Summary
In human longevity studies, single nucleotide polymorphism (SNP) analysis identified a large number of genetic variants with small effects, yet not easily replicable in different populations. New insights may come from the combined analysis of different SNPs, especially when grouped by metabolic pathway. We applied this approach to study the joint effect on longevity of SNPs belonging to three candidate pathways, the insulin/insulin‐like growth factor signalling (IIS), DNA repair and pro/antioxidant. We analysed data from 1,058 tagging SNPs in 140 genes, collected in 1825 subjects (1,089 unrelated nonagenarians from the Danish 1905 Birth Cohort Study and 736 Danish controls aged 46–55 years) for evaluating synergic interactions by SNPsyn. Synergies were further tested by the multidimensional reduction (MDR) approach, both intra‐ and interpathways. The best combinations (FDR<0.0001) resulted those encompassing IGF1R‐rs12437963 and PTPN1‐rs6067484, TP53‐rs2078486 and ERCC2‐rs50871, TXNRD1‐rs17202060 and TP53‐rs2078486, the latter two supporting a central role of TP53 in mediating the concerted activation of the DNA repair and pro‐antioxidant pathways in human longevity. Results were consistently replicated with both approaches, as well as a significant effect on longevity was found for the GHSR gene, which also interacts with partners belonging to both IIS and DNA repair pathways (PAPPA,PTPN1,PARK7, MRE11A). The combination GHSR‐MREA11, positively associated with longevity by MDR, was further found influencing longitudinal survival in nonagenarian females (p = .026). Results here presented highlight the validity of SNP‐SNP interactions analyses for investigating the genetics of human longevity, confirming previously identified markers but also pointing to novel genes as central nodes of additional networks involved in human longevity.
Keywords: aging, epistasis, genetic component of human longevity, pathway‐based analysis, SNP, synergic interaction
1. INTRODUCTION
Association studies can identify the association of individual gene variants to a given phenotype. Nevertheless, such analysis is unable to explain the biological complexity of several diseases and complex phenotypes such as human aging and longevity.
Recent advances in DNA technology and association studies have uncovered a very large number of common susceptibility variants for complex traits; however, the translation into practice of their role is complicated by the evidence that such variants operate together to influence the final phenotype. The major challenge is represented by the relatively small genetic contribution to the variation in phenotype, which in human lifespan was estimated to be approximately 25% (Herskind et al., 1996) determined by many, mostly still uncharacterized, genes (Deelen et al., 2013), possibly belonging to conserved pathways (Johnson, Dong, Vijg & Suh, 2015). Single‐SNP analyses may miss such a complexity, primarily because if a genetic factor operates through a mechanism involving multiple genes, and is also affected by environmental factors, the single investigation may not examine statistical interactions between loci that are informative about the biological and biochemical pathways underpinning the phenotype. On the contrary, SNP interactions may carry more information about the phenotype than those observed from individual SNPs alone (Gerke, Lorenz & Cohen, 2009; Su et al., 2015). Assessing SNP‐SNP interactions at the gene level co‐occurring within a specific phenotype and not due to linkage disequilibrium (LD) can overcome this problem, possibly finding specific subprocesses more strongly associated with the phenotype than single‐SNP analysis (Cordell, 2009). However, the search of all possible SNP‐SNP interactions is challenging from an analytical point of view, because of the number of pairwise statistical tests to be performed. Only recently, significant advances in statistical approaches make it possible to analyse multiple SNPs in large genetic data sets, considering the main pathway to which the gene belongs and possible covariates, as required in the analysis of complex traits (Curk, Rot & Zupan, 2011).
Several SNPs in genes belonging to distinct pathways have been associated with the longevity phenotype (Soerensen et al., 2012; Dato et al., 2013 and references therein, Rose et al., 2015; Crocco, Montesanto, Passarino & Rose, 2016; De Luca, Crocco, De Rango, Passarino & Rose, 2016). GWAS of human longevity in worldwide samples (North America, Europe and very recently China) generally failed to give new insights into genetic determinants of human longevity: only the TOMM40/APOE/APOC1 locus, associated with longevity, was replicated in different populations (Deelen et al., 2014; Lin et al., 2016; Newman et al., 2010; Sebastiani et al., 2012), while rs2149954 on 5q33.3 (Deelen et al., 2014; Zeng et al., 2016) and the FOXO3A locus (Broer et al., 2015 and references therein) are the other signals showing population‐specific associations. Some authors attempted to analyse epistatic intragenic (Tan, Soerensen, Kruse, Christensen & Christiansen, 2013; Zeng et al., 2010) or intergenic effects (Napolioni, Giannì, Carpi, Predazzi & Lucarini, 2011a; Napolioni et al., 2011b), however comprising a limited number of genetic variants. Recently, the analysis of the joint effect of a SNP set grouped by pathway or gene region (pathway/group‐based analysis) on human longevity was carried out by Deelen and co‐authors (Deelen et al., 2013), who analysed GWAS data of 1,021 SNPs in 68 genes from the insulin‐Igf1 signalling (IIS) pathway and 88 SNPs in 13 telomere maintenance (TM) pathway genes. The comparison of genotype data between 403 unrelated nonagenarians from the Leiden Longevity Study and 1,670 younger population controls confirmed the role of genetic variation in IIS and TM pathways in the predisposition to longevity. However, while IIS had many genes associated with the trait, the association of TM with survival was mainly determined by the POT1 gene. Similarly, we previously performed a pathway analysis of 592 SNPs in the DNA repair pathway in the same in the same study population as investigated in the present study indicating association of subprocesses of this pathway in human longevity (Debrabant et al., 2014). Here, we wanted to explore synergies inside a large SNP data set, by applying statistical methodologies allowing us to test the interaction both inside and among SNP variants belonging to three main candidate pathways of human longevity such as IIS, DNA damage signalling and repair, and pro/antioxidant response. To this aim, we analysed 1,058 SNPs from 140 genes in 1,825 subjects (1,089 cases aged 92–93, 736 controls aged 46–55) of Danish origin, previously analysed for single‐SNP associations with longevity (Soerensen et al., 2012).
