Abstract
Nicotianamine aminotransferase (NAAT), the key enzyme involved in the biosynthesis of mugineic acid family phytosiderophores (MAs), catalyzes the amino transfer of nicotianamine (NA). MAs are found only in graminaceous plants, although NA has been detected in every plant so far investigated. Therefore, this amino transfer reaction is the first step in the unique biosynthesis of MAs that has evolved in graminaceous plants. NAAT activity is dramatically induced by Fe deficiency and suppressed by Fe resupply. Based on the protein sequence of NAAT purified from Fe-deficient barley (Hordeum vulgare) roots, two distinct cDNA clones encoding NAAT, naat-A and naat-B, were identified. Their deduced amino acid sequences were homologous to several aminotransferases, and shared consensus sequences for the pyridoxal phosphate-binding site lysine residue and its surrounding residues. The expression of both naat-A and naat-B is increased in Fe-deficient barley roots, while naat-B has a low level of constitutive expression in Fe-sufficient barley roots. No detectable mRNA from either naat-A or naat-B was present in the leaves of either Fe-deficient or Fe-sufficient barley. One genomic clone with a tandem array of naat-B and naat-A in this order was identified. naat-B and naat-A each have six introns at the same locations. The isolation of NAAT genes will pave the way to understanding the mechanism of the response to Fe in graminaceous plants, and may lead to the development of cultivars tolerant to Fe deficiency that can grow in calcareous soils.
The Gramineae family, with about 9,000 species, is economically the most important family of flowering plants. Graminaceous plants are found throughout the world and comprise about 20% of the earth's vegetation. Primarily important as a basic food source, graminaceous plants provide us with cereals bred from selected wild species. The second most important use of the graminaceous plants is as pasture and hay for livestock. Increasing the productivity of graminaceous plants will be an important strategy for dealing with the rapid population increase predicted for developing countries in the 21st century.
Even though there is a large amount of Fe in soils, plants growing in soils with a high pH, such as calcareous soils, develop Fe deficiency that is observable as chlorosis. Under these adverse conditions, graminaceous plants secrete Fe chelators called mugineic acid family phytosiderophores (MAs) (Takagi, 1976) from their roots to acquire sparingly soluble Fe as Fe(III)-MAs through the use of Fe(III)-MA transporter(s). This mechanism was named strategy II by Römheld and Marschner (1986). The chemical properties (Takemoto et al., 1978; Nomoto et al., 1981; Sugiura et al., 1981; Mino et al., 1983) and physiological significance (Takagi et al., 1984; Römheld and Marschner, 1986; Marschner et al., 1987; Mori et al., 1987, 1991; Kawai et al., 1988; Mihashi and Mori, 1989) of MAs have already been established.
The amount of MAs secreted increases under Fe deficiency stress and is correlated with a plant's tolerance to Fe deficiency. Of the graminaceous plants, barley (Hordeum vulgare) is the most tolerant to Fe deficiency and secretes the largest amount of MAs, while rice is the most susceptible to Fe deficiency and secretes very little MAs (Sugiura et al., 1981; Takagi et al., 1984). MAs are synthesized from l-Met (Mori and Nishizawa, 1987). Nicotianamine synthase (NAS) catalyzes the bonding of three S-adenosyl Met molecules to form nicotianamine (NA) (Higuchi et al., 1994, 1999), which is then converted to deoxymugineic acid by an amino-group transfer catalyzed by nicotianamine aminotransferase (NAAT) (Kanazawa et al., 1995) and subsequent reduction at the 3′-carbon of the keto acid (Nomoto et al., 1987). The other MAs, including mugineic acid, hydroxymugineic acid, epihydroxymugineic acid, avenic acid (Mori and Nishizawa, 1989; Mori et al., 1990; Shojima et al., 1990; Ma and Nomoto, 1993), and epihydroxydeoxymugineic acid (Ma et al., 1999), are produced by the subsequent hydroxylation of deoxymugineic acid.
Of the enzymes participating in the biosynthesis of MAs, NAS and NAAT are strongly induced by Fe deficiency. The increase in the activity of these enzymes after the onset of Fe deficiency is well correlated with the amount of MAs secreted. Recently, several NAS genes (the nas series) have been cloned from barley (Higuchi et al., 1999), Arabidopsis (DDBJ/EMBL/GenBank accession nos. AB021934, AB021935, and AB021936), and rice (DDBJ/EMBL/GenBank accession no. AB021746). Although NA has been detected in all plants so far surveyed, including monocots and dicots (Noma and Noguchi, 1976; Fushiya et al., 1982; Rudolph et al., 1985), MAs have been found only in graminaceous plants. Therefore, the amino-group transfer of NA catalyzed by NAAT in the biosynthesis of MAs is a crucial step that distinguishes graminaceous plants from the other members of the plant kingdom (Kanazawa et al., 1994). Because of its important role in the biosynthesis of MAs under Fe-deficient conditions, many attempts have been made to purify NAAT to homogeneity, but these have achieved only partial success (Ohata et al., 1993; Kanazawa et al., 1994, 1995). In our previous work, the activity of two NAAT isozymes was seen on DEAE chromatography eluted with a linear KCl gradient, and these isozymes were named NAAT-I and NAAT-II (Kanazawa et al., 1995). In barley, NAAT-I is present in the roots of Fe-deficient plants, but is absent in the roots of Fe-sufficient plants. In contrast, a small amount of NAAT-II is present in Fe-sufficient roots, although its activity is also induced by Fe deficiency. In the present study, a third NAAT isozyme (NAAT-III) was found in the non-binding fraction from DEAE chromatography. Based on the partial amino acid sequences obtained from purified NAAT-III, we succeeded in isolating two distinct cDNAs and one genomic DNA clone encoding NAAT.
MATERIALS AND METHODS
Plant Materials
Barley (Hordeum vulgare L. cv Ehimehadakamugi No.1) was grown hydroponically according to the method of Kanazawa et al. (1994) in a greenhouse under natural light. When the third leaf emerged (20 d after germination), plants were transferred to either Fe-free culture solution (Fe-deficient plants) or culture medium containing 10−4 m Fe-EDTA (Fe-sufficient plants). Two weeks after transplanting, the roots were harvested, dipped in liquid nitrogen, crushed, and stored at −80°C until use.
Chemicals
Nicotianamine was synthesized using the method of Shioiri et al. (1997).
Preparation of NA-Affinity Columns
NA was bound to EAH-Sepharose 4B (Amersham-Pharmacia Biotech, Uppsala, Sweden) as follows: NA solution (20 μmol mL−1, pH 4.5) was added to the EAH-Sepharose 4B gel (NA solution:gel, 2:1) and mixed gently and thoroughly. The carbodiimide aqueous solution was added to the suspension to a final concentration of 0.1 m. The suspension was mixed gently, and the pH was adjusted to between 4.5 and 6.0 during the 1st h by the addition of 0.1 m NaOH. After that, the product was washed at least three times with 0.1 m acetate buffer (pH 4.0) containing 0.5 m NaCl. There was a final wash with distilled water.
