-171 | 5 | 167 | 3.79 | 6.16 | 0.09 | − 0.03 | ZM00147844 | ZM00150817 | 154700000 | 171900000 | CML444 | ||
| qMLN_06-06 | 6 | 202 | 8.15 | 14.15 | − 0.13 | − 0.01 | S6_5159730 | S6_6270908 | 5159730 | 6270908 | CML539 | ||
| qMLN_06-16 | 6 | 215 | 5.68 | 9.51 | 0.00 | − 0.16 | ZM00150684 | S6_16291106 | 12700000 | 16291106 | CML444 | ||
| qMLN_09-100 | 9 | 230 | 3.51 | 7.27 | 0.01 | − 0.13 | ZM00149322 | ZM00145937 | 96500000 | 116800000 | CML444 | ||
| Mo37 × CML144 | |||||||||||||
| MLN-early | qMLN_03-130 | 3 | 129.6 | 5.6 | 16.63 | 0.11 | 0.03 | 38.76 | PHM1745_16 | PZA00920_1 | 129095914 | 131969810 | CML144 |
| qMLN_09-100 | 9 | 112.5 | 5.33 | 16.32 | − 0.05 | − 0.43 | PZA00060_2 | PZB00221_3 | 95769540 | 113201792 | Mo37 | ||
| MLN-late | qMLN_03-129 | 3 | 128.6 | 5.58 | 13.97 | 0.18 | − 0.04 | 39.13 | PZA00413_20 | PHM1745_16 | 113820730 | 129095914 | CML144 |
| qMLN_06-19 | 6 | 14.2 | 3.18 | 9.61 | − 0.14 | − 0.02 | PZA00158_2 | PZA00440_1 | 13254840 | 21599925 | Mo37 | ||
| qMLN_08-157 | 8 | 156.4 | 4.32 | 13.94 | − 0.15 | 0.12 | PZA00460_3 | PHM4786_9 | 156320002 | 157402090 | Mo37 | ||
MLN-early indicates MLN score 21 days after first post inoculation; MLN-late indicates MLN score 42 days after first post inoculation; fav parent indicates parental genotype from where favorable allele for MLN resistance is contributing. Markers with italicized letters are the QTL consistent across MLN-early and MLN-late
LOD logarithm of odds, Add additive effect, Dom dominance effect, PVE phenotypic variance explained
aQTL name composed by the trait code followed by the chromosome number in which the QTL was mapped and a physical position of the QTL
The first two principal components explained 24% of the total variation (Fig. S2). The PCA revealed a population structure of the four parents with three clusters. JLAM analyses with three biometric models together revealed 16 and 10 main effect QTL for MLN-early and MLN-late, respectively (Table 3). With model A, six QTL each were associated with MLN-early and MLN-late and were identified, which together explained 38.27 and 16.44% of the total phenotypic variance, respectively. With model B, by including population effect, nine QTL were detected for MLN-early whereas only one QTL was detected for MLN-late. Model C by considering the nested effect of SNPs in each population, three and four different QTL were identified for MLN-early and MLN-late, respectively. Across these models, one QTL each was overlapped between models A and B and models A and C for MLN-early, whereas only one overlapped QTL was detected between models A and B for MLN-late.
Table 3.
Analysis of trait-associated markers, allele substitution (α) effects, and the total phenotypic variance (R2) of the joint linkage association mapping in multiple segregating F3 populations based on three different biometrical models
| MLN_early | QTL namea | chr | Position (Mbp) | Model A | Model B | Model C | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α-Effect | P value | PVE (%) | α-Effect | P value | PVE (%) | α-Effect | P value | PVE (%) | ||||
| S1_35581569 | qMLN_01-36 | 1 | 35.58 | – | – | – | 0.21 | 1.18E−13 | 7.50 | – | – | – |
| S1_293055726 | qMLN_01-293 | 1 | 293.06 | 0.12 | 1.85E−10 | 5.90 | – | – | – | – | – | – |
| S3_56468658 | qMLN_03-56 | 3 | 56.46 | 0.21 | 1.08E−20 | 21.70 | 0.27 | 1.38E−38 | 21.70 | – | – | – |
| S3_138773656 | qMLN_03-139 | 3 | 138.77 | − 0.15 | 5.64E−22 | 8.50 | – | – | – | − 0.17 | 5.64E−22 | 19.90 |
| S4_5150195 | qMLN_04-05 | 4 | 5.15 | 0.11 | 8.72E−09 | 2.50 | – | – | – | – | – | – |
| S6_21886770 | qMLN_06-21 | 6 | 21.88 | 0.20 | 2.30E−18 | 10.10 | – | – | – | – | – | – |
| S6_39371783 | qMLN_06-39 | 6 | 39.37 | – | – | – | – | – | – | − 0.06 | 1.57E−07 | 3.50 |
