Abstract
The archive of the Universidad de Costa Rica maintains a nineteenth-century French collection of drawings and lithographs in which the biodeterioration by fungi is rampant. Because of nutritional conditions in which these fungi grew, we suspected that they possessed an ability to degrade cellulose. In this work our goal was to isolate and identify the fungal species responsible for the biodegradation of a nineteenth-century art collection and determine their cellulolytic activity. Fungi were isolated using potato-dextrose-agar (PDA) and water-agar with carboxymethyl cellulose (CMC). The identification of the fungi was assessed through DNA sequencing (nrDNA ITS and α-actin regions) complemented with morphological analyses. Assays for cellulolytic activity were conducted with Gram’s iodine as dye. Nineteen isolates were obtained, of which seventeen were identified through DNA sequencing to species level, belonging mainly to genera Arthrinium, Aspergillus, Chaetomium, Cladosporium, Colletotrichum, Penicillium and Trichoderma. For two samples that could not be identified through their ITS and α-actin sequences, a morphological analysis was conducted; they were identified as new species, named Periconia epilithographicola sp. nov. and Coniochaeta cipronana sp. nov. Qualitative tests showed that the fungal collection presents important cellulolytic activity.
Introduction
Variations in the composition and appearance of a material as a consequence of the action of microorganisms is known as biodeterioration1. This phenomenon becomes evident with the presence of reddish-brown or yellowish-brown patches, microfungal structures and textural changes, which are commonly found in ancient documents2. These conditions apply to a nineteenth-century French collection of drawings and lithographs by Bernard Romain Julien (1802–1871) that is held in the archive of the School of Plastic Arts of Universidad de Costa Rica. The damage due to the microbial proliferation in these works of art is related to the storage conditions, especially to the damp and warm environments3. To design an effective and specific treatment according to the species growing in the laminae led to the isolation and identification of the fungal species responsible for the foxing of the lithographs.
Previous investigations of the microbiota in antique documents reported the presence of fungi that belong mainly to genera Alternaria, Aspergillus, Chaetomium, Cladosporium, Penicillium, and Trichoderma1,2,4–7. For instance, El Bergadi et al. (2013) isolated, identified and characterized the microbiota of manuscripts from an ancient collection of the Medina of Fez and found Aspergillus niger, Aspergillus oryzae, Mucor racemus, and Penicillium chrysogenum, as the most frequent species from a total of 31 fungal isolates8.
Because of nutritional limitations in which these fungi grew and where cellulose of the laminae was the only source of carbon, the species responsible for the biodeterioration were believed to possess cellulolytic activity8,9. This cellulolytic activity is of interest for multiple biotechnological processes, such as treatment of agroindustrial residues10,11 or production of cellulases12. This condition was first deduced and published in 1903 by van Iterson in “La décomposition del la cellulose pas les microrganismes”7. An investigation of the microbial diversity in a nineteenth-century Islamic and Koranic book led to the discovery of nine fungal species with the ability to degrade carboxymethylcellulose (CMC), including Aspergillus niger, A. oryzae and Hypocrea lixii8. Michaelsen et al. (2009) and Pinzari et al. (2006) described Aspergillus versicolor, A. nidulans, A. terreus and Chaetomium globosum as agents in the microbiological damage of old documents5,6.
Given the interest in the developing methods for protecting and preserving ancient documents from microbial degraders13 and the importance of obtaining microorganisms or enzymes with the capacity to degrade lignocellulosic wastes14, the aim of the present work was to isolate and identify the fungal species responsible for the biodegradation of a nineteenth-century art collection and to determine their cellulolytic activity. We found 19 fungal isolates belonging mainly to genera Arthrinium, Aspergillus, Chaetomium, Cladosporium, Colletotrichum, Penicillium and Trichoderma. Two samples not identified through their DNA sequences were identified through morphological analysis as new fungal species, namely Periconia epilithographicola sp. nov. and Coniochaeta cipronana sp. nov. Qualitative tests showed that the fungus collection presents important cellulolytic activity.
Methods
Sampling and isolation of cellulolytic fungi
A total of 13 laminae from a nineteenth-century French collection of lithographs belonging to Universidad de Costa Rica with signs of biodeterioration were sampled in areas of critical damage (colored or discolored areas, microfungal structures or other observable textural changes in the paper) with sterile cotton swabs, which were subsequently submerged in Phosphate Buffered Saline (PBS, 100 µL). Samples (50 µL) were cultured onto potato dextrose agar (PDA; Difco Potato Dextrose agar, BD company, France), and onto water agar with carboxymethyl cellulose (CMC, 1%, Sigma-Aldrich) with kanamycin (km, 50 µg/mL, Sigma-Aldrich). Morphologically distinct colonies were isolated and purified onto plates with the same culture media2,15–17.