Purification of NAAT
NAAT was purified according to the method of Kanazawa et al. (1995) until the hydroxylapatite column step (except for the addition of protease inhibitor E-64 to the extraction buffer). All of the experimental processes were conducted at 4°C except the enzyme assay. The frozen roots (150 g) from Fe-deficient plants were homogenized in 225 mL of extraction buffer using an electric juicer. The extraction buffer consisted of 0.2 m Tris/HCl (pH 8.0), 10 mm EDTA, 0.1 mm p-APMSF, 0.1 mm E-64, 10 mm DTT, 5% (v/v) glycerol, and 5% (w/v) insoluble PVP. The homogenate was centrifuged at 8,000g for 30 min, and the supernatant was applied to a column packed with gel (Butyl Toyopearl 650M, TOSOH, Tokyo), and hydroxylapatite gel (100–350 mesh, Nacalai Tesque, Kyoto). The active fraction eluted from the hydroxylapatite gel was concentrated using MOLCUT (Millipore, Tokyo) and the buffer was exchanged with 20 mm Tris/HCl (pH 8.0), 10 mm KCl, and 10 mm DTT. The sample was applied to a column packed with DEAE Sephacel (Pharmacia Biotech). The non-binding fraction from the DEAE Sephacel column was applied to a NA-Sepharose 4B column after buffer exchange. The buffer used for the NA-Sepharose 4B column consisted of 20 mm Tris/HCl (pH 8.0), 10 mm KCl, 1 mm DTT, and 5 mm EDTA. After washing the column with the same buffer, the absorbed proteins were eluted with a specific elution buffer (1 mm NA, 10 mm KCl, 20 mm Tris/HCl [pH 6.0]). The eluted sample was separated by two-dimensional gel electrophoresis (2D-SDS-PAGE) as described below.
NAAT Enzyme Assay
Protein was assayed using a kit (Bio-Rad Laboratories, Hercules, CA). Aliquots of samples containing 5 to 50 μg of protein were assayed for NAAT activity according to the method of Kanazawa et al. (1994). Four microliters of 0.25 m NaBH4 was then added to reduce the reaction product to deoxymugineic acid (Ohata et al., 1993), which was then analyzed by HPLC (Mori et al., 1987).
Gel Electrophoresis
The proteins were separated by 2D-SDS-PAGE following the method of O'Farrell (1975). To cover the pI range from 5.0 to 8.0, the gel contained both 1.6% (v/v) (pH 5.0—8.0) and 0.4% (v/v) (pH 3.0—10.0) Ampholines (Daiichi Chemicals, Tokyo). Protein extracts (100 μg) were subjected to IEF at 400 V for 15 h and then at 800 V for 1 h. After the gels equilibrated for 15 min in SDS-PAGE sample buffer (2.3% [w/v] SDS, 10% [w/v] glycerol, 5% [v/v] 2-ME, 62.5 mm Tris-HCl [pH 6.8], and 0.1% [w/v] bromphenol blue), they were loaded onto slab gels (10%, w/v) for SDS-PAGE in the second dimension. The gels were stained with 0.25% (w/v) Coomassie Brilliant Blue R-250 in a mixture of 50% (v/v) methanol and 10% (v/v) acetate, and destained in a solution of 50% (v/v) methanol and 10% (v/v) acetate.
Analysis of Partial Amino Acid Sequences
The amino acid sequences of the amino terminus and the CNBr digests of several proteins detected on the two-dimensional gels were determined. Chemical digestion with CNBr followed the method of Gross (1976) with slight modifications. Each protein was cut from the two-dimensional gels. Isolated proteins were eluted from the gels by soaking in a 10-fold volume of 70% (v/v) formic acid containing 1% (v/v) CNBr in a 1.5-mL microtube and incubated overnight at 4°C. The supernatant was collected, dried under reduced pressure, resuspended in SDS-PAGE sample buffer, and incubated overnight at room temperature. The peptides were separated by electrophoresis using Tricine SDS-PAGE (Schägger and Von Jagow, 1987) in 16.5% (w/v) acrylamide gels. The peptides were transferred onto a PVDF membrane by electroblotting and stained with Coomassie Brilliant Blue. Each band on the PVDF membrane was cut out, and the amino acid sequence was determined by automated Edman degradation on a gas-phase sequencer (model 477A protein sequencer and model 120A PTH analyzer, Applied Biosystems, Tokyo).
Amplifying cDNA Encoding NAAT-III Using Degenerate Primers
Oligonucleotides derived from the sequences of four peptides were synthesized and used as primers for PCR. cDNA was synthesized from poly(A+) RNA prepared from Fe-deficient barley roots by using a dT17 adaptor primer, 5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′, and reverse transcriptase. The amplification was successfully carried out using two-step PCR with Taq polymerase (Sawady, Tokyo) as described below. The PCR reaction buffer (50 μL) consisted of 1 mm Tris-HCl (pH 8.8), 50 mm KCl, 1.5 mm MgCl2, 0.2 mm dNTP, and 1.25 units of Taq polymerase. The first PCR was conducted using a sense degenerate primer mix (5′-GCIGTIGARTGGAAYTTYGCIMG-3′) derived from the amino-terminal amino acid sequence (AVEWNFAR) and 3′ adaptor primer (5′-GACTCGAGTCGACATCG-3′) using the cDNA as a template. The second PCR was conducted using the degenerate primer mix derived from the amino-terminal amino acid sequence and a degenerate antisense primer mix (5′-GCDATRTGICCRAAIACICC-3′) derived from one of the internal amino acid sequences (AIHGFVG) of the template that was in the reaction solution obtained from the first PCR. The resulting PCR products were separated on a 0.8% (v/v) agarose gel, eluted with a GeneClean II kit (Funakoshi, Tokyo), and subcloned into the pT7Blue T-vector (Novagen, Madison, WI). The DNA insert from the clones was sequenced by the dideoxynucleotide chain-termination method using a dideoxy sequencing kit (BcaBEST, TaKaRa Shuzo, Shiga, Japan), and was also used as a probe to screen a cDNA library.
Isolating cDNA Encoding NAAT-III
A pYH23 cDNA library was prepared from poly(A+) RNA from Fe-deficient barley roots. pYH23 is an expression vector for yeast, and has the ADH (alcohol dehydrogenase) promoter driving expression of the introduced cDNAs. The cDNA library was screened by colony hybridization using the PCR product (probe I, see Fig. 2). The probe was labeled with [α-32P]dATP using a random-primer labeling kit (version 2, TaKaRa). The labeled DNA was purified in a nick column (Pharmacia). The cloned cDNAs were sequenced with a DNA sequencer (A373, Applied Biosystems) using the following three kits: a dye terminator cycle sequencing kit (Ready Reaction, Perkin Elmer, Foster City, CA), a fluorescent labeled primer cycle sequencing kit (Thermo Sequence, Amersham Life Science, Tokyo), and a dideoxy sequencing kit (BcaBEST, TaKaRa).
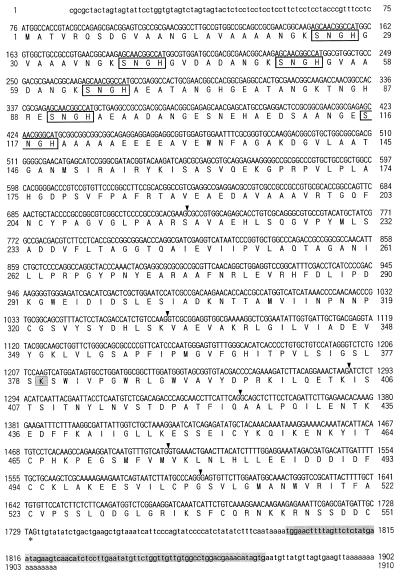
Figure 2.
Nucleotide and deduced amino acid sequences of the cDNA encoding naat-A (a in Fig. 1) from barley. The predicted amino acid sequence starting with the putative initiator Met is shown below the nucleotide sequence. The stop codon is indicated by an asterisk (*). The directly determined amino acid sequences of the amino-terminal and CNBr digests of protein a are underlined. Arrows indicate the positions of the PCR primers. Probe I used for additional studies is the sequence between the two primers indicated by arrows. The specific probe (probe-II) for the 3′-terminal sequence used for genomic Southern hybridization and the putative PLP-binding Lys residue are shaded. There are three core SNGH sequences in the N-terminal region (boxed). Arrowheads show the six introns deduced from the genomic DNA sequence in Figure 8.
Northern-Blot Analysis
Total RNA was isolated from the roots and leaves of both Fe-sufficient and Fe-deficient barley according to the procedure of Naito et al. (1988). The RNA was denatured and electrophoresed on 1.2% (v/v) agarose gels containing 5% (v/v) formaldehyde, blotted onto nylon membranes (Hybond-N+, Amersham), and hybridization was conducted using total naat-A cDNA or naat-B cDNA as probes following the instruction manual for the direct nucleic acid labeling and detection systems (LCA, Amersham Life Science).
Southern-Blot Analysis
Genomic DNA was prepared from barley leaves (Murray and Thompson, 1980). DNA samples were digested with BamHI and HindIII and separated by electrophoresis on 0.8% (v/v) agarose gels. The gels were blotted to Hybond-N+ membranes, prehybridized for 1 h, and hybridized overnight with 32P-labeled 3′-specific probes for naat-A or naat-B (see Figs. 3 and 4). Prehybridization and hybridization were conducted at 65°C in a mixture of 5× SSPE, 4× Denhardt's solution, 0.1% (v/v) SDS, and 100 μg mL−1 denatured salmon-sperm DNA. After hybridization, the filters were washed with 2× SSPE and 0.1% (v/v) SDS at 65°C for 10 min and then twice at 42°C for 5 min, and visualized with a bio-imaging analyzer (BAS 2000, Fujix, Tokyo).
Figure 3.
Nucleotide and deduced amino acid sequences of the cDNA encoding naat-B (NAAT-A isozyme). The stop codon is indicated by an asterisk (*). The specific probe (probe III) for the 3′-terminal sequence used for genomic Southern hybridization and the putative PLP-binding Lys residue are shaded. There are six core SNGH sequences in the N-terminal region (boxed). Arrowheads show the six introns deduced from the genomic DNA sequence in Figure 8.
Figure 4.
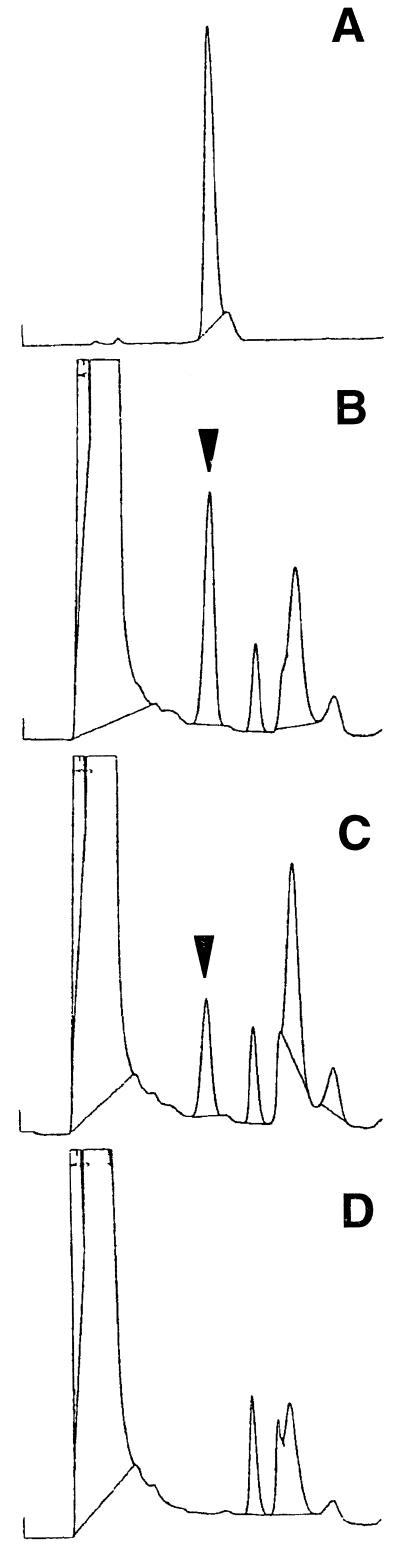
NAAT activity of naat-A and naat-B expressed in the yeast. naat-A and naat-B were introduced into wild-type S. cerevisiae, and NAAT activity was determined from the amount of deoxymugineic acid detected with HPLC. The arrowhead indicates the retention time for deoxymugineic acid. A, Deoxymugineic acid as a standard; B, naat-A; C, naat-B; D, pYH23 (control). The specific activities (pkat mg−1 protein) of NAAT-A, NAAT-B, and pYH23 expressed in yeast are 0.36 ± 0.01, 0.13 ± 0.03, and not detected, respectively. Each value represents the mean of four replicates.
Construction of Yeast Expression Vector pYH23
The yeast expression vector (pYH23) was constructed in the following manner. The plasmid YEplac181 (Gietz and Sugino, 1988) was cleaved with XbaI and EcoRI, blunted with T4 DNA polymerase, and re-ligated to remove most of the multiple cloning sites. The HindIII site was similarly deleted. To construct pYH23, the plasmid was digested with SphI and the 728-bp fragment from pVT100U (Vernet et al., 1987) containing the ADH1 expression cassette was inserted. To insert a NotI site in the multicloning site of the cassette, the plasmid was cleaved with BamHI and blunted with T4 DNA polymerase, and the phosphorylated linker AGCGGCCGCT (TaKaRa Shuzo) was inserted. The new plasmid, pYH23, has HindIII, PvuII, PstI, XhoI, SstI, XbaI, and NotI sites in the ADH1 expression cassette. Although the ADH promoter is generally considered to be constitutive, expression from this promoter is actually repressed as much as 10-fold on non-fermentable carbon sources (Denis et al., 1983).
Yeast Transformation and NAAT Assay
Yeast (Saccharomyces cerevisiae wild type CM3260, MATalpha trp1-63 leu2-3, 112 gcn4-101 his3-60 ura3-52 FRE1-HIS3::URA3 ctr1-3 [Dancis et al., 1994]) were transformed as described by Schiestl and Gietz (1989) using naat-A cDNA or naat-B cDNA in pYH23 vector. The empty vector was used as a control. All experiments were carried out in synthetic defined medium containing Glc (Sherman, 1991), so that expression of NAAT protein in yeast cells was constitutive.
Transformed yeast was cultured at 30°C for 24 h, centrifuged at 2,000 rpm for 5 min, and the pellet was washed in sterile distilled water. The protein was extracted from the washed pellet of transformed yeast by crushing the cells in extraction buffer (0.2 m Tris/HCl [pH 8.0], 10 mm EDTA, 5% [v/v] glycerol, 50 mm KCl, 1 mm p-APMSF, 3 mm DTT, 0.1 μg mL−1 chymostatin, 0.5 μg mL−1 leupeptin, and 7.2 μg mL−1 E-64) using glass beads. The extract was centrifuged at 2,500 rpm for 5 min, and the supernatant was centrifuged again at 14,500 rpm for 30 min. The yeast extracts were assayed for NAAT activity using the method described above.
Isolation of Genomic Clones of naat-A and naat-B from the λFIX II Barley Genomic Library
A λFIX II genomic barley (var Igri) library was obtained commercially (Stratagene, La Jolla, CA). Plaques (200,000) were screened using naat-A cDNA as a probe and three clones were selected by plaque hybridization. According to the restriction enzyme mapping, these three clones contained almost the same genomic DNA fragments. The insert (approximately 11.2 kb) was separated into four fragments by EcoRI and NotI, and subcloned into pBluescriptII SK(+) vector. All fragments were sequenced using automated DNA sequencers (DSQ-2000L, Shimadzu Scientific Instruments, Tokyo; ABI PRISM 310 genetic analyzer, Perkin Elmer).
RESULTS
Purification of NAAT
The enzyme purification procedure is summarized in Table I. Two NAAT isozymes have already been isolated from Fe-deficient barley (NAAT-I and NAAT-II) using DEAE chromatography with a linear KCl gradient elution (Kanazawa et al., 1995). In addition to these two isozymes, we noticed that NAAT activity was also present in the fraction passing through the DEAE column from the roots of Fe-deficient barley only. Initially, we thought that the NAAT activity in this fraction was NAAT-I and NAAT-II that failed to bind to the DEAE Sephacel gel. However, after the fraction passing through the DEAE column was re-applied to the DEAE column, NAAT activity was again detected in the non-binding fraction. In addition, the specific enzyme activity of the NAAT in this fraction (non-bound fraction of DEAE in Table II) was increased by the NA-affinity chromatography step with various ligands more efficiently than the binding fraction (Table II). Therefore, we assumed that this fraction contained another NAAT isozyme and named it NAAT-III.
Table I.
Summary of enzyme purification
| Purification Step | Total Protein | Specific Activity | Yield | Purification |
|---|---|---|---|---|
| mg | pkat mg−1 | % | -fold | |
| Crude extract | 523 | 9.98 | 100 | 1 |
| Butyl Toyopearl | 160 | 34.3 | 105 | 3.44 |
| Hydroxylapatite | 33.8 | 254 | 164 | 25.5 |
| DEAE Sephacel | 7.09 | 261 | 35.3 | 26.2 |
| NA-Sepharose | 0.84 | 1298 | 20.8 | 130 |
Table II.
Increase in the specific activity of NAAT-III by affinity chromatography
| Binding Fraction | Non-Bound Fraction | |||||||
|---|---|---|---|---|---|---|---|---|
| Ligand | NA | α-KG | α-KG | A-2-C | NA | α-KG | α-KG | A-2-C |
| Solute of specific elution buffer | NA | NA | α-KG | A-2-C | NA | NA | α-KG | A-2-C |
| pkat mg−1 protein | ||||||||
| Before affinity chromatography | 519 | 261 | ||||||
| After affinity chromatography | 96 | 599 | 706 | 575 | 1298 | 601 | 926 | 868 |
Binding and non-bound fractions of DEAE were applied to affinity chromatography columns with three ligands and eluted by four specific elution buffers. For the ligands, NA was used as a substrate, 2-oxoglutarate (α-KG) as an amino group acceptor, and azetidine-2-carboxylic acid (A-2-C) as a substrate analog. The concentration of the solute in the specific elution buffers was 1 mm.
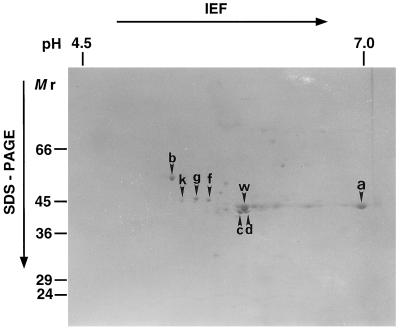
In the NA-affinity chromatography step, the specific activity of NAAT-III was increased more efficiently when using NA to elute the protein rather than other ligands. Even after NA-affinity chromatography, however, several protein spots were still detected on 2D-SDS-PAGE (Fig. 1). Eight spots (a, b, c, d, f, g, k, and w, in Fig. 1) that were candidates for NAAT protein were extracted from the gel and their partial amino acid sequences were determined. The N termini of proteins c and d were blocked. Of the spots for which the N-terminal sequences could be analyzed, proteins b, f, and g did not match any N-terminal amino acid sequences in the database. Protein k was identified as enolase. The partial amino acid sequence of the peptides obtained by CNBr digestion of protein w was highly homologous to formate dehydrogenase (EC 1.21.2.). The gene encoding barley formate dehydrogenase has been identified, isolated, and characterized previously (Suzuki et al., 1998).
Figure 1.
2D-SDS-PAGE of the eluted fraction from NA-affinity chromatography. The arrowheads indicate candidate proteins for NAAT.
With protein a, a homology search failed to find any matches for the N-terminal sequence GHAAAAAVEWNFARG, and an internal sequence obtained with CNBr digestion, VKLNLHLLEEIHDDIDF, but two other internal peptides, VIINPNNP?G and GVFGHIA, showed a high homology to Ala aminotransferase and Glu dehydrogenase, respectively. Both Ala aminotransferase and Glu dehydrogenase are pyridoxal phosphate (PLP)-dependent enzymes. Since NAAT is also a PLP-dependent enzyme (Shojima et al., 1990), we thought that protein a was most likely NAAT-III.
Isolation of cDNA Clones for NAAT
Based on the N-terminal and internal amino acid sequences of protein a, one sense amino-terminal oligonucleotide and three sense and three antisense primers for each internal amino acid sequence were synthesized. These primers were used for PCR using cDNA from Fe-deficient barley roots as the template. The combination of the two primers used in the second PCR amplified a 722-bp DNA fragment encoding the partial amino acid sequence (VIINPNNP) of protein a (Fig. 2). To isolate a full-length cDNA clone, a cDNA library prepared from poly(A+) RNA from Fe-deficient barley roots was screened with this 722-bp fragment. Two clones were isolated after screening 5 × 104 colonies. The predicted amino acid sequence of one clone (naat-A) contained all of the partial amino acid sequences of protein a, while another clone (naat-B) encoded a protein with very similar amino acid sequences. The nucleotide and deduced amino acid sequences of naat-A and naat-B cDNA are shown in Figures 2 and 3, respectively (DDBJ accession nos. D88273 [naat-A] and AB005788 [naat-B]).
The naat-A cDNA was 1,660 nucleotides long, including 26 adenines in a poly(A+) tail. We assume that the start codon corresponded to the first Met codon located at nucleotide positions 62 to 64 of the naat-A cDNA. The nucleotide sequence around the initiation codon matches the consensus sequence for eukaryotic translation initiation sites (Kozak, 1987). The naat-A cDNA contains 61 nucleotides in the 5′-noncoding region, 1,386 nucleotides within the coding region, and 187 nucleotides in the 3′-noncoding region that precedes the poly(A+) tail. The open reading frame (ORF) encodes a polypeptide of 461 amino acid residues. Direct amino acid sequences of CNBr digests of protein a completely matched segments of the amino acid sequence deduced from the naat-A cDNA nucleotide sequence. However, protein a lacked 32 amino acids from the N-terminal end of the predicted amino acid sequence. The molecular mass of the polypeptide (from the actual N-terminal amino acid, G-33, to the last amino acid residue) was calculated to be 46.6 kD and the pI 6.8. These values were similar to those estimated from the position by 2D-SDS-PAGE (Fig. 1). The N-terminal 32 amino acid residues after the first Met residue are likely a signal peptide that is cleaved just before G-33 during protein maturation (Fig. 2).
The naat-B cDNA is 1,910 nucleotides long, including 15 adenines in a poly(A+) tail (Fig. 3). We assume that the first Met codon located at nucleotide positions 76 to 78 is the start codon, because there was no other Met codon upstream from this position in the genomic DNA sequence (Fig. 7A). The naat-B cDNA contained an ORF of 1,653 nucleotides encoding a polypeptide of 551 amino acid residues. The molecular mass of the polypeptide (from the first Met to the last Cys residue) was calculated to be 58.1 kD and the pI 5.5.
Figure 7.
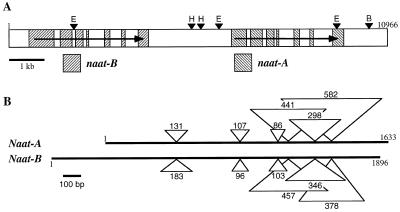
Genomic DNA sequence of naat-A and naat-B in a tandem array. A, Organization of the genomic clone (11.0 kb) that contains both the naat-B and naat-A genes (arrow points from the 5′ to the 3′ end). The naat-A and naat-B genes each contain seven exons (hatched boxes). Arrowheads show the restriction sites for (E) EcoRI, (H) HindIII, and (B) BamHI. B, Organization of the naat-A and naat-B genes. Triangles show the six intron insertion positions in each gene. The numbers of nucleotides in each intron are indicated.
NAAT Activity in Yeast Transformed with naat-A or naat-B
To confirm that naat-A and naat-B actually encode NAAT enzymes, they were introduced into S. cerevisiae and the NAAT activity was measured (Fig. 4). NAAT activity was detected in the yeast transformed with naat-A or naat-B, while NAAT activity was not detected in yeast transformed with the empty vector (pYH23). In the yeast, the specific enzyme activity of naat-A was greater than that of naat-B, although these specific activities were far less than that of the crude extract of Fe-deficient barley roots (see Table I). These results demonstrate that both naat-A and naat-B encode NAAT enzymes and that NAAT-A is NAAT-III (protein a in Fig. 1).
Expression of naat-A and naat-B in Barley Roots
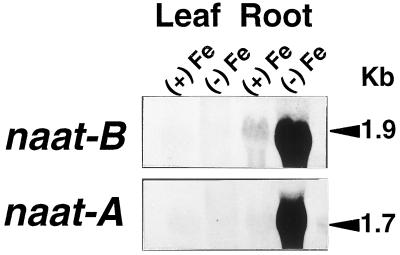
To examine the expression of naat-A and naat-B, we performed northern-blot analysis on total RNA from Fe-deficient and Fe-sufficient leaves and roots, as shown in Figure 5. When naat-A cDNA or naat-B cDNA were used as probes for northern analysis, the naat-A probe hybridized preferentially with a 1.7-kb mRNA and the naat-B probe preferentially hybridized with 1.9-kb mRNA. In Fe-deficient roots, transcripts of both naat-A and naat-B were abundant, while in Fe-sufficient roots, naat-A was not detectable and a trace amount of the 1.9-kb band of naat-B was detected. The results indicate that Fe deficiency induces the expression of both naat-A and naat-B, although there is a basal level of naat-B expression in Fe-sufficient roots (Fig. 5).
Figure 5.
Northern analysis of naat-A and naat-B in barley roots. RNA was extracted from Fe-deficient (−Fe) or Fe-sufficient (+Fe) roots and leaves. Each lane was loaded with 20 μg of RNA. The RNA was blotted onto a nylon membrane and hybridized with a total length of naat-A cDNA or naat-B cDNA.
Southern-Blot Analysis
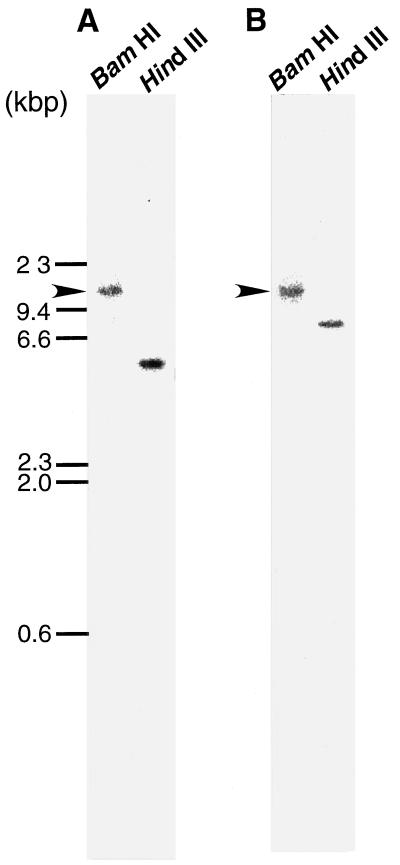
Total genomic DNA was digested with two restriction enzymes, BamHI and HindIII, and the resulting DNA fragments were hybridized with a probe common to naat-A and naat-B (probe-I in Fig. 2). However, we only obtained a smeared image (data not shown; discussed below). We then hybridized the same membrane with the probes specific to naat-A cDNA (probe-II in Fig. 2) or naat-B cDNA (probe-III in Fig. 3). As shown in Figure 6, hybridization with these specific probes yielded a single band in both the BamHI and HindIII digests. The BamHI band in both genes seemed to be the same size. These results suggest that one copy each of naat-A and naat-B are localized close together on the same chromosome.