| S6_82022555 | qMLN_06-82 | 6 | 82.02 | – | – | – | 0.12 | 7.88E−07 | 2.70 | – | – | – |
| S6_99946471 | qMLN_06-100 | 6 | 99.94 | – | – | – | − 0.08 | 2.00E−07 | 3.00 | – | – | – |
| S6_120159068 | qMLN_06-120 | 6 | 120.16 | – | – | – | 0.11 | 7.36E−07 | 2.70 | – | – | – |
| S6_158478115 | qMLN_06-158 | 6 | 158.48 | – | – | – | 0.10 | 1.48E−07 | 1.60 | – | – | – |
| S7_3671560 | qMLN_07-037 | 7 | 3.67 | 0.08 | 1.57E−07 | 3.20 | – | – | – | – | – | – |
| S8_147097693 | qMLN_08-147 | 8 | 147.09 | – | – | – | 0.14 | 1.13E−10 | 4.70 | – | – | – |
| S9_94515942 | qMLN_09-95 | 9 | 94.51 | – | – | – | – | – | – | 0.07 | 8.45E−07 | 2.70 |
| S9_137154420 | qMLN_09-137 | 9 | 137.15 | – | – | – | 0.08 | 8.45E−07 | 3.10 | – | – | – |
| S10_145280961 | qMLN_10-145 | 10 | 145.28 | – | – | – | 0.11 | 1.47E−11 | 6.00 | – | – | – |
| Total PVE (%) | 38.27 | 30.81 | 25.5 | |||||||||
| MLN-late | ||||||||||||
| S1_7162859 | qMLN_01-071 | 1 | 7.16 | − 0.13 | 1.34E−06 | 2.80 | – | – | – | – | – | – |
| S2_30361545 | qMLN_02-30 | 2 | 30.36 | 0.16 | 8.08E−09 | 2.90 | 0.16 | 6.14E−08 | 4.20 | – | – | – |
| S3_119323182 | qMLN_03-119 | 3 | 119.32 | − 0.13 | 9.58E−08 | 3.60 | – | – | – | – | – | – |
| S3_133187288 | qMLN_03-133 | 3 | 133.19 | – | – | – | – | – | – | − 0.11 | 2.30E−18 | 15.00 |
| S3_188926823 | qMLN_03-189 | 3 | 188.93 | 0.11 | 1.29E−09 | 4.80 | – | – | – | – | – | – |
| S5_205155852 | qMLN_05-205 | 5 | 205.16 | – | – | – | – | – | – | 0.01 | 9.58E−08 | 3.60 |
| S6_5441847 | qMLN_06-05 | 6 | 5.44 | – | – | – | – | – | – | − 0.02 | 1.29E−09 | 4.80 |
| S6_38273901 | qMLN_06-39 | 6 | 38.27 | 0.09 | 3.52E−07 | 4.30 | – | – | – | – | – | – |
| S7_19623847 | qMLN_07-19 | 7 | 19.62 | 0.15 | 1.23E−10 | 5.60 | – | – | – | – | – | – |
| S7_123880597 | qMLN_07-123 | 7 | 123.88 | – | – | – | – | – | – | 0.09 | 3.52E−07 | 4.10 |
| Total PVE (%) | 16.44 | 4.19 | 7.91 | |||||||||
MLN-early indicates MLN score 21 days after first post inoculation; MLN-late indicates MLN score 42 days after first post inoculation; R2 indicates proportion of phenotypic variance explained
Chr chromosome, MLM mixed linear model, MAF minor allele frequency
aQTL name composed by the trait code followed by the chromosome number in which the QTL was mapped and a physical position of the QTL
JLAM is expected to increase the resolution within QTL intervals detected by individual population linkage analyses. Therefore, in this study, we tried to identify the QTL that fell within the confidence interval of MLN resistance QTL identified through the biparental approach. On chromosome 3, JLAM identified five SNP markers associated with MLN resistance QTL, among them, one SNP (S3_119323182) fell within the confidence interval of QTL identified in population 4 (113 to 129 Mbp) (Tables 2 and 3) whereas the same SNP fell just outside the confidence intervals of the major QTL in other three populations. SNP marker detected on chromosome 5 (S5_205155852) was located just outside the confidence interval of QTL detected on population 2. On chromosome 6, JLAM detected eight MLN resistance QTL. One of them located in bin 6.00 (S6_5441847) resided within the confidence interval of QTL identified in the population 3 (S6_5159730-S6_6270908). Other SNPs fell just outside the confidence interval of the QTL detected on chromosome 6 (Tables 2 and 3). On the chromosome 9, JLAM discovered one QTL that resided in bin 9.03 fell under the QTL detected on populations 3 and 4 (95.7 to 116.8 Mbp) (Tables 2 and 3). This SNP marker (S9_94515942) can serve as an anchor landmark to delimit the confidence interval for these QTL. A set of putative candidate genes associated with significant SNPs/QTL was identified (Table S2). All the QTL detected for individual populations and JLAM were mapped on one integrated map (Fig. S3).