Molecular identification
To identify the various fungal isolates, DNA extraction was performed using the method described by Lodhi et al. (1994) with modifications18. First, two disks (diameter 0.8 cm) from each fungal colony were introduced into Eppendorf tubes (1.5 mL). An extraction buffer (750 µL) was added, followed by the vortex of the sample and an incubation period (30 min at 67 °C). DNA was then precipitated with the addition of a CHCl3:octanol mixture (24:1, 750 µL), separation of the supernatant, and addition of isopropanol (600 µL, Sigma-Aldrich) and ethanol (500 µL, 70% v/v, Sigma-Aldrich). The DNA was eventually resuspended in AE buffer (50 µL, Qiagen) containing RNAse (1 µL, Fermentas). PCR reactions were performed to amplify the ITS (ITS 4 and ITS 5) and actin (Act-512F and Act-783R) regions using a reaction mix (PCR Master Mix, 10 µL, 2X, Thermo Scientific), water (7 µL), primers (0.5 µL, 10 μM) and DNA (2 µL, 50 ng/μL)19,20. All PCR reactions were performed in a thermocycler (Applied Biosystems 9902, Norwalk, USA) according to conditions described by Carbone & Kohn (1999) and White et al. (1990) for actin and ITS primers, respectively19,20.
The amplified products were purified with a clean-up kit (EXO-AP, Thermo Scientific, USA) and sequenced with a genetic analyzer (ABI 3130xl) and a reaction kit (Big Dye v.3 Terminator Cycle Sequencing Ready Reaction Kit, Applied Biosystems, USA), using ITS and actin primers (1 μM). Sequences were analyzed with software (MEGA 7), and were run through a Standard Nucleotide BLAST (Genbank, NCBI nucleotide database) to assess the similarity with reported sequences of fungal species. The BLAST searches were run excluding uncultured/environmental samples in the database. To corroborate the results, the BLAST search was repeated limiting the search to sequences only from type material. All sequences have been deposited in the GenBank database under the accession numbers that appear in Supplementary Table S1.
Morphological identification
Two species that did not have a close match to anything in Genbank, were examined in more detail to determine their morphological characteristics. Morphological analyses followed recommendations and techniques described by Ellis (1971) for hyphomycetous fungi and common methods in mycology21,22. Fungal isolates were cultured in CMD (BBL Corn Meal Dextrose agar, BD Company, France) and PDA (Difco Potato Dextrose agar, BD company, France) for 7 to 10 days near 25 °C. An optical microscope (Olympus BX-40, Japan) was used with an attached camera (18 megapixels, OMAX, Korea); software (ToupView, ToupTek Photonics, China) was used to measure structures.
Screening of cellulolytic activity
Cellulase-producing microorganisms were screened on agar plates enriched with only CMC as a source of carbon, with Gram’s iodine as indicator (Prelab)23–26. This qualitative determination is based on the interaction of iodine with cellulose and its components in its degraded form, such that the integral biopolymer holds Gram’s iodine dye; whereas areas with cellulose hydrolyzed by enzymes result in clear zones or the appearance of a pale halo15,27. The halo was measured for the subsequent calculation of the enzymatic index (EI), a semi-quantitative estimate of the enzyme activities, according to this formula15.
| 1 |
For this purpose, fungal discs (diameter 0.8 cm) were grown in a solid medium composed of water agar (1.6%), CMC (1%) and kanamycin (km, 50 µg/mL). After cultures were incubated (7 days, 30 °C), plates were flooded with Gram’s iodine stain (10 mL, 10 min) and washed with water to enable the observation, photographing and measurement of the clear zone around the fungal growth23–26. Software (ImageJ, version 1.51j8) was used to measure the diameters28. The experiment was repeated twice (on separate days) with duplicates of each isolate. Pleurotus ostreatus served as a positive control29.
Results and Discussion
Isolation and identification of fungi isolated from drawings and lithographs
Through the screening of the lithographs, the total count of fungi isolated was 19, of which eight grew directly in water agar with CMC-km and eleven were first isolated from PDA and then recultivated in water agar with CMC as the sole source of carbon. The proliferation of fungi in the latter culture medium is in accordance with the environment in which they were isolated (limited sources of carbon, with cellulose as sole nutrient). Laminae #5 was the most contaminated, with ten isolations; followed by laminae #7 and #10, with 3 isolations each (see Supplementary Figure S1). The fungal isolates showed diverse forms, sizes, elevations, borders, surfaces, opacity, color and growth rates, as shown in Fig. 1.
Figure 1.
Fungal diversity in ancient lithographs. Several fungi were isolated from stained and degraded areas from nineteenth-century drawing laminae. On the top row from left to right are isolates #5, #10, #19, #9, #8, and #15. On the bottom row from left to right are isolates #16, #22, #21, #11, #23 and #26. Samples in the image were grown in PDA during 6 days. We thank Dr. Salomón Chaves (Instituto de Investigaciones en Arte) for authorizing the use of images from the collection of drawings by Bernard Romain Julien in this manuscript.
BLAST searches in GenBank database resulted in the classification of nineteen isolates into fifteen species and nine genera (Table 1). These nineteen isolates had at least a 98% similarity with known species. The most prevalent genus was Cladosporium. Of the nine identified genera, Aspergillus, Chaetomium, Cladosporium, Penicillium, and Trichoderma are reported as common microbiota in ancient works of art1,2,17,30,31. The actin region was sequenced to confirm the results obtained with the ITS region, and to classify to species level some samples that could not be done with ITS. For all cases in which both ITS and actin sequences were obtained, the fungi were classified within the same species, except isolate #9 in which the actin region denied conclusive results obtained with ITS.
Table 1.
Molecular identification of isolated fungi using ITS and actin regions.
| Isolate# | Identification | ITS and closest accesion number | Actin and closest accesion number | ||||
|---|---|---|---|---|---|---|---|
| Accession | Identity | Coverage | Accession | Identity | Coverage | ||
| 4 | Cladosporium sphaerospermum | KP701988.1 | 100% | 100% | EU570272.1 | 98% | 99% |
| 5 | Penicillium chrysogenum a | KC009774.1 | 100% | 100% | AM920435.1 | 97% | 100% |
| 6 | Penicillium westlingii b | JN617668.1 | 100% | 100% | AM920435.1 | 83% | 55% |
| 7 | Cladosporium tenuissimum c |
KP701937.1* KJ596320.1* |
100% | 100% | LN834582.1 | 100% | 99% |
| 8 | Aspergillus niger | KJ365316.1 | 100% | 100% | AM270331.1 | 99% | 99% |
| 9 | Cladosporium sp. | KP701937.1*KJ596320.1* | 100% | 100% | — | — | — |
| 10 | Arthrinium arundinis b | KF144889.1 | 100% | 100% | AY951865.1 | 76% | 76% |
| 11 | Cladosporium angustisporum c |
MG250413.1*MG199960.1* KP701978.1* KP701964.1* KP701938.1* KP701935.1* KP701930.1* KP701908.1* |
100% | 100% | LN834540.1 | 100% | 100% |
| 12 | Aspergillus versicolor | NR_131277.1 | 100% | 95% | — | — | — |
| 13 | Chaetomium cf. subglobosumb | NR_144826.1 | 96% | 99% | KF545191.1 | 99% | 100% |
| 15 | Cladosporium angustisporum c |
MG250413.1*MG199960.1* KP701978.1* KP701964.1* KP701938.1* KP701935.1* KP701930.1* KP701908.1* |
100% | 100% | LN834540.1 | 100% | 100% |
| 16 | Cladosporium cladosporioides c |
MG250413.1*MG199960.1* KP701978.1* KP701964.1* KP701938.1* KP701935.1* KP701930.1* KP701908.1* |
100% | 100% | KT600582.1 | 99% | 96% |
| 17 | Chaetomium cf. subglobosumb | NR_144826.1 | 100% | 99% | KF545191.1 | 99% | 100% |
| 19 | Periconia sp.d | HQ608027.1 | 99% | 100% | KP184118.1 | 83% | 95% |
| 20 | Chaetomium cf. subglobosum b | NR_144826.1 | 99% | 99% | KF545191.1 | 99% | 100% |
| 21 | Coniochaeta sp.d | KX869958.1 | 99% | 100% | AY579255.1 | 71% | 100% |
| 22 | Aspergillus niger | KJ365316.1 | 100% | 100% | AM270331.1 | 99% | 99% |
| 23 | Trichoderma cf. Longibrachiatumb | KT336509.1 | 100% | 100% | JQ238613.1 | 98% | 99% |
| 26 | Colletotrichum kahawae c | NR_144787.1 | 100% | 98% | JX009431.1 | 99% | 100% |
aIsolates had homology with two fungi of different species with the ITS region analysis, but through sequencing of the actin region it was possible to confirm the identification.
bNo register in the NCBI GenBank database for actin sequencing regions.
cITS region sequencing allowed to identify only the isolates at genus level; the actin region enabled an identification at specie level.
dNo register in the NCBI GenBank database for either ITS or actin sequencing regions.
*Accessions with same similarity.
Two isolates were only identified to genus or class levels using both ITS and actin regions. Specifically, isolate #19 was classified within the genus Periconia, and isolate #21 was classified within the class Sordariomycetes, both in the phylum Ascomycota. Since these two isolates did not have a close match to any sequence in the Genbank, traditional morphological analyses and descriptions (e.g. microscopy and use of taxonomic literature) were done to elucidate the identity of these isolates.
Description of two new fungal species
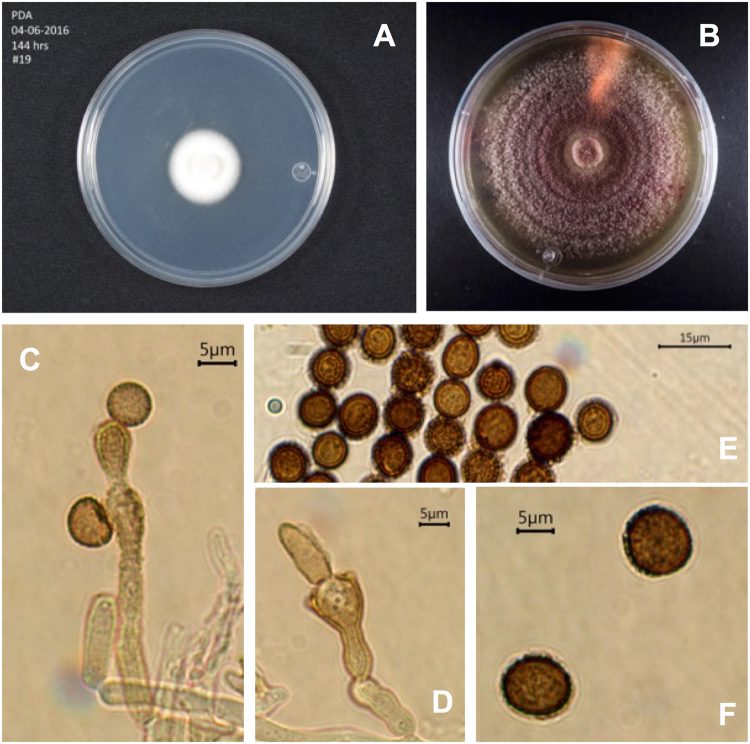
Periconia epilithographicola C
Coronado-Ruiz, R. Avendaño, E. Escudero-Leyva, G. Conejo-Barboza, P. Chaverri & M. Chavarría sp. nov. Fig. 2. Mycobank: MB825093 GenBank: MF422162 (ITS) & MF422179 (actin). Etymology: epilithographicola, because it was found growing over art lithographs. Holotype: Costa Rica, San José, San Pedro de Montes de Oca, Universidad de Costa Rica; on art lithographs; May 19th, 2016; collected by Avendaño R.; extype culture CBS 144017, a permanently preserved, metabolically inactive culture (=#19). Diagnosis: Periconia species producing a pinkish to reddish pigment. Straight conidiophores; globose, echinulated, golden-brown conidia. Colonies: At 25 °C after three weeks, on CMD, attaining 25 mm diam., colony white, cottony. On PDA, attaining 60 mm diam., colony effuse, pinkish (similar to OAC486), with creeping hyphae; conidiophores visible, forming small agglutinated black sticky drop-like structures. Conidiophores: macronematous, with creeping hyphae forming stipes 251.6–270 · 3.6–6.1 µm, straight, branched singly near the base, seven or more septate, grayish to black. Conidiogenous cells: holoblastic, (5.1−) 7 · 10 (−11.5) µm (n = 15), sub-globose to ellipsoid, finely roughened, yellowish to brown (slightly more brilliant than OAC757). Conidia: globose, (7.8−) 9.2 (−10.7) µm diam. (n = 30), golden to brown (similar to OAC705), echinulated, catenated, sometimes forming long chains. Habitat: Growing on aged lithographs of Instituto de Investigaciones en Arte (Universidad de Costa Rica). Notes: Several Periconia species share similar characteristics of the conidiophore, differing mainly in the conidia size. Periconia pseudobyssoides conidia are larger, (12−) 15–17 (−20) µm diam. and brown-reddish32; P. byssoides conidia are 10–15 µm in diam21.; P. saraswatiurensis conidia are 9–12 µm diam., also secreting dark green to purple pigments in culture21. The only species with a similar conidial size is P. jabalpurensis but it lacks septa in the conidiophores; P. macrospinosa shows conidia of up to 35 µm diam. with long spines (<2 µm)21, which does not fit Periconia epilithographicola.
Figure 2.
Periconia epigraphicola. (A) PDA Culture ca 6 days. (B), PDA Culture ca 20 days. (C) Conidiogenous cell forming conidia. (D) Conidiogenous cell. (E) Catenated spinulose conidia. (F) Spinulose conidia.
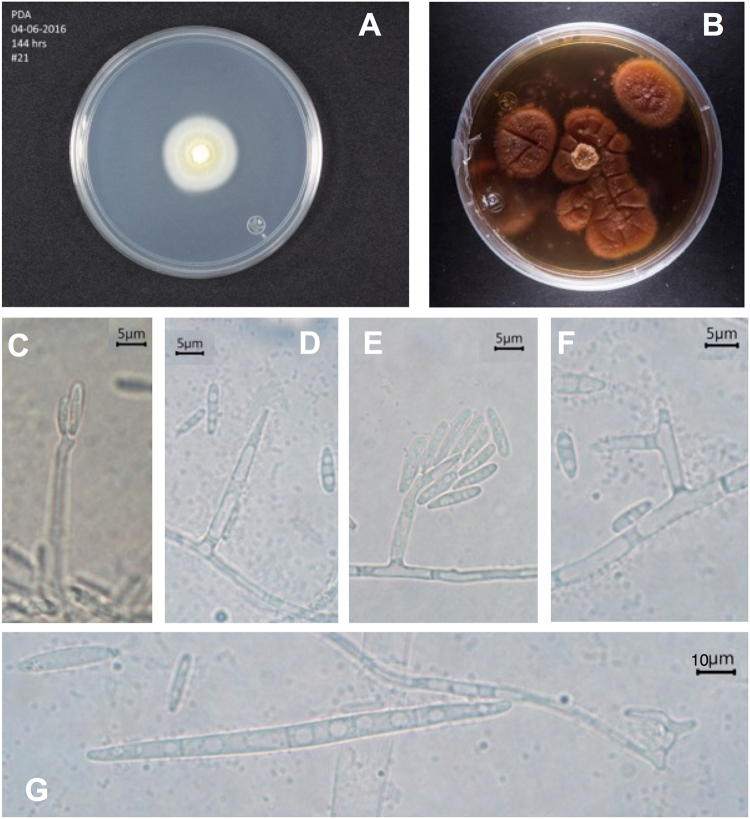
Coniochaeta cipronana C
Coronado-Ruiz, R. Avendaño, E. Escudero-Leyva, G. Conejo-Barboza, P. Chaverri & M. Chavarría sp. nov. Fig. 3. Mycobank: MB825094 GenBank: MF422164 (ITS) & MF422181 (actin). Etymology: as a reference to Centro de Investigaciones en Productos Naturales (CIPRONA, Universidad de Costa Rica) for the impact and transcendence of the research in the field of natural products over 38 years. Holotype: Costa Rica, San José, San Pedro de Montes de Oca, Universidad de Costa Rica; from art lithograph; May 19th, 2016; collected by Avendaño R.; extype culture CBS 144016, a permanently preserved, metabolically inactive culture (=#21). Diagnosis: Nodulisporium-like conidiophore, with macro- and microconidia, hyaline, macroconidia 5–7-septate slightly curved, fusiform, microconidia cylindrical 1–2-septate. Colonies: At 25 °C after 3 weeks on CMD, reaching 20 mm diam., hyaline to white. On PDA attaining 25 mm diam., colony white, then turning purple (similar to OAC555), cracking and turning the media dull orange (lighter than OAC789). Conidiophores: Nodulisporium-like. Conidiogenous cells: Simple, mainly straight or sometimes curled, cylindrical, (5.8−) 21.8 (−28.8) · (2.2−) 2.8 (−3) μm (n = 15), arising directly from hyphae and stretching toward the apex, sometimes dichotomously branched, dimorphic, without collarete, hyaline. Short conidiophores (3−) 4.8 (−5) · 2 μm (n = 15). Conidia: macroconidia fusiform, (40.1−) 60.6 (−74.6) · (3−) 3.5 (−5) μm (n = 30), 5–7-septate, slightly curved, hyaline, smooth; microconida cylindrical (11.6−) 16.5 (−25) · (2.2−) 2.8 (−3.6) μm (n = 30), 1–2-septate, hyaline, smooth. Habitat: Growing on aged lithographs of Instituto de Investigaciones en Arte (Universidad de Costa Rica). Notes: This species, because of the Nodulisporium-like conidiophore, is similar to Coniochaeta ershadii, especially in the size of the conidiogenous cells. The conidia produced by C. ershadii are prominently smaller33 than those present in C. cipronana; the presence of macro- and microconidia is also a distinguishing character.
Figure 3.
Coniochaeta cipronana. (A) PDA Culture ca 6 days. (B) PDA Culture ca 20 days. (C–E) Conidiogenous cell with small conidia. (F) Dicotomic conidiophore. (G) Macroconidia 5–7 septate.
The new fungal species described belong to Periconia Tode and Coniochaeta (Sacc.) Cooke genera (see Supplementary Figures S2 and S3). Periconia is a polyphyletic genus Pleosporales (Dothideomycetes, Ascomycota), with a complicated taxonomy and a poorly understood phylogeny32. This genus has been widely reported as a common endophyte from the roots of several plants, like a Periconia species isolated from Piper longum producing metabolites with a high pharmacological potential34 and the melanized hyphae are believed to protect the fungi from environmental oxidation35. Some species have been reported as parasites in leaves of Xanthium strumarium and Ipomea muricara in India and others as decomposers in bamboo statches36. Coniochaeta (Coniochaetaceae, Coniochaetales, Sordariomycetes, Ascomycota) was introduced as a subgenus of Rosellinia De Not. for species with hairy perithecia but differing by the absence of amyloid asci in their sexual stages33. Many Coniochaeta conidiophores produce Lecytophora-like structures. Like Periconia, Coniochaeta requires further taxonomic and phylogenetic studies37. About 70 species and six synonyms are included in the genus Coniochaeta and most of the isolates are reported from dung, necrotic wood, soil and plant surfaces38.
Cellulase activity of the fungal isolates
Assay of the cellulase activity showed that 95% of the samples produce extracellular enzymes that break down cellulose into smaller oligosaccharides or monosaccharides, as evident from the clear zone observed after staining the plates with Gram’s iodine (see Table 2 and Supplementary Figure S4). This fraction that includes the two new species (sample #19: Periconia epilithographicola and sample #21: Coniochaeta cipronana) also comprehends species of Arthrinium, Aspergillus, Chaetomium, Cladosporium, Colletotrichum, Penicillium, and Trichoderma, being the first four commonly reported with cellulolytic activity31,39–45. These observations are congruent with the habitat of restricted carbon sources, in which sheets or laminae made of fibers of cellulose pulp were the support material for the growth of microorganisms.
Table 2.
Enzymatic indices of the isolates on CMC agar stained with Gram Iodine after incubation for seven days.
| Isolate# | Enzymatic index |
|---|---|
| 4 | No activity |
| 5 | 3,3 ± 0,2 |
| 6 | 1,91 ± 0,07 |
| 7 | 2,87 ± 0,05 |
| 8 | 0,92 ± 0,02 |
| 9 | 1,6 ± 0,1 |
| 10 | 1,62 ± 0,08 |
| 11 | 2,74 ± 0,02 |
| 12 | 1,47 ± 0,09 |
| 13 | 1,243 ± 0,005 |
| 15 | 2,87 ± 0,03 |
| 16 | 1,47 ± 0,03 |
| 17 | 1,199 ± 0,004 |
| 19 | 1,861 ± 0,002 |
| 20 | 1,17 ± 0,08 |
| 21 | 1,57 ± 0,06 |
| 22 | 1,086 ± 0,006 |
| 23 | 1,39 ± 0,03* |
| 26 | 0,80 ± 0,06 |
| Control: P. ostreatus | 1,8 ± 0,1 |
*EI was measured after 24 h.
Importantly, 32% of the total isolates had a significantly superior enzymatic index relative to a positive control (P. ostreatus), i.e., isolates #5 (Penicillium chrysogenum), #7 (Cladosporium tenuissimum), #11 (Cladosporium angustisporum) and #23 (Trichoderma cf. longibrachiatum). Other studies have characterized these species as effective cellulase producers16,40. Isolates #5 (Penicillium chrysogenum) and #23 (Trichoderma cf. longibrachiatum) had an outstanding performance relative to the positive control and the rest of the isolated fungi. Specifically, isolate #5 presented an enzymatic index for cellulose activity almost twice of that of the positive control. Isolates of these species not only have presented important cellulase activity but also have been the object of study for their capacity to produce xylanases46, or tanases47.
The case of isolate # 23 (Trichoderma cf. longibrachiatum) was even more striking. For this fungus, EI is reported for 24 h (see Table 2) because after 7 days (the period in which the other isolates were measured) the microorganism had covered the entire Petri plate, evidence of an accelerated growth and a large capacity to use the CMC as the sole source of carbon. The result (1.39 ± 0.03) was slightly smaller than the positive control (1.8 ± 0.1, measured after seven days). However, as previously mentioned, isolate # 23 was measured at 24 h. This result implies a large rate of enzymatic (cellulase) production from fungus #23 in a medium rich in cellulose, relative to the rest of the fungi studied, which is important for the development of biotechnological applications and industry. Many studies have featured this species as a fungus with great cellulase activity48–51. Many commercial cellulases can be purchased in purified form after production with this species (e.g. C9748 Sigma-Aldrich or E-CELTR from Megazyme). Investigations with isolation # 23 will continue to evaluate its potential to degrade lignocellulosic residues from agricultural activity in Costa Rica (e.g., wastes from pineapple production).
In summary, in isolating, identifying and characterizing the cellulolytic activity of the fungi responsible for the biodegradation of a nineteenth-century collection, several species of fungi were found to have the ability to produce cellulases. In addition, two new species of fungi were identified and named Periconia epilithographicola sp. nov. and Coniochaeta cipronana sp. nov., which also have cellulolytic activity. A knowledge of the microorganisms that colonized the Bernard Romain Julien collection belonging to Universidad de Costa Rica will allow the development of strategies directed to the conservation of these ancient lithographs. This work also contributes to the knowledge of new species with cellulolytic activity, which is a topic of perennial interest for biotechnology because of the important role of fungal cellulolytic enzymes in commercial food processing, performing the hydrolysis of cellulose during drying of beans, in the textile industry and laundry detergents, in the conversion of biomass into industrially important solvents or fuels, and their potential application for the bioremediation of wastes.
Electronic supplementary material
Acknowledgements
We are grateful to Instituto de Investigaciones en Arte (II Arte) of Universidad de Costa Rica and Dr. Salomon Chaves Badilla for the access and support for the study and selection of the collection of drawings. This work was supported by resources of the Vice-rectory of Research of Universidad de Costa Rica (project number 726-B6-115), Centro Nacional de Innovaciones Biotecnológicas (CENIBiot) and Reintegra (Red en Ciencia y Conservación del Patrimonio Cultural Nacional).
Author Contributions
M.C. conceived and designed the experiments; C.C.-R., R.A., G.C.-B., E.E.-L. performed the experiments; C.C.-R., R.A., E.E.-L., P.C., M.C. analyzed the data; P.C., M.C. contributed reagents or materials or analytical tools; C.C.-R., E.E.-L., P.C., M.C. wrote the paper. All authors reviewed and approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24934-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sterflinger K, Piñar G. Microbial deterioration of cultural heritage and works of art — tilting at windmills. Appl. Microbiol. Biotechnol. 2013;97:9637–9646. doi: 10.1007/s00253-013-5283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mesquita N, et al. Fungal diversity in ancient documents. a case study on the archive of the University of Coimbra. Int. Biodeter. Biodegr. 2009;63:626–629. doi: 10.1016/j.ibiod.2009.03.010. [DOI] [Google Scholar]
- 3.Rakotonirainy MS, Heude E, Lave B. Isolation and attempts of biomolecular characterization of fungal strains associated to foxing on a 19th century book. J. Cult. Herit. 2007;8:126–133. doi: 10.1016/j.culher.2007.01.003. [DOI] [Google Scholar]
- 4.Garg KL, Kamal K, Mishra AK. Role of fungi in the deterioration of wall paintings. Sci. Total Environ. 1995;167:255–271. doi: 10.1016/0048-9697(95)04587-Q. [DOI] [Google Scholar]
- 5.Michaelsen A, Piñar G, Montanari M, Pinzari F. Biodeterioration and restoration of a 16th-century book using a combination of conventional and molecular techniques: A case study. Int. Biodeter. Biodegr. 2009;63:161–168. doi: 10.1016/j.ibiod.2008.08.007. [DOI] [Google Scholar]
- 6.Pinzari F, Pasquariello G, De Mico A. Biodeterioration of paper: a SEM study of fungal spoilage reproduced under controlled conditions. Macromol. Symp. 2006;238:57–66. doi: 10.1002/masy.200650609. [DOI] [Google Scholar]
- 7.Sterflinger K, Pinzari F. The revenge of time: fungal deterioration of cultural heritage with particular reference to books, paper and parchment. Environ. Microbiol. 2012;14:559–566. doi: 10.1111/j.1462-2920.2011.02584.x. [DOI] [PubMed] [Google Scholar]
- 8.El Bergadi, F., Laachari, F., Elabed, S., Mohammed, I. & Ibnsouda, S. Cellulolytic potential and filter paper activity of fungi isolated from ancients’ manuscripts from the Medina of Fez. Ann. Microbiol. 64, 815–822 (2013).
- 9.Montegut, D., Indictor, N. & Koestler, R. J. Fungal deterioration of cellulosic textiles: a review. Int. Biodeterior. 28, 209–226 (1991).
- 10.Anwar Z, Gulfaz M, Irshad M. Agro-industrial lignocellulosic biomass a key to unlock the future bio-energy: A brief review. J. Radiat. Res. Appl. Sci. 2014;7:163–173. doi: 10.1016/j.jrras.2014.02.003. [DOI] [Google Scholar]
- 11.Berlemont R. Distribution and diversity of enzymes for polysaccharide degradation in fungi. Sci. Rep. 2017;7:222. doi: 10.1038/s41598-017-00258-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiranjeevi T, et al. Optimization of holocellulolytic enzymes production by Cladosporium Cladosporioides using Taguchi-L’16 orthogonal array. J. Biobased Mater. Bioenergy. 2012;6:1–10. doi: 10.1166/jbmb.2012.1201. [DOI] [Google Scholar]
- 13.Sequeira S, Phillips A, Cabrita E, Macedo M. Antifungal treatment of paper with calcium propionate and parabens: Short-term and long-term effects. Int. Biodeterior. Biodegradation. 2017;120:203–215. doi: 10.1016/j.ibiod.2017.03.005. [DOI] [Google Scholar]
- 14.Montella S, et al. Discovery of genes coding for carbohydrate-active enzyme by metagenomic analysis of lignocellulosic biomasses. Sci. Rep. 2017;7:42623. doi: 10.1038/srep42623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florencio C, Couri S, Farinas C. Correlation between agar plate screening and solid-state fermentation for the prediction of cellulase production by Trichoderma strains. Enzyme Res. 2012;2012:793708–793715. doi: 10.1155/2012/793708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makeshkumar V, Mahalingam PU. Isolation and characterization of rapid cellulose degrading fungal pathogens from compost of agro wastes. Int. J. Pharm. Biol. Arch. 2011;2:1695–1698. [Google Scholar]
- 17.Sanmartín P, DeAraujo A, Vasanthakumar A. Melding the old with the new: trends in methods used to identify, monitor, and control microorganisms on cultural heritage materials. Microb. Ecol. 2016;74:1–17. doi: 10.1007/s00248-016-0770-4. [DOI] [PubMed] [Google Scholar]
- 18.Lodhi A, Ye G, Weeden N, Reisch B. A simple and efficient method for DNA extraction from Grapevine Cultivars and Vitis species. Plant Mol. Biol. Rep. 1994;12:6–13. doi: 10.1007/BF02668658. [DOI] [Google Scholar]
- 19.White, T., Bruns, S., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 315–322 (1990).
- 20.Carbone I, Kohn L. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycology. 1999;91:553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- 21.Ellis, M. Dematiaceous hyphomycetes (CABI Publishing, 1971).
- 22.Sutton, B.C. The Coelomycetes. Fungi Imperfecti with Pycnidia, Acervuli and Stromata (CABI Publishing, 1980).
- 23.Bakar N, Abd-Aziz S, Hassan M, Ghazali F. Isolation and selection of appropriate cellulolytic mixed microbial cultures for cellulases production from oil palm empty fruit bunch. Biotech. 2010;9:73–78. doi: 10.3923/biotech.2010.73.78. [DOI] [Google Scholar]
- 24.Gohel HR, Contractor CN, Ghosh SK, Braganza VJ. A comparative study of various staining techniques for determination of extra cellular cellulase activity on Carboxy MethylCellulose (CMC) agar plates. Int. J. Curr. Microbiol. App. Sci. 2014;3:261–266. [Google Scholar]
- 25.Johnsen HR, Krause K. Cellulase activity screening using pure carboxymethylcellulose: Application to soluble cellulolytic samples and to plant tissue prints. I. J. Mol. Sci. 2014;15:830–838. doi: 10.3390/ijms15010830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meddeb-Mouelhi F, Moisan JK, Beauregard M. A comparison of plate assay methods for detecting extracellular cellulase and xylanase activity. Enzyme Microb. Technol. 2014;66:16–19. doi: 10.1016/j.enzmictec.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Easteal A. Interaction between iodine and ethyl cellulose. J. Appl. Polym. Sci. 1999;71:1303–1314. doi: 10.1002/(SICI)1097-4628(19990222)71:8<1303::AID-APP10>3.0.CO;2-T. [DOI] [Google Scholar]
- 28.Schneider C, Rasband W, Eliceiri K. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valásková V, Baldrian P. Estimation of bound and free fractions of lignocellulose-degrading enzymes of wood-rotting fungi Pleurotus ostreatus, Trametes versicolor and Piptoporus betulinus. Research in Microbiology. 2006;157:119–124. doi: 10.1016/j.resmic.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Dunca S, et al. Study of the contaminating microbiota of old paper supports. Eur. Sci. J. 2014;3:237–251. [Google Scholar]
- 31.Naji K, Abdullah Q, Al-Zaqri A, Alghalibi S. Evaluating the biodeterioration enzymatic activities of fungal contamination isolated from some ancient Yemeni mummies preserved in the national museum. Biochem. Res. Int. 2014;2014:1–9. doi: 10.1155/2014/481508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markovskaja S, Kacergius A. Morphological and Molecular Characterization pf Periconia pseudobyssoides sp. nov. and a closely related P. byssoides. Mycol. Progress. 2014;13:291–302. doi: 10.1007/s11557-013-0914-6. [DOI] [Google Scholar]
- 33.Asgari B, Zare R, Gams W. Coniochaeta ershadii, a new species from Iran and a key to well-documented Coniochaeta species. Nova Hedwigia. 2007;84:175–187. doi: 10.1127/0029-5035/2007/0084-0175. [DOI] [Google Scholar]
- 34.Verma VC, Lobkovsky E, Gange AC, Singh SK, Prakash S. Piperine production by endophytic fungus Periconia sp. isolated from Piper longum L. J. Antibiot. 2011;64:427–431. doi: 10.1038/ja.2011.27. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Z, Zhang C, Lin, Kubicek C. Identity, diversity, and molecular phylogeny of the endophytic mycobiota in the roots of rare wild rice (Oryza granulate) from a nature reserve in Yunnan, China. Appl. Environ. Microbiol. 2010;76:1642–1652. doi: 10.1128/AEM.01911-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao R, Rao D. The genus Periconia from India. Mycopathol. Mycol. Appl. 1964;22:285–310. doi: 10.1007/BF02049062. [DOI] [PubMed] [Google Scholar]
- 37.Khan Z, et al. Coniochaeta polymorpha, a new species from endotracheal aspirate of a preterm neonate, and transfer of Lecytophora species to Coniochaeta. Antonie van Leeuwenhoek. 2013;104:243–252. doi: 10.1007/s10482-013-9943-z. [DOI] [PubMed] [Google Scholar]
- 38.Xie J, et al. An endophytic Coniochaeta velutina producing broad spectrum antimycotics. J. Microbiol. 2015;53:390–397. doi: 10.1007/s12275-015-5105-5. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, et al. The different roles of Penicillium oxalicum LaeA in the production of extracellular cellulase and β-xylosidase. Front. Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.02091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe C, et al. Fungi isolated from maize (Zea mays L.) grains and production of associated enzyme activities. Int. J. Mol. Sci. 2015;16:15328–15346. doi: 10.3390/ijms160715328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciferri O. Microbial degradation of paintings. Appl. Environ. Microbiol. 1999;65:879–855. doi: 10.1128/aem.65.3.879-885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Said A, Saleem A, Maghraby T, Hussein M. Cellulase activity of some phytopathogenic fungi isolated from diseased leaves of broad bean. Int. J. Curr. Microbiol. App. Sci. 2014;3:883–900. [Google Scholar]
- 43.Ghose TK. Measurement of cellulase activities. Pure Appl. Chem. 1987;59:257–268. doi: 10.1351/pac198759020257. [DOI] [Google Scholar]
- 44.Gopinath S, Anbu P, Hilda A. Extracellular enzymatic activity profiles in fungi isolated from oil-rich environments. Mycoscience. 2005;46:119–126. doi: 10.1007/S10267-004-0221-9. [DOI] [Google Scholar]
- 45.Wang M, Lu X. Exploring the synergy between cellobiose dehydrogenase from Phanerochaete chrysosporium and cellulase from Trichoderma reesei. Front. Microbiol. 2016;7:1–10. doi: 10.3389/fmicb.2016.00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haas H, Herfurth E, Stoffler G, Redl B. Purification, characterization and partial amino acid sequences of a xylanase produced by Penicillium chrysogenum. Biochim. Biophys. Acta. 1992;1117:279–286. doi: 10.1016/0304-4165(92)90025-P. [DOI] [PubMed] [Google Scholar]
- 47.Rajakumar G, Nandy S. Isolation, purification, and some properties of Penicillium chrysogenum Tannase. Appl. Environ. Microbiol. 1983;46:525–527. doi: 10.1128/aem.46.2.525-527.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandhu D, Kalra M. Production of cellulase, xylanase and pectinase by Trichoderma longibrachiatum on different substrates. T. Brit. Mycol. Soc. 1982;79:409–413. doi: 10.1016/S0007-1536(82)80034-X. [DOI] [Google Scholar]
- 49.Roger J, Nacas J. Interrelationship of Xylanase Induction and Cellulase Induction of Trichoderma longibrachiatum. Appl. Environ. Microbiol. 1990;56:2535–2539. doi: 10.1128/aem.56.8.2535-2539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalra M, Sidhu M, Sandhu D. Partial purification, characterization and regulation of cellulolytic enzymes from Trichoderma longibrachiatum. J. Appl. Bacteriol. 1986;61:73–80. doi: 10.1111/j.1365-2672.1986.tb03760.x. [DOI] [Google Scholar]
- 51.Chutani P, Sharma K. Concomitant production of xylanases and cellulases from Trichoderma longibrachiatum MDU-6 selected for the deinking of paper waste. Bioprocess Biosyst. Eng. 2016;39:747–758. doi: 10.1007/s00449-016-1555-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.