Figure 6.
Southern hybridization analysis of naat-A and naat-B. Genomic DNA from barley was digested with BamHI and HindIII and then blotted onto a nylon membrane and hybridized with 32P-labeled specific probes: probe II in Figure 3 and probe III in Figure 4 for the 3′-terminal sequences of naat-A (A) and naat-B (B), respectively. Arrows indicate bands of the same size of BamHI restriction fragments of naat-A and naat-B.
Genomic DNA Sequence of naat-A and naat-B
Using naat-A cDNA as a probe, a barley genomic library was screened. Positive clones were obtained and an 11.2-kb DNA fragment was completely sequenced. This 11.2-kb fragment contained both naat-B and naat-A in this order (Fig. 7A), separated by 3.0 kb. Compared with the cDNA, both naat-A and naat-B have six introns at the same positions (Figs. 2, 3, and 7B). The positions of the inserted introns are indicated with arrows in the cDNA sequences of naat-A (Fig. 2) and naat-B (Fig. 3). The genomic DNA data will appear in the DDBJ/EMBL/GenBank under the accession no. AB024006.
DISCUSSION
After several column chromatography steps, we purified a third isoform of NAAT, NAAT-III, which was induced only in Fe-deficient barley roots (Table I). Based on the partial amino acid sequences of protein a, a candidate for NAAT-III seen on 2D-SDS-PAGE (in Fig. 1), we isolated two distinct cDNAs, naat-A (Fig. 2) and naat-B (Fig. 3). The expression of these cDNAs in transformed yeast confirmed that they both encoded NAATs (Fig. 4).
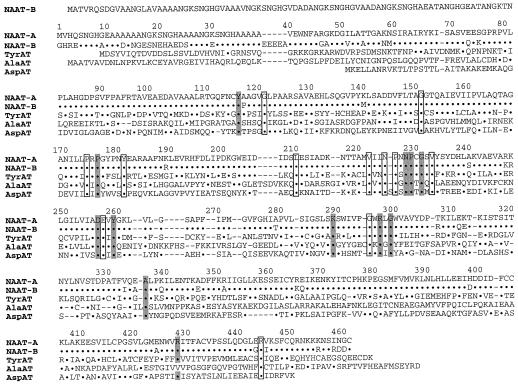
The deduced amino acid sequence of naat-A is 88.1% identical to the deduced amino acid sequence of naat-B (Fig. 8). The N-terminal of NAAT-B is longer than that of NAAT-A, and their homology is low at both the 5′- and 3′-terminal ends. The cDNAs of both naat-A and naat-B have repeated SNGH amino acid sequences at their N termini; there are three repeats in naat-A and six in naat-B. The N-terminal amino acid of the mature NAAT-A protein is G-33, which is located in the third SNGH sequence from the 5′-terminal of the coding region. The repeated SNGH sequence has not been reported previously and its function is not known.
Figure 8.
Aligned amino acid sequences of NAAT-A, NAAT-B, TyrAT (P04694; Tyr aminotransferase from rat) (Ernest et al., 1977), AlaAT (P52894; Ala aminotransferase from barley) (Good and Crosby, 1989), and Asp aminotransferase (M59430; Asp aminotransferase from Bacillus sp.) (Taniguchi et al., 1995). The residues are numbered according to the deduced amino acid sequence of naat-A (NAAT-A). Functionally or structurally important residues following the results in Mehta et al. (1989) are shadowed. Residues identical to NAAT-A are represented with dots. Identical residues in the five aminotransferase sequences are boxed. Gaps, represented by dashes, have been inserted to achieve maximum homology.
The presence of a signal peptide at the N terminus of NAAT-A suggests that NAAT-A is targeted to the rough endoplasmic reticulum (ER). Although the actual N terminus of mature NAAT-B protein was not determined, the hydropathy profile of the deduced amino acid sequence of NAAT-B predicted the presence of a signal peptide at the N terminus. In addition, NAAT-A and NAAT-B contain two Asn residues (N-217 and N-324 in Fig. 2 and N-148 and N-307 in Fig. 3) that are potential glycosylation sites (Keller et al., 1992). Therefore, both NAATs may be imported into the rough ER. However, they do not have an ER retention signal (KDEL) in their C termini.
Previously, we reported the appearance of distinct vesicles in the root cells of Fe-deficient barley before sunrise, just before the diurnal secretion of MAs. These vesicles were covered with ribosomes and were thought to originate from the rough ER (Nishizawa and Mori, 1987). An electron microscope-autoradiography study showed that the 14C-labeled Met supplied to the Fe-deficient barley after sunset (when MA secretion has ceased but MA production continues) was incorporated into these vesicles. Based on these results, we postulated that MAs are produced in these distinct vesicles and stored until they are secreted. Of the enzymes that participate in MA biosynthesis, NAS works efficiently at pH 9.0 (Higuchi et al., 1994) and the optimum pH of NAAT is between 8.5 and 9.0 (Kanazawa et al., 1994). These results also suggest that neither NAS and NAAT is present in the cytoplasm but, rather, in some compartment within the cells. It is conceivable that NAATs are sorted in the rough ER and catalyze the amino transfer of NA in these particular vesicles derived from the rough ER. Interestingly, the deduced amino acid sequence of NAS does not have a signal peptide at the N-terminal that would allow it to be sorted into the rough ER, but it does have two potential transmembrane domains, and computer research predicts that it would be sorted into the ER (Higuchi et al., 1999). Therefore, two enzymes involved in MA synthesis might be present in the specific vesicles derived from the rough ER. The precise localization of NAS and NAAT should be clarified by immunocytochemistry in future studies.
The deduced amino acid sequence of naat-A cDNA (NAAT-A) was 33.3% similar to the amino acid sequence of Tyr aminotransferase from rat liver (accession no. P04694) (Fig. 8). In addition, NAAT-A showed 25.5% and 21.7% similarity to the amino acid sequences of Ala aminotransferase from barley (accession no. P52894) and Asp aminotransferase from Bacillus sp. (accession no. M59430), respectively. None of these similarities to the three aminotransferases is very high. However, a comparison between NAAT-A, NAAT-B, and the other three aminotransferases shows that they have conserved regions that may be functionally or structurally important residues. Asp aminotransferase is the most extensively examined pyridoxal enzyme. X-ray crystallographic characterization of Asp aminotransferase, along with comparison of the amino acid sequences of Asp aminotransferase from various species and those of other aminotransferases, have revealed important conserved amino acid residues (Mehta et al., 1989).
The important residues are as follows: Lys-289 is a Schiff base with PLP. Tyr-116, Asn-228, Asp-256, Tyr-260, and Arg-297 form hydrogen bonds to PLP. Pro-176 and Pro-229 are both in the cis conformation. Gly-231 participates in a turn of the polypeptide chain located at the domain interface. Arg-428 forms a salt bridge/hydrogen bond with the α-carboxylate group of the substrate. Gly-299 and Ala-336 are conserved, but their significance is unknown. Comparison of the amino acid homology between the five enzymes in Figure 8 identifies Gly-121, Gly-151, Pro-174, Tyr-181, Leu-210, Val-222, Asn-225, Pro-226, Val-233, Glu-257, Gly-295, and Arg-444 as potentially important amino acid residues for transamination or conformation. NAAT is a PLP-dependent enzyme (Shojima et al., 1990), and it is thought that NAAT binds PLP to the lysyl residue (Lys-289) via an aldimine bond, as is the case with other pyridoxal enzymes (Pfleiderer et al., 1968). Thus, the conservation of these important residues in NAAT-A and NAAT-B is consistent with NAAT being a PLP-dependent enzyme.
Although we have not presented the data, we isolated a 5′-terminal truncated cDNA clone that encoded a protein missing 163 amino acid residues at the N-terminus of NAAT-A. This clone did not show NAAT activity when introduced into yeast, although its deduced amino acid sequence contained the putative PLP-binding Lys residue and the surrounding amino acid residues. The lack of NAAT activity may be due to a missing amino acid residue (Tyr-116). These assumptions of the functional or structural importance of the conserved residues should be confirmed by x-ray characterization of NAAT-A and NAAT-B.
We confirmed that naat-A encodes NAAT-III. On the other hand, it is likely that naat-B encodes an isoform of NAAT-III, possibly one of the two isoforms (NAAT-I or NAAT-II) described by Kanazawa et al. (1995). naat-A was only expressed in Fe-deficient roots, whereas naat-B had a low basal level of expression in Fe-sufficient roots and expression was also induced under Fe deficiency. Kanazawa et al. (1995) reported that both NAAT-I and NAAT-II are induced by Fe deficiency, although NAAT-II is also detectable in Fe-sufficient plants. Therefore, naat-B is a candidate as the gene for NAAT-II (NAAT-B). NAAT-B may contribute to the production of a basal level of MAs under non-Fe-stress conditions, whereas the induction of both NAAT-A and NAAT-B may increase the level of MAs under Fe starvation. Takagi et al. (1984) reported that even under Fe-sufficient hydroponic conditions, barley always secretes a low amount of MA with some perturbed pattern.
There are several reports that Zn deficiency also induces MA production in wheat (Triticum aestivum cv Aroona and cv Durati) (Cakmak et al., 1994; Walter et al., 1994), although the induction after Zn deficiency is delayed, and less phytosiderophore is produced than under Fe deficiency. Although we did not measure the release of MAs with Zn-deficiency treatment in barley, Zn may regulate naat-A and naat-B as well as Fe. Therefore, a time-course study of the expression of naat genes under various nutritional stress conditions is also necessary.
The smeared image (data not shown) in the Southern analysis using the common probe I (Fig. 2) might have been caused by the existence of other related genes (possibly encoding other PLP-requiring enzymes) in the genome. Because the washings of the Southern blot were not very stringent, it did not allow detection of any such genes. The single positive band in the BamHI column (arrows in Fig. 6) was the same size for both naat-A and naat-B, suggesting that the two genes are very near to each other. This was confirmed by cloning a genomic DNA that contained the full-length naat-A and naat-B genes in a tandem arrayed with only one BamHI site (Fig. 7).
We cloned two naat genes as an important step toward our ultimate objective of developing rice, sorghum, and corn that are tolerant to Fe-deficient conditions. The gene encoding NAAT-I has not yet been cloned. Identification of the cis element(s) responsive to Fe deficiency in naat genes is the next step in our study.
ACKNOWLEDGMENT
We thank Dr. Emmanuel Delhaize of Commonwealth Scientific and Industrial Research Organization, Australia, for the critically review and editing for English of this manuscript.
LITERATURE CITED
- Cakmak I, Gülüt KY, Marschner H, Graham RD. Effect of zinc and iron deficiency on phytosiderophore release in wheat genotypes differing in zinc deficiency. J Plant Nutr. 1994;17:1–17. [Google Scholar]
- Dancis A, Yuan DS, Haile D, Askwith C, Eide D, Moehl EC, Kaplan J, Klausner RD. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell. 1994;76:393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- Denis CL, Ferguston J, Young ET. mRNA levels for the fermentable alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. J Biol Chem. 1983;25:1165–1171. [PubMed] [Google Scholar]
- Ernest MJ, Chen CL, Feigelson P. Induction of tyrosine aminotransferase synthesis in isolated liver cell suspensions. J Biol Chem. 1977;252:6783–6791. [PubMed] [Google Scholar]
- Fushiya S, Takahashi K, Nakatsuyama S, Sato Y, Nozoe S, Takagi S. Co-ocurrence of nicotianamine and avenic acid in Avena sativa and Oryza sativa. Phytochemistry. 1982;21:1907–1908. [Google Scholar]
- Gietz RD, Sugino A. New yeast and Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;72:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Good AG, Crosby WL. Anaerobic induction of alanine aminotransferase in barley root tissue. Plant Physiol. 1989;90:1305–1309. doi: 10.1104/pp.90.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross E. The cyanogen bromide reaction. Methods Enzymol. 1976;11:238–255. [Google Scholar]
- Higuchi K, Kanazawa K, Nishizawa NK, Chino M, Mori S. Purification and characterization of nicotianamine synthase from Fe-deficient barley roots. Plant Soil. 1994;165:173–179. [Google Scholar]
- Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S. Cloning of nicotianamine synthase, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 1999;119:471–480. doi: 10.1104/pp.119.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa K, Higuchi K, Nishizawa NK, Fushiya S, Chino M, Mori S. Nicotianamine aminotransferase activities are correlated to the phytosiderophore secretions under Fe-deficient conditions in Gramineae. J Exp Bot. 1994;45:1903–1906. [Google Scholar]
- Kanazawa K, Higuchi K, Nishizawa NK, Fushiya S, Mori S. Detection of two distinct isozymes of nicotianamine aminotransferase in Fe-deficient barley roots. J Exp Bot. 1995;46:1241–1244. [Google Scholar]
- Kawai S, Takagi S, Sato Y. Mugineic acid-family phytosiderophores in root-secretions of barley, corn and sorghum varieties. J Plant Nutr. 1988;11:633–642. [Google Scholar]
- Keller DJ, Kreibich G, Gilmore R. Oligosaccharyltransferase activity is associated with a protein complex composed of ribophorins I and II and a 48 kD protein. Cell. 1992;69:55–65. doi: 10.1016/0092-8674(92)90118-v. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Nomoto K. Two related biosynthetic pathway of mugineic acids in gramineous plants. Plant Physiol. 1993;102:373–378. doi: 10.1104/pp.102.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Takeda S, Chong YC, Iwashita T, Matsumoto H, Takeda K, Nomoto K. Genes controlling hydroxylations of phytosiderophores are located on different chromosomes in barley (Hordeum vulgare L.) Planta. 1999;270:16549–16554. [Google Scholar]
- Marschner H, Römheld V, Kissel M. Localization of phytosiderophore release and of iron uptake along intact barley roots. Physiol Plant. 1987;71:157–172. [Google Scholar]
- Mehta PK, Hale TI, Christen P. Evolutionary relationships among aminotransferases—tyrosine aminotransferase, histidinol-phosphate aminotransferase, and aspartate aminotransferase—are homologous proteins. Eur J Biochem. 1989;186:246–253. doi: 10.1111/j.1432-1033.1989.tb15202.x. [DOI] [PubMed] [Google Scholar]
- Mihashi S, Mori S. Characterization of mugineic-acid-Fe transporter in Fe-deficient barley roots using the multi-compartment transport box method. Biol Metals. 1989;2:146–154. [Google Scholar]
- Mino Y, Ishida T, Ota N, Inoue M, Nomoto K, Takemoto T, Tanaka H, Sugiura Y. Mugineic acid-iron (III) complex: characterization and implication for absorption and transport of iron in gramineous plants. J Am Chem Soc. 1983;105:4671–4676. [Google Scholar]
- Mori S, Nishizawa N. Methionine as a dominant precursor of phytosiderophores in Gramineae plants. Plant Cell Physiol. 1987;28:1081–1092. [Google Scholar]
- Mori S, Nishizawa N. Identification of barley chromosome no. 4, possible encoder of genes of mugineic acid synthesis from 2′-deoxymugineic acid using wheet-barley addition lines. Plant Cell Physiol. 1989;30:1057–1060. [Google Scholar]
- Mori S, Nishizawa NK, Fujigaki J. Identification of rye chromosome 5R as a carrier of the genes for mugineic acid synthase and hydroxymugineic acid synthase using wheat-rye addition lines. Jpn J Genet. 1990;65:343–352. [Google Scholar]
- Mori S, Nishizawa NK, Hayashi H, Chino M, Yoshimura E, Ishihara J. Why are young rice plants highly susceptible to iron deficiency? Plant Soil. 1991;130:143–156. [Google Scholar]
- Mori S, Nishizawa N, Kawai S, Sato S, Takagi S. Dynamic state of mugineic acid and analogous phytosiderophores in Fe-deficient barley. J Plant Nutr. 1987;10:1003–1011. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito S, Dube P-H, Beachy R-N. Differential expression of conglycinin alpha′ and beta subunit genes in transgenic plants. Plant Mol Biol. 1988;11:109–124. doi: 10.1007/BF00015664. [DOI] [PubMed] [Google Scholar]
- Nishizawa NK, Mori S. The particular vesicle appearing in barley root cells and its relation to mugineic acid secretion. J Plant Nutr. 1987;10:1012–1020. [Google Scholar]
- Noma M, Noguchi M. Occurrence of nicotianamine in higher plants. Phytochemistry. 1976;15:1701–1702. [Google Scholar]
- Nomoto K, Sugiura Y, Takagi S. Mugineic acid, studies on phytosiderophores: microbes, plants and animals. In: Winkelmann G, Van der Helm D, Nielands JB, editors. Iron Transport in Microbes, Plants and Animals. Weinheim, Germany: VCH Publishers; 1987. pp. 401–425. [Google Scholar]
- Nomoto K, Yoshioka H, Arima M, Fushiya S, Takagi S, Takemoto T. Structure of 2′-deoxymugineic acid, a novel amino acid possessing an iron-chelating activity. Chimia. 1981;7:249–250. [Google Scholar]
- O'Farrell PH. High resolution two-dimentional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ohata T, Kanazawa K, Mihashi S, Nishizawa NK, Fushiya S, Nozoe S, Chino M, Mori S. Biosynthetic pathway of phytosiderophores in iron-deficient graminaceous plants: development of assay system for the detection of nicotianamine aminotransferase activity. Soil Sci Plant Nutr. 1993;39:745–749. [Google Scholar]
- Pfleiderer G, Stock A, Ortanderl F, Mella K. Uber die coenzymbindung in der glutamat-pyruvat-transaminase aus schweineherz. Eur J Biochem. 1968;5:18–23. doi: 10.1111/j.1432-1033.1968.tb00330.x. [DOI] [PubMed] [Google Scholar]
- Römheld V, Marschner H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol. 1986;70:175–180. doi: 10.1104/pp.80.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph A, Becker R, Scholz G, Prochazka Z, Toman J, Macel T, Herout V. The occurrence of the amino acid nicotianamine in plants and microorganisms: a re-investigation. Biochem Physiol Pflanzen. 1985;180:557–563. [Google Scholar]
- Schägger H, Von Jagow G. Tricine-sodium dodecyl sulfatepolyacrylamide gel electrophoresis for the separation of proteins in the range from 1 kDa to 100kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schiestl PH, Gietz RD. High efficiency transformation of intact yeast cells using single standard nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shioiri T, Irako N, Sakakibara S, Matsuura F, Hamada Y. A new efficient synthesis of nicotianamine and 2′-deoxy mugineic acid. Heterocycles. 1997;44:519–530. [Google Scholar]
- Shojima S, Nishizawa NK, Fushiya S, Nozoe S, Irifune T, Mori S. Biosyntheisis of phytosiderophores. Plant Physiol. 1990;93:1497–1503. doi: 10.1104/pp.93.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura Y, Tanaka H, Mino Y, Ishida T, Ota N, Inoue M, Nomoto K, Yoshioka H, Takemoto T. Structure, properties, and transport mechanism of iron (III) complex of mugineic acid, a possible phytosiderophore. J Am Chem Soc. 1981;103:6979–6982. [Google Scholar]
- Suzuki K, Itai R, Suzuki K, Nakanishi H, Nishizawa NK, Yoshimura E, Mori S. Formate dehydrogenase, an enzyme of anaerobic metabolism, is induced by Fe deficiency in barley roots. Plant Physiol. 1998;116:725–732. doi: 10.1104/pp.116.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S. Naturally occurring iron-chelating compounds in oat- and rice-root washing. I. Activity measurement and preliminary characterization. Soil Sci Plant Nutr. 1976;22:423–433. [Google Scholar]
- Takagi S, Nomoto K, Takemoto S. Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. J Plant Nutr. 1984;7:469–477. [Google Scholar]
- Takemoto T, Nomoto K, Fushiya S, Ouchi R, Kusano G, Hikino H, Takagi S, Matsuura Y, Kakudo M. Structure of mugineic acid, a new amino acid possessing an iron-chelating activity from root washing of water-cultured Hordeum vulgare. Proc Jpn Acad. 1978;54:469–473. [Google Scholar]
- Taniguchi M, Kobe A, Kato M, Sugiyama T. Aspartate aminotransferase isozymes in Planicum miliaceum L., an NAD-Malic enzyme-type C4 plant: structures, and expression patterns. Arch Biochem Biophys. 1995;318:295–306. doi: 10.1006/abbi.1995.1233. [DOI] [PubMed] [Google Scholar]
- Vernet T, Dignard D, Thomas DV. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- Walter A, Römheld V, Marschner H, Mori S. Is the release of phytosiderophores in zinc-deficient wheat plants a response to impaired iron utilization? Physiol Plant. 1994;92:493–500. [Google Scholar]