Abstract
Unraveling the molecular mechanisms of nitrate regulation and deciphering the underlying genetic network is vital for elucidating nitrate uptake and utilization in plants. Such knowledge could lead to the improvement of nitrogen-use efficiency in agriculture. Here, we report that the FIP1 gene (factor interacting with poly(A) polymerase 1) plays an important role in nitrate signaling in Arabidopsis thaliana. FIP1 encodes a putative core component of the polyadenylation factor complex. We found that FIP1 interacts with the cleavage and polyadenylation specificity factor 30-L (CPSF30-L), which is also an essential player in nitrate signaling. The induction of nitrate-responsive genes following nitrate treatment was inhibited in the fip1 mutant. The nitrate content was also reduced in fip1 seedlings due to their decreased nitrate uptake activity. Furthermore, the nitrate content was higher in the roots but lower in the roots of fip1, which may result from the downregulation of NRT1.8 and the upregulation of the nitrate assimilation genes. In addition, qPCR analyses revealed that FIP1 negatively regulated the expression of CIPK8 and CIPK23, two protein kinases involved in nitrate signaling. In the fip1 mutant, the increased expression of CIPK23 may affect nitrate uptake, resulting in its lower nitrate content. Genetic and molecular evidence suggests that FIP1 and CPSF30-L function in the same nitrate-signaling pathway, with FIP1 mediating signaling through its interaction with CPSF30-L and its regulation of CIPK8 and CIPK23. Analysis of the 3′-UTR of NRT1.1 showed that the pattern of polyadenylation sites was altered in the fip1 mutant. These findings add a novel component to the nitrate regulation network and enhance our understanding of the underlying mechanisms for nitrate signaling.
Keywords: Arabidopsis, FIP1, CPSF30-L, nitrate signaling, nitrate uptake and assimilation, CIPK8, CIPK23
Introduction
Nitrogen is an essential macronutrient, and its availability in soil is a major limiting factor for plant growth and development. N fertilizers are routinely used to increase agricultural productivity; however, the low NUE of many crops means that a large portion of the N application cannot be absorbed by plants and is lost to the environment, leading to various environmental and ecological problems, such as eutrophication and soil acidification, as well as increased economic costs for farmers (Canfield et al., 2010). Improving NUE and understanding how plants regulate their growth and development in response to different levels and forms of N available in the rhizosphere are essential for addressing these problems and improving the sustainability of agriculture.
Nitrate is the main source of inorganic N for terrestrial plants, which have evolved sophisticated regulatory mechanisms to withstand changing nitrate concentrations in the environment (Crawford and Glass, 1998). In the Arabidopsis thaliana genome, four gene families (NRT1/PTR, NRT2, CLC, and SLAC1/SLAH) are responsible for nitrate absorption and distribution (Krapp et al., 2014). Once absorbed into the cells, a portion of the nitrate is reduced to nitrite and then to ammonium by the action of NR and NiR, respectively (Crawford and Glass, 1998). The ammonium is then assimilated into glutamine by glutamine synthetase (Stitt, 1999).
Nitrate serves not only as a nutrient, but also as a potent signal regulating the long-term and short-term development and physiology of plants. In the long term, nitrate affects the metabolism, growth, and development of Arabidopsis (Wang et al., 2007; Gutiérrez, 2012; Vidal et al., 2013). Several essential genes have been found to be involved in regulating the effects of nitrate on root architecture, including ANR1, NRT1.1 (NITRATE TRANSPORTER 1.1, also called NPF6.3 and CHL1), AFB3-miR393, NLP7, TCP20, HRS1, and HHO1 (Zhang and Forde, 1998; Castaings et al., 2009; Ho et al., 2009; Wang et al., 2009; Vidal et al., 2010, 2013; Guan et al., 2014, 2017; Medici et al., 2015). Oligopeptide signals have also been shown to mediate N-dependent root architecture via the CLE-CLAVATA1 module (Araya et al., 2014) and via leucine-rich repeat receptor kinases (LRR-RKs) (Tabata et al., 2014).
In the short term, nitrate induces the PNR in roots and shoots, during which the expression of more than 1000 genes can be rapidly altered (Wang et al., 2003, 2007; Scheible et al., 2004; Krouk et al., 2010a; Alvarez et al., 2012). Important nitrate regulatory genes have been identified over the last decade that play crucial roles in regulating nitrate-responsive genes such as NRT1.1, NRT2.1, NRT2.2, NITRATE REDUCTASE 1 (NIA1), NIA2, and NiR (Castaings et al., 2009; Alvarez et al., 2014; Guan et al., 2014, 2017; Xu et al., 2016; Li et al., 2017). The only nitrate sensor to have been identified thus far, NRT1.1, triggers nitrate responses, regulating the expression of CIPK8 as a positive factor and CIPK23 as a negative factor during PNR (Ho et al., 2009; Hu et al., 2009). The CIPK23-CBL9 protein complex has been implicated in the switch between the dual affinities of NRT1.1, through the phosphorylation of a threonine residue (Thr101) (Ho et al., 2009). NLP6 and NLP7 act as key activators of nitrate assimilatory genes (Castaings et al., 2009; Konishi and Yanagisawa, 2013; Marchive et al., 2013). NRG2, another recently identified regulator, was found to act upstream of NRT1.1 and interact with NLP7 in the nucleus (Xu et al., 2016). LBD37/38/39 function as negative regulators that repress the expression of a subset of genes involved in nitrate uptake and assimilation (Rubin et al., 2009). TARGET (transient assay reporting genome-wide effects of transcription factors) and ChIP-sequencing analyses were used to identify roles for SPL9 and bZIP1, respectively, in the PNR (Krouk et al., 2010b; Para et al., 2014; Vidal et al., 2015). Both TGA1 and TGA4 are induced by nitrate treatments and involved in nitrate transport and metabolic functions (Alvarez et al., 2014; O’Brien et al., 2016). Furthermore, TGA1 can interact with CIPK23, suggesting that phosphorylation may be important for TGA1 activation (Yazaki et al., 2016).
Recently, a 65-kDa subunit of the CPSF CPSF30-L was found to function upstream of NRT1.1 in nitrate signaling, where it affects nitrate uptake and assimilation (Li et al., 2017); however, the structure of the regulatory network modules and the underlying molecular mechanisms remain uncharacterized. CPSF30 has two spliced forms, a larger one (CPSF30-L) and a smaller one (CPSF30-S) (Delaney et al., 2006). CPSF30-S interacts with FIP1 (factor interacting with poly(A) polymerase 1), an important regulator of the nuclease activity of CPSF30 (Addepalli and Hunt, 2007); however, whether CPSF30-L can interact with FIP1 has yet to be reported. Here, we demonstrate that FIP1 interacts with CPSF30-L and plays an important role in nitrate signaling. Our results show that FIP1 also modulates the nitrate content in plants by regulating their nitrate transport, allocation, and assimilation. Moreover, FIP1 negatively regulates the expression of CIPK8 and CIPK23. Molecular and genetic analyses revealed that FIP1 and CPSF30-L function in the same nitrate signaling pathway.
Materials and Methods
Plant Materials
Arabidopsis thaliana (Columbia-0 ecotype) and homozygous transgenic seeds containing the NRP-YFP construct (SS204-9) (Wang et al., 2009) were used as the wild types (WTs). The mutant lines chl1-13 (original name: Mut21; containing a NRT-YFP construct) (Wang et al., 2009), cipk8-1 (Hu et al., 2009), cipk23-3 (Ho et al., 2009), nrg2-3 and nlp7-4 (Xu et al., 2016), and cpsf30 (original name: Mut65; containing a NRT-YFP construct) (Li et al., 2017) were described previously. The fip1 mutant (Salk_087117), containing a T-DNA insertion in the sixth exon of FIP1, was obtained from ABRC and used for further analysis (Alonso et al., 2003). The construct p35S::FIP1 in destination vector pMDC43 (Thermo Fisher Scientific) was transformed into the fip1 mutant using the Agrobacterium-mediated floral dip method (Xu et al., 2016). Homozygous transgenic lines were isolated as complementation line (FIP1/fip1) for further investigation.