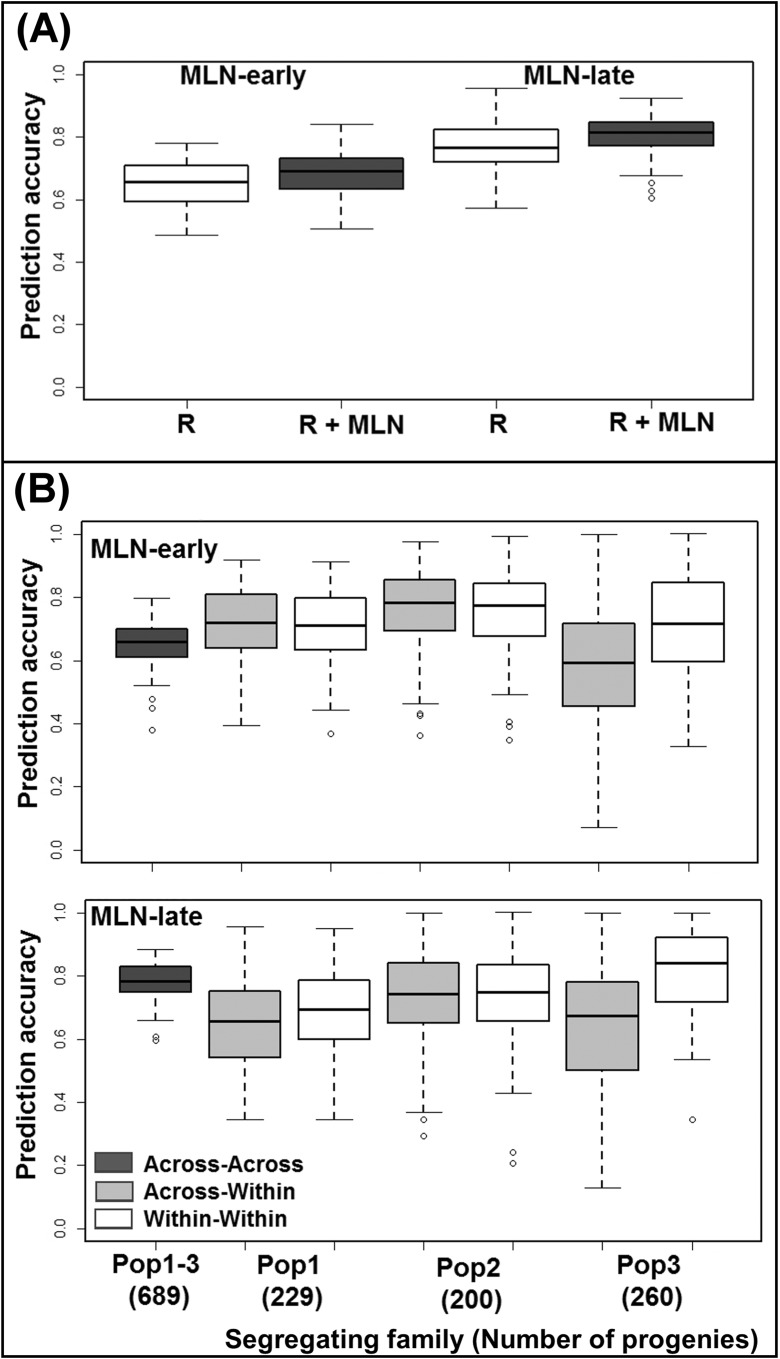
We used five-fold cross-validation to assess the accuracy of genomic predictions for resistance to MLN-early and MLN-late traits by combining the data from the first three populations and within each population (Fig. 2a, b). The cross-validated prediction accuracy when both the training and estimation sets were formed across populations was high with 0.65 and 0.77 for MLN-early and MLN-late, respectively. For both MLN-early and MLN-late, the prediction accuracy was slightly improved by 2% with inclusion of the MLN resistance-associated markers into the prediction model (Fig. 2a). When the training set was derived from across populations and the prediction set was within population, the prediction accuracy was high and varied from 0.58 to 0.72 for MLN-early and 0.65 to 0.73 for MLN-late. Prediction accuracy of genomic breeding values within each biparental population ranged from 0.71 to 0.76 for MLN-early and 0.68 to 0.82 for MLN-late scores (Fig. 2b).
Fig. 2.
Genome-wide prediction accuracies based on random markers (a) (R) and random + MLN resistance associated significant markers (R + MLN), and prediction accuracy based on three different scenarios (b). Scenario 1a—estimation and prediction across families; scenario 1b—estimation across and prediction within families; and scenario 2—both estimation and prediction within biparental segregating families, with five-fold cross-validation for MLN disease severity at early and late stages of infection
Discussion
MLN is a complex challenge that has to be effectively addressed through several simultaneously implemented strategies, including development and deployment of MLN-resistant germplasm (Prasanna 2016). Over the last 4 years, CIMMYT has screened more than 120,000 germplasm entries against MLN under artificial inoculation at the centralized MLN screening facility established in Naivasha, Kenya (http://mln.cimmyt.org/). Although a substantial proportion of pre-commercial and commercial maize germplasm in SSA is susceptible to MLN, these intensive efforts enabled identification of promising CIMMYT maize germplasm with tolerance/resistance to MLN, including the individual viruses (MCMV and SCMV).
Phenotype-based selection strategies are often resource-intensive and time-consuming. Identifying and validating MLN resistance-associated molecular markers which are stable across diverse genetic backgrounds could potentially enable pre-selection of genomic regions in Africa-adapted sub-tropical maize germplasm, thereby contributing to enhanced genetic gains. In this study, we performed linkage mapping, JLAM, and GS to understand the genetic architecture of MLN resistance and validate earlier findings in sub-tropical maize germplasm.