2. RESULTS
In this work, we used a combination of different approaches for understanding the relationships between SNP variants in the predisposition to become long‐lived. First, we computed case–control based SNP‐SNP interaction without pathway constraints using the whole data set, and we plotted interaction networks from significant SNP combinations. Then, we performed pathway‐based case–control analysis, looking for epistatic interactions inside original assigned pathways and among different pathways by applying a MDR approach. Finally, we analysed those pairs of SNPs significantly enriched in cases respect to controls for their influence on survival in the oldest old.
2.1. SNPsyn analysis in the entire data set
By testing all possible combinations among the 1,058 available variants, assuming a significance level p < .0001 and correcting for multiple comparisons (FDR <0.005), we found a set of 73 best‐interacting SNP pairs (Table S1). Figure 1 shows the interaction network, composed by 33 intrapathway network (11 IIS; 17 DNA repair; 5 pro‐antioxidant) and 40 interpathway network (22 IIS‐DNA repair; 11 pro‐antioxidant‐DNA repair; 7 pro‐antioxidant‐IIS). Sixteen of 73 SNP‐SNP interactions were inside the same gene: these interactions could be due to noncasual association among markers (LD), however modest (r 2 < 0.8), as indicated by the positive values of synergy (Syn) among variants (all interactions reported significant showed a Syn >0). In any case, the single contributions (I1 and I2) of SNP1 and SNP2 to the synergic interaction can give information about the more influencing SNP of the pairs and thus help in prioritizing the SNP in future analysis, so that no one SNP was a priori removed (Tables 1 and S1).
Figure 1.
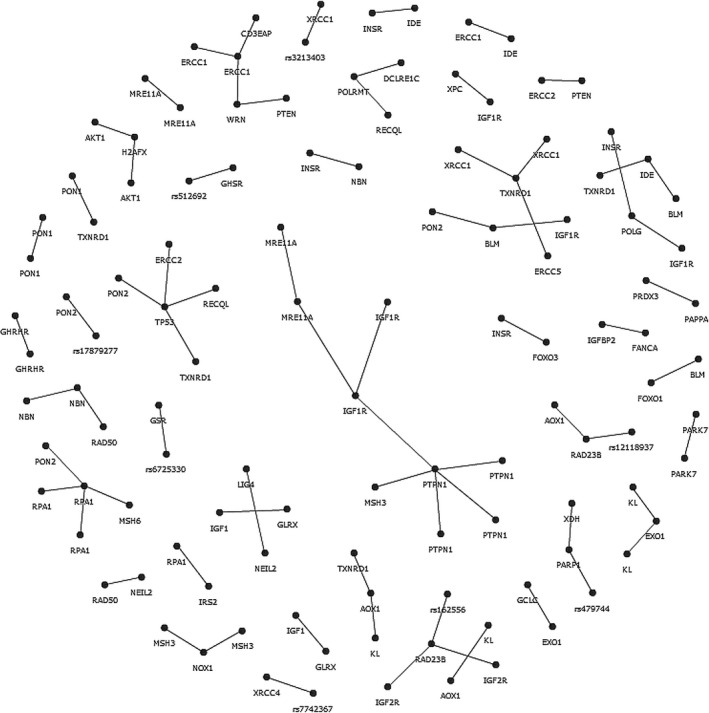
Interaction network obtained by the analysis of SNP‐SNP interactions by SNPsyn in the whole data set, comprising the all three analysed pathways (IIS, DNA repair and pro‐antioxidant). Each dot represents a SNP, indicated by the corresponding gene, when univocally assigned or by dbSNP notation, when present in a regulatory region, and not assigned by the SNPsyn output
Table 1.