Growth and Treatment Conditions
Seeds were germinated on nylon mesh floating in a 2.5-mM ammonium succinate [(NH4)2Suc] solution for 7 days. To detect the expression of the nitrate-responsive genes, the roots were treated with 10 mM KNO3 or KCl for 2 h and then harvested. For fluorescence microscopy, seedlings were grown on the KNO3 medium for 4 days before being observed using a Nikon Eclipse Ti-S microscope (Nikon, Tokyo, Japan). The fluorescence intensity was quantified using Image J (Schneider et al., 2012).
To determine expression profiles, various tissues were harvested either from plants grown for 7 days in ½ MS medium (pH = 5.7, 10 mM KNO3, and 10 mM NH4NO3) or for 6 weeks in soil. Roots and shoots were harvested from seedlings grown in ½ MS solution for 7 days and used to determine their nitrate concentration, NR activity, and amino acid content. The expression levels of the genes involved in nitrate transportation and assimilation were determined in the roots and shoots of these seedlings, whereas the expression of the nitrate regulatory genes was measured in seedlings grown in 2.5 mM (NH4)2Suc, 10 mM KNO3, and 10 mM NH4NO3 for 7 days.
qPCR Analysis
Total RNA was isolated from Arabidopsis roots and shoots using a Total RNA Miniprep Kit (CWBIO, Beijing, China). cDNA synthesis was carried out using the RevertAid first-stand Synthesis System Kit (Thermo Fisher Scientific, Waltham, MA, United States). An UltraSYBR Green Mixture qPCR Kit (CWBIO) was used for the qPCR reaction, following the manufacturer’s protocol. Gene expression was determined by real-time PCR using an ABI7500 Fast Real-Time PCR System (Thermo Fisher Scientific). TUB2 (At5g62690) was used as the internal reference gene.
Expression Profile Analysis
A GUS assay was performed according to Xu et al. (2016). A 381-bp genomic sequence located upstream of the FIP1 start codon was cloned into the pMDC163 destination vector (Thermo Fisher Scientific). Transgenic plants containing the ProFIP1::GUS construct were grown on ½ MS for 7 days or in soil for 6 weeks before their GUS activity was assessed.
Nitrate, NR Activity, and Amino Acid Content Assays
Plant nitrate content was determined using the salicylic acid method, as described previously (Zhao and Wang, 2017). The amino acid content and NR activity of the seedlings were tested using a Micro Amino Acid Content Assay Kit and a Micro Nitrate Reductase (NR) Assay Kit (Solarbio, Beijing, China), respectively.
Yeast Two-Hybrid Assays
Full-length cDNA fragments of CPSF30-L or CPSF30-L containing a point mutation in nucleotide 376 (G to A; mCPSF30-L) were introduced into the pGBKT7 vector (Clontech Laboratories, Mountain View, CA, United States), while a full-length cDNA fragment of FIP1′ a 1335-bp sequence encoding the N-terminal of FIP1 (FIP1-S1), a 2196-bp sequence encoding the C-terminal of FIP1 (FIP1-S2), were ligated into the pGADT7 vector (Clontech Laboratories). The two-hybrid interaction was performed following the instructions provided by the manufacturer (Clontech Laboratories).
GST Pull-Down Assays
Full-length cDNA of CPSF30-L or mCPSF30-L was cloned into pGEX4T-1 (GE) to produce a GST-CPSF30-L or GST-mCPSF30-L product as bait protein, respectively. FIP1 or FIP1-S1 was cloned into pET28a (Novagen) to produce His-FIP1 or His-FIP1-S1 product as prey protein, respectively. The constructs were introduced into Escherichia coli strain BL21. The preparation and immobilization of the bait protein, the preparation and capture of the prey protein, and bait-prey elution were performed using a GST protein interaction pull-down kit (Thermo). The prepared eluent was loaded into wells and electrophoresis was run in the stacking and separating gel, respectively. Following SDS-PAGE, the proteins were transferred onto blotting membrane and blocked, then adding anti-HIS mAb (Zoonbio) as primary antibody and HRP-conjugated secondary antibody (Zoonbio). The Chemiluminescent (ECL) was used to visualize protein bands as recommended by the manufacturer (Thermo).
BiFC Analysis
Transient bimolecular fluorescence complementation (BiFC) assays in Arabidopsis mesophyll protoplast were performed as described (Xu et al., 2016). Full-length cDNA of CPSF30-L and mCPSF30-L were cloned into the Gateway compatible binary vectors pSITE-NEYFP vectors containing the N-terminal fragments of YFP (YFPN). Full-length cDNA of FIP1 and FIP1-S1 were cloned into pSITE-CEYFP containing the C-terminal fragments of YFP (YFPC), respectively. These vectors CPSF30-L-YFPN and FIP1-YFPC, CPSF30-L-YFPN and FIP1-S1-YFPC, and mCPSF30-L-YFPN and FIP1-YFPC were cotransfected into protoplast and the empty vectors YFPN and YFPC were used as negative controls. The fluorescence of transfected protoplast was observed using confocal microscope (Leica TCS SP5II).
Analysis of Polyadenylation in NRT1.1 3′-UTR
For analysis of NRT1.1 3′-UTR, the nested PCR technique was used as described (Li et al., 2017). The seedlings of WT, fip1, cpsf30, and fip1cpsf30 were grown on 10 mM KNO3 for 7 days and total RNA were extracted, and then reverse transcription polymerase chain reaction (RT-PCR) was performed. The 3′-UTR of NRT1.1 after two rounds of PCR was analyzed by polyacrylamide gel electrophoresis (PAGE).
Results
The Interaction Between FIP1 and CPSF30-L
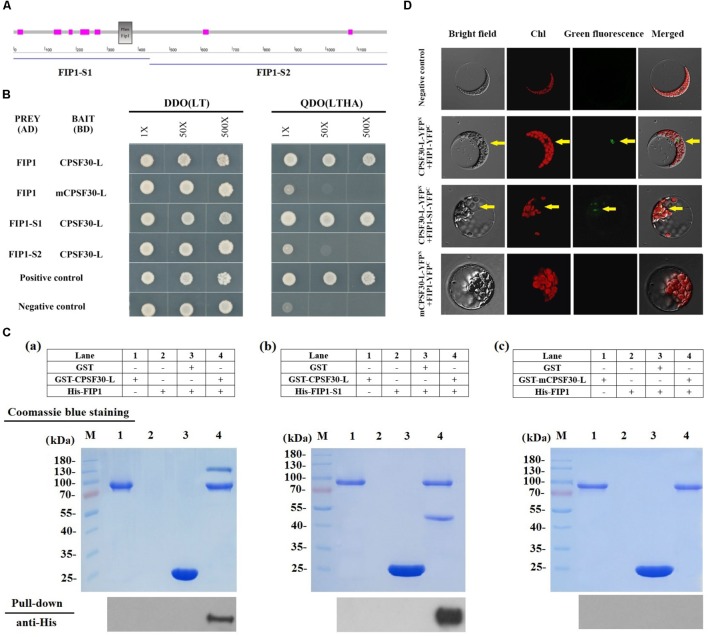
We first tested if FIP1 can interact with CPSF30-L using a yeast two-hybrid assay. Full length FIP1 and FIP1 segments were used for the assays: segment 1 comprised the N-terminal 445 amino acid residues while segment 2 contained the C-terminal 731 amino acid residues (Figure 1A). The CPSF30-L protein and a CPSF30-L variant containing a point mutation resulting in a conversion of Gly to Arg at the 126th amino acid in the third zinc finger, mCPSF30 (Li et al., 2017), were used as bait proteins. We found that CPSF30-L and FIP1 interacted with each other; however, mCPSF30 did not interact with FIP1 (Figure 1B). Specifically, CPSF30-L interacted with the N-terminus but not the C-terminus of FIP1.