QTL analyses in each of the four populations identified three major QTL genomic regions on chromosome 3, between 113 and 131 Mbp (bin 3.04) and 145 and 160 Mbp (bin 3.05). Major QTL detected on chromosome 6 are also mapped in three genomic regions, between 2 and 6 Mbp, 15 and 21 Mbp, and 85 and 96 Mbp (bin 6.00/01). On chromosome 9, we found major consistent QTL on bin 9.03, between 95 and 116 Mbp. Interestingly, the major QTL on chromosome 3 consistently expressed at both early and late stages of MLN infection; however, most of the QTL on chromosome 6 showed stage-specific expression. Genomic regions particularly on bin 3.04 and bin 6.00/01 in chromosomes 3 and 6, respectively, are known as the regions rich in resistant genes to multiple maize viruses, including SCMV, Maize dwarf mosaic virus (MDMV), and Johnson grass mosaic virus (JGMV; Xia et al. 1999; Jones et al. 2007; Ingvardsen et al. 2010; Ding et al. 2012; Stewart et al. 2013; Tao et al. 2013; Zambrano et al. 2014), Wheat streak mosaic virus (WSMV; Jones et al. 2011), Maize mosaic virus (MMV; Zambrano et al. 2014), and maize chlorotic dwarf virus (MCDV; Jones et al. 2004; Zambrano et al. 2014). The results of the current study also indicate the importance of the same regions having QTL with major effects. Nevertheless, whether the same region/s are contributing for resistance to both SCMV and MCMV or SCMV alone warrants further study.
The major effect QTL on chromosomes 3, 6, and 9 are interesting targets for either marker-assisted backcrossing (MABC) or marker-assisted recurrent selection (MARS) to introgress into different genetic backgrounds particularly on highly susceptible, widely using elite lines. These QTL can also be potentially used in maize breeding with the aim to enrich target alleles in F2 populations prior to producing DH lines from such populations. In this study, we also found a few new major QTL on chromosomes 1, 5, and 8; however, these QTL were expressed in specific populations and at specific stages of MLN infection.
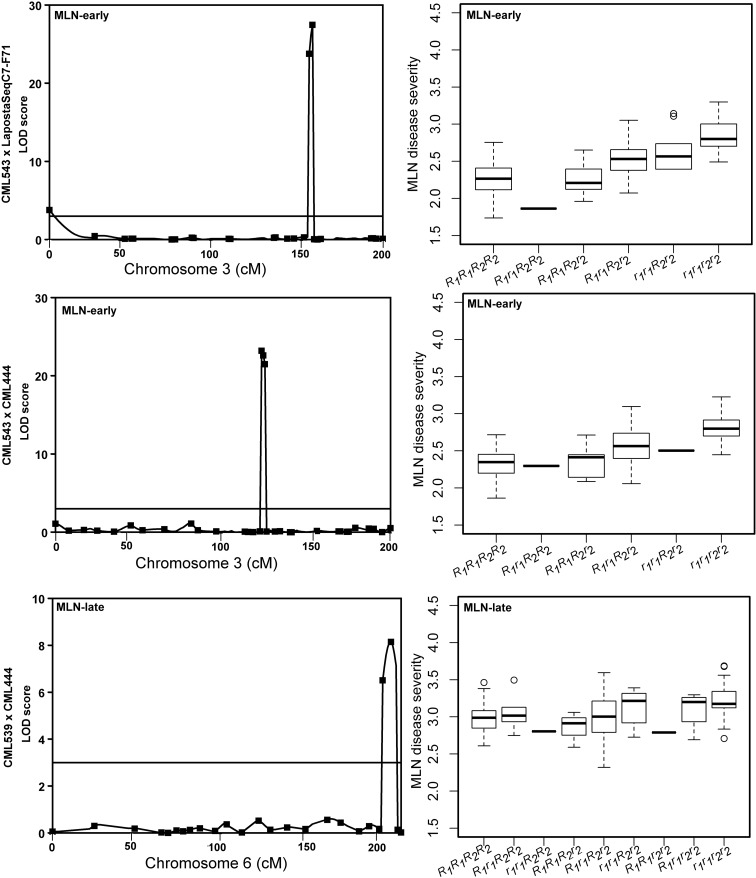
In population 1, major QTL identified on chromosome 3 with a LOD score of 27.48 and explaining 37% of the phenotypic variation revealed that CML 543 is the source of favorable alleles. The segregation alleles from two tightly linked markers for this major QTL reveal that F3 plants with low disease severity score were strongly associated with alleles from CML543, the resistant parent (Fig. 1) for both MLN-early and MLN-late. Similar association was also found for other major QTL observed on population 2, with strong association between alleles from two closely linked markers with low disease severity data (Fig. 1). This suggests CML 543 can be used as a potential donor to introgress the major QTL identified on chromosome 3. In contrast, the distribution of MLN-late disease severity in population 3 with markers linked to QTL on chromosome 6 showed some differences in the action of genes for controlling MLN tolerance. The population 3 phenotypes for MLN-late were skewed toward the susceptible parent. F3 plants having homozygous dominant alleles from one marker locus and homozygous recessive allele from other marker loci showed strong association with low disease severity score. This warrants further study to clarify on whether the identified QTL on chromosome 6 carries one gene or more than one gene before concluding on the type of gene/s involved in MLN resistance (Fig. 1).
Fig. 1.
Major QTL for MLN resistance in three F3 populations. A likelihood of odds (LOD) scan showing the QTL identified on chromosomes 3 and 6 in three F3 populations. Box-whisker plots display the level of disease resistance or severity for different allele combinations at resistance gene loci explaining > 10% of the phenotypic variation for MLN-early and MLN-late as determined by two strongly associated SNP markers
JLAM was implemented with the aim to take the advantages of both the high detection power of linkage mapping and improved resolution of association mapping to robustly identify the MLN resistance QTL. We applied three biometric models to increase the possibility to capture maximum number of QTL associated with MLN resistance. In this study, with three biometric models, we found 16 and 10 main effect QTL significantly associated with resistance to MLN at early and late stages of disease infection, respectively. The QTL identified on chromosomes 3, 5, 6, and 9 were consistent with the QTL found by single-population-based linkage analyses. Further, JLAM also drastically increased the resolution within the confidence intervals in some of MLN resistance QTL on chromosomes 3, 5, 6, and 9 (Tables 2 and 3). Moreover, we also found a few new QTL associated with MLN resistance that were not detected by linkage mapping but found only with JLAM. This could be attributed to the higher power and resolution offered by combined linkage and association mapping by exploiting both the variations across and within populations.