List of top‐ranked 22 SNP‐SNP interactions, sorted by FDR <= 0.001, organized according to the synergy value, as resulting from SNPsyn analysis on the whole genotypic data set [1825 samples (1089 long‐lived, 736 younger controls), 1059 SNPs]. SNP‐SNP interactions inside the same gene captured by the algorithm could be due to a modest LD between markers, below the r 2 = 0.8 used as cutoff. Syn: synergy value, FDR: false discovery rate; I: information, I1 and I2: contribution of SNP1 and SNP2 to the synergic interaction, respectively
| Syn | I | p‐value | FDR | Snp 1 | Pathway | Chr 1 | Snp 2 | Y37 | Chr 2 | I 1 | I 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.0205 | 0.0325 | <.0001 | <0.0001 | GHSR (rs512692) | IIS | 3 | GHSR (rs572169) | IIS | 3 | 0.0006 | 0.0115 |
| 0.0199 | 0.0246 | <.0001 | <0.0001 | ERCC1 (rs3212961) | DNA repair | 19 | ERCC1 (rs762562) | DNA repair | 19 | 0.0034 | 0.0013 |
| 0.0181 | 0.0297 | <.0001 | <0.0001 | GHRHR (rs2267723) | IIS | 7 | GHRHR (rs4988505) | IIS | 7 | 0.0112 | 0.0004 |
| 0.0159 | 0.0214 | <.0001 | <0.0001 | PTPN1 (rs2038526) | IIS | 20 | PTPN1 (rs6067484) | IIS | 20 | 0.0001 | 0.0054 |
| 0.0143 | 0.0178 | <.0001 | <0.0001 | ERCC1 (rs3212961) | DNA repair | 19 | ERCC1 (rs3212964) | DNA repair | 19 | 0.0034 | 0.0001 |
| 0.0140 | 0.0182 | <.0001 | <0.0001 | NBN (rs12680687) | DNA repair | 8 | NBN (rs2735385) | DNA repair | 8 | 0.0041 | 0.0001 |
| 0.0126 | 0.0182 | <.0001 | <0.0001 | PTPN1 (rs2426164) | IIS | 20 | PTPN1 (rs6067484) | IIS | 20 | 0.0002 | 0.0054 |
| 0.0125 | 0.0165 | <.0001 | <0.0001 | XRCC1 (rs1799782) | DNA repair | 19 | XRCC1 (rs3213403) | DNA repair | 19 | 0.0005 | 0.0035 |
| 0.0120 | 0.0189 | <.0001 | <0.0001 | TXNRD1 (rs17202060) |
Pro/Anti oxidant |
12 | TP53 (rs2078486) | DNA repair | 17 | 0.0007 | 0.0062 |
| 0.0112 | 0.0196 | <.0001 | <0.0001 | IGF1R (rs12437963) | IIS | 15 | PTPN1 (rs6067484) | IIS | 20 | 0.0030 | 0.0054 |
| 0.0112 | 0.0186 | <.0001 | <0.0001 | TP53 (rs2078486) | DNA repair | 17 | ERCC2 (rs50871) | DNA repair | 19 | 0.0062 | 0.0012 |
| 0.0102 | 0.0165 | <.0001 | 0.0006 | PTPN1 (rs6063534) | IIS | 20 | PTPN1 (rs6067484) | IIS | 20 | 0.0008 | 0.0054 |
| 0.0098 | 0.0157 | <.0001 | 0.0006 | RPA1 (rs11656253) | DNA repair | 17 | RPA1 (rs17292175) | DNA repair | 17 | 0.0058 | 9,61E‐01 |
| 0.0097 | 0.0155 | <.0001 | 0.0006 | MRE11A (rs512150) | DNA repair | 11 | MRE11A (rs592068) | DNA repair | 11 | 0.0041 | 0.0017 |
| 0.0091 | 0.0156 | <.0001 | 0.0006 | AOX1 (rs2465661) |
Pro/Anti oxidant |
2 | RAD23B (rs11573709) | DNA repair | 9 | 0.0024 | 0.0041 |
| 0.0087 | 0.0161 | <.0001 | 0.0009 | WRN (rs11574218) | DNA repair | 8 | ERCC1 (rs3212961) | DNA repair | 19 | 0.0040 | 0.0034 |
| 0.0084 | 0.0158 | <.0001 | 0.0009 | AOX1 (rs2256977) |
Pro/Anti oxidant |
2 | KL (rs2283368) | IIS | 13 | 0.0039 | 0.0035 |
| 0.0081 | 0.0154 | <.0001 | 0.0009 | PARP1 (rs1136410) | DNA repair | 1 | FOXO3A (rs479744) | IIS | 6 | 0.0048 | 0.0024 |
| 0.0127 | 0.0147 | <.0001 | 0.0012 | PRDX3 (rs1553850) |
Pro/Anti oxidant |
10 | PAPPA (rs449807) | IIS | 9 | 0.0010 | 0.0011 |
| 0.0122 | 0.0139 | <.0001 | 0.0012 | MRE11A (rs10831227) | DNA repair | 11 | MRE11A (rs604845) | DNA repair | 11 | 0.0005 | 0.0013 |
| 0.0126 | 0.0129 | <.0001 | 0.0014 | INSR (rs2252673) | IIS | 19 | IDE (rs7078413) | IIS | 10 | 0.0001 | 0.0002 |
| 0.0120 | 0.0127 | <.0001 | 0.0014 | EXO1 (rs1635518) | DNA repair | 1 | KL (rs9527026) | IIS | 13 | 0.0003 | 0.0004 |
Further reducing the significance level, by selecting a p < .0001, we prioritized 22 top‐ranked synergies among SNPs, significantly enriched in longevity (Table 1). The best combination (FDR <0.0001) between SNPs in different genes was between TXNRD1‐rs17202060 and TP53‐rs2078486, respectively, belonging to the pro/antioxidant and DNA repair pathway. The second‐best interaction was found among IGF1R‐rs12437963 and PTPN1‐rs6067484, both belonging to the IIS pathway, and the third among genes TP53‐rs2078486 and ERCC2‐rs50871, both belonging to the DNA repair pathway.
Less significant interactions were found between TP53 and two genes from the same DNA repair pathway: TP53 (rs2078486)‐PON2 (rs2299267) and TP53 (rs2078486)‐RECQL (rs2284392) (FDR = 0.0032 and 0.0038, respectively); although not holding an FDR<0.005, they suggest a central role of TP53 in mediating the concerted activation of DNA repair and pro‐antioxidant pathways, as for example in stressful conditions.
2.2. MDR analysis
SNPsyn gave us a whole picture of the significantly interacting SNPs in the whole data set available for our study; however, that approach does not include covariates in the analysis, such as sex or age, simply dividing the data in cases and controls. Thus, to improve the case–control analysis of SNP‐SNP interactions, a MDR approach was applied. Table 2 reports the list of two‐ and three‐loci interactions prioritized by MDR analysis as the most significant for consistency value (minimum 2/10 replicates) and accuracy level (>0.5), between genes belonging to the same pathway and in the whole data set (last rows).