FIGURE 1.
FIP1 interacts with CPSF30-L. (A) Secondary structure of FIP1 defined using SMART software. FIP1-S1, N-terminal of FIP1, comprising 445 amino acids with a Fip1 conserved domain (indicated by the gray box); FIP1-S2, C-terminal of FIP1, comprising 731 amino acids. The seven fuchsia boxes indicate regions of low complexity. (B) Yeast two-hybrid (Y2H) assay of the FIP1 and CPSF30-L interaction. CPSF30-L, a BD vector containing cDNA encoding CPSF30-L (a 65-kDa transcript of At1g30460.2); mCPSF30-L, a BD vector containing CPSF30-L cDNA with a point mutation at nucleotide 376 (G to A); FIP1, an AD vector containing FIP1 cDNA (a transcript of At5g58040); FIP1-S1, an AD vector containing cDNA comprising a 1335-bp sequence encoding the N-terminal of FIP1; FIP1-S2, an AD vector containing cDNA comprising a 2196-bp sequence encoding the C-terminal of FIP1; DDO (LT), SD medium lacking leucine and tryptophan used to verify the co-transformation of AD and BD plasmids; QDO (LTHA), SD medium lacking leucine, tryptophan, histidine, and adenine used to test for autoactivation or interaction. Positive controls (BD-pGBKT7-53 and AD-pGADT7-T) and negative controls (BD-pGBKT7-Lam and AD-pGADT7-T) are also shown. (C) GST pull-down assay to detect the in vitro interaction of FIP1 and CPSF30-L. The tagged protein GST-CPSF30-L, GST-mCPSF30-L, His-FIP1, and His-FIP1-S1 were synthesized in Escherichia coli. His-FIP1 (a) or His-FIP1-S1 (b) was pulled down using glutathione agarose beads with GST-CPSF30-L, (c) His-FIP1 was not pulled down with GST-mCPSF30-L, as detected using an anti-His antibody. (D) BiFC analysis for interaction between FIP1 and CPSF30-L. CPSF30-L and mCPSF30-L were fused to N-terminal fragments of YFP, FIP1 and FIP1-S1 were fused to C-terminal fragments of YFP, respectively. Different combinations of expression vectors and negative control (indicated on the left of the panel) were transfected into Arabidopsis mesophyll protoplast. Presence of YFP signal indicates reconstitution of YFP through protein interaction of the tested pairs. Yellow arrows indicate the nucleus.
To confirm the interaction between FIP1 and CPSF30-L, in vitro GST pull-down assay and in vivo test with BiFC assays in Arabidopsis mesophyll protoplast were performed. In pull-down assay, the interaction of GST tagged CPSF30-L and His-tagged FIP1 was inspected. His-FIP1 or His-FIP1-S1 was readily pulled down by glutathione agarose beads with GST-CPSF30-L, as detected using an anti-His antibody (Figure 1Ca,b). When mCPSF30-L was used, His-FIP1 could not be pulled down (Figure 1Cc). In BiFC assay, a direct interaction was observed between FIP1 and CPSF30-L as well as FIP1-S1 and CPSF30-L in the nucleus of the protoplast when CPSF30-L-YFPN was coexpressed with FIP1-YFPC and FIP1-S1-YFPC. But no interaction was found when mCPSF30-L-YFPN and FIP1-YFPC were coexpressed (Figure 1D). These results suggest that CPSF30-L interacts with the N-terminus of FIP1, and this interaction depends on a Gly at the 126th amino acid of CPSF30-L.
The fip1 Mutant Is Defective in the PNR
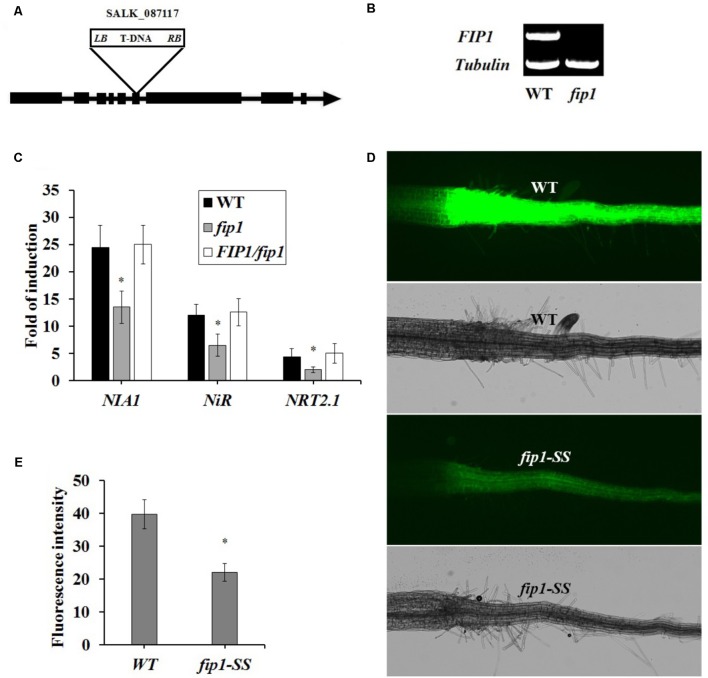
To explore the function of FIP1, we obtained a fip1 mutant (Salk_087117), containing a T-DNA insertion in the sixth exon (Figure 2A), from ABRC. FIP1 expression was undetectable in this mutant (Figure 2B). To determine whether FIP1 is involved in the PNR, we determined the expression levels of three known nitrate-responsive genes, NRT2.1 (encoding a high-affinity nitrate transporter), NIA1, and NiR, using qPCR. As shown in Figure 2C, the induction of these three genes by nitrate treatment was significantly decreased in the fip1 mutant, but was restored to WT levels in the FIP1/fip1 complementation line, indicating that FIP1 plays an important role in nitrate signaling. In addition, we previously constructed a transgenic Arabidopsis line containing a nitrate-responsive reporter (SS204-9) that exhibited strong YFP fluorescence in the roots of Arabidopsis plants in the presence of nitrate (Wang et al., 2009). We introduced the nitrate-responsive reporter into the fip1 mutant by crossing it with SS204-9 to generate fip-SS. The fip1-YFP line had a significantly reduced fluorescence in the presence of nitrate in comparison with the WT (Figures 2D,E). These results show that FIP1 functions as an important regulatory gene in the PNR.
FIGURE 2.
FIP1 regulates the expression of nitrate-responsive genes. (A) Schematic map of the T-DNA insertion site in the fip1 mutant. Exons and introns are represented by black boxes and lines, respectively. The location of the T-DNA insertion in FIP1 is indicated by a triangle. (B) RT-PCR analysis of FIP1 mRNA levels in the wild type (WT) and fip1. Total RNA was isolated from 7-days-old seedlings grown on ½ MS. TUB2 serves as the internal control. (C) The expression of nitrate-responsive genes in the roots of the fip1 mutant and complementation line (FIP1/fip1). Seedlings were grown on medium with 2.5 mM ammonium succinate as the sole nitrogen source for 7 days, and then treated with 10 mM KNO3 or KCl as a control for 2 h. The transcripts of the nitrate-responsive genes were quantified using qPCR. Error bars represent the SD of the biological replicates (n = 4). Asterisks indicate significant differences (p < 0.05, U-test). (D) Nitrate responsiveness in 4-days-old fip1-SS seedlings grown on a medium containing KNO3, revealed using a fluorescent reporter system (NRP-YFP). (E) Quantification of root fluorescence in the WT and the fip1-SS mutant described in (D). Error bars represent the SD of the biological replicates (n = 60), asterisks indicate significant differences to WT (p < 0.05, U-test).
To test whether FIP1 expression can be induced by nitrate, the seedlings were grown for 7 days on medium containing 2.5 mM ammonium succinate as the sole N source, and then treated with 10 mM KNO3. We found that the expression of FIP1 was not significantly altered in the roots following nitrate treatment (Supplementary Figure 1), indicating that FIP1 expression is not induced by nitrate.