The results obtained in this study revealed some common genetic loci with previous large GWAS on MLN resistance (Gowda et al. 2015) and SCMV resistance (Zambrano et al. 2014; Li et al. 2016). QTL on chromosome 3 at 113, 133, and 189 Mbp were identified in both the association panel and in population 4 and JLAM panel. Similarly, QTL reported on chromosomes 5 (199 Mbp) and 6 (85 Mbp) in the association panel were also found in population 2 and population 3. Taken together, these results indicate that there is common genomic regions particularly across populations on chromosomes 3, 6, and 9 which contributing significantly on resistance to MLN.
The ability to predict and select best disease-resistant lines without phenotyping in biparental populations based on genomic-estimated breeding values is an important application of GS in maize breeding (Zhang et al. 2017). The primary method for doing this is through GS models (Meuwissen et al. 2001; Lorenz et al. 2011), a strategy that is well established in large commercial seed companies but still in its infancy among public sector breeding programs. The potential and limits of GS-based predictions have been examined in maize for several traits (Albrecht et al. 2011; Riedelsheimer et al. 2012; Zhao et al. 2012; Bernardo 2014; Crossa et al. 2017). Although MLN resistance is relatively complex (because of combination of two viruses), we observed high prediction accuracy of > 0.65 across three populations, which is comparable with the previously reported prediction accuracy for MLN (Gowda et al. 2015) and NCLB (Technow et al. 2013) (Fig. 2a). We observed small improvements in prediction accuracy by including MLN resistance-associated markers suggesting the possibility of considerable contribution from several minor effect QTL which were not detected by linkage mapping studies.
Success of GS in maize breeding depends on the type of training populations used and their genetic relationships with the prediction set. In this study, we tested predictions within and across biparental populations. The results clearly suggested that for MLN, the accuracy is not affected significantly when training populations are based on either a single biparental population or multiple populations or even when a selected panel of breeding lines (Gowda et al. 2015) is used. QTL mapping and JLAM results suggest that the genetic architecture of MLN is perhaps much less complex compared to traits like grain yield. Therefore, for comprehensive improvement of MLN resistance in breeding materials, we suggest to incorporate GS in breeding programs, as GS allows to capture contributions of even small effect QTL along with the major effect QTL. The prediction accuracy for MLN was slightly higher when the training set was derived from the same population than from a combination of many populations. This might be attributed to varying levels of relatedness and confounding population structure. Nevertheless, the prediction accuracy is still promising and encouraging to apply GS as one option to select the best MLN-resistant lines by reducing the phenotyping efforts.
High-throughput and cost-effective genotyping platforms are required to implement GS routinely in the breeding programs. Recent advances in sequencing technologies like GBS provide the capacity to genotype substantial number of breeding lines at low costs (Elshire et al. 2011). The cost per sample for GBS is comparable with the low-density SNPs obtained from the single-plex arrays. In this study, for across populations, phenotypic selection accuracy which is estimated as h (square root of heritability) was moderate for early and late stages of MLN infection. Whereas for GS, selection accuracy was slightly higher for MLN-early and MLN-late. By considering the possibility to complete up to three maize cycles per year (Lorenzana and Bernardo 2009), GS is more efficient in terms of genetic gain per year. With rapid reduction in genotyping cost, it is possible to effectively apply GS for MLN resistance routinely in maize breeding programs in SSA.
Conclusion
In this study, we used four biparental populations to understand the genetic architecture of MLN resistance and validate the earlier findings in CIMMYT-derived sub-tropical maize germplasm. Two major QTL were identified on chromosomes 3 and 6 across different genetic backgrounds; these could be potential candidates for genomic-assisted breeding. JLAM scan also identified 26 main effect QTL significantly associated with resistance to MLN. The genomic regions identified on chromosomes 1, 3, 6, and 9 are consistently detected in both linkage mapping and JLAM. Further validation could lead to development of production markers for MLN resistance. Introgressing these major QTL on chromosomes 3, 6, and 9 into elite inbred lines could improve the level of resistance to MLN. GS results revealed higher genetic gain per year for marker-based selection. These results suggest that integration of GS in maize breeding even with small training population sizes is an attractive complement to phenotypic selection to improve resistance to MLN. Overall, the study confirmed that MLN resistance is controlled by a few major genes and several minor genes.
Electronic supplementary material
(DOCX 101 kb)
(DOCX 137 kb)
(DOCX 1.73 mb)
(DOCX 12.6 kb)
(DOCX 17.4 kb)
Acknowledgements
The authors wish to thank CIMMYT field technicians at the different experiment stations in Kenya for managing trials; the management of Kenya Agricultural and Livestock Research Organization (KALRO) for giving us access to the experiment station; and CIMMYT laboratory technicians in Kenya for preparing samples for genotyping. We want to thank ARC South Africa for providing one population for this study under IMAS project. We also want to thank Dr. Edward S. Buckler and the Institute of Genomic Diversity, Cornell University, for the high-density genotyping (GBS) and imputation service and the Monsanto Company for low-density genotyping and also providing marker database that links the genetic positions of the proprietary SNP markers with public markers.
Author contributions
YB, DM, BMP, and MG—conceived the experiment; YB, MG, and SLM—conducted the field evaluations and phenotyping; MG and KS coordinated the GBS experiments; MG—carried out the linkage, GS, and GWAS analyses; MG, YB, DM, BD, MO, BMP, JMB, KS, SM and SLM—interpreted the results and drafted the manuscript.
Funding information
This study was supported by various projects, especially the STMA, WEMA, and MLND-Africa projects funded by the Bill & Melinda Gates Foundation, USAID, and Syngenta Foundation for Sustainable Agriculture, besides the CGIAR Research Program on MAIZE.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Key message
Analysis of genetic architecture of MLN resistance in four biparental populations revealed QTL with major effects on chromosomes 3, 6, and 9 that were consistent across genetic backgrounds and environments.
Electronic supplementary material
The online version of this article (10.1007/s11032-018-0829-7) contains supplementary material, which is available to authorized users.
References
- Adams IP, Harju VA, Hodges T, Hany U, Skelton A, Rai S, Deka MK, Smith J, Fox A, Uzayisenga B, Ngaboyisonga C, Uwumukiza B, Rutikanga A, Rutherford M, Ricthis B, Phiri N, Boonham N. First report of maize lethal necrosis disease in Rwanda. New Dis Rep. 2014;29:22. doi: 10.5197/j.2044-0588.2014.029.022. [DOI] [Google Scholar]
- Albrecht T, Wimmer V, Auinger H-J, Erbe M, Knaak C, Ouzunova M, Simianer H, Schön C-C. Genome-based prediction of testcross values in maize. Theor Appl Genet. 2011;123:339–350. doi: 10.1007/s00122-011-1587-7. [DOI] [PubMed] [Google Scholar]
- Bernardo R. Genomewide selection when major genes are known. Crop Sci. 2014;54(1):68–75. doi: 10.2135/cropsci2013.05.0315. [DOI] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Cairns JE, Crossa J, Zaidi PH, Grudloyma P, Sanchez C, Araus JL, Thaitad S, Makumbi D, Magorokosho C, Bänziger M, Menkir A. Identification of drought, heat, and combined drought and heat tolerant donors in maize. Crop Sci. 2013;53(4):1335–1346. doi: 10.2135/cropsci2012.09.0545. [DOI] [Google Scholar]
- Crossa J, de los Campos G, Perez P, Gianola D, Burgueno J, Araus JL, et al. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics. 2010;186:713–724. doi: 10.1534/genetics.110.118521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J, Beyene Y, Kassa S, Perez P, Hickey JM, Chen C, et al. Genomic prediction in maize breeding populations with genotyping-by-sequencing. G3 Genes Genom Genet. 2013;3:1903–1926. doi: 10.1534/g3.113.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J, Pérez-Rodríguez P, Cuevas J, Montesinos-López O, Jarquín D, de los Campos G, Burgueño J, González-Camacho JM, Pérez-Elizalde S, Beyene Y, Dreisigacker S, Singh R, Zhang X, Gowda M, Roorkiwal M, Rutkoski J, Varshney RK. Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 2017;22(11):961–975. doi: 10.1016/j.tplants.2017.08.011. [DOI] [PubMed] [Google Scholar]
- De Groote H, Oloo F, Tongruksawattana S, Das B. Community-survey based assessment of the geographic distribution and impact of maize lethal necrosis (MLN) disease in Kenya. Crop Prot. 2016;82:30–35. doi: 10.1016/j.cropro.2015.12.003. [DOI] [Google Scholar]
- Dekkers JCM. Prediction of response to marker-assisted and genomic selection using selection index theory. J Anim Breed Genet. 2007;124:331–341. doi: 10.1111/j.1439-0388.2007.00701.x. [DOI] [PubMed] [Google Scholar]
- Ding J, Li H, Wang Y, Zhao R, Zhang X, Chen J, Xia Z, Wu J. Fine mapping of Rscmv2, a major gene for resistance to sugarcane mosaic virus in maize. Mol Breed. 2012;30:1593–1600. doi: 10.1007/s11032-012-9741-8. [DOI] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;14:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One. 2014;9(2):e90346. doi: 10.1371/journal.pone.0090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda M, Das B, Makumbi D, Babu R, Semagn K, Mahuku G, Olsen MS, Bright JM, Beyene Y, Prasanna BM. Genome-wide association and genomic prediction of resistance to maize lethal necrosis disease in tropical maize germplasm. Theor Appl Genet. 2015;128(10):1957–1968. doi: 10.1007/s00122-015-2559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective Bonferroni test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Ingvardsen CR, Xing Y, Frei UK, Lübberstedt T. Genetic and physical fine mapping of Scmv2, a potyvirus resistance gene in maize. Theor Appl Genet. 2010;120:1621–1634. doi: 10.1007/s00122-010-1281-1. [DOI] [PubMed] [Google Scholar]
- Jensen SG, Wysong DS, Ball EM, Higley PM. Seed transmission of maize chlorotic mottle virus. Plant Dis. 1991;75:497–498. doi: 10.1094/PD-75-0497. [DOI] [Google Scholar]