Table 2.
Significant results of SNP‐SNP associations with longevity, resulting by MDR analysis. Two‐ and three‐loci interactions are shown, with respective pathways, prioritized for CV consistency and training balanced accuracy (> 0.5)
| SNP combination | Genes included in the best combination | Pathways involved | High‐risk combination for longevity | OR (95% C.I.) | p‐value of interaction analysis (case–control study) by MDR | p‐value of survival analysis (Danish 1905 cohort) | |
|---|---|---|---|---|---|---|---|
| Males | Females | ||||||
| rs572169(1)/rs512692(2) | GHSR/GHSR | IIS | SNP1‐GG/SNP2‐T carriers | 0.61 (0.50–0.97) | .001 | .961 (N = 285) | .070 (N = 715) |
| rs572169(1)/rs6067484(2) | GHSR/PTPN1 | IIS | SNP1‐GG/SNP2‐AG | 0.52 (0.42–0.66) | .001 | .192 (N = 291) | .431 (N = 727) |
| rs572169(1)/rs512692(2)/rs4837525(3) | GHSR/GHSR/PAPPA | IIS | SNP1‐GG/SNP2‐TT/SNP3‐AG | 0.86 (0.68–1.09) | .001 | .716 (N = 282) | .262 (N = 709) |
| rs50871(1)/rs2078486(2) | ERCC2/TP53 | DNA repair | SNP1‐C carriers/SNP2‐G carriers | 0.80 (0.64–1.0) | .001 | .746 (N = 308) | .946 (N = 757) |
| rs170548(1)/rs2436514(2) | ATM/LONP1 | DNA repair | SNP1‐A carriers/SNP2‐AG | 0.67 (0.56–0.82) | .002 | .722 (N = 300) | .912 (N = 728) |
| rs2236270(1)/rs162557(2)/rs3756704(3) | GSS/ CYP1B1/GLRX | Pro/Antioxidant | SNP1‐AC/SNP2‐GG/SNP3‐AG | 0.69 (0.52–0.91) | .001 | .514 (N = 299) | .508 (N = 747) |
| rs572169(1)/rs533984(2) | GHSR‐MRE11A | IIS‐DNA repair | SNP1‐GG/SNP2‐G carriers | 0.55 (0.45–0.67) | .001 | .636 (N = 288) | .026 (N = 724) |
| rs572169(1)/rs533984(2)/rs225119(3) | GHSR/MRE11A/PARK7 | IIS‐DNA repair‐pro/antioxidant | SNP1‐GG/SNP2‐G carriers/SNP3‐G carriers | 0.53 (0.43–0.65) | .001 | .340 (N = 285) | .096 (N = 717) |
High‐risk genetic combinations for longevity resulting from MDR approach was tested for involvement in survival (in the oldest cohort), by analysing carriers of specific candidate combinations, respect to the other genotypes, for the sample stratified by sex. Odds ratio with 95% confidence interval (C.I.) was determined by logistic regression applied to candidate SNP combinations that were reported. p‐value of MDR was calculated by permutation analysis, while those reported for survival are referred to the log‐rank (Mantel–Cox) test.
2.2.1. MDR: Analysis intrapathways
First, the MDR confirmed the SNPsyn approach for what concerns the most significant SNP‐SNP interactions. The SNP‐SNP combination rs572169‐rs512692 inside the gene GHSR significantly interact with rs4837525 in the PAPPA gene inside the IIS pathway. rs572169‐GHSR also showed an interesting interaction with rs606748‐PTPN1. As indicated by the interaction graph (a), interaction dendrogram (b) and histograms (c) in Figure 2, these SNP‐SNP combinations jointly explain the most entropy, with the rs572169 SNP having the largest univariate effect (1.53% of explained entropy in the network), and the GG genotype driving the high‐risk combinations for longevity. The performance evaluation of each model (Table 2) shows that the two‐locus epistatic interaction performs better in terms of modelling (higher CV) compared to three‐locus interactions in these pathways. In the DNA repair sub‐data set (Figure 3), two high‐level epistatic interactions were found: one between rs50871 (ERCC2 gene) and rs2078486 (TP53), and another between rs170548 (ATM) and rs2436514 (LONP1). Although the two indicated SNP‐SNP interactions reported low CV (Table 2), they were both significant by permutation analysis (p < .002). Interestingly, the ERCC2‐TP53 interaction was highly significant also in SNPsyn analyses (FDR <.001 and p < .001), thus indicating a consistent association across methods of this SNP‐SNP combination with longevity. The largest univariate effect on longevity for the DNA repair pathway was recognized to rs13251813 (WRN gene), with 1.44% of explained entropy in the network (Figure 3). In the pro‐antioxidant data set, a three‐level SNP combination was found significantly associated with longevity among rs2236270 (GSS gene)‐ rs162557 (CYP1B1 gene)‐rs3756704‐GLRX (p < .001) (Table 2 and Figure 4).
Figure 2.