FIP1 Is Mainly Expressed in the Vascular Tissues of the Leaves and Roots
To further explore the regulation of the nitrate response by FIP1, its expression pattern was investigated using qPCR. FIP1 was found to be expressed in all tissues investigated, but was particularly highly expressed in the roots and leaves (Figure 3A). Histochemical analyses using transgenic lines harboring the GUS gene driven by the FIP1 promoter revealed that FIP1 was predominantly expressed in the vascular tissues of the roots and leaves (Figure 3B). GUS staining was also observed in the trichomes and stomata on the leaves, as well as in the flowers (Figure 3B). The expression profile of FIP1 suggests that it may function in the nitrate signaling pathway of several tissues.
FIGURE 3.
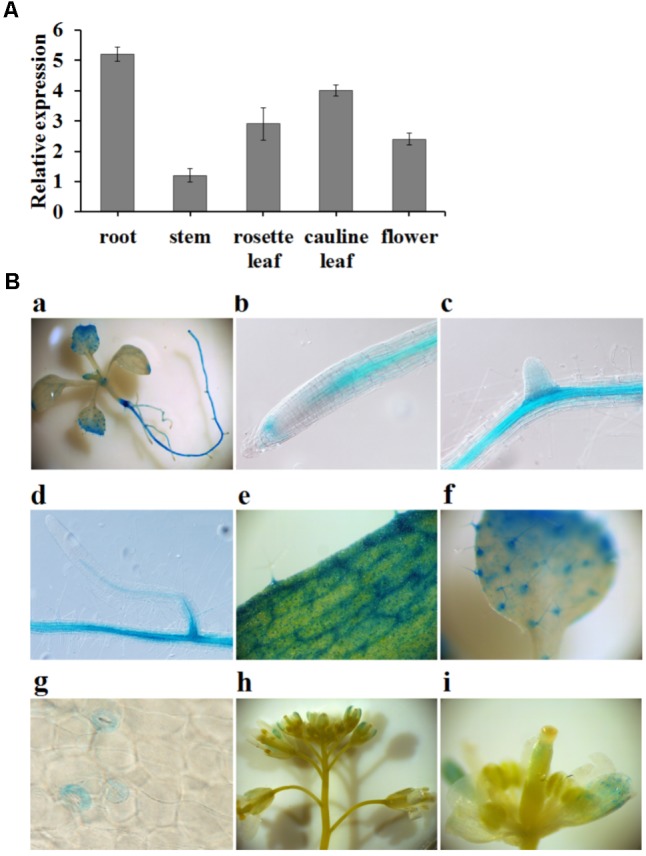
The expression pattern of FIP1. (A) Relative expression levels of FIP1 in different tissues quantified using qPCR. Tissues were harvested either from 7-days-old plants (seedlings and roots) grown in 1/2 MS solution or 6-week-old plants (leaves, stems, and flowers) grown in soil. Error bars represent the SD of the biological replicates (n = 4). (B) Histochemical staining of GUS activity in transgenic plants expressing ProFIP1::GUS. GUS activity was detectable in the (a) whole seedling, (b) root tip, (c,d) lateral roots, (e) rosette leaves, (f) cotyledons, (g) stomata, and (h,i) flowers.
FIP1 Regulates the Uptake and Allocation of Nitrate Between the Shoot and Root
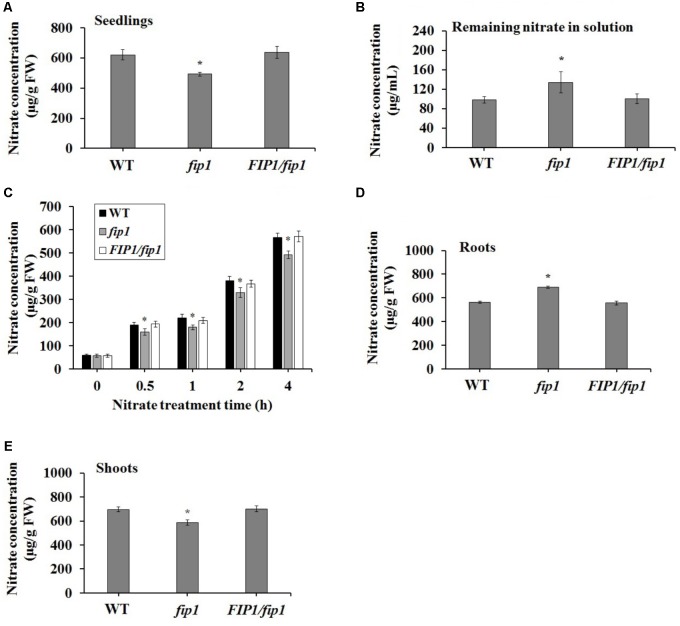
Some nitrate regulators affect the accumulation of nitrate within the plant (Castaings et al., 2009; Wang et al., 2009; Xu et al., 2016; Li et al., 2017). To test the physiological effects of FIP1, we measured the nitrate content of plants grown on ½ MS medium. Nitrate accumulation was significantly decreased in the fip1 seedlings compared with the WT (Figure 4A). This decreased nitrate content may result from defects in nitrate uptake and/or increased nitrate assimilation; therefore, we determined the remaining nitrate concentration in the solution after 7 days of seedling growth. The remaining nitrate concentration in the solution was significantly higher for fip1 than for WT (Figure 4B). We tested the nitrate uptake of seedlings grown in the presence of 2.5 mM ammonium succinate for 7 days and then treated with 5 mM KNO3 for different durations, and found that the nitrate content was reduced significantly in the fip1 mutant at the time points tested (Figure 4C). Taken together, these results indicate that FIP1 affects nitrate uptake. To further investigate the distribution of nitrate in the plants, we quantified the accumulation of nitrate in both the shoots and roots. The nitrate content was higher in the roots but lower in the shoots of fip1, and this phenotype was recovered in the FIP1/fip1 complementation line (Figures 4D,E), suggesting that the allocation of nitrate within the plant is altered in the fip1 mutant. Taken together, these findings show that FIP1 regulates nitrate uptake and allocation.
FIGURE 4.
FIP1 affects nitrate uptake and accumulation. Seedlings were grown in ½ MS solution for 7 days and their nitrate content was quantified. (A) The nitrate concentration in the seedlings. (B) The nitrate concentration remaining in the solution after 7 days of seedlings growth. (C) Nitrate uptake in seedlings grown for 7 days in 2.5 mM ammonium succinate and then treated with 5 mM KNO3 for the indicated periods. The nitrate concentration in the roots (D) and shoots (E). Error bars represent the SD of the biological replicates (n = 5). Asterisks indicate significant differences in the transgenic lines in comparison with the WT (p < 0.05, U-test). FW, fresh weight.
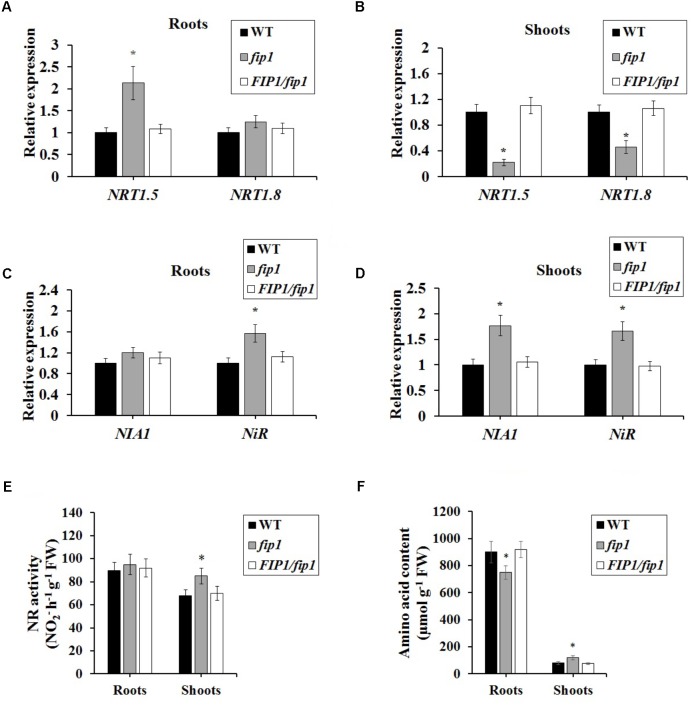
The decreased nitrate content in fip1 led us to investigate the expression of genes known to be involved in nitrate transport and assimilation. For this assay, steady state levels of mRNA after a week of growth on nitrate were determined by growing seedlings on ½ MS for 7 days then harvesting roots and shoots separately for RNA extraction. Our qPCR analysis showed that NRT1.5 expression was increased in the roots of fip1 compared with the WT, while the expression of NRT1.5 and NRT1.8 was decreased in the fip1 shoots in comparison with the WT (Figures 5A,B). These phenotypes were recovered in the FIP1/fip1 complementation line. No significant differences in expression were found for the other genes tested (Supplementary Table 1). NRT1.8 functions in nitrate unloading from the xylem to the shoots (Li et al., 2010). Therefore, the decreased expression of NRT1.8 in the shoots may result in the lower nitrate content in the shoots but higher in the roots of fip1.
FIGURE 5.
FIP1 regulates the expression of genes involved in nitrate transport and assimilation in the roots and shoots of seedlings grown for 7 days in ½ MS solution. (A,B) The relative expression levels of nitrate transport genes NRT1.5 and NRT1.8 were determined in the roots (A) and shoots (B) using qPCR. (B,C) The expression levels of the nitrate assimilation genes NIA1 and NiR were detected in the roots (C) and shoots (D) using qPCR. (E) Nitrate reductase (NR) activity and (F) amino acid content in fip1 roots and shoots. Error bars represent the SD of the biological replicates (n = 5). Asterisks indicate significant differences in the transgenic lines in comparison with the WT (p < 0.05, U-test).
We also determined the expression levels of the nitrate assimilation-related genes. The expression levels of NIA1 and NiR were much higher in fip1 than in the WT (Figure 5C), while in the roots only the expression of NiR was higher in fip1 (Figure 5D and Supplementary Table 2). Furthermore, we detected the NR activity and amino acid content of the plants. The NR activity in the shoots of fip1 was higher than that of the WT (Figure 5E), while the amino acid content was lower in the roots and higher in the shoots of fip1 compared with the WT (Figure 5F). These results demonstrate that FIP1 also affects the expression of the genes involved in nitrate assimilation, which may be another reason for the lower nitrate content in the shoots of fip1.
FIP1 Regulates the Expression of CIPK8 and CIPK23 in the Presence of Nitrate and Functions in the Same Nitrate-Signaling Pathway as CPSF30
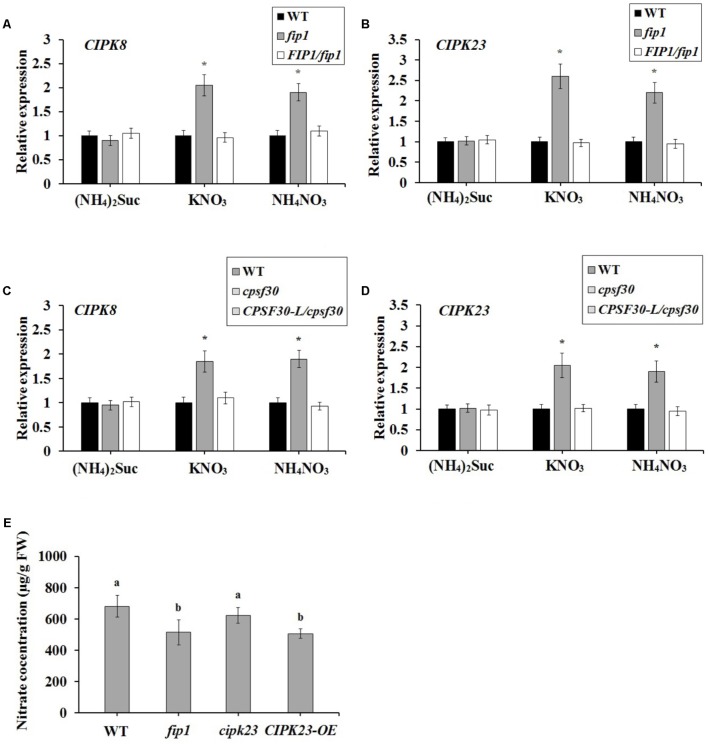
To further explore the relationship between FIP1 and the previously characterized nitrate regulatory genes, we grew nitrate regulation mutant plants, lacking CPSF30, NRT1.1, NLP7, CIPK8, CIPK23, or NRG2, for 7 days under three N sources (2.5 mM ammonium succinate, 10 mM KNO3, or 10 mM NH4NO3), and then quantified their expression of FIP1. There was no significant difference in the expression of FIP1 between the WT and each mutant under the different N sources (Supplementary Figure 2), indicating that the above nitrate regulatory genes do not regulate its expression. We also detected the expression of these genes in fip1, revealing that the expression of CIPK8 and CIPK23 in fip1 was significantly increased in comparison with the WT in the presence of nitrate, and that these changes were recovered in the FIP1/fip1 complementation line (Figures 6A,B). No significant differences were found for the other genes tested (Supplementary Table 3). CIPK8 and CIPK23 are important regulatory genes involved in nitrate signaling (Ho et al., 2009; Hu et al., 2009); therefore, our results suggest that FIP1 may play an essential role in the PNR by regulating the expression of the nitrate regulatory genes CIPK8 and CIPK23.
FIGURE 6.
FIP1 regulates the expression of CIPK8 and CIPK23 and affects seedling nitrate content in the presence of nitrate. RNA was extracted from seedlings grown for 7 days with various sources of nitrogen [(NH4)2Suc, KNO3, or NH4NO3 media]. The expression levels of CIPK8 (A) and CIPK23 (B) in the fip1 mutant, determined using qPCR. The expression levels of CIPK8 (C) and CIPK23 (D) in the cpsf30 mutant, determined using qPCR. Error bars represent the SD of the biological replicates (n = 4). (E) Nitrate content in the CIPK23 overexpression line (CIPK23-OE) grown in ½ MS medium for 7 days. Error bars represent the SD of the biological replicates (n = 5). Asterisks and different letters indicate significant differences to the WT (p < 0.05, U-test).
The expression of CIPK8 and CIPK23 is also known to be controlled by NRT1.1 (Ho et al., 2009; Hu et al., 2009). As CPSF30-L regulates the expression of NRT1.1, we investigated whether CPSF30-L affected the expression of CIPK8 and CIPK23. We found that CIPK8 and CIPK23 expression was significantly higher in the cpsf30 mutant than in the WT in the presence of nitrate, a phenotype that was recovered in the CPSF30-L/cpsf30 complementation line (Figures 6C,D). These results demonstrate that, like FIP1, CPSF30-L can modulate the expression of CIPK8 and CIPK23.
CIPK23 was previously reported to phosphorylate Thr101 of NRT1.1, thereby facilitating a shift to its high-affinity status (Ho et al., 2009). In theory, the increased expression of CIPK23 may strengthen the phosphorylation of NRT1.1, enhancing its high-affinity nitrate transport activity and reducing nitrate uptake in the presence of sufficient nitrate. We therefore quantified the nitrate content of the CIPK23 overexpression line (CIPK23-OE). The nitrate content of this line was reduced, whereas no significant difference was observed between the nitrate contents of cipk23 and the WT (Figure 6E). These results suggest that the increased expression of CIPK23 in fip1 may enhance the phosphorylation of NRT1.1, resulting in the lower nitrate accumulation of this mutant.
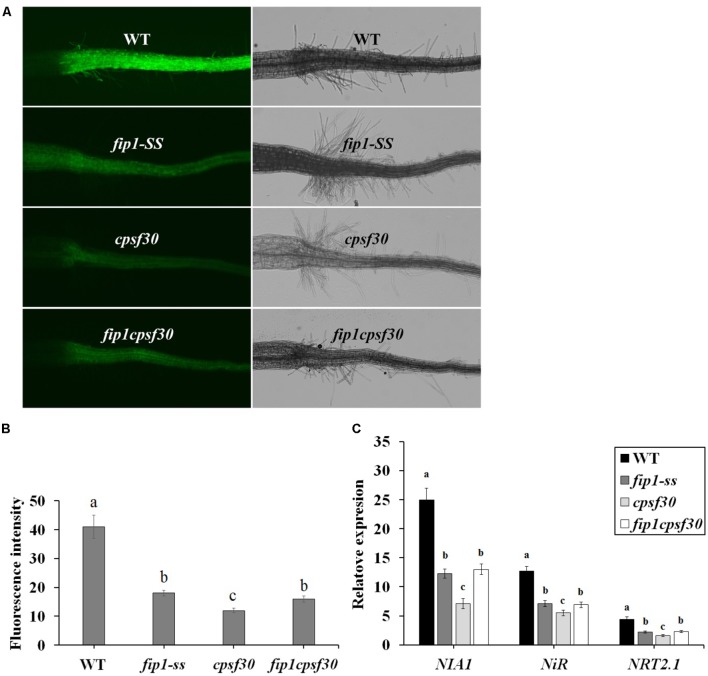
To further investigate the relationship between FIP1 and CPSF30-L, we constructed a double mutant (fip1cpsf30) by crossing fip1-SS and cpsf30. RT-PCR results showed that the expression of FIP1 was undetectable and sequencing results exhibited that the point mutation of the cpsf30 mutant was present in the double mutant (Supplementary Figure 3). The fluorescence intensity, indicating nitrate responsiveness, was significantly lower in fip1 than in the WT but higher than in cpsf30, while no significant difference was found between fip1 and the fip1cpsf30 double mutant (Figures 7A,B), suggesting that FIP1 and CPSF30-L may regulate the nitrate response via the same pathway. The expression levels of the nitrate-responsive genes NIA1, NiR, and NRT2.1 were found to be higher in the fip1cpsf30 double mutant than in cpsf30, while no significant difference in expression was observed between the fip1cpsf30 double mutant and the fip1 single mutant (Figure 7C). This further confirms that FIP1 and CPSF30-L function in the same nitrate-signaling pathway. The similar phenotype of fip1 mutant and the double mutant may be explained by the possibility that FIP1 and CPSF30-L may be part of a larger complex. Loss of FIP1 can results in a change in the complex that reduces the complex’s activity by a certain amount. Loss of CPSF30-L may result in a larger change that reduces activity more than loss of FIP1. Loss of both genes results in a change that mimics loss of FIP1 alone. This would happen if CPSF30-L binding requires FIP1 binding. Loss of FIP1 prevents CPSF30-L binding so that the phenotype of double mutant looks like that of the fip1 single mutant.
FIGURE 7.
FIP1 and CPSF30 function in the same nitrate signaling pathway. (A) Nitrate responsiveness in 4-days-old WT, fip1, cpsf30, and fip1cpsf30 seedlings grown in KNO3 medium, revealed using a fluorescent reporter system (NRP-YFP). (B) Quantification of root fluorescence in the plants described in (A). Error bars represent the SD of the biological replicates (n = 60). (C) The expression of nitrate-responsive genes in the roots of WT, fip1, cpsf30, and fip1cpsf30 plants. The seedlings were grown for 7 days on the medium containing 2.5 mM ammonium succinate as the sole nitrogen source, and then treated with 10 mM KNO3 or 10 mM KCl as a control for 2 h. The transcripts of the nitrate-responsive genes were quantified using qPCR. Error bars represent the SD of the biological replicates (n = 4). Asterisks and different letters indicate significant differences (p < 0.05, U-test).
Previous study has shown that CPSF30-L can affect APA of 3′-UTR in NRT1.1 mRNA (Li et al., 2017). Since FIP1 and CPSF30 function in the same nitrate signaling pathway, it’s possible that FIP1 also affects the APA of the NRT1.1. Therefore, 3′-UTR of NRT1.1 was amplified by nested PCR from the WT, fip1, cpsf30, and fip1cpsf30 seedlings grown on KNO3 medium. The results showed that there was a band (marked as a in Figure 8) in the WT, but this fragment was almost invisible in fip1, cpsf30, and fip1cpsf30 double mutant while a lower band (marked as b) was more obvious in fip1 and cpsf30 mutants compared to that in WT (Figure 8). In addition, the fip1 and cpsf30 mutants contained a single band (marked as c) which was almost invisible in the WT. These results indicate that FIP1 and CPSF30-L can similarly affect the APA of the 3′-UTR of NRT1.1 mRNA. However, the bands b and c were almost invisible in fip1cpsf30 double mutant, implying that the APA of 3′-UTR in NRT1.1 mRNA in the double mutant may be more complicated.
FIGURE 8.
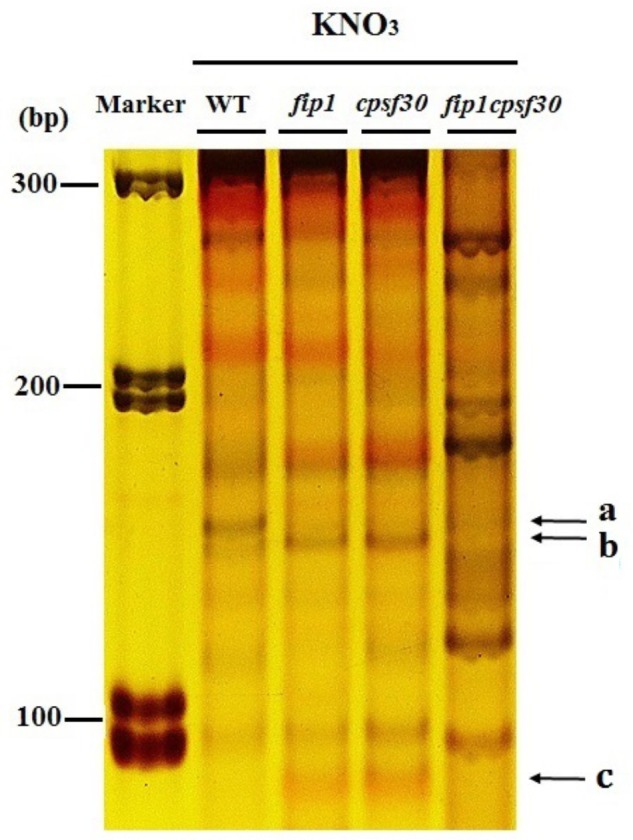
FIP1 can affect the APA in 3′-UTR of NRT1.1 mRNA. Polyacrylamide gel electrophoresis (PAGE) was run to separate the amplified bands of different 3′-UTR polyadenylated forms of NRT1.1 in WT, fip1, cpsf30, and fip1cpsf30. The seedlings were grown on 10 mM KNO3 for 7 days and total RNA was extracted for RT-PCR.
Discussion
In Arabidopsis, CPSF30 has two spliced forms, a smaller one (CPSF30-S) and a larger one (CPSF30-L) (Delaney et al., 2006). CPSF30-S plays a role in a number of distinct developmental processes and physiological responses, partly resulting from a global shift in the poly(A) site choice of numerous responding genes (Zhang et al., 2008; Bruggeman et al., 2014; Liu et al., 2014); however, the function of CPSF30-L was largely unknown (Delaney et al., 2006). Recently, we reported that a nitrate regulatory mutant, Mut65, lacked CPSF30 function (Li et al., 2017). Further investigation demonstrated that CPSF30-L and not CPSF30-S was responsible for the nitrate signaling (Li et al., 2017). It is known that CPSF30-S can interact with FIP1, a putative core component of a pre-mRNA processing complex (Addepalli and Hunt, 2007) and that this interaction regulates the nuclease activity of CPSF30. However, whether CPSF30-L could interact with FIP1 and whether FIP1 was involved in nitrate signaling were not known. The data we present here indicate that the N-terminus of FIP1 does indeed interact with CPSF30-L (Figure 1) and plays an important role in nitrate signaling.