- Jones MW, Redinbaugh MG, Anderson RJ, Louie R. Identification of quantitative trait loci controlling resistance to Maize chlorotic dwarf virus. Theor Appl Genet. 2004;110:48–57. doi: 10.1007/s00122-004-1757-y. [DOI] [PubMed] [Google Scholar]
- Jones MW, Redinbaugh MG, Louie R. The Mdm1 locus and maize resistance to Maize dwarf mosaic virus. Plant Dis. 2007;91:185–190. doi: 10.1094/PDIS-91-2-0185. [DOI] [PubMed] [Google Scholar]
- Jones MW, Boyd EC, Redinbaugh MG. Responses of maize (Zea mays L.) near isogenic lines carrying Wsm1, Wsm2, and Wsm3 to three viruses in the Potyviridae. Theor Appl Genet. 2011;123:729–740. doi: 10.1007/s00122-011-1622-8. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugenic. 1944;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- Li Z, Chen J, Han L, Wen J, Chen G, Li H, Wang Y, Zhao R, Zhang X, Xia Z, Yan J. Association mapping resolving the major loci Scmv2 conferring resistance to sugarcane mosaic virus in maize. Europ J Plant Pathol. 2016;145(2):385–391. doi: 10.1007/s10658-015-0852-z. [DOI] [Google Scholar]
- Liu W, Gowda M, Steinhoff J, Maurer HP, Würschum T, Longin CFH, Cossic F, Reif JC. Association mapping in an elite maize breeding population. Theor Appl Genet. 2011;123:847–858. doi: 10.1007/s00122-011-1631-7. [DOI] [PubMed] [Google Scholar]
- Lorenz AJ, Chao S, Asoro FG, et al. Genomic selection in plant breeding: knowledge and prospects. Adv Agron. 2011;110:77–123. doi: 10.1016/B978-0-12-385531-2.00002-5. [DOI] [Google Scholar]
- Lorenzana R, Bernardo R. Accuracy of genotypic value predictions for marker-based selection in biparental plant populations. Theor Appl Genet. 2009;120:151–161. doi: 10.1007/s00122-009-1166-3. [DOI] [PubMed] [Google Scholar]
- Louie R. Sugarcane mosaic virus in Kenya. Plant Dis. 1980;64:944–947. doi: 10.1094/PD-64-944. [DOI] [Google Scholar]
- Lukanda M, Owati A, Ogunsanya P, Valimunzigha K, Katsongo K, Ndemere H, Kumar PL. First report of maize chlorotic mottle virus infecting maize in the Democratic Republic of the Congo. Plant Dis. 2014;98(10):1448–1448. doi: 10.1094/PDIS-05-14-0484-PDN. [DOI] [PubMed] [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10(8):565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Mahuku G, Lockhart BE, Wanjala B, Jones MW, Kimunye JN, Stewart LR, Cassone BJ, Sevgan S, Nyasani JO, Kusia E, Kumar PL, Niblett CL, Kiggundu A, Asea G, Pappu HR, Wangai A, Prasanna BM, Redinbaugh M. Maize lethal necrosis (MLN), an emerging threat to maize-based food security in sub-Saharan Africa. Phytopathology. 2015;105(7):956–965. doi: 10.1094/PHYTO-12-14-0367-FI. [DOI] [PubMed] [Google Scholar]
- Mahuku G, Wangai AW, Sadessa K, Teklewold A, Wegary D, Adams I, Smith J, Braidwood L, Feyissa B, Regassa B, Wanjala B, Kimunye JN, Mugambi C, BoTtomley E, Bryce S, Ayalneh D, Prasanna BM. First report of maize chlorotic mottle virus and maize lethal necrosis on maize in Ethiopia. Plant Dis. 2015;99(12):1870–1870. doi: 10.1094/PDIS-04-15-0373-PDN. [DOI] [Google Scholar]
- Meng L, Li H, Zhang L, Wang J. QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015;3(3):269–283. doi: 10.1016/j.cj.2015.01.001. [DOI] [Google Scholar]
- Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanna BM (2016) Developing and deploying abiotic stress-tolerant maize varieties in the tropics: challenges and opportunities. In Molecular Breeding for Sustainable Crop Improvement. Springer, New York, pp 61–77 Springer International Publishing. (pp. 61–77)
- R Development Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- Rawlings JO, Pantula SG, Dickey DA. Applied regression analysis: a research tool. 2. New York, NY: Springer; 1998. [Google Scholar]
- Redinbaugh MG, Pratt RC. Virus resistance. In: Bennetzen JL, Hake SC, editors. Handbook of maize: its biology. New York: Springer; 2009. pp. 251–268. [Google Scholar]
- Reif JC, Liu W, Gowda M, Maurer HP, Möhring J, Fischer S, Schechert A, Würschum T. Genetic basis of agronomically important traits in sugar beet (Beta vulgaris L.) investigated with joint linkage association mapping. Theor Appl Genet. 2010;121:1489–1499. doi: 10.1007/s00122-010-1405-7. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C, Czedik-Eysenberg A, Grieder C, Lisec J, Technow F, Sulpice R, Altmann T, Stitt M, Willmitzer L, Melchinger AE (2012) Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet. 10.1038/ng.1033 [DOI] [PubMed]
- SAS Institute Inc . SAS 9.2 Intelligence Platform: System Administration Guide. Cary: SAS Institute Inc; 2010. [Google Scholar]
- Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. doi: 10.1214/aos/1176344136. [DOI] [Google Scholar]
