Abstract
Linear mixed models (LMM) are widely used to estimate narrow sense heritability explained by tagged single-nucleotide polymorphisms (SNPs). However, those estimates are valid only if large sample sizes are used. We propose a Bayesian covariance component model (BCCM) that takes into account the genetic correlation among phenotypes and genetic correlation among individuals. The use of the BCCM allows us to circumvent issues related to small sample sizes, including overfitting and boundary estimates. Using expression of genes in breast cancer pathway, obtained from the Multiple Tissue Human Expression Resource (MuTHER) project, we demonstrate a significant improvement in the accuracy of SNP-based heritability estimates over univariate and likelihood-based methods. According to the BCCM, except CHURC1 (h2 = 0.27, credible interval = (0.2, 0.36)), all tested genes have trivial heritability estimates, thus explaining why recent progress in their eQTL identification has been limited.
Introduction
For many phenotypes, there is a substantial difference between estimates of narrow sense heritability from family studies and variance explained by discovered single-nucleotide polymorphisms (SNPs) from genome-wide association studies (GWAS) [1, 2]. This gap is a key component of the missing heritability problem [3]. Existing genotyping technologies have allowed narrow sense heritability to be estimated from unrelated individuals using all SNPs in the genotyping platform (typically most common with a minor allele frequency >0.05) [4]. However, given that we cannot exclude the possibility of existing rare variants with large effects that have not been detected by genotyping arrays, this SNP-specific heritability is only a lower bound of the true narrow sense heritability. Nevertheless, we do not yet fully understand the gap between SNP heritability and the variance explained by replicated SNPs.
Several hypotheses have been investigated to explain this problem. Recent attempts suggest that previous estimates are biased and that large sample sizes are required to obtain accurate results [5–7]. Naturally, violation of model assumptions can result in biased estimates. For example, using a model that does not capture existing epistatic effects will risk biasing the SNP heritability estimates [6]. Moreover, linear mixed models (LMM) implicitly assume that all SNPs have an effect on the phenotype as part of the infinitesimal assumption. Violation of this assumption was thought to be a possible source of bias given the widespread belief that the majority of SNPs are null [8]; however, recent studies found that the effect of this assumption is negligible on SNP heritability estimates [7, 9]. Furthermore, in twin studies, the phenotypic variation due to any shared environment might be significant. Therefore, a model that accounts for only a unique environment can inflate heritability estimates.
Biased estimates are not necessarily caused by model assumptions violations; they can also be a result of the assumptions of the estimation procedure itself. For example, the “set to zero” convention, a numerical adjustment used to ensure that variance estimates are positive, will upwardly bias the heritability. Additionally, in many cases, when small sample sizes are used, the variance components are inaccurately estimated taking boundary values. These potential sources of bias are all associated with heritability estimates from variance components models.
The restricted maximum likelihood (REML) method [10] is the mainstream method for estimating variance components. It is implemented in a variety of genome-wide software packages, such as the genome-wide complex trait analysis GCTA [11], efficient mixed-model association EMMA eXpedited [12], FAST LMM [13], and the genome-wide efficient mixed-model association GEMMA [14]. All these methods are equivalent in the sense that they are all based on the same classical univariate LMM. Indeed, some of these methods—e.g., EMMA, FAST LMM, and GEMMA—even produce identical p-values in genetic association testing applications [14]. However, these methods differ in their computational complexity, with GEMMA being the most efficient in this regard [14].
REML produces unbiased estimates of the variance components if they are allowed to be negative [15]; otherwise, its estimates are very likely to degenerate in the boundary of the parameter space when small samples are used. When a variance parameter is estimated as zero, this should not imply that it is close to zero. Instead, it commonly indicates a large amount of uncertainty about it [16–18]. In multiple-phenotype models, the problem with estimates extends to another class of degeneracy, namely, non-positive definite estimates of the covariance matrices. Such estimates not only are uninterpretable but also can result in underestimated standard errors for the fixed-effect part of an LMM [19]. This feature is misleading in GWAS because an SNP of interest is typically tested by modeling its effect as fixed; therefore, an underestimated standard error will lead to overconfidence about the estimated effect.
Multivariate LMM have recently emerged as a tool to increase statistical power by incorporating correlations among multiple phenotypes. Such models can be fitted using, for example, multi-trait mixed-model MTMM [20] and GCTA [21], both of which are limited to bivariate phenotypes. A popular multivariate method that extended the number of phenotypes to more than two was recently proposed by Zhou and Stephens [22] and implemented in GEMMA software. Both the univariate and multivariate versions of GEMMA are widely used in genetic epidemiology. Therefore, we use them as our benchmark given that they have one advantage over the aforementioned methods: speed [14, 22]. GEMMA relies on the maximum likelihood (ML) method including its restricted version. In this study, however, we propose that an improved estimation is obtained using a full Bayes approach, specifically, the use of inverse Wishart (IW) prior for the covariance matrices and a diffuse normal distribution on the covariate coefficients.
The outline of this article is as follows. First, we provide some definitions and notations about the matrix-normal distribution. Second, we discuss the widely used model for multiple phenotypes and subsequently state the definition of marginal SNP heritability. Third, we unravel the equivalence between the multivariate model under study and multivariate ridge regression. This equivalence indicates that the model has the advantage of including all tagged SNPs while accommodating inevitable correlations among them (linkage disequilibrium). The ridge representation is used further to (a) explain the degeneracy problem associated with estimates of the covariance matrices in genome-wide studies and (b) provide a fast evaluation of the posterior distribution of the SNP effect sizes, which can subsequently be used for predictions as model checking. Next, we present the Bayesian covariance component model (BCCM) and its simplified form, which facilitates the use of many “off-the-shelf” Bayesian software. Via simulations, we show that the BCCM can accurately retrieve the real SNP heritability value under different structures of genetic correlations. The simulated data are used further to evaluate the SNP heritability estimates from GEMMA. The benefits of our model are shown further using expression of genes involved in a breast cancer (BC) pathway. Finally, a scaled version of the inverse Wishart (SIW) is used to assess for prior sensitivity.
Methods
Definitions and notations
The matrix-normal distribution is a generalization of the multivariate normal distribution, which allows us to model correlations among and within subjects [23]. The probability density function for the random matrix X (d × n) that follows the matrix-normal distribution with mean matrix M (d × n) column covariance matrix A (n × n) and row covariance matrix B (d × d) denoted by X ~ MNn,d(M,A,B) has the following form:
| 1 |
Its expected value and second-order expectations are given by E[X] = M, and , respectively.
One way to understand how the matrix normal generalizes the multivariate normal distribution is to assume we have n one-dimensional variates that are independent and identically distributed as normal with zero mean and variance σ2, i.e., xi ~ N(0, σ2). This can be written equivalently as a multivariate normal distribution Xn×1 ~ Nn(0,σ2In). Now, assume we have n d-dimensional variates that are independent and identically distributed as multivariate normal with zero mean and covariance matrix B, i.e., the vectors Xi ~ Nd(0,B). Because these variates are independent, concatenating them will result in a vector with a block diagonal covariance matrix , which is itself equivalent to [X1,…, Xn] ~ MNn,d(0, In, B).
Multiple-phenotype model
We consider the matrix-variate model given by
| 2 |
where n and d are the number of individuals and phenotypes, respectively. Here, Y is a d × n phenotypic matrix; X is a k × n matrix of covariates, such as age and sex; and β is a d × k matrix of corresponding coefficients. η is a d × n matrix of random effects that is independent of the d × n matrix of errors ∈. The random effect term is used to model any correlation between and within individuals. The n × n relatedness matrix K represents the genetic covariance between individuals and is typically estimated in advance using the genotype data of p SNPs and n individuals. In other words, it is the sample covariance matrix based on the genotype matrix Z (p × n) with rows pre-processed to have zero mean and unit variance, K = ZtZ/p. The d × d matrix Σ represents the genetic covariance matrix within individuals. Σ∈ and In specify the environmental covariance matrices within and between individuals, respectively.
Below, we state the SNP heritability definition under this model and discuss problems hindering its estimation.
Marginal SNP heritability
SNP heritability is defined as the proportion of additive phenotypic variance explained by tagged SNPs. The diagonal elements of the genetic covariance matrix Σ represent the polygenic variances of the d phenotypes. Therefore, the SNP heritability of the ith phenotype according to the multivariate model is defined as follows:
| 3 |
Estimation of Σ and Σ∈ requires estimation of d(d + 1) different parameters. Clearly, this number increases rapidly with the number of phenotypes. Such a large number of parameters can make existing algorithms unstable, e.g., by producing covariance matrices that are not positive definite and standard error matrices with large or sometimes uninterpretable entries (NAN). To explain these issues in more detail, it is instructive to first describe the nature of these covariance matrices or, in other words, their relation to SNP effect sizes. To this end, we proceed by writing the matrix-variate model in Eq. (2) in terms of SNP effect sizes. In statistics, this is referred to as ridge regression.
Generalized Bayesian interpretation of ridge regression
The Bayesian interpretation of ridge regression assumes that the regression coefficients of a multiple regression model are independent and identically normally distributed [24]. Here, we aim to provide a broader Bayesian interpretation of ridge regression in the context of matrix-normal distribution. Consider the matrix-normal regression model of p SNP effects on d phenotypes:
| 4 |
with matrix-normal prior to the effect sizes1
| 5 |
where the Ip (p × p) and Σβ (d × d) represent the effect size covariances between and within SNPs, respectively. Thus, we are assuming that effect sizes are correlated within SNPs and independent across SNPs. Exploiting the multivariate normal equivalence of matrix-normal distribution, model 4 can be rewritten as follows:
| 6 |
Similarly, the prior on the effect sizes is written as follows:
| 7 |
which is itself equivalent to [βz]:j ~ Nd(0,Σβ/p) j = 1, …, p.
Here, vec refers to matrix vectorization. Now,
The multivariate normal equivalence of model (2) without βX is given as follows:
| 8 |
Noting that both vec(η) and Zt ⊗ Id vec(βz) have the same probability model, namely Nnd(0,K ⊗ Σ), it becomes clear that when the relatedness matrix is estimated using , the multivariate ridge regression (Eq. (4) with (5)) is equivalent to the multiple-phenotype model in Eq. (2). This equivalence shows that the multiple-phenotype model has the advantage of handling linkage disequilibrium in an integrative manner, i.e., without the need for an initial LD pruning step (see [25] for relevant discussion on how ridge regression handles LD).
Boundary estimation problem
To understand the causes of non-positive definite estimates of the genetic covariance matrix Σ, it is easier to think of model (2) using its equivalent form from the ridge regression representation as follows:
| 9 |
where βz is the D×P matrix of effect sizes for the d phenotypes and p SNPs. A natural estimator of the genetic covariance matrix would be . An estimate of this form that is not positive definite may signal a phenotype with zero genetic variance. This occurs when all SNP effect sizes for a particular phenotype are equal. Another common cause is the existence of perfect linear dependency between the SNP effect sizes of different phenotypes. However, given that we know that these conditions are implausible a priori, we address the non-positive definiteness problem that corresponds to these factors by taking a Bayesian approach via the assignment of an IW prior to the covariance matrix.
The scaling matrix V of the IW distribution (IW(V,v)) and its degrees of freedom v determine how informative the prior is. For example, the least informative IW is formed by taking the scaling matrix to be the identity matrix and the degrees of freedom to be the least such that the distribution remains proper. Assigning an IW to the covariance matrix is equivalent to assigning an inverse gamma distribution to the variances. To show the effect of the choice of the degrees of freedom on the correlations, 50,000 d-dimensional matrices from the IW(Id,v) with v = d, d + 1 and d + 2, were randomly generated (supplementary note). The least informative IW corresponds to v = d+1, and use of this parameter combination has the effect of setting an approximate uniform distribution on the genetic correlations.
Simplified full Bayes model
Using univariate LMM, Lippert et al. [13] showed that a spectrally transformed model using a spectral decomposition of the relatedness matrix significantly reduces computational complexity. Similar approaches were subsequently adopted by Zhou and Stephens [14, 22] and Pirinen et al. [26]. Following these developments, we spectrally decompose the relatedness matrix, which allows us to write the matrix-variate model in Eq. (2) as a multivariate normal model on the transformed data for each individual independently as follows:
| 10 |
where U is an n × n orthogonal matrix of normalized eigenvectors, and rj is the corresponding n eigenvalues. Here, [A]:j is the jth column of the matrix A. Eventually, the full Bayes model will have the following hierarchical structure:
Here, diag(10,000, k) is a k × k diagonal matrix. We implemented the simplified full Bayes model as a module for the existing and widely used Bayesian analysis software rjags [27], R2jags [28], and coda [29]. The relevant BUGS code is provided in the supplementary note. Next, we provide an algorithm to predict unobserved phenotypic values, which can be used as a model-checking technique.
SNP-based prediction
Prediction can be performed first by estimating the effect sizes using the computationally efficient formula for the mode of their posterior distribution,
| 11 |
where M = pΣ∈Σ−1. Second by incorporating the estimated effect sizes to obtain the predicted phenotypic values of the new sample based on its genotypes Znew:
| 12 |
Here, y=vec(Y) and the effect sizes b = vec(βz) are the values estimated from the original data. Steps are delineated in the supplementary note.
Multiple Tissue Human Expression Resource (MuTHER) project
One of the main aims of the MuTHER consortium [30] is to quantify the variation in gene expression that is due to genetic factors and ultimately provides insights into the mechanisms underlying the disease susceptibility of associated SNPs. To this end, genome-wide expression profiles (Illumina HT-12v3 Chip) and genome-wide association data (Illumina 610k or 1 M chip) were previously obtained from three tissues adipose, skin and lymphoblastoid cell lines (LCL) from blood samples of 856 Caucasian female twins aged 38.7–86.6 years and living throughout the United Kingdom [30]. All recruited females were from the TwinsUK adult registry [31, 32].
In this work SNPs from the original study [30], which were filtered at an MAF >0.01 and IMPUTE info value >0.6 were used (p = 2,238,276). The filtered list of probes given by Grundberg et al. [30], which excluded polymorphic probes and probes mapping to multiple genes or to genes of uncertain function, was also used.
BCCM is not tailored for twins, in the sense that it does not consider the shared environment, which can result in inflated heritability estimates. Accordingly, only a twin of each pair was combined with the available singletons, resulting in 446 samples suitable for the analysis. There were no differences in batch effects after removing the twin structure; therefore, only age was included as a non-genotype covariate.
In this study, gene expression measurements obtained from LCL tissues were used. The expression data were downloaded directly from ArrayExpress, and access to the genotypes and covariates was granted from the TwinsUK Steering Committee.
Results
Simulation study
We simulated 50 bivariate phenotypes using relatedness from a publicly available genotype data, which can be found in the “example” folder that is part of the GEMMA software [33]. Given the genotypes of p = 12,226 SNPs across n = 1940 subjects, with missing genotypes replaced by the mean genotype, we simulated bivariate phenotypes, controlling their SNP heritability and their genetic correlation. Six distinct scenarios were used to simulate bivariate phenotypes from the matrix-variate model given in equation (2). First, assuming both weak genetic correlation and low SNP heritability h2 = 0.25. Second, assuming weak genetic correlation r = 0.3 but moderate SNP heritability h2 = 0.5. Third, assuming weak genetic correlation r = 0.3 but high SNP heritability h2 = 0.7. Fourth, assuming strong genetic correlation r = 0.7 but weak SNP heritability h2 = 0.25. Fifth, assuming strong genetic correlation r = 0.7 but moderate SNP heritability h2 = 0.5. Finally, assuming both strong genetic correlation r = 0.7 and high SNP heritability h2 = 0.7.
The simplified full Bayes model (BCCM) was fitted to the simulated data sets using the BUGS code (supplementary note), via the R2jags package [28], as its function “autojags”, has the capacity to automatically run Markov chain Monte Carlo (MCMC) models till convergence to the equilibrium state is believed to be achieved, based on the Gelman-Rubin diagnostic [34] (see supplementary note for more information on this diagnostic). This makes R2jags a handy tool for simulations, as it saves the user having to inspect convergence at each replicate. Equilibrium here refers to the situation under which states are believed to be stochastically independent of each other.
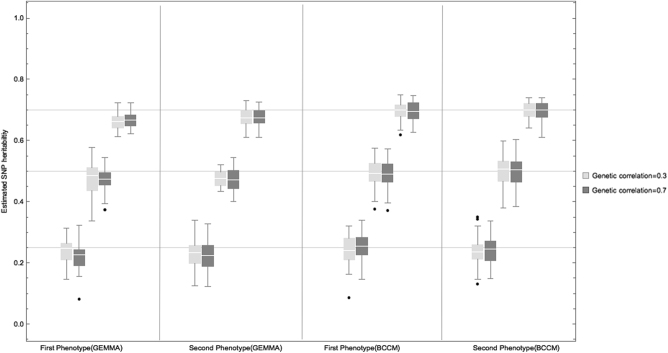
The results from the six scenarios are shown in Fig. 1, which suggest that the proposed BCCM can indeed retrieve the marginal SNP heritability regardless of the strength of genetic correlation between the two phenotypes.
Fig. 1.
Performance of BCCM and GEMMA for SNP heritability estimation. For a given genetic correlation, the SNP heritability of the two phenotypes was estimated, and results across 50 replicates are shown in two separate box plots each correspond to a different phenotype. This was done using different magnitudes of true heritability (0.25, 0.5, 0.7). We can see that the BCCM works well under all scenarios, as average estimates almost coincide with the true heritability values (horizontal lines). There are few outliers (dots), which are expected to disappear as the number of MCMC iterations tends to infinity. On the other hand, using the same type and number of simulated data used in assessing the BCCM, GEMMA seems to slightly underestimate the true heritability (horizontal lines). The estimation accuracy of both methods will increase as the sample size tends to infinity
In parallel, GEMMA was applied to the same type of simulated data, i.e. phenotypes simulated under the same assumptions that were used for the assessment of the BCCM. Overall, in most scenarios, both methods produced similar interval widths; however, in all scenarios (except when both real heritability and correlation are small) the average estimates from the BCCM are noticeably closer to the true SNP heritability values compared to those of the GEMMA’s type (Fig. 1).
Heritability estimation using MuTHER consortium data
To illustrate the benefits of our method (BCCM), we applied it to a pre-defined gene set from the MuTHER project, namely, genes in a BC pathway [35]. We chose 20 filtered genes comprising d = 30 filtered probes. We performed two types of comparisons to characterize the variability in the heritability estimates: Bayesian versus frequentist approaches and univariate versus multivariate approaches. In each scenario, confidence and credible intervals of the heritability estimates were inspected to assess their uncertainty.
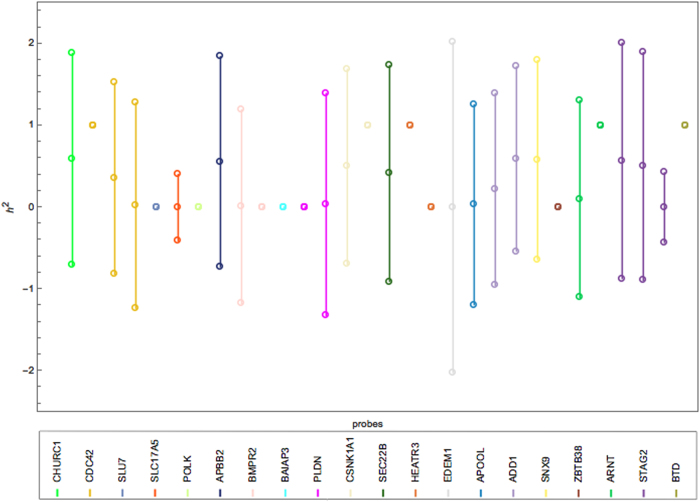
For univariate ML-based analysis, GEMMA software was used to obtain heritability estimates, which also facilitates the use of Wald’s method to compute confidence intervals. The model was separately fitted to each probe when d = 1 using its gene expression as a response variable and the genotypes as explanatory variables modeled via a random term. Figure 2 presents the Wald’s confidence intervals for the univariate likelihood-based heritability of each probe. In almost half of the cases, the estimates are indeed at the boundary. The remaining confidence intervals are wide, and in many cases, they spread beyond the parameter space (e.g., including negative values for heritability), which make them difficult to interpret.
Fig. 2.
Interval plot of the heritability of 20 BC genes (30 filtered probes) in the LCL tissue using univariate LMM in GEMMA
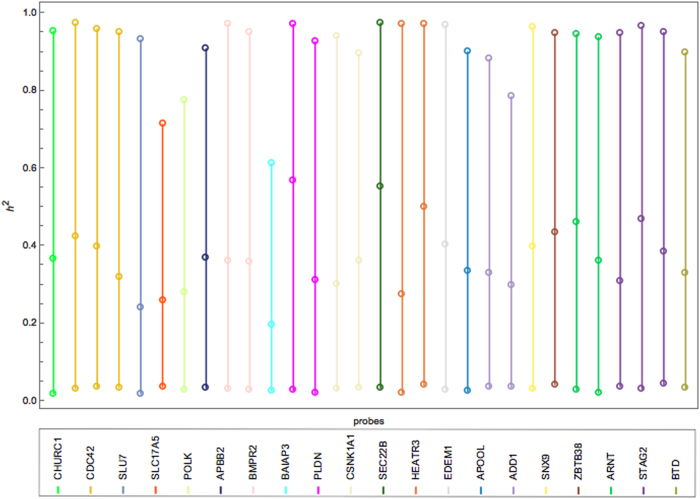
For a better characterization of the variability in the heritability estimates, a Bayesian univariate model was used. The model is based on a diffuse gamma prior for the scalar precisions: G(0.001, 0.001) with a unity mean and variance of 1000. The Bayesian estimates from the univariate analysis differ significantly from their REML counterparts (Figs. 2 and 3) as both remain susceptible to variability. Thus, the variance/uncertainty remained large despite the very large number of iterations (see [36] for relevant discussion). Convergence is further discussed below.
Fig. 3.
Interval plot of the heritability of 20 BC genes (30 filtered probes) in the LCL tissue using Bayesian univariate analysis with a diffuse gamma prior on the variance components
Although GEMMA can theoretically be applied to any number of phenotypes, when we attempted to fit five probes from the BC pathway using the filtered set of individuals (N = 446), the numerical algorithm failed to produce valid standard error matrices. In addition, the covariance matrices were not positive definite because of the small sample size and large covariance matrices. We overcame this problem by assigning the IW prior, as described above.
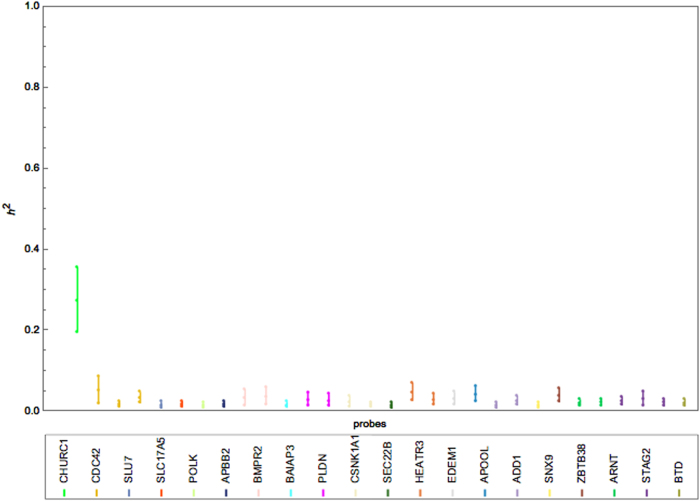
The credible intervals of the heritability of each probe are much narrower under the BCCM (Fig. 4) than under its univariate counterpart (Fig. 3), suggesting very little uncertainty about the heritability estimates. Our examination of convergence (supplementary note) showed that convergence is not only satisfactorily achieved under the multivariate analysis (BCCM) but also achieved with a shorter MCMC run than under the univariate model (supplementary note).
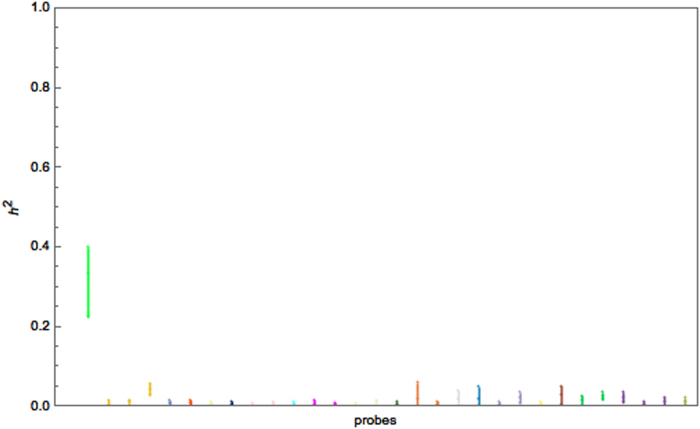
Fig. 4.
Interval plot of the heritability of 20 BC genes (30 filtered probes) in the LCL tissue using the BCCM with a non- informative IW prior on the covariance components
Finally, in contrast to both types of univariate analysis, the BCCM provided an insight into the SNP relevancy in explaining the variation in expression. Specifically, most tested BC genes [35] have negligible heritability (average ~ 0.03). The exception is CHURC1, which has a relatively high heritability of 0.27 with a credible interval (0.2, 0.36). These results were recorded after a 150,000 burn-in period using a sample of 5000 resulting from 25,000 iterations with a thinning interval of 5.
Model assessment
Based on credible intervals, the results from the BC pathway support the idea that BCCM can produce more accurate estimates of SNP heritability than classical multivariate and univariate models. It is also clear that the genetic architectures of complex phenotypes are miscellaneous, and no single method will be the most efficient in capturing all of them. The “gold standard” for model assessment and comparison is therefore an improved phenotype prediction using a new data set (see SNP-based prediction above).
In our example data, phenotype prediction as a model checking technique has its pitfalls. In the BC pathway, our method produced expression heritability estimates that are close to zero. Therefore, any attempt to predict the expression using SNPs is expected to fail because the heritability (variance explained by genotyped SNPs) is very low. Accordingly, we had to resort to alternative approaches in an attempt to provide evidence in support of our SNP heritability estimates. The approaches were (a) finding literature that could support or refute the results and (b) using more flexible prior specifications.
Real data example
To trace back the heritability estimates, we looked at previously reported cis eQTLs for the 20 genes from the MuTHER study tested here. According to Grundberg et al. [30], there are 196 SNPs associated with CHURC1 expression, i.e., with p-values <10−8; however, the other 19 had no reported eQTL, supporting the finding that their expression has limited SNP heritability. To determine whether the same conclusion can be drawn using a different analysis, we used univariate GEMMA to scan chromosome 14 for association with CHURC1 expression and identified 180 SNPs with p-values <10−8. Given that GEMMA fits an LMM with the fixed-effect part being the genotypes of the tested SNP, proximal contamination [37]; that is the situation when the tested SNP is assumed both fixed and random, can incur power loss. To eliminate this issue, we followed the GCTA approach and excluded chromosome 14 from the computations of the relatedness matrix. In addition to the 180 SNPs identified before the exclusion, only one additional significant SNP was detected because of a slight decrease in most of the p-values after the exclusion. Overall, there is a significant overlap between the SNPs from our analysis and those obtained by Grundberg et al. [30].
We determined whether there is an effect of eQTLs on heritability estimates.
To this end, we repeated the analysis, excluding the SNPs previously associated with CHURC1 expression from the model, and the trace plots for its heritability appeared unstable using the same burn-in period and thinning interval that was used before the exclusion. We therefore explored additional convergence diagnostics, leading to the choice of a million iterations with a thinning interval of 1000 after a similar burn-in period of 150,000. Nevertheless, convergence remained an issue for the heritability of CHURC1. However, the heritability of each of the remaining genes remained close to zero with acceptable convergence.
Sensitivity analysis of prior distributions
Possible arguments against the use of the IW are (a) the uncertainty for all variance parameters are controlled by a single degree of freedom, namely v=d+1, and (b) the lack of independence between the variances and the correlations. To overcome the first issue and alleviate the second, we used a scaled version of the IW.
The scaled IW was first introduced by O’Malley and Zaslavsky [38], and its relative advantage over the unscaled IW was discussed by Gelman and Hill [39]. The basic idea is to decompose the covariance matrix into a diagonal matrix and a matrix V distributed as (Id, d + 1); Σ = AV A. This idea implies that Σ ~ IW(AIdA, d + 1), AId A = diag (a1,…,ad). This prior will not shrink the correlations, so their marginal prior will remain uniform when v = d + 1. However, the variances now can be estimated more freely from the data. To determine the scaling needed, we added another level of variability by assigning a uniform prior U(0, 100) for each scaling parameter (see supplementary for the relevant BUGS code). The SNP heritability estimates using the SIW are very similar to those obtained using the unscaled method (Figs. 4 and 5), implying that the above concerns that plague the IW do not affect the posterior distributions of the heritability.
Fig. 5.
Interval plot of the heritability of 20 BC genes (30 filtered probes) in the LCL tissue using the BCCM with a non- informative SIW prior on the covariance components and a uniform prior on its scale parameters
Discussion
Motivated by the boundary estimation problem that frequently arise when attempting to estimate SNP heritability using ML-based methods, we developed a BCCM model. Two key problems are addressed by our model. First, it takes into account the tendency of the ML or REML estimates of the covariance matrices to be non-positive definite even when the number of phenotypes is not very large. Our approach overcame this problem by adding an extra level of variability to existing multivariate models through the assignment of a non-informative IW prior that allows the data to dominate while guarding against positive definiteness problems. Second, and as a result, the BCCM produced heritability estimates with less uncertainty.
The advancements offered by the BCCM were shown in comparison with state of the art methods. Specifically, using real data, we pinpointed the convergence issues in Bayesian univariate analyses (Fig. 3), the boundary estimation problem in classical univariate analyses, which occurred in 40% of the times (Fig. 2), the extremely wide confidence intervals of the heritability from classical univariate analyses, which occurred in 60% of the times (Fig. 2), and finally the boundary estimation problem in classical multivariate analysis. Using the same example, we showed how BCCM overcame these problems by improving chain convergence, producing results that are interpretable, prior insensitive, and in agreement with the literature.
We argue that the significant enhancement in convergence pertains to the extra amount of information used in the BCCM, that is the multiple phenotypic values for every individual in the study. Further, the genetic correlation matrix from the MUTHER study does possess useful information. Although 50% of pairwise correlations lie between 0.05 and 0.25, there are pairs of genes with high genetic correlation reaching −0.5 and 0.8 (supplementary note). This means that there are some probes exhibiting shared genetic etiology, that is when alleles affecting both phenotypes tend to be found within the same individual. Accordingly, at first, heritability results from BCCM might seem counterintuitive, given the strong genetic correlation between few pairs of probes. For example, one might expect another heritably expressed gene to come up in addition to CHURC1. However, even though high correlation indicates shared genetic bases, there is no reason to expect a shared variant to explain expression variation in two correlated genes by the same magnitude. In other words, the strength of the variant’s effect on a pair of correlated phenotypes can be different.
Although simulations were used to illuminate the performance of BCCM under a single instance (infinitesimal assumption) of the whole spectrum of genetic architectures, caution should be taken when performing method comparison based on simulations, as conclusions from generated phenotype data are assumption-specific. Important assumptions that can affect the interpretation of simulation-based method comparison include model and estimation assumptions as well as the chosen number of replicates. In our comparison, the model assumptions used to generate the phenotypes are of no concern, since BCCM and multivariate GEMMA (under the null hypothesis of no association) differ only in the estimation method. Accordingly, the number of iterations used in the underlying numerical algorithm (MCMC iterations in BCCM and the combined iterations of Newton–Raphson and the expectation-maximization (EM) algorithm in GEMMA) is an important determinant of our simulation results. In this occasion, it is important to recall that, by definition the EM algorithm does not guarantee to find the ML estimate; however, the theoretical results associated with it, such as the increase in the likelihood with each iteration, makes it a numerically stable procedure, when combined with Newton–Raphson method [40]. On the other hand, true convergence of a Markov chain to the equilibrium occurs only at infinity; therefore, there cannot exist any method that can guarantee MCMC convergence. However, the available diagnostic tools are all heuristic windows that can suggest whether the chain is close enough to the equilibrium. Finally, the number of replicates, which is generally not well defined, was kept fixed for the method comparison, but increasing it further, could improve the accuracy of the average SNP heritability estimate.
The BCCM can be built upon to advance its use. For example, although gene expression phenotypes were used in this study, the BCCM can be applied to any normalized quantitative phenotypes; however, for binary phenotypes, e.g. case–control status, a liability threshold model that correct for both, measurements scale and ascertainment bias should be used. This represents a useful extension of the univariate ML-based method for SNP heritability estimation of binary phenotypes [41]. Further, the BCCM might be extended to more than two covariance components, e.g. to estimate phenotypic variation due to shared environment between twins, or to partition heritability into the contributions of genomic regions [42].
Although by definition, the BCCM can tackle any number of phenotypes, in practice this number cannot be increased indefinitely. The main reason would be the formidable computational complexity, which scales about cubically with the number of phenotypes. Pathways are ideal applications of BCCM, as the number of phenotypes will be moderate and thus computational complexity will be tractable. Also, phenotypes within pathways are expected to be correlated, and accordingly the non-informative IW prior will remain a reasonable choice. In contrast, with large number of phenotypes that are not necessarily within a pathway, we might not expect all of them to be correlated, in which case, it may be desirable to enforce sparsity on the covariance matrices, representing a promising avenue for future research.
From a computational perspective, the simplified form of the BCCM allowed the posterior distribution of SNP heritability to be determined at a feasible computational cost using rjags [27]. This feature is advantageous because it saves users from having to write their own MCMC code. However, it should be noted that alternative software will be needed for genome-wide association scan, as rjags [27] will be intrinsically slow owing to the number of iterations required for convergence. Finally, the multivariate model used herein is based on the infinitesimal assumption, which might not be favored by all real data. Since in reality, the genetic architectures of complex phenotypes are unknown, a more flexible prior that fits a wide range of settings may be desirable. A mixture of matrix-variate normal distributions for the effect size matrix is likely to provide a gain in estimation accuracy and ultimately in phenotype prediction, representing another avenue for future research.
Extrapolating from the impact that SNP heritability has had on the genetic of complex phenotypes, we believe that the BCCM has the potential to pave the way for a major reshuffle in our understanding of the missing heritability problem; in particular, the gap between estimates of SNP heritability and variance explained by replicated SNPs. For example, instead of taking current estimates for granted, then approaching the missing heritability problem from an SNP discovery perspective, one can first reevaluate such estimates, ideally by means of predictions. In other words, the estimate that improves the prediction of unobserved phenotypic values would be the most accurate. The prediction algorithm provided in the method section could easily be implemented for this task.
Various consortia can benefit from implementing the BCCM. For example, estimates of genetic variances among the five psychiatric disorders discussed here [43] can be improved by considering measurements of additional disorders, such as obsessive compulsive disorder, generalized anxiety disorder, tic disorder, etc. However, this could compromise the performance of the multivariate LMM used to obtain the previous estimates [43] due to potentially large covariance matrices. In this case, the BCCM augmented by a liability threshold model that correct for both scale and bias holds significant potential in providing more accurate estimates of both SNP heritability and correlations among large number of disorders. Further afield, prediction results discussed in the same paper [43] could also be improved by applying our algorithm to a larger number of disorders.
Highly heritable gene expression could serve as candidate genes in other studies. For example, the statistical power to detect genotype–phenotype associations depends, to a non-trivial extent, on SNP heritability. Indeed, in our example data, we have shown the intertwined between eQTLs and the SNP heritability of gene expression. In particular, the lack of convergence for the heritability of CHURC1 after excluding its eQTLs, which also recapitulates the role of the prior distribution. In other words, the expected polygenic variance of CHURC1 expression is zero after removing its eQTLs (assuming neither epistatic nor very small effects); however, given that we are sampling from a family of positive definite matrices (IW), the resulting estimate will not be zero, which probably caused the lack of convergence. Given this important observation, efforts to detect eQTLs could be directed to genes whose expression are indeed influenced by genotypes.
The above application has an overarching importance in advancing our understanding of the functionality of non-coding variants discovered from GWAS. For example, once heritably expressed genes have been identified in a disease-relevant tissue using BCCM, a functional follow-up of GWAS associations can be carried by looking for an overlap between the GWAS associations and variants within or associated with the identified heritably expressed genes (eQTLs). Repeating the process could eventually lead to a network of genes involved in the pathogenesis of the disease.
Finally, the results from the SNP heritability estimation of the expression of the tested BC pathway genes, specifically CHURC1, and its eQTLs are provocative and underscore the need to investigate CHURC1 effect on BC status. We have made the first promising steps toward designing a method orthogonal to the one in this paper to investigate the extent to which significant heritability estimates of the expression of BC-related genes will translate into improved predictive accuracy of BC status.
Code implementation
BUGS codes used in this work are provided in the supplementary materials.
Electronic supplementary material
Acknowledgements
The work described in this paper was funded by the Saudi Government as part of the author’s PhD scholarship. The TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. Many thanks to Wicher Bergsma, Nick Dand, and Doug Speed for their useful comments. I am also grateful to the authors of the MuTHER study for sharing their list of filtered probes.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Note that and are different. The first value corresponds to the effect sizes of any covariates other than SNP genotypes—e.g., sex and age—whereas the second value is specifically for the SNP genotypes, which are stored in Z.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0100-z) contains supplementary material, which is available to authorized users.
References
- 1.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maher B. The case of the missing heritability. Nature. 2008;456:18. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 3.Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright FA, Sullivan PF, Brooks AI, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 2014;46:430–7. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109:1193–8. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaitlen N, Kraft P. Heritability in the genome-wide association era. Hum Genet. 2012;131:1655–64. doi: 10.1007/s00439-012-1199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods. 2013;9:1. doi: 10.1186/1746-4811-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X, Carbonetto P, Stephens M. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 2013;9:e1003264. doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patterson HD, Thompson R. Recovery of inter-block information when block sizes are unequal. Biometrika. 1971;58:545–54. doi: 10.1093/biomet/58.3.545. [DOI] [Google Scholar]
- 11.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HM, Sul JH, Service SK, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–54. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–5. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verbeke G, Molenberghs G. Linear mixed models for longitudinal data. Springer Science & Business Media, New York, United States; 2009.
- 16.Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian data analysis. Boca Raton, FL, USA: Chapman & Hall CRC; 2014. [Google Scholar]
- 17.Gelman A. Prior distributions for variance parameters in hierarchical models. Bayesian Anal. 2006;1:515–34. doi: 10.1214/06-BA117A. [DOI] [Google Scholar]
- 18.Chung Y, Rabe-Hesketh S, Dorie V, Gelman A, Liu J. A nondegenerate penalized likelihood estimator for variance parameters in multilevel models. Psychometrika. 2013;78:685–709. doi: 10.1007/s11336-013-9328-2. [DOI] [PubMed] [Google Scholar]
- 19.Chung Y, Gelman A, Rabe-Hesketh S, Liu J, Dorie V. Weakly informative prior for point estimation of covariance matrices in hierarchical models. J Educ Behav Stat. 2015;40:136–57. doi: 10.3102/1076998615570945. [DOI] [Google Scholar]
- 20.Korte A, Vilhjálmsson BJ, Segura V, Platt A, Long Q, Nordborg M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat Genet. 2012;44:1066–71. doi: 10.1038/ng.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Stephens M. Efficient algorithms for multivariate linear mixed models in genome-wide association studies. Nat Methods. 2014;11:407. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawid AP. Some matrix-variate distribution theory: notational considerations and a Bayesian application. Biometrika. 1981;68:265–74. doi: 10.1093/biomet/68.1.265. [DOI] [Google Scholar]
- 24.Hoerl AE, Kennard RW. Ridge regression: biased estimation for nonorthogonal problems. Technometrics. 1970;12:55–67. doi: 10.1080/00401706.1970.10488634. [DOI] [Google Scholar]
- 25.Malo N, Libiger O, Schork NJ. Accommodating linkage disequilibrium in genetic-association analyses via ridge regression. Am J Hum Genet. 2008;82:375–85. doi: 10.1016/j.ajhg.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirinen M, Donnelly P, Spencer CC. Efficient computation with a linear mixed model on large-scale data sets with applications to genetic studies. Ann Appl Stat. 2013;7:369–90. doi: 10.1214/12-AOAS586. [DOI] [Google Scholar]
- 27.Plummer M. JAGS: a program for analysis of Bayesian graphical models using Gibbs sampling. Proceedings of the 3rd international workshop on distributed statistical computing, March 20-22, Vienna, Austria 2003.
- 28.Su Y, Yajima M. R2jags: using R to run “JAGS” 2015. https://cran.r-project.org/web/packages/R2jags/index.html
- 29.Plummer M, Best N, Cowles K, Vines K. CODA: convergence diagnosis and output analysis for MCMC. R News 2006;6:7–11.
- 30.Grundberg E, Small KS, Hedman AringK, et al. Mapping cis-and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–9. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moayyeri A, Hammond CJ, Hart DJ, Spector TD. The UK adult twin registry (TwinsUK resource) Twin Res Hum Genet. 2013;16:144–9. doi: 10.1017/thg.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spector TD, Williams FM. The UK adult twin registry (TwinsUK) Twin Res Hum Genet. 2006;9:899–906. doi: 10.1375/twin.9.6.899. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X. GEMMA user manual 2016. USA: University of Chicago.
- 34.Gelman A, Donald R. Inference from iterative simulation using multiple sequences. Stat Sci. 1992;1:457–72. doi: 10.1214/ss/1177011136. [DOI] [Google Scholar]
- 35.Ginestier C, Cervera N, Finetti P, et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006;12:4533–44. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- 36.Furlotte NA, Heckerman D, Lippert C. Quantifying the uncertainty in heritability. J Hum Genet. 2014;59:269–75. doi: 10.1038/jhg.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nat Genet. 2014;46:100–6. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Malley AJ, Zaslavsky AM. Cluster-level covariance analysis for survey data with structured nonresponse. Technical report, Department of Health Care Policy, Harvard Medical School, Boston, United States; 2005.
- 39.Gelman A, Hill J. Data analysis using regression and multilevel/hierarchical models. Cambridge University Press, New York, United States; 2006.
- 40.Pawitan Y: In all likelihood: statistical modelling and inference using likelihood. Oxford University Press, New York, United States; 2001.
- 41.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kostem E, Eskin E. Improving the accuracy and efficiency of partitioning heritability into the contributions of genomic regions. Am J Hum Genet. 2013;92:558–64. doi: 10.1016/j.ajhg.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maier R, Moser G, Chen GB, et al. Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder, and major depressive disorder. Am J Hum Genet. 2015;96:283–94. doi: 10.1016/j.ajhg.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.