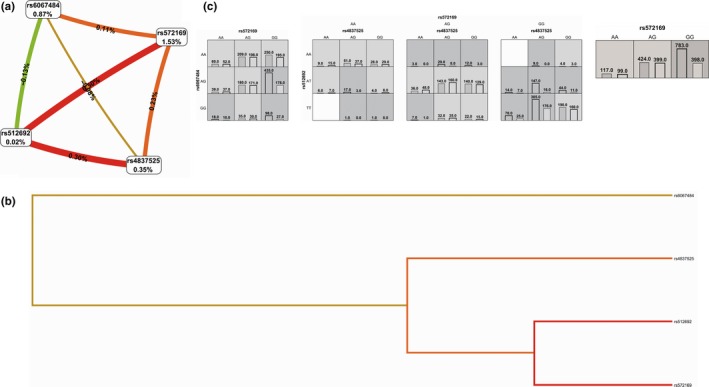
Interaction graph (a) and interaction dendrogram (b) for the IIS data set, resulting from MDR analysis. In a, for each SNP is reported in per cent the value of information gain (IG), while numbers in the connections indicate the entropy‐based IG for the SNP pairs. Red bar and orange bar indicate the high‐level synergies on the phenotype, while the brown indicate a medium‐level interaction, green and blue connections with negative IG values indicate redundancy or lack of synergistic interactions between the markers. Histograms in (c) reports the distributions of cases (left bars) and controls (right bars) for three‐locus and two‐locus genotype combinations of SNPs. The effect of rs572169(GHSR gene) is showed alone, because of its univariate effect. Dark‐shaded cells are considered “high risk,” while light‐shaded cells are considered “low risk” for longevity. White cells indicate no subjects with those genotype combinations that were observed in the data set
Figure 3.
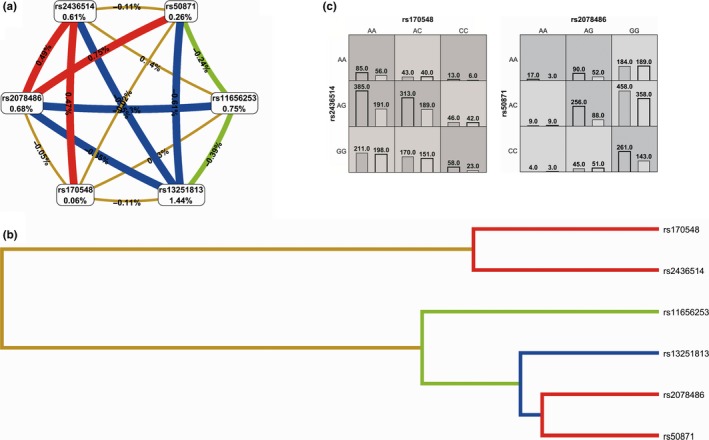
Interaction graph (a) and interaction dendrogram (b) for the DNA repair data set, resulting from MDR analysis. In a, for each SNP is reported in per cent the value of information gain (IG), while numbers in the connections indicate the entropy‐based IG for the SNP pairs. Red bar and orange bar indicate the high‐level synergies on the phenotype, while the brown indicate a medium‐level interaction, green and blue connections with negative IG values indicate redundancy or lack of synergistic interactions between the markers. Histograms in (c) report the distributions of cases (left bars) and controls (right bars) for three‐locus and two‐locus genotype combinations of SNPs. The effect of rs13251813 (WRN gene) is shown alone, because of its univariate effect. Dark‐shaded cells are considered “high risk,” while light‐shaded cells are considered “low risk.” White cells indicate no subjects with those genotype combinations that were observed in the data set
Figure 4.
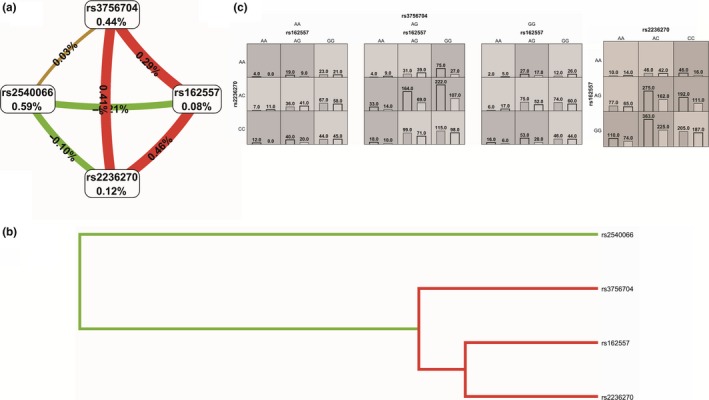
Interaction graph (a) and interaction dendrogram (b) for the pro/antioxidant data set, resulting from MDR analysis. In a, for each SNP is reported in per cent the value of information gain (IG), while numbers in the connections indicate the entropy‐based IG for the SNP pairs. Red bar and orange bar indicate the high‐level synergies on the phenotype, while the brown indicate a medium‐level interaction, green and blue connections with negative IG values indicate redundancy or lack of synergistic interactions between the markers. Histograms in (c) reports the distributions of cases (left bars) and controls (right bars) for three‐locus and two‐locus genotype combinations of SNPs. Dark‐shaded cells are considered “high‐risk” while light‐shaded cells are considered “low risk.” White cells indicate no subjects with those genotype combinations that were observed in the data set
2.2.2. MDR: Analysis interpathways
Figure 5 shows the interaction graph (a) and interaction dendrogram (b) for the whole genotypic data set, as resulting from MDR analysis. In this case, we can see whether there are significant epistatic interactions between genes belonging to different metabolic biological pathways, among those investigated in this article. A significant epistasis was found between rs572169 (GHSR gene) belonging to the IIS pathway, which still has the largest univariate effect on the longevity phenotype (1.53% explained entropy), and rs533984 (MRE11A gene), from the DNA repair pathway. rs533984 positively interacts with rs225119 (PARK7), which belongs to the pro‐antioxidant pathway. Both the two‐locus interaction GHSR‐MRE11A and the three‐locus combination with PARK7 were found to be significant (p = .001, Table 2); this may still depend on the higher univariate effect on longevity of the rs572169 SNP at the GHSR gene, with homozygotes GG favoured for longevity compared to the other genotypic combinations both in three‐ and two‐locus models.
Figure 5.
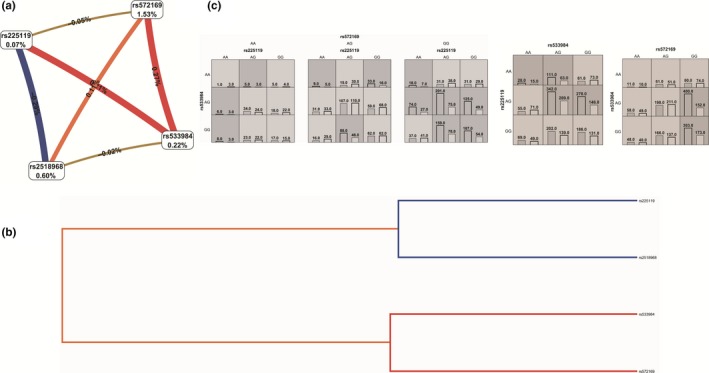
Interaction graph (a) and interaction dendrogram (b) for the DNA whole genetic data set, resulting from MDR analysis. In a, for each SNP is reported in per cent the value of Information Gain (IG), while numbers in the connections indicate the entropy‐based IG for the SNP pairs. Red bar and orange bar indicate the high‐level synergies on the phenotype, while the brown indicate a medium‐level interaction, green and blue connections with negative IG values indicate redundancy or lack of synergistic interactions between the markers. Histograms in (c) reports the distributions of cases (left bars) and controls (right bars) for three‐locus and two‐locus genotype combinations of SNPs. Dark‐shaded cells are considered “high‐risk” while light‐shaded cells are considered “low risk.” White cells indicate no subjects with those genotype combinations that were observed in the data set
2.3. Survival analysis
Based on the information about postsurvey mortality available for the oldest cohort, the Danish 1905, we tested two‐ and three‐locus interactions significantly associated with longevity status in the previous analyses. With this analysis, we aimed to test whether those combinations, associated with an advantage in older individuals compared to younger subjects, were also favourable for survival at very old ages. Thus, we classified the subjects from the 1905 Danish cohort as carriers or noncarriers of a candidate risk combination for longevity and tested their survival as number of months survived from the recruitment. We performed survival analysis separately in male and female cohorts, considering the great importance of sex in survival differences inside a human population (reviewed in Alberts et al., 2014). As indicated in Table 2, among all relevant combinations tested, only that between rs572169 (GHSR) and rs533984 (MREA11) was found significantly associated with extreme survival in females (Figure 6) (p = .026, log‐rank Mantel–Cox test). In males, no effect on survival was observed for any of the candidate risk combinations of different SNPs.
Figure 6.
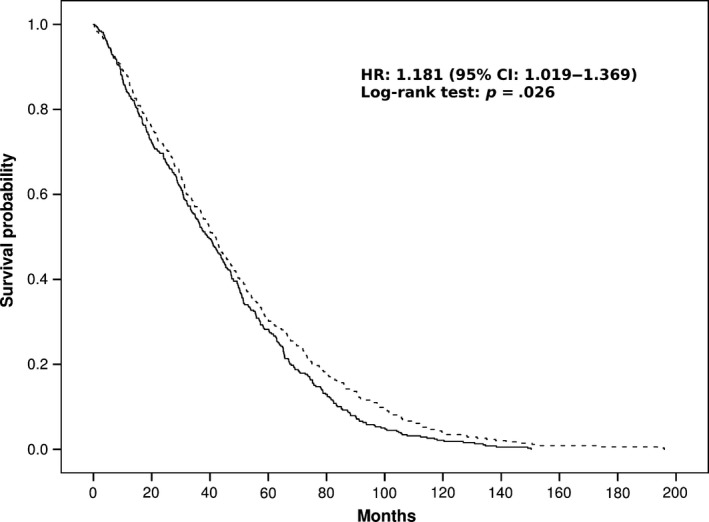
Survival function of female carriers of the protective combination rs572169(GG)‐rs533984(G/‐) (dashed line) vs. the cumulative survival function of all those carrying the other genotypic combinations at this loci (solid line). Time is expressed in months, where 0 is considered the time of recruitment, and each individual is followed up for his/her survival status till death. p‐value refers to log‐rank (Mantel–Cox) test. HR value and its confidence interval from the Cox regression analysis are reported inside the figure
3. DISCUSSION
Many reasons for the missing replicability in genetic association studies of survival to ages ≥90 years have been suggested, as it was recently reviewed by Sebastiani and coworkers, who indicated demographical reasons, that is the definition of the age windows for survival for birth cohort, as the main bias source when comparing different association studies on human longevity (Sebastiani, Nussbaum, Andersen, Black & Perls, 2016). Another possible explanation of the missing replicability and missing heritability may be related to the complexity of longevity (Dato et al., 2017), which harbours many heterogeneity sources, including the effect of rare variants, not captured by standard genomewide genotyping, and interactions between different loci, an often‐cited reason for the lack of success in genetic studies of complex diseases (Moore, 2003). With the aim of exploring this poorly investigated genetic effect, we re‐analysed a genetic data set previously described and used for single‐SNP and gene set analyses (Debrabant et al., 2014; Soerensen et al., 2012), for SNP‐SNP interactions. The findings obtained indeed indicate that an epistatic analysis approach is very much applicable for candidate gene/pathway data and hence might contribute to the knowledge concerning the genetics of human longevity.
First, we carried out SNP‐SNP association analysis by a classical case–control approach; that is, we compared the oldest‐old and youngest individuals for the frequency of relevant SNP pairs. We chose the SNPsyn software among the various existing tools reviewed in Cordell (2009), because it directly calculates the amount of synergy, also reflecting a balanced effect of two variants with equal importance. The most interesting findings of this analysis were the epistatic interactions intergenes (TP53‐DNA repair pathway/TXNRD1‐pro‐antioxidant pathway and TP53‐DNA repair pathway/ERCC2‐DNA repair pathway). Furthermore, while the interaction analysis confirmed the association of rs572169 of GHSR reported in a previous study using single‐SNP‐analysis (Soerensen et al., 2012), single variants in TP53, TXNRD1 or ERCC2 here identified were not previously found. In particular, a central role of TP53 might not seem surprising as TP53 is a very well‐known tumour suppressor in DNA damage response which balances tumour surveillance and maintenance of stem cell pools, finally resulting in beneficial effects both for cancer protection and longevity (Reinhardt & Schumacher, 2012). In addition, the strong interaction with TXNRD1, recently found associated with the quality of aging (Dato, De Rango, Crocco, Passarino & Rose, 2015), suggests a central role of TP53 in mediating the concerted activation of DNA repair and the pro‐antioxidant pathway. TP53 has been suggested to exert longevity assurance functions by impacting on the conserved IGF‐1 system (Feng & Levine, 2010), yet we did not find interactions at genetic level among TP53 and genes belonging to the IIS pathway in this study population.
According to the MDR analysis, the GHSR gene has an important univariate effect on longevity in the Danish study population; the analyses suggest synergistic interactions, favourable for longevity, with both the PAPPA and PTPN1 genes from the same IIS pathway in longevity, but also with the MRE11A gene from the DNA repair and PARK7 gene from the pro‐antioxidant pathway. Survival analysis in the oldest‐old cohort demonstrated that the interaction between GHSR and MRE11A (rs572169/rs533984) has a positive effect on female survival (p = .026), with carriers of the combination rs572169‐GG /rs533984‐G showing a less mortality. Interestingly, MRE11A, according to SNPsyn analysis, is part of a network with IGFIR, PTPN1 and MSH3. This multiple level interaction highlights the interaction between the IIS pathway (IGF1R and PTPN1) and DNA repair (MRE11A and MSH3). The link between the IGF1R‐PTPN1 proteins is known: literature data indicate a crucial role of PTPN1 as a tumour suppressor, able to negatively regulate multiple pathways of cell growth directly through IGF1R and IR, or indirectly through leptin receptor GHR (Neel & Tonks, 2016). Experimental data report that the protein interaction IGF1R‐PTPN1 is regulated by insulin levels (Bandyopadhyay et al., 1997). Furthermore, in experimental organisms, PTPN1 is involved in inflammatory mechanisms and insulin resistance associated with diabetes and obesity during aging (González‐Rodríguez et al., 2012). Thus, although still not studied in relation to successful aging, the observed interaction among genetic variants of IGF1R and PTPN1 in Danish long‐lived could reflect a conserved mechanism of cellular signalling, involved in the development of metabolic diseases and tumorigenesis.
In conclusion, the results here presented illustrate the validity of interaction approach and network analyses for investigating the genetic contribution in human longevity. The analyses not only confirm a gene previously identified by single‐SNP‐analysis in the same data set (GHSR), but found novel interaction partners of such gene (PAPPA, PTPN1, MRE11A and PARK7). Furthermore, this methodology identifies novel genes as central nodes of additional networks (e.g., TP53) involved in human longevity. Obviously, these interesting findings will need further validation, possibly by the confirmation of top SNP‐SNP interactions found in this work independently replicated in additional study populations. Moreover, functional analyses (expression studies and protein interaction testing) should be implemented, to determine the biological relevance of the statistical interactions.
4. EXPERIMENTAL PROCEDURES
4.1. Study population and genetic data set
The characteristics of the study population were elsewhere reported (Soerensen et al., 2012) and summarized in Table S2 (Supplementary materials). Briefly, the sample studied here included 1,089 cases (mean age: 93.1 years, age range 92.2–93.8 years, 29% males and 71% females) drawn from eligible members of the Danish 1905 Birth Cohort Study (Nybo et al., 2001) and 736 controls (mean age 50.6, age range 46.0–55.0, gender distribution 50% males and 50% females) from the Study of Middle Aged Danish Twins (MADT) (Skytthe et al., 2013). Information on survival status per 1st of January 2015 for the oldest‐old was retrieved from the Danish Central Population Register (Pedersen, Gotzsche, Moller & Mortensen, 2006). Permission to collect blood samples and to use survey information was granted by The Danish Regional Committees on Biomedical Research Ethics.
The genotype data set and genotyping methodology by the Illumina GoldenGate platform has been described in previous papers (Soerensen et al., 2010, 2012). To exclude the rare variants, a minimum allele frequency (MAF) level of 5% was chosen for performing the analyses, leaving 1,058 SNPs from the original data set to test SNP‐SNP associations with longevity. A table reporting all SNPs investigated can be found in the Supplementary material (Table S3), classified by gene and pathway. In detail, 317 SNPs were belonging to the IIS pathway, 260 to pro‐antioxidant response, 481 to DNA damage signalling and repair. Furthermore, functional prediction of most significant SNPs in SNP pairs found to be associated with longevity, based on Genome Browser information (Haploreg, http://archive.broadinstitute.org/mammals/haploreg/haploreg.php) was reported in Table S4.
4.2. Statistical methodology
Genotype counts for testing HWE were performed in the control group sample (Li & Li, 2008; Schaid, Batzler, Jenkins & Hildebrandt, 2006) by Plink (Purcell et al., 2007), choosing a cutoff of 0.001 for excluding SNPs not in HWE in the control sample. Minor allele frequency (MAF) was computed for each SNP, and differences in genotype frequency between cases controls were calculated by RobustSNP (So & Sham, 2011) in the R environment (https://wwwr-project.org/). As reported in Soerensen et al., 2012; tagging SNPs were prioritized by establishing a cutoff of r 2 = .8, LOD = 3, minor allele frequency (MAF)>5% and a minimum distance between SNPs = 60 bp criteria. The selection of common variants (MAF ≥ 5) and a LD r 2 < .8 is widely used in genomewide association studies (Fadista, Manning, Florez & Groop, 2016). Furthermore, we tested LD levels by genomic region in the control sample (data not shown).
4.3. SNPsyn analysis of SNP‐SNP interactions
SNP‐SNP interactions significantly differing in frequency between cases and controls were computed by SNPsyn, according to the method described in Curk et al., 2011; which allows the discovery of synergistic pairs of SNPs from large data sets in association studies with complex traits. Synergy (Syn) among SNPs was estimated by an information‐theoretic approach, without specifying interaction models, but assuming an additive model, where the expected amount of information on the phenotype for a combination of SNPs is equal to the sum of information of the individual SNPs. For each SNP‐SNP combination, information gain (IG) and entropy were calculated, which corresponds to the level of association between markers and phenotype. Pairs of SNPs with positive synergy (Syn>0) were called synergistic; negative synergy (Syn<0) indicates that the two SNPs carry redundant information, an effect typically observed among highly correlated SNPs in LD. Information about pathway membership by Gene Ontology was used for performing enrichment analysis on subpathways, for genes associated with the phenotype. Obtained significance scores were corrected for multiple testing using the false discovery rate (FDR) method described by Benjamini and Yekutieli (2001). In this paper, significant associations were considered those reporting a FDR< 0.005 (p‐value <.0001). Because FDR is a measure of the per cent of false‐positive results, and it is inversely proportional to the number of SNPs tested, we chose a stringent level to minimize the false‐positive combinations in our analysis.
Interaction analyses were computed online, at the website http://snpsyn.biolab.si/.
4.4. Gene set enrichment analysis
Gene set enrichment analysis was performed by SNPsyn on the whole data set to find biological processes in which represented genes were involved in human longevity (based on the known information about their gene products (data driven by Gene Ontology)). Which gene sets to test were determined by SNPsyn based on significant association with the phenotype (as previously explained). Subsequently, the gene sets were used as a cluster (query) set in this gene enrichment analysis, with the goal to determine whether members of a gene set tend to occur towards the top (or bottom) of the list. In such case, the gene set was correlated with the phenotypic class distinction. Fold enrichment score reflects the degree to which a set is overrepresented at the extremes (top or bottom) of the entire ranked list.
4.5. MDR analysis of epistatic interactions
For testing the epistatic interaction between pairs of SNPs, multifactor dimensionality reduction (MDR) was applied (Moore, 2004; Ritchie et al., 2001). This methodology can reveal high‐order interactions among genes collaborating with respect to a given phenotype, with the aim of determining multilocus genotype combinations associated with high or low risk of disease. The entropy‐based clustering algorithm used by MDR sets a contingency table for k SNPs and calculates case–control ratios for each of the possible multilocus genotypes. Hence, a genotype combination is considered high‐level if more present in cases respect to controls. The MDR interaction model describes percentage of entropy (information gain or IG) by each factor (values in the nodes indicate independent main effect) or 2‐way interaction. Graphical visualization through connections among the markers help to interpret additive and nonadditive interactions effects on phenotype: positive values of entropy indicate synergistic or nonadditive interaction while negative entropy values indicate redundancy between the markers or lack of any synergistic interaction between the markers. As for colours, connections in red and orange indicate nonlinear interaction, connections in green and brown indicate independence or additivity and redundancy (blue lines). In a data set of more than two interacting factors, the best model is considered the one found more consistent in different replicates (expressed as CV, consistency values and accuracy, i.e. training balanced accuracy level). For calculating significance, permutation testing is applied, dividing the data set into 10 portions, and using nine portions as a training data set, and the remaining as a testing data set. One thousand permutations were performed, to determine a cutoff threshold for an α = 0.05 significance level. Missing genotypes were imputed with the MDR data tool software (version 0.4.3), which allows the global replacement of unknown values and the imputation of the data from a model constructed by the software from the existing data set.
MDR analyses were implemented in the open‐source MDR software package version 3.0.2 (available on http://www.multifactordimensionalityreduction.org/).
4.6. Survival analysis of relevant SNP‐SNP interactions
Kaplan–Meier survival curves with the Mantel–Cox log‐rank test were used to compare survival of oldest‐old subjects with different SNP‐SNP combinations (Therneau & Grambsch, 2000). Survival analyses were performed using the survival package in R, as described in Therneau (2005). A significance level of 0.05 was chosen in all tests.
AUTHOR CