Abstract
Objective
Changes in progranulin (GRN) expression have been hypothesized to alter risk for Alzheimer's disease (AD). We investigated the relationship between GRN expression in peripheral blood and clinical diagnosis of AD and mild cognitive impairment (MCI).
Methods
Peripheral blood progranulin gene expression was measured, using microarrays from Alzheimer's (n = 186), MCI (n = 118), and control (n = 204) subjects from the University of California San Francisco Memory and Aging Center (UCSF‐MAC) and two independent published series (AddNeuroMed and ADNI). GRN gene expression was correlated with clinical, demographic, and genetic data, including APOE haplotype and the GRN rs5848 single‐nucleotide polymorphism. Finally, we assessed progranulin protein levels, using enzyme‐linked immunosorbent assay, and methylation status using methylation microarrays.
Results
We observed an increase in blood progranulin gene expression and a decrease in GRN promoter methylation in males (P = 0.007). Progranulin expression was 13% higher in AD and MCI patients compared with controls in the UCSF‐MAC cohort (F 2,505 = 10.41, P = 3.72*10−5). This finding was replicated in the AddNeuroMed (F 2,271 = 17.9, P = 4.83*10−8) but not the ADNI series. The rs5848 SNP (T‐allele) predicted decreased blood progranulin gene expression (P = 0.03). The APOE4 haplotype was positively associated with progranulin expression independent of diagnosis (P = 0.04). Finally, we did not identify differences in plasma progranulin protein levels or gene methylation between diagnostic categories.
Interpretation
Progranulin mRNA is elevated in peripheral blood of patients with AD and MCI and its expression is associated with numerous genetic and demographic factors. These data suggest a role in the pathogenesis of neurodegenerative dementias besides frontotemporal dementia.
Introduction
The 88 kDa progranulin and its 6 kDa processed products (granulins) represent a class of secreted proteins with diverse functions peripherally and in the brain. Granulins and progranulin have been implicated as potent immunomodulators and cell‐cycle regulators. In the CNS, progranulin expression increases with age in both neurons and microglia, and plays a role in neurite outgrowth, synapse modification, and the prevention of neuronal apoptosis.1, 2, 3, 4 This neuroprotective function is highlighted by the relationship between progranulin and neurodegenerative disease; heterozygous loss‐of‐function mutations in the gene encoding progranulin (GRN) cause frontotemporal dementia (FTD),5, 6 and a common rs5848 allele in the 3′UTR of GRN has been associated with both decreased serum and brain progranulin expression levels and increased risk of developing Alzheimer's disease (AD).7, 8, 9 Additionally, misregulation of progranulin expression has been implicated in parkinsonism, neuronal ceroid lipofuscinosis, and other neuropsychiatric disorders.10, 11
Mutations in the GRN gene may contribute to the risk of developing AD.8, 12 Additionally, progranulin localizes at the margins of amyloid plaques in both mouse and human postmortem brain tissue, and increased progranulin mRNA levels have been reported in the brains of multiple AD‐mouse models.13 Overexpression of progranulin in these models has been shown to slow plaque deposition and cognitive decline.14, 15 Taken together, these data suggest a direct role of progranulin in AD pathogenesis. Previous studies examining protein levels in peripheral blood failed to detect a relationship between peripheral progranulin protein levels and AD status.16 We first observed a relationship between GRN mRNA levels and AD status in a small patient series,17 but no study to date has conclusively shown a connection between progranulin expression and sporadic AD.
Early detection of AD has increasingly become a focus of the biomedical community, as future treatment modalities will likely hinge on slowing or preventing neurodegeneration before it has occurred. As such, mild cognitive impairment (MCI)–defined by focal memory or executive function deficits not explained by age, without dementia or loss of day‐to‐day function – has become a major focus of study.18 MCI patients convert to AD at a rate of 10–15% per year.19, 20 However, the majority of MCI patients will not transition to dementia, and some may display spontaneous improvement.19 Currently, estimates of hippocampal or entorhinal cortex volume coupled with cognitive function testing are the best predictors of disease transition.20, 21 However, reconstructive MRI imaging is expensive and not easily transferable to community hospitals. Thus, further work is needed to uncover peripheral blood markers of MCI.
We studied GRN expression levels in peripheral blood in a large patient series with AD and MCI, and asymptomatic controls, as well as in multiple datasets from the literature. We correlated GRN mRNA levels with demographic characteristics, disease status, genetic risk factors, methylation at the GRN locus, and progranulin protein levels as assayed by ELISA.
Material and Methods
Subjects and samples
This study received prior approval from the Institutional Review Board at the University of California San Francisco, and informed consent was obtained from subjects prior to study enrollment and sample collection.
UCSF‐MAC cohort
In this study, 530 patients clinically diagnosed as either AD, MCI, or unaffected controls were enrolled at the UCSF‐MAC between 2006 and 2016. MRI and Amyloid PET imaging were not uniformly performed to support diagnosis.
Peripheral blood from each subject was collected in Paxgene tubes and kept in ice prior to total RNA isolation. RNA extraction was performed, using the RNeasy QIAcube extraction kit (Qiagen) and RNA quantity was determined, using a Nanodrop instrument (Nanodrop Technologies). RNA quality was assessed with the Agilent Bioanalyzer (Agilent Technology) and samples with an RNA Integrity Number (RIN) <7 were excluded. RNA libraries were hybridized to Illumina HumanHT‐12 V4.0 microarrays at the UCLA Neuroscience Genomics Core. Microarray slides were scanned and signal processed using Illumina BeadStation and the BeadStudio software package in preparation for subsequent analysis. Here, 22 samples were ultimately excluded due to poor RIN or were detected as outliers (described below) for a final cohort size of n = 508.
AddNeuroMed cohort
The AddNeuroMed cohort 1 is publicly available in the Gene Expression Omnibus (GEO) repository (Accession: GSE63060).22 Briefly, this patient cohort is composed of 329 samples diagnosed as AD, MCI, or control with RNA hybridized to Illumina HumanHT‐12 V3.0 microarrays. Ultimately 21 outliers were removed and an additional 34 samples were dropped to correct for an age confound (described below) for a final cohort size of n = 274.
ADNI
Data used in this manuscript were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 under the direction of Principal Investigator Dr. Michael W. Weiner, MD to interrogate whether biological markers and clinical assessment can be combined to measure the progression of MCI and early AD.
The ADNI_Gene_Expression_Profile dataset is made available from the Alzheimer's Disease Neuroimaging Initiative (ADNI) repository. Samples were prepared as described (http://adni.loni.usc.edu/wp-content/uploads/2008/07/ADNI_GO_Procedures_Manual_06102011.pdf). Briefly, 811 patients in the ADNI cohort were categorized as Control, Early MCI, Late MCI, or AD using published clinical criteria. Total RNA was extracted and hybridized to an Affymetrix Human Genome U219 array and scanning and signal extraction were performed, using the Affymetrix GeneTitan system. After quality control, outlier removal, and stratification for an age confound (described below), the final processed cohort was n = 617.
San Antonio family heart study
The San Antonio Family Heart Study (SAFHS) dataset23 is publicly available through ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under the accession E‐TABM‐305. This cohort is composed of 1240 peripheral blood lymphocyte samples hybridized to Illumina Human‐6 v1 Expression BeadChip microarrays. Following outlier removal, we stratified this dataset to remove collinearity between sex and smoking status (described below). We then excluded all samples below 30 years old to match the age of our other datasets. The final processed cohort was n = 543.
Genotyping
Subjects in the UCSF‐MAC cohort had genomic DNA isolated from peripheral blood following standard procedures. APOE and GRN rs5848 genotyping were carried out by real‐time PCR on an Applied Biosystems 7900HT Real Time PCR machine (Applied Biosystems, Foster City, CA), using Taqman SNP Genotyping Assays (#C___7452046_20, C___3084793_20, and C____904973_10 for rs5848, rs429358, and rs7412, respectively). Assays were run in triplicate. The SDS version 2.3 software was used to analyze the raw data and to call the genotypes.
Array processing
Microarray raw signal processing for the UCSF‐MAC cohort was performed using the lumi package.24 First, within‐sample raw gene expression intensities were normalized using variance‐stabilized transformation (VST)25 and interarray normalization was performed with robust spline normalization. Probes with a detection score below standard threshold (P = 0.01) for all samples were dropped along with probes not annotated within the lumiHumanAll.db database. Next, ComBat from the sva package26 was used to perform batch correction. Outliers were removed using a connectivity Z‐score (threshold > 2) calculated using the fundamentalNetworkConcepts function from the WGCNA package.27 The AddNeuroMed cohort was processed using the same pipeline except that log2 normalization was used instead of VST because some quality control information was missing from the raw data.
The raw data for the ADNI dataset is not publicly available. Therefore, we performed analysis on preprocessed array data which were normalized, using standard Robust‐Multi‐Array Averaging from the affy package.28 We subsequently excluded 33 outliers using connectivity Z‐scores (threshold > 2) and 11 samples with a RIN < 7, and performed batch correction using ComBat. Probes were annotated using the Affymetrix hgu219.db database from BioConductor. The SAFHS cohort was analyzed using the same pipeline as the UCSF‐MAC cohort except for batch correction, which was not performed as batch information was not available.
Removal of confounding covariates
We used linear modeling and G‐tests29 to determine if age or sex were collinear with diagnosis using a significance threshold of P < 0.05. When significant collinearity was observed, samples were stratified, using a randomized nonbiased approach to drop samples until collinearity was no longer observed. Thus, final analysis was run on cohorts with age, sex, and diagnosis verified as independent variables. For the SAFHS dataset, sample stratification was used to remove collinearity between sex and smoking status.
Statistical analysis
Intensity values for probes querying GRN expression were collected from each platform. Each GRN probe was verified for every cohort by ensuring that average expression intensity was greater than the 60th percentile of all detected probes – no probes were dropped. For Illumina platforms, two progranulin probes were identified: AAGGCTCGATCCTGCGAGAAGGAAGTGGTCTCTGCCCAGCCTGCCACCTT (Probe 1, mapping to exon 11) and GGCCTTCCCTGTCAGAAGGGGGTTGTGGCAAAAGCCACATTACAAGCTGC (Probe 2, mapping to the 3′UTR). Neither location harbors known SNPs or InDels at a high population frequency (>1:1000). As probes were correlated (r 2 = 0.75), we reported only probe 1, which also had higher mean expression values. Linear models were used to assess significance of diagnosis or genotype with gender, age, and interactions (when appropriate) included as covariates. An F‐test was used to assess significance of categorical predictors with more than two groups, and Welch's T‐test was used for continuous predictors or categorical predictors with two groups. Post hoc pairwise testing was done using Tukey's test with a significance threshold of P < 0.05. The Wilcoxon rank sum test was used in a case of small, nonnormal sample.
Quantification of progranulin protein by ELISA
Progranulin protein in human plasma and cerebrospinal fluid (CSF) was quantified by A&G Pharmaceutical Inc. (Columbia, MD) using their Progranulin (GP88/PGRN) ELISA, which detects full‐length progranulin in both biofluids. The detection limit of this assay is 100 pg/mL with a working range up to 20 ng/mL. Assay details have been described previously.30, 31, 32 Briefly, a subset of patients enrolled in the UCSF‐MAC cohort underwent peripheral blood draws (n = 266) and/or lumbar puncture (n = 80) during multiple follow‐up visits. Each sample was run in duplicate and normalized against a recombinant progranulin standard and two reference serum samples. A coefficient of variation score (SD/mean*100) was calculated for every sample. The 11 plasma samples with a coefficient of variation >15% were dropped as a quality control measure. Patients who underwent multiple draws had their plasma or CSF PGRN values averaged. Outliers were removed using the R boxplot function. Statistical analysis was done using the same models and tests as described for gene expression.
Methylation
DNA was extracted from peripheral blood using standard methods. DNA methylation was quantified using the Illumina Human Methylation 450K microarray. Preprocessing was run with the RnBeads package33, using the default options for quality control. Background correction was performed with the normal exponential convolution, using out‐of‐band probes (noob) method,34 and arrays were normalized using beta mixture quantile dilation (BMIQ).35 The normalized data was corrected for batch effect using the parametric empirical Bayes method from ComBat.26 The RnBeads package was also used to extract GRN promoter methylation β‐values, which were computed by averaging the β‐values of all 10 probes from 1.5 kb upstream to 0.5 kb downstream of the transcription start site. RNBeads was similarly used to extract gene body methylation, again by averaging the β‐values of all 10 probes from the transcription start site to the end of the gene.
Results
Demographic and diagnostic determinants of GRN expression
General characteristics of all processed cohorts (postquality control and confounder stratification) are described in Table 1. We first analyzed the UCSF‐MAC cohort (n = 508).
Table 1.
Summary characteristics of four patient cohorts analyzed
| Cohort | Diagnosis | Sample (n) | Male (n) | Female (n) | Mean Age | SD Age |
|---|---|---|---|---|---|---|
| UCSF‐MAC | Combined | 508 | 240 | 268 | 69.8 | 10.4 |
| Control | 204 | 95 | 109 | 70.9 | 11.2 | |
| MCI | 118 | 61 | 57 | 69.7 | 10.1 | |
| AD | 186 | 84 | 102 | 68.8 | 9.5 | |
| AddNeuroMedd | Combined | 274 | 107 | 167 | 72.9 | 5.7 |
| Control | 95 | 38 | 57 | 71.8 | 5.8 | |
| MCI | 66 | 32 | 34 | 73.1 | 4.9 | |
| AD | 113 | 37 | 76 | 73.7 | 5.9 | |
| ADNI | Combined | 617 | 326 | 291 | 73.8 | 6.5 |
| Control | 240 | 114 | 126 | 74.3 | 5.6 | |
| eMCI | 159 | 91 | 68 | 73.7 | 5.8 | |
| lMCI | 178 | 96 | 82 | 73.0 | 7.2 | |
| AD | 40 | 25 | 15 | 75.6 | 9.8 | |
| SAFHS | Combined | 543 | 252 | 291 | 49.7 | 13.5 |
| Smoker | 158 | 85 | 73 | 48.1 | 12.0 | |
| Nonsmoker | 385 | 167 | 218 | 50.2 | 13.9 |
First, we assessed the relationship between gene expression, sex, and age, after ensuring no confounding between our predictors. We verified no collinearity between sex and diagnosis in our dataset (G‐test, G‐statistic(2) = 1.3, P = 0.52) nor between age and diagnosis (F 2,505 = 2.04, P = 0.13). We next fit a linear model for our covariates. We observed that GRN mRNA expression levels were significantly higher in males than in females (log2 fold‐change [log2FC] = 0.09, corresponding to a 6% increase, T‐statistic = 6.26, P = 0.013, Fig. 1A) across diagnostic categories, and that age was significantly but trivially correlated with progranulin mRNA levels (Pearson's r 2 = 0.008, P = 0.04; Fig. 1B) consistent with previous reports.11 We next compared GRN levels between diagnostic categories (F 2,505 = 10.41, P = 3.7*10−5). Patients diagnosed with AD had a statistically significant increase (log2FC = 0.17, corresponding to +13%, Tukey's test P = 0.00019) in GRN mRNA expression compared with controls. We also observed a similar increase (log2FC = 0.17, P = 0.0012) in the MCI group compared with controls (Fig. 1C). We observed no significant differences in GRN mRNA levels between AD and MCI. When we modeled interactions between sex and diagnosis, as well as age and diagnosis, we did not observe significant effects, arguing against a synergistic relationship between these covariates.
Figure 1.
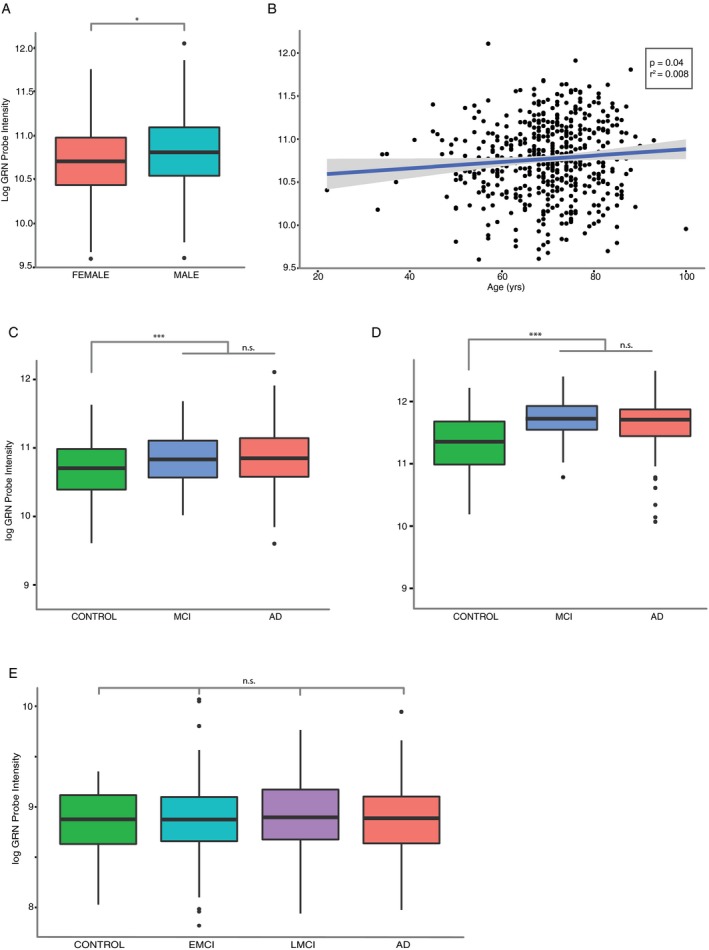
Effect of demographic characteristics and disease status on peripheral blood GRN gene expression. (A) Progranulin gene expression is higher in males than females. UCSF‐MAC cohort, Welch's t‐test. (B) Progranulin gene expression increases with age across diagnostic categories. UCSF‐MAC cohort, linear regression. (C) Progranulin expression is higher in AD and MCI patients compared with controls. UCSF‐MAC cohort, F‐test and analysis of variance (AOV) with Tukey's post hoc test. (D) same as (C) in the AddNeuroMed cohort. (E) lack of difference in progranulin expression in AD, early MCI (EMCI), and later MCI (LMCI) patients compared with controls. ADNI cohort, F‐test and aov with Tukey's post hoc test. General cohort characteristics are described in Table 1. (A, C, D, E) standard boxplot representing median and interquartile range (IQR), whiskers represent 1.5 IQR greater or less than the upper and lower quartile. (B) scatterplot with line of best‐fit. *P < 0.05, ***P < 0.001, n.s. P > 0.05.
We next sought to replicate our findings. We first analyzed the AddNeuroMed Cohort (n = 274; Table 1). We initially observed collinearity between age and diagnosis (F 2,305 = 5.81, P = 0.003) and therefore stratified and discarded 34 samples until collinearity was no longer significant (F 2,271 = 3.0, P = 0.051). Sex remained independent after stratification (G(2) = 4.38, P = 0.11). We again fit a linear model with all covariates and relevant interaction terms. Progranulin mRNA expression again was higher in males compared with females (log2FC = 0.12, +9%, T‐statistic = 4.4, P = 0.037), but age was no longer predictive (T‐statistic = 2.3, P = 0.13). We observed a significant effect based on diagnosis (F 2,271 = 17.9, P = 4.8*10−8) but no interaction effects. Similar to the UCSF‐MAC cohort, we observed a significant increase (log2FC = 0.28, corresponding to +21%, TukeyHSD; P = 5.6*10−6) in progranulin mRNA expression for AD patients compared with controls, and an increase (log2FC = 0.35, +27%, P = 6.4*10−7) for MCI patients compared to controls (Fig. 1D).
We next validated these sex and age findings in a third dataset, the San Antonio Family Heart Study cohort (diagnosis was not relevant for this dataset). After stratifying to correct for collinearity between sex and smoking status, we excluded all samples under 30 years of age to more appropriately match mean cohort age with our other cohorts while still maintaining statistical power (n = 543, mean age = 49.7 years). We ensured no collinearity between age (T‐statistic = 1.06, P = 0.3) or smoking status (G(2) = 4.48, P = 0.08) and sex and fit a linear model. We again found that progranulin was higher in males than females (log2FC = 0.07, +5%, T‐statistic = 4.8, P = 0.03) and found no correlation with age (T‐statistic = 0.04, P = 0.8). Interestingly, smoking status was an independent predictor of blood progranulin mRNA expression in this cohort, with a 6% GRN increase in smokers (log2FC = 0.08, T‐statistic = 4.5, P = 0.03).
Finally, we analyzed the ADNI patient cohort (n = 617), which is divided into asymptomatic controls, early MCI (eMCI), late MCI (lMCI, with regard to clinical disease progression), and AD. This cohort was initially confounded with significant collinearity between age (F 3,696 = 10.9, P = 5*10‐7), sex (G(3) = 11.1, P = 0.01) and diagnosis. We stratified and excluded samples (described in methods) until age (F 3,613 = 2.49, P = 0.06) and sex (G(3) = 5.58, P = 0.13) were no longer collinear with diagnosis. However, a plate batch effect remained (G(24) = 38.9, P = 0.03), which we corrected with the ComBat package. In this cohort, neither age nor sex were predictive of progranulin expression, and we did not observe differences in progranulin expression levels between diagnostic categories (F 3,611 = 0.31, P = 0.82, TukeyHSD post hoc P > 0.05 for all comparisons; Fig. 1E).
Effects of genotype on GRN expression
Next, we genotyped samples from the UCSF‐MAC cohort for the risk‐associated rs5848 GRN variant. Homozygotes for the risk‐associated allele (T:T, n = 25) had a 12.3% decrease in progranulin mRNA expression compared with C:C carriers (log2FC = −0.19, Wilcoxon Rank sum test, P = 0.02) supporting an earlier report of decreased GRN mRNA levels in postmortem brain tissue of T:T genotype carriers.36 We also found that T‐allele carriers had significantly lower progranulin levels than C:C homozygotes (log2FC = −0.1, Welch, T506 = 2.24, P = 0.03; Fig. 2A). After ensuring there was no collinearity between the rs5848 genotype and sex (G‐statistic1 = 0.21, P = 0.65), we fit a linear model using sex and haplotype as an interaction term, but did not observe any significant interaction in our data. Additionally, using a Fisher's exact test, we observed an enrichment of T‐genotype carriers in the AD population compared with controls, which did not reach statistical significance (OR = 1.46, 95% CI = 0.95–2.38, P = 0.09). Thus, it is noteworthy that AD patients on average still have elevated progranulin gene expression despite the overrepresentation of rs5848 T‐allele carriers (associated with lower progranulin levels) in the AD patient population.
Figure 2.
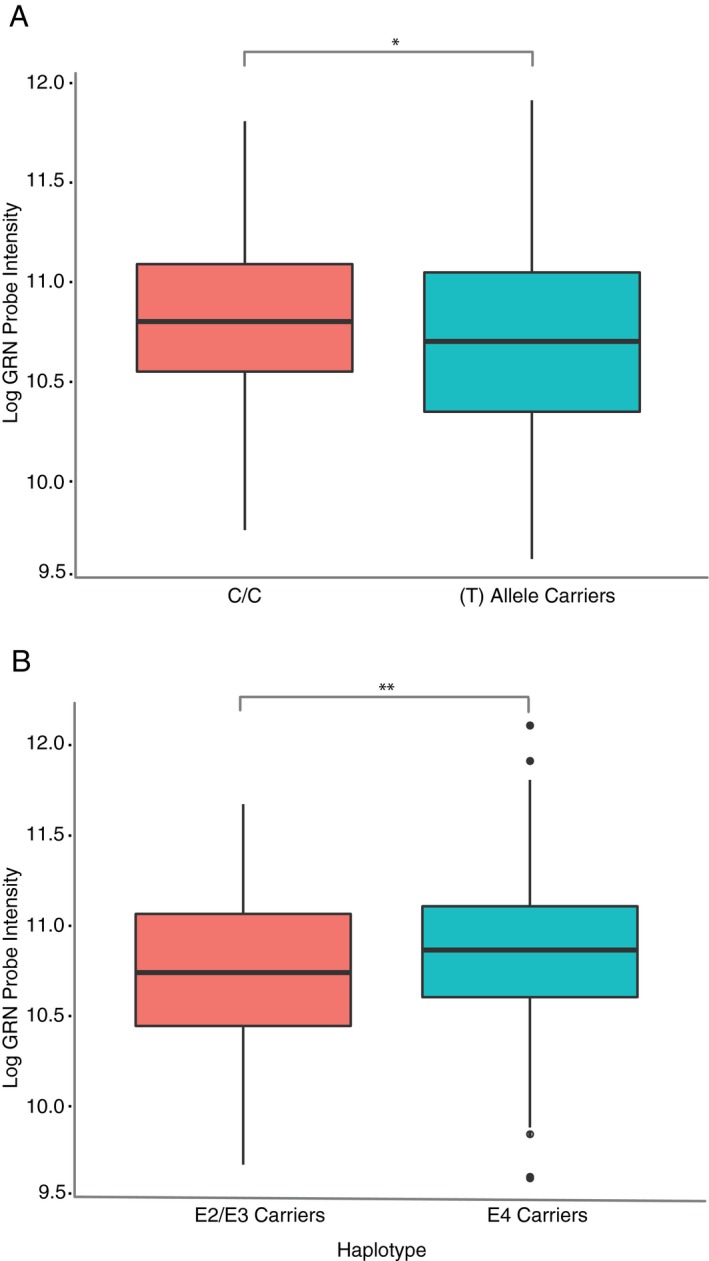
Common AD‐associated genetic variants and GRN mRNA expression in peripheral blood. (A) Progranulin gene expression is lower in rs5848 risk allele carriers (C:T or T:T genotypes) compared with low‐risk genotype (C:C). UCSF‐MAC cohort, Welch's t‐test. (B) Progranulin gene expression is higher in APOE risk allele (E4) carriers (homozygote or heterozygote) compared with low‐risk haplotypes (E2 or E3 carriers). UCSF‐MAC cohort, Welch's t‐test. (A‐B) standard boxplot representing median and IQR, whiskers represent 1.5 IQR greater or less than the upper and lower quartile. *P < 0.05, **P < 0.01.
We also studied the relationship between GRN gene expression levels and the AD risk‐associated APOE haplotype.37 We observed a significant effect of the APOE haplotype on progranulin levels in the UCSF‐MAC cohort across diagnostic categories, with progranulin mRNA levels significantly higher in E4 risk allele carriers compared with E2/E3 haplotypes (log2FC = 0.12, +9%, Welch, T (441) = 2.82 P = 0.005, Fig. 2B). However, there was also significant collinearity between APOE haplotype and disease status; the E4 haplotype was significantly enriched in AD patients (Fisher's exact test, OR = 4.83, P = 6.3*10−11) compared with controls, confounding our results.
Therefore, we attempted to estimate the independent relationship between APOE haplotype and GRN expression. After removing the effects of diagnosis as a confound using a parametric empirical Bayesian estimator (ComBat), we still observed significantly higher progranulin gene expression levels in ApoE4 haplotype carriers (log2FC = 0.08, Welch T‐test, P = 0.04), suggesting that APOE4 genotype might function as a trans expression quantitative trait locus (eQTL) controlling GRN expression. We validated this finding in the Genotype‐Tissue Expression consortium dataset (GTEx).38 Analysis of rs429358 SNP (minor C‐allele tags the APOE4 haplotype) and RNA‐seq data from 328 samples revealed a significant association between expression and genotype (effect size = 0.12, P = 0.004) in whole blood. We checked the relationship between APOE4 carrier status and GRN expression in the ADNI dataset but failed to detect a difference in GRN expression between APOE4 carriers and controls (T (613) = 1.43, P = 0.15). We also performed the converse analysis in the UCSF‐MAC cohort. Using ComBat to remove the effect of APOE haplotype, we then modeled the effect of diagnosis on progranulin expression. We found that AD samples still had significantly higher progranulin expression (log2FC = 0.17, T (450) = 3.9, P = 0.0001) compared with controls suggesting that diagnosis predicts progranulin expression independent of APOE4 sample overrepresentation.
GRN protein levels in CSF and plasma
We next utilized ELISA to quantify progranulin protein levels in plasma (n = 266) and CSF (n = 80) in a subset of patients from the UCSF‐MAC cohort. After ensuring no collinearity with age (F 2,263 = 1.84, P = 0.16) and sex (G‐statistic2 = 2.32, P=0.31), we observed no correlation between plasma progranulin protein and gene expression in peripheral blood (Pearson's r = 0.065, df = 206, P = 0.35; Fig. 3A). However, we observed a sex dimorphism in our plasma data, with females across diagnostic categories having significantly higher progranulin protein levels than males (fold‐change = 0.07, Welch's T 264 = 2.14, P = 0.03, Fig. 3B) which recapitulates an earlier study,16 and is the opposite finding from our gene expression data, where males had higher GRN mRNA expression. We identified no correlation between age and plasma GRN protein levels (Pearson's r = 0.09, df = 264, P = 0.16). We next assessed the relationship between disease status and progranulin protein levels. In plasma, we found no difference between GRN protein levels across all disease categories (F 2,263 = 1.72, P = 0.18, Tukey's post hoc P > 0.05 all comparisons; Fig. 3C) again replicating previous findings.16 We observed no correlation between progranulin levels in the CSF and in plasma (Pearson's r = 0.13, df = 63, P = 0.31; Fig. 3E). AD patients had statistically lower CSF progranulin levels than control subjects (F 2,66 = 3.45, P = 0.04, TukeyHSD P = 0.03 for AD vs. Control, Fig. 3D), confirming an earlier report.39 We observed no relationship between age or sex and progranulin levels in CSF.
Figure 3.
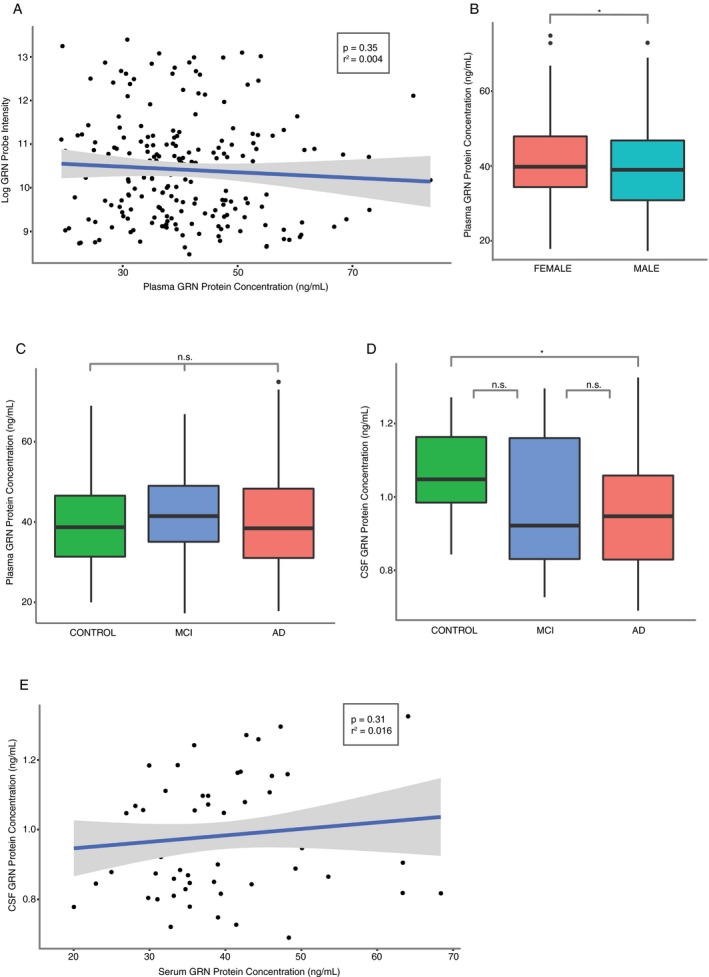
Progranulin protein levels by ELISA in plasma and cerebrospinal fluid (CSF). (A) Plasma progranulin protein levels are not correlated with gene expression within patients. n = 266, linear regression. (B) Females have significantly higher plasma GRN protein levels than males. Welch's t‐test. (C) Lack of difference in plasma GRN protein levels between AD, MCI, and control patients. Analysis of variance (AOV), Tukey's post hoc test. (D) AD patients have significantly lower CSF GRN protein levels than MCI and control patients. n = 80, AOV, Tukey's post hoc test. (E) Lack of correlation between plasma and CSF GRN protein levels. n = 80, linear regression. All analysis performed with the UCSF‐MAC cohort. (B–D) standard boxplot representing median and IQR, whiskers represent 1.5 IQR greater or less than the upper and lower quartile. (A,E) scatterplot with line of best‐fit. *P < 0.05, n.s. P > 0.05.
Analysis of GRN methylation status
Finally, we analyzed DNA methylation at the GRN locus in a subset of subjects (AD patients [n=128] and controls [n=227]) from the UCSF‐MAC cohort. We found no correlation between GRN promoter CpG methylation and GRN expression (T (91) = 0.16, r = 0.02, P = 0.8) nor GRN gene body methylation and GRN gene expression (T (91) = 1.52, r = −0.16, P = 0.13). We next fit a linear model predicting methylation status, using all relevant predictors. Males had lower GRN promoter CpG methylation than females (Log2FC = 0.004, T (351) = 7.32, P = 0.007; Fig. 4C) but no differences in gene body methylation T (351) = 1.06, P = 0.3). Conversely, there was a negative correlation between age and GRN gene body methylation; methylation decreased significantly with age (T (351) = 62.8, r = ‐0.39, P = 3*10−14; Fig. 4D). We identified no correlation between GRN promoter methylation and age (T (351) = 1.11, r = 0.05, P = 0.29) in agreement with Galimberti and colleagues.40 We identified no significant difference in methylation β‐values for either the GRN promoter (T (351) = 0.07, P = 0.79) or gene body (T (351) = 0.17, P = 0.67) between AD and control patients (Fig. 4A,B).
Figure 4.
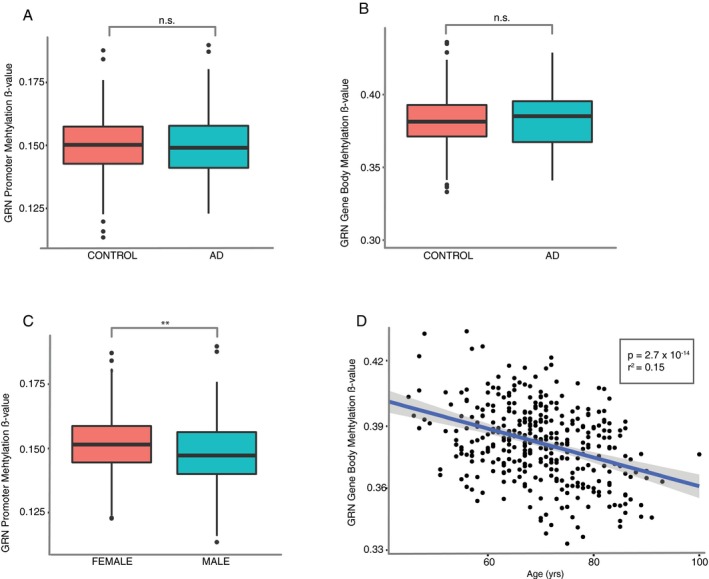
Analysis of GRN promoter and gene body DNA (CpG) methylation in DNA from peripheral blood. (A–B) lack of difference in promoter (A) or gene body (B) methylation between AD and control patients. n = 355, Welch's t‐test. (C) GRN promoter methylation is significantly lower in males compared with females across diagnostics categories. Welch's t‐test. (D) GRN gene body methylation decreases with age. Linear regression. All analysis performed with the UCSF‐MAC cohort. (A‐C) standard boxplot representing median and IQR, whiskers represent 1.5 IQR greater or less than the upper and lower quartile. (D) scatterplot with line of best‐fit. **P < 0.01, n.s. P>0.05.
Discussion
We present here data indicating that patients with sporadic Alzheimer's disease have significantly increased progranulin mRNA in peripheral blood compared with controls. Additionally, we find that progranulin expression is significantly increased in patients with mild cognitive impairment. While we have replicated these findings in an independent patient cohort (AddNeuroMed), we failed to replicate them in the ADNI cohort. Of note, both our cohort and the AddNeuroMed cohort used Illumina microarray platforms while the ADNI gene expression data was run on an Affymetrix platform. We therefore hypothesize that this discrepancy may be a feature of Affymetrix probes, which may not sensitively or reliably detect changes in progranulin expression levels. This is supported by our additional failure to detect any correlation between progranulin expression and age, sex, or genotype in the ADNI dataset.
While we identified differences in mean progranulin expression between groups, our results indicate that progranulin cannot be used on its own as a sensitive or specific biomarker of disease. However, our results suggest that it should be possible to identify further differentially expressed genes that could perhaps then be incorporated into a diagnostic panel. Because peripheral blood can be drawn from living patients in the most basic clinical settings, this has broader clinical utility. Most saliently, this study represents the first association between peripheral progranulin expression and MCI to our knowledge, suggesting that progranulin may play an early role in AD pathogenesis.
Previous studies in mouse models of AD have found that the artificial increase in progranulin levels can slow disease progression including disrupting Aβ plaque deposition and neurotoxicity.14 Furthermore, loss‐of‐function mutations in the progranulin gene are risk factors for developing AD as well as other neurodegenerative disorders.4, 6, 8 Together these findings suggest that progranulin plays a primarily neuroprotective role, reactively modifying or guarding against neurodegenerative processes. Our data support this hypothesis. The small subset of patients with the rs5848 (T:T) haplotype had significantly lower progranulin levels and were also more likely to have AD, as expected.7, 9 However, AD patients in general had higher progranulin levels, especially when excluding haplotypes that directly lower progranulin expression and cause disease. This suggests that progranulin expression might increase as an effect rather than a cause of disease. It is also conceivable that undiscovered cis or trans eQTLs that control progranulin expression exist, and therefore modify risk for AD. For example, we identify the APOE haplotype as a possible trans eQTL that influences progranulin expression, independent of AD diagnosis.
If progranulin plays a functional role in AD pathogenesis, we would expect to ultimately observe changes in protein levels. However, analysis of our ELISA data failed to show a difference in progranulin protein levels between AD, MCI, and control patients, consistent with earlier reports.16 There was also no correlation between progranulin protein levels in plasma and CSF and no correlation between protein and gene expression.39 Our ELISA findings are entirely consistent with and replicate previous independent reports, indicating that our results are not the result of operational error or a technical artifact. One likely interpretation is that the current ELISA methodology fails to accurately detect subtle changes in plasma progranulin levels. The progranulin protein has complex posttranslational regulation, including glycosylation and variable cleavage into a variety of intermediaries as well as any of 8 final granulin products. These various protein configurations have myriad and often contradictory functions.41 In fact, the current ELISA used is specific for unprocessed progranulin.30 Given high antibody specificity and the diversity of progranulin end products, it is unlikely that ELISA can sensitively detect overall changes in specific forms of progranulin being expressed in AD, MCI, and controls; rather it is likely reflective of a particular subset of progranulin products.
Additionally, progranulin and granulins undergo complex spatial regulation with progranulin being shuttled to both the lysosome or excreted into the extracellular space. As such, the biological fluid and cellular components assayed have a large impact on progranulin measurements.42 This might explain the sex dimorphism discrepancy between our gene expression and ELISA data in addition to differences in progranulin cleavage and processing. ELISA, which measures extracellular fluid (plasma), may not be directly comparable to intracellular gene expression profiles. Nevertheless, the functional significance of possibly increased progranulin secretion in females versus relative intracellular retention in males requires further study.
Finally, our data suggest that blood progranulin gene expression is higher overall in males than in females. Our GRN methylation data also supports this finding, as males have reduced promoter methylation, suggesting a derepressed state primed for transcriptional activation. These data are intriguing considering that progranulin expression in rat hippocampus was found to be under the control of estrogen.43 Although there have been numerous studies describing sex dimorphisms in gene methylation in peripheral blood, these studies have failed to specifically identify progranulin.44, 45 Nevertheless, it is especially interesting to consider this sex dimorphism as women are twice as likely to develop AD as men. If progranulin indeed plays a protective role, increased endogenous progranulin expression in males might partly explain this phenomenon, and this might be mechanistically mediated by differences in progranulin promoter methylation.
Author Contributions
Study design and conception: YC and GC. Patient recruitment and trial design: AK, AB, and BM. Sample and data acquisition: AB, BM, DD, YZ, GS, BY. Data analysis: YC, DN, and GC. Manuscript preparation: YC and GC. All authors provided critical feedback.
Conflicts of Interest
Ginette Serrero and Binbin Yue are employees of A&G Pharmaceutical Inc., which has a proprietary PGRN antibody used for the ELISA portion of this work. Ginette Serrero holds issued patents related to the measurement of Progranulin in biological fluids. No other authors report conflicts of interest or financial stakes in this work.
Acknowledgments
This work was supported by the NIH (RC1 AG035610, R01 AG26938, P50 AG023501, P01 AG019724), the John Douglas French Alzheimer's Foundation, and the Tau Consortium. Samples from the National Cell Repository for Alzheimer's Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study. We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691). We thank contributors who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible. The Genotype‐Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 08/07/2017.
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH‐12‐2‐0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol‐Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann‐La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Funding Information
This work was supported by the NIH (RC1 AG035610, R01 AG26938, P50 AG023501, P01 AG019724), the John Douglas French Alzheimer's Foundation, and the Tau Consortium. Samples from the National Cell Repository for Alzheimer's Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study. We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691). We thank contributors who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible. The Genotype‐Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 08/07/2017.
Funding Statement
This work was funded by NIH grants RC1 AG035610, R01 AG26938, P50 AG023501, P01 AG019724, and U01 AG024904; John Douglas French Alzheimer's Foundation grant ; Tau Consortium grant ; National Cell Repository for Alzheimer's Disease (NCRAD) grant ; National Institute on Aging (NIA) grant U24 AG21886; NINDS Informatics Center for Neurogenetics and Neurogenomics grant P30 NS062691; Common Fund of the Office of the Director of the National Institutes of Health grant ; NCI grant ; NHGRI grant ; NHLBI grant ; NIDA grant ; NIMH grant ; DOD ADNI grant W81XWH‐12‐2‐0012.
References
- 1. Gao X, Joselin AP, Wang L, et al. Progranulin promotes neurite outgrowth and neuronal differentiation by regulating GSK‐3β . Protein Cell 2010;1:552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Luo L, Lü L, Lu Y, et al. Effects of hypoxia on progranulin expression in HT22 mouse hippocampal cells. Mol Med Rep 2014;9:1675–1680. [DOI] [PubMed] [Google Scholar]
- 3. Lui H, Zhang J, Makinson SR, et al. Progranulin deficiency promotes circuit‐specific synaptic pruning by microglia via complement activation. Cell 2016;165:921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olney NT, Alquezar C, Ramos EM, et al. Linking tuberous sclerosis complex, excessive mTOR signaling, and age‐related neurodegeneration: a new association between TSC1 mutation and frontotemporal dementia. Acta Neuropathol 2017;134:813–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baker M, Mackenzie IR, Pickering‐Brown SM, et al. Mutations in progranulin cause tau‐negative frontotemporal dementia linked to chromosome 17. Nature 2006;442:916–919. [DOI] [PubMed] [Google Scholar]
- 6. Yu C‐EE, Bird TD, Bekris LM, et al. The spectrum of mutations in progranulin: a collaborative study screening 545 cases of neurodegeneration. Arch Neurol 2010;67:161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsiung G‐YY, Fok A, Feldman HH, et al. rs5848 polymorphism and serum progranulin level. J Neurol Sci 2011;300:28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perry DC, Lehmann M, Yokoyama JS, et al. Progranulin mutations as risk factors for Alzheimer disease. JAMA Neurol 2013;70:774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheng J, Su L, Xu Z, Chen G. Progranulin polymorphism rs5848 is associated with increased risk of Alzheimer's disease. Gene 2014;542:141–145. [DOI] [PubMed] [Google Scholar]
- 10. Kittel‐Schneider S, Weigl J, Volkert J, et al. Further evidence for plasma progranulin as a biomarker in bipolar disorder. J Affect Disord 2014;157:87–91. [DOI] [PubMed] [Google Scholar]
- 11. Petkau TL, Leavitt BR. Progranulin in neurodegenerative disease. Trends Neurosci 2014;37:388–398. [DOI] [PubMed] [Google Scholar]
- 12. Brouwers N, Sleegers K, Engelborghs S, et al. Genetic variability in progranulin contributes to risk for clinically diagnosed Alzheimer disease. Neurology 2008;71:656–664. [DOI] [PubMed] [Google Scholar]
- 13. Pereson S, Wils H, Kleinberger G, et al. Progranulin expression correlates with dense‐core amyloid plaque burden in Alzheimer disease mouse models. J Pathol 2009;219:173–181. [DOI] [PubMed] [Google Scholar]
- 14. Minami SS, Min S‐W, Krabbe G, et al. Progranulin protects against amyloid β deposition and toxicity in Alzheimer's disease mouse models. Nat Med 2014;20:1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Kampen JM, Kay DG. Progranulin gene delivery reduces plaque burden and synaptic atrophy in a mouse model of Alzheimer's disease. PLoS ONE 2017;12:e0182896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Piscopo P, Rivabene R, Galimberti D, et al. Gender effects on plasma PGRN levels in patients with Alzheimer's disease: a preliminary study. J Alzheimers Dis 2013;35:313–318. [DOI] [PubMed] [Google Scholar]
- 17. Coppola G, Karydas A, Rademakers R, et al. Gene expression study on peripheral blood identifies progranulin mutations. Ann Neurol 2008;64:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petersen RC, Smith GE, Waring SC, et al. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol 1999;56:303–308. [DOI] [PubMed] [Google Scholar]
- 19. Mitchell AJ, Shiri‐Feshki M. Rate of progression of mild cognitive impairment to dementia–meta‐analysis of 41 robust inception cohort studies. Acta Psychiatr Scand 2009;119:252–265. [DOI] [PubMed] [Google Scholar]
- 20. Risacher SL, Saykin AJ, West JD, et al. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res 2009;6:347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gainotti G, Quaranta D, Vita MG, Marra C. Neuropsychological predictors of conversion from mild cognitive impairment to Alzheimer's disease. J Alzheimers Dis 2014;38:481–495. [DOI] [PubMed] [Google Scholar]
- 22. Sood S, Gallagher IJ, Lunnon K, et al. A novel multi‐tissue RNA diagnostic of healthy ageing relates to cognitive health status. Genome Biol 2015;16:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Göring HH, Curran JE, Johnson MP, et al. Discovery of expression QTLs using large‐scale transcriptional profiling in human lymphocytes. Nat Genet 2007;39:1208–1216. [DOI] [PubMed] [Google Scholar]
- 24. Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics 2008;24:1547–1548. [DOI] [PubMed] [Google Scholar]
- 25. Lin SM, Du P, Huber W, Kibbe WA. Model‐based variance‐stabilizing transformation for Illumina microarray data. Nucleic Acids Res 2008;36:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–127. [DOI] [PubMed] [Google Scholar]
- 27. Dong J, Horvath S. Understanding network concepts in modules. BMC Syst Biol 2007;1:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of affymetrix genechip data at the probe level. Bioinformatics 2004;20:307–315. [DOI] [PubMed] [Google Scholar]
- 29. Hoey J. The two‐way likelihood ratio (G) test and comparison to two‐way chi squared test. arXiv 2012.
- 30. Tkaczuk KR, Yue B, Zhan M, et al. Increased circulating level of the survival factor GP88 (Progranulin) in the serum of breast cancer patients when compared to healthy subjects. Breast (Auckl) 2011;5:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edelman MJ, Feliciano J, Yue B, et al. GP88 (progranulin): a novel tissue and circulating biomarker for non–small cell lung carcinoma. Hum Path 2014;45:1893–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kimura A, Takemura M, Saito K, et al. Increased cerebrospinal fluid progranulin correlates with interleukin‐6 in the acute phase of neuromyelitis optica spectrum disorder. J Neuroimmunol 2017;305:175–181. [DOI] [PubMed] [Google Scholar]
- 33. Assenov Y, Müller F, Lutsik P, et al. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods 2014;11:1138–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. TricheTJ Jr, Weisenberger DJ, Van Den Berg D, et al. Low‐level processing of illumina infinium DNA methylation beadarrays. Nucleic Acids Res 2013;41:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Teschendorff AE, Marabita F, Lechner M, et al. A beta‐mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013;29:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fenoglio C, Galimberti D, Cortini F, et al. Rs5848 variant influences GRN mRNA levels in brain and peripheral mononuclear cells in patients with Alzheimer's disease. J Alzheimers Dis 2009;18:603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van Cauwenberghe C, Van Broeckhoven C, Sleegers K. The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet Med 2018;18:421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. GTEx Consortium . The Genotype‐Tissue Expression (GTEx) project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilke C, Gillardon F, Deuschle C, et al. Cerebrospinal fluid progranulin, but not serum progranulin, is reduced in GRN‐negative frontotemporal dementia. Neurodegener Dis 2016;17:83–88. [DOI] [PubMed] [Google Scholar]
- 40. Galimberti D, D'Addario C, Dell'osso B, et al. Progranulin gene (GRN) promoter methylation is increased in patients with sporadic frontotemporal lobar degeneration. Neurol Sci 2013;34:899–903. [DOI] [PubMed] [Google Scholar]
- 41. Abella V, Pino J, Scotece M, et al. Progranulin as a biomarker and potential therapeutic agent. Drug Discov Today 2017;22:1557–1564. [DOI] [PubMed] [Google Scholar]
- 42. Kao A, McKay A, Singh PP, et al. Progranulin, lysosomal regulation and neurodegenerative disease. Nat Rev Neuro 2017;18:325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suzuki M, Lee HC, Kayasuga Y, et al. Roles of progranulin in sexual differentiation of the developing brain and adult neurogenesis. J Reprod Dev 2009;55:351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu J, Morgan M, Hutchison K, Calhoun VD. A study of the influence of sex on genome wide methylation. PLoS ONE 2010;5:e10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hall E, Volkov P, Dayeh T, et al. Sex differences in the genome‐wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol 2014;15:522. [DOI] [PMC free article] [PubMed] [Google Scholar]