To determine whether FIP1 participates in nitrate signaling, we quantified the expression of the nitrate-responsive genes in fip1 and found that this mutant was defective in the PNR (Figure 2). Our histochemical assay and qPCR analysis showed that FIP1 was mainly expressed in the stele of the primary root, basal lateral root, vascular tissue of the leaf, and stomata, and was not induced by nitrate (Figure 3 and Supplementary Figure 1). The expression profile of FIP1 was coincident with that of CPSF30 (Li et al., 2017). The common expression sites of both genes further support the suggestion that FIP1 interacts with CPSF30-L.
CPSF30-L is known to regulate nitrate uptake and the expression of the nitrate transporter gene, NRT1.1 (Li et al., 2017). Here, we demonstrated that the nitrate content was lower in the fip1 mutant seedlings because of its defective nitrate uptake (Figures 4A–C); however, the expression levels of the known genes involved in nitrate uptake, such as NRT1.1 and NRT1.2, were not affected in fip1 (Supplementary Table 1). Furthermore, the expression of CIPK23 was increased in fip1 (Figure 6B). As CIPK23 can phosphorylate NRT1.1 to switch its activity to have a high affinity for nitrate, the increased expression of CIPK23 may upregulate the phosphorylation and nitrate transport activity of NRT1.1, reducing nitrate uptake under conditions with sufficient nitrate. Indeed, the nitrate concentration in CIPK23-OE was decreased (Figure 6E), suggesting that the increased expression of CIPK23 might result in the lower nitrate content observed in the fip1 mutant.
In addition, FIP1 affects the distribution of nitrate between the roots and shoots. The fip1 mutant accumulated more nitrate than WT in the roots, but less in the shoots (Figures 4D,E), in contrast to the cpsf30 mutant, which accumulates less nitrate than the WT in both its roots and shoots (Li et al., 2017). The lower nitrate content in the shoots and higher in the roots of the fip1 mutant may be due to the following possibilities: (1) the lower expression of NRT1.8 in the shoots may decrease nitrate unloading from the xylem to the shoot tissues (Figure 5B); (2) the increased expression of the nitrate assimilation genes may cause more nitrate to be reduced in the shoots (Figure 5D). The NR activity and amino acid contents in the fip1 mutant were indeed higher than in the WT (Figures 5E,F). In the cpsf30 mutant, the decreased expression of NRT1.8 and the increased expression of the nitrate assimilation genes in the shoots also led to a lower nitrate content in the shoots (Li et al., 2017). These results indicate that FIP1 plays an important role in regulating nitrate uptake, transport, and assimilation.
We investigated the relationships between FIP1 and the nitrate regulatory genes, revealing that FIP1 functions as a negative regulator to modulate the expression of CIPK8 and CIPK23 in the PNR, similar to the role of CPSF30-L (Figure 6). It has been reported that both CIPK8 and CIPK23 are involved in regulating the nitrate response (Ho et al., 2009; Hu et al., 2009); thus, our results suggest that FIP1 may play an essential role in the PNR by regulating the expression of CIPK8 and CIPK23. Moreover, our genetic and molecular findings suggest that FIP1 and CPSF30-L work in the same nitrate-signaling pathway (Figure 7), FIP1 also alter the 3′-UTR of NRT1.1, similar with CPSF30-L (Figure 8); however, the fip1 and cpsf30 mutants have some phenotypic differences, such as their nitrate distributions and their regulation of NRT1.1 expression, reflecting the complexity of nitrate regulation.
FIP1 is a core component of a pre-mRNA processing complex that is conserved between plants, yeast, and humans (Preker et al., 1995; Kaufmann et al., 2004; Forbes et al., 2006). In plants, FIP1 is an RNA-binding protein and its N-terminus (containing 137 amino acids) can interact with poly(A) polymerase and stimulate its activity. In addition, the N-terminus of FIP1 interacts with other protein machinery involved in the 3′-end processing of pre-mRNAs such as CstF77, CFIm-25, PabN1, and CPSF30 (Forbes et al., 2006). Furthermore, it has been reported that three distinct hubs centered around FIP1, CPSF100, and CLPS are involved in poly(A) processing in Arabidopsis (Hunt et al., 2008). The biological function of FIP1 is unknown; however, the findings presented here indicate that FIP1 plays an important role in nitrate signaling and the regulation of nitrate uptake, transport, and assimilation.
In Arabidopsis, the 28-kDa CPSF30-S subunit contains three zinc finger motifs and functions in polyadenylation, while the 65-kDa CPSF30-L subunit contains a YTH (YT521 homology) RNA-binding domain in addition to three zinc fingers (Delaney et al., 2006). In both splicing forms, the first zinc finger binds RNA and the third zinc finger has nuclease activity, but the function of the second zinc finger domain is unknown. FIP1 was previously shown to interact with the third zinc finger of CPSF30-S, thereby regulating its nuclease activity (Addepalli and Hunt, 2007). Furthermore, a disulfide bond can be formed between the side chains of two cysteine residues in the third zinc finger domain, which, when reduced, results in the loss of endonuclease activity in CPSF30-S (Addepalli et al., 2010). CPSF30-S plays a variety of roles in a number of distinct developmental and physiological responses, partly resulting from a global shift in the poly(A) site choice of the corresponding genes (Zhang et al., 2008; Bruggeman et al., 2014; Liu et al., 2014); however, whether CPSF30-L functions in the same way to influence APA remains unknown. Interestingly, a mutation in the third zinc finger motif of CPSF30-L affects the poly(A) processing of NRT1.1 mRNA, resulting in the altered expression of NRT1.1 (Li et al., 2017). These findings suggest that CPSF30-L mediates APA function in a similar manner to CPSF30-S; therefore, the function of CPSF30-L in regulating nitrate signaling may depend on its endonuclease activity. Here, we found that FIP1 could interact with CPSF30-L and also alter the APA in the 3′-UTR of NRT1.1. FIP1 and CPSF30-L function in the same pathway to regulate nitrate signaling. There results suggest that FIP1 and CPSF30-L may modulate the expression of nitrate related genes through affecting their APA process.
Author Contributions
YW and NC designed the project. CW, WZ, ZhL, and YB performed the experiments. YW, CW, and ZeL analyzed the data. YW, NC, and CW wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Weihua Wu for providing the CIPK23-OE line and Dr. Fei Ni for the useful discussion.
Abbreviations
- APA
alternative polyadenylation
- CPSF
cleavage and polyadenylation specificity factor
- GUS
β-glucuronidase
- N
nitrogen
- NiR
nitrite reductase
- NR
nitrate reductase
- NUE
nitrogen use efficiency
- PNR
primary nitrate response
- YFP
yellow fluorescent protein
Footnotes
Funding. This work was supported by the National Key R&D Program of China (Grant No. 2016YFD0100701), the NSFC (Grant No. 31670247), and funding from the Funds of Shandong “Double Tops” Program (Grant No. SYL2017YSTD01) awarded to YW.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00593/full#supplementary-material
References
- Addepalli B., Hunt A. G. (2007). A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kDa subunit of cleavage and polyadenylation specificity factor. Nucleic Acids Res. 35 4453–4463. 10.1093/nar/gkm457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addepalli B., Limbach P. A., Hunt A. G. (2010). A disulfide linkage in a CCCH zinc finger motif of an Arabidopsis CPSF30 ortholog. FEBS Lett. 584 4408–4412. 10.1016/j.febslet.2010.09.043 [DOI] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Alvarez J. M., Riveras E., Vidal E. A., Gras D. E., Contreras-López O., Tamayo K. P., et al. (2014). Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80 1–13. 10.1111/tpj.12618 [DOI] [PubMed] [Google Scholar]
- Alvarez J. M., Vidal E. A., Gutiérrez R. A. (2012). Integration of local and systemic signaling pathways for plant N responses. Curr. Opin. Plant Biol. 15 185–191. 10.1016/j.pbi.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Araya T., Miyamoto M., Wibowo J., Suzuki A., Kojima S., Tsuchiya Y. N., et al. (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111 2029–2034. 10.1073/pnas.1319953111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q., Garmier M., de Bont L., Soubigou-Taconnat L., Mazubert C., Benhamed M., et al. (2014). The polyadenylation factor subunit cleavage and polyadenylation specificity factor 30: a key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol. 165 732–746. 10.1104/pp.114.236083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield D. E., Glazer A. N., Falkowski P. G. (2010). The evolution and future of earth’s nitrogen cycle. Science 330 192–196. 10.1126/science.1186120 [DOI] [PubMed] [Google Scholar]
- Castaings L., Camargo A., Pocholle D., Gaudon V., Texier Y., Boutet-Mercey S., et al. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57 426–435. 10.1111/j.1365-313X.2008.03695.x [DOI] [PubMed] [Google Scholar]
- Crawford N. M., Glass A. D. M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3 389–395. 10.1016/S1360-1385(98)01311-9 [DOI] [Google Scholar]
- Delaney K. J., Xu R. Q., Zhang J. X., Li Q. Q., Yun K. Y., Falcone D. L., et al. (2006). Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 140 1507–1521. 10.1104/pp.105.070672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes K. P., Addepalli B., Hunt A. G. (2006). An Arabidopsis Fip1 homolog interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J. Biol. Chem. 281 176–186. 10.1074/jbc.M510964200 [DOI] [PubMed] [Google Scholar]
- Guan P., Ripoll J. J., Wang R., Vuong L., Bailey-Steinitz L. J., Ye D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U.S.A. 114 2419–2424. 10.1073/pnas.1615676114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P., Wang R., Nacry P., Breton G., Kay S. A., Pruneda-Paz J. L., et al. (2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 111 15267–15272. 10.1073/pnas.1411375111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez R. A. (2012). Systems biology for enhanced plant nitrogen nutrition. Science 336 1673–1675. 10.1126/science.1217620 [DOI] [PubMed] [Google Scholar]
- Ho C. H., Lin S. H., Hu H. C., Tsay Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138 1184–1194. 10.1016/j.cell.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Hu H. C., Wang Y. Y., Tsay Y. F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57 264–278. 10.1111/j.1365-313X.2008.03685.x [DOI] [PubMed] [Google Scholar]
- Hunt A. G., Xu R. Q., Addepalli B., Rao S., Forbes K. P., Meeks L. R., et al. (2008). Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genomics 9:220. 10.1186/1471-2164-9-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann I., Martin G., Friedlein A., Langen H., Keller W. (2004). Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23 616–626. 10.1038/sj.emboj.7600070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. commun. 4:1617. 10.1038/ncomms2621 [DOI] [PubMed] [Google Scholar]
- Krapp A., David L. C., Chardin C., Girin T., Marmagne A., Leprince A. S., et al. (2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 65 789–798. 10.1093/jxb/eru001 [DOI] [PubMed] [Google Scholar]
- Krouk G., Crawford N. M., Coruzzi G. M., Tsay Y. F. (2010a). Nitrate signaling: adaptation to fluctuating environments. Curr. Opin. Plant Biol. 13 266–273. 10.1016/j.pbi.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Krouk G., Mirowski P., LeCun Y., Shasha D. E., Coruzzi G. M. (2010b). Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 11:R123. 10.1186/gb-2010-11-12-r123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Y., Fu Y. L., Pike S. M., Bao J., Tian W., Zhang Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 2010 1633–1646. 10.1105/tpc.110.075242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang R., Gao Y., Wang C., Zhao L., Xu N., et al. (2017). The Arabidopsis CPSF30-L gene plays an essential role in nitrate signaling and regulates the nitrate transceptor gene NRT1.1. New Phytol. 216 1205–1222. 10.1111/nph.14743 [DOI] [PubMed] [Google Scholar]
- Liu M., Xu R., Merrill C., Hong L., Von Lanken C., Hunt A. G., et al. (2014). Integration of developmental and environmental signals via a polyadenylation factor in Arabidopsis. PLoS One 9:e115779. 10.1371/journal.pone.0115779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C., Roudier F., Castaings L., Bréhaut V., Blondet E., Colot V., et al. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4:1713. 10.1038/ncomms2650 [DOI] [PubMed] [Google Scholar]
- Medici A., Marshall-Colon A., Ronzier E., Szponarski W., Wang R., Gojon A., et al. (2015). AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 6:6274. 10.1038/ncomms7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J. A., Vega A., Bouguyon E., Krouk G., Gojon A., Coruzzi G., et al. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9 837–856. 10.1016/j.molp.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Para A., Li Y., Marshall-Colón A., Varala K., Francoeur N. J., Moran T. M., et al. (2014). Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111 10371–10376. 10.1073/pnas.1404657111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P. J., Lingner J., Minvielle-Sebastia L., Keller W. (1995). The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell 81 379–389. 10.1016/0092-8674(95)90391-7 [DOI] [PubMed] [Google Scholar]
- Rubin G., Tohge T., Matsuda F., Saito K., Scheible W. R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21 3567–3584. 10.1105/tpc.109.067041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W. R., Morcuende R., Czechowski T., Fritz C., Osuna D., Palacios-Rojas N., et al. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136 2483–2499. 10.1104/pp.104.047019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 671–755. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M. (1999). Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2 178–186. 10.1016/S1369-5266(99)80033-8 [DOI] [PubMed] [Google Scholar]
- Tabata R., Sumida K., Yoshii T., Ohyama K., Shinohara H., Matsubayashi Y. (2014). Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346 343–346. 10.1126/science.1257800 [DOI] [PubMed] [Google Scholar]
- Vidal E. A., Alvarez J. M., Moyano T. C., Gutiérrez R. A. (2015). Transcriptional networks in the nitrate response of Arabidopsis thaliana. Curr. Opin. Plant Biol. 27 125–132. 10.1016/j.pbi.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Vidal E. A., Araus V., Lu C., Parry G., Green P. J., Coruzzi G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107 4477–4482. 10.1073/pnas.0909571107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E. A., Moyano T. C., Krouk G., Katari M. S., Tanurdzic M., McCombie W. R., et al. (2013). Integrated RNA-seq and sRNA-seq analysis identifies novel nitrate-responsive genes in Arabidopsis thaliana roots. BMC Genomics 14:701. 10.1186/1471-2164-14-701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Okamoto M., Xing X., Crawford N. M. (2003). Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 132 556–567. 10.1104/pp.103.021253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xing X., Crawford N. M. (2007). Nitrate acts as a transcriptome signal at micromolar concentration in Arabidopsis root. Plant Physiol. 145 1735–1745. 10.1104/pp.107.108944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xing X., Wang Y., Tran A., Crawford N. M. (2009). A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 151 472–478. 10.1104/pp.109.140434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Wang R., Zhao L., Zhang C., Li Z., Lei Z., et al. (2016). The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 28 485–504. 10.1105/tpc.15.00567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki J., Galli M., Kim A. Y., Nito K., Aleman F., Chang K. N., et al. (2016). Mapping transcription factor interactome networks using Halo Tag protein arrays. Proc. Natl. Acad. Sci. U.S.A. 113 E4238–E4247. 10.1073/pnas.1603229113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Forde B. G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279 407–409. 10.1126/science.279.5349.407 [DOI] [PubMed] [Google Scholar]
- Zhang J., Addepalli B., Yun K. Y., Hunt A. G., Xu R., Rao S., et al. (2008). A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS One 3:e2410. 10.1371/journal.pone.0002410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L. F., Wang Y. (2017). Nitrate assay for plant tissues. Bio Protoc. 7:e2029 10.21769/BioProtoc.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.