- Semagn K, Beyene Y, Warburton M, Tarekegne A, Mugo S, Meisel B, Sehabiague P, Prasanna B. Meta-analyses of QTL for grain yield and anthesis silking interval in 18 maize populations evaluated under water-stressed and well-watered environments. BMC Genomics. 2013;14:313. doi: 10.1186/1471-2164-14-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiferaw B, Prasanna BM, Hellin J, Bänziger M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Sec. 2011;3(3):307–327. doi: 10.1007/s12571-011-0140-5. [DOI] [Google Scholar]
- Stewart LR, Haque MA, Jones MW, Redinbaugh MG. Response of maize (Zea mays L.) lines carrying Wsm1, Wsm2, and Wsm3 to the potyviruses Johnsongrass mosaic virus and Sorghum mosaic virus. Mol Breed. 2013;31:289–297. doi: 10.1007/s11032-012-9789-5. [DOI] [Google Scholar]
- Tao YF, Jiang L, Liu QQ, Zhang Y, Zhang R, Ingvardsen CR, Frei UK, Wang BB, Lai JS, Lubberstedt T, Xu ML. Combined linkage and association mapping reveals candidates for Scmv1, a major locus involved in resistance to sugarcane mosaic virus (SCMV) in maize. BMC Plant Biol. 2013;13:162. doi: 10.1186/1471-2229-13-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technow F, Bürger A, Melchinger AE. Genomic prediction of northern corn leaf blight resistance in maize with combined or separated training sets for heterotic groups. G3. 2013;3:197–203. doi: 10.1534/g3.112.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utz HF, Melchinger AE, Schön CC. Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics. 2000;154:1839–1849. [PMC free article] [PubMed] [Google Scholar]
- Vivek BS, Krishna GK, Vengadessan V, Babu R, Zaidi PH, Kha LQ, Mandal SS, Grudloyma P, Takalkar S, Krothapalli K, Singh IS, Ocampo ETM, Xingming F, Burgueño J, Azrai M, Singh RP, Crossa J. Use of genomic estimated breeding values results in rapid genetic gains for drought tolerance in maize. Plant Genome. 2017;10:1–8. doi: 10.3835/plantgenome2016.07.0070. [DOI] [PubMed] [Google Scholar]
- Wangai AW, Redinbaugh MG, Kinyua ZM, Miano DW, Leley PK, Kasina M, Mahuku G, Scheets K, Jeffers D. First report of maize chlorotic mottle virus and maize lethal necrosis in Kenya. Plant Dis. 2012;96:1582–1582. doi: 10.1094/PDIS-06-12-0576-PDN. [DOI] [PubMed] [Google Scholar]
- Whittaker JC, Thompson R, Denham MC. Marker-assisted selection using ridge regression. Genet Res. 2000;75:249–252. doi: 10.1017/S0016672399004462. [DOI] [PubMed] [Google Scholar]
- Wisser RJ, Balint-Kurti PJ, Nelson RJ. The genetic architecture of disease resistance in maize: a synthesis of published studies. Phytopathology. 2006;96(2):120–129. doi: 10.1094/PHYTO-96-0120. [DOI] [PubMed] [Google Scholar]
- Würschum T, Liu W, Gowda M, Maurer HP, Fischer S, Schechert A, Reif JC. Comparison of biometrical models for joint linkage association mapping. Heredity. 2012;108:332–340. doi: 10.1038/hdy.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XC, Melchinger AE, Kuntze L, Lubberstedt T. Quantitative trait loci mapping of resistance to Sugarcane mosaic virus in maize. Phytopathology. 1999;89:660–667. doi: 10.1094/PHYTO.1999.89.8.660. [DOI] [PubMed] [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES. Genetic design and statistical power of nested association mapping in maize. Genetics. 2008;178:539–551. doi: 10.1534/genetics.107.074245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano JL, Jones MW, Brenner E, Francis DM, Tomas A, Redinbaugh MG. Genetic analysis of resistance to six virus diseases in a multiple virus-resistant maize inbred line. Theor Appl Genet. 2014;127(4):867–880. doi: 10.1007/s00122-014-2263-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao W, Li M, Chen H, Zhu S, Fan Z. Real-time TaqMan RT-PCR for detection of Maize chlorotic mottle virus in maize seeds. J Virol Methods. 2011;171:292–294. doi: 10.1016/j.jviromet.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang X, Perez-Rodriguez P, Semagn K, Beyene Y, Babu R, Lopez-Cruz MA, et al. Genomic prediction in biparental tropical maize populations in water-stressed and well-watered environments using low-density and GBS SNPs. Heredity. 2015;114:291–299. doi: 10.1038/hdy.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wang H, Beyene Y, Semagn K, Liu Y, Cao S, Cui Z, Ruan Y, Burgueño J, San Vicente F, Olsen M, Prasanna BM, Crossa J, Yu H, Zhang X. Effect of trait heritability, training population size and marker density on genomic prediction accuracy estimation in 22 bi-parental tropical maize populations. Front Plant Sci. 2017;8:1916. doi: 10.3389/fpls.2017.01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Gowda M, Liu W, Würschum T, Maurer HP, Longin FH, Ranc N, Reif JC. Accuracy of genomic selection in European maize elite breeding populations. Theor Appl Genet. 2012;124:769–776. doi: 10.1007/s00122-011-1745-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 101 kb)
(DOCX 137 kb)
(DOCX 1.73 mb)
(DOCX 12.6 kb)
(DOCX 17.4 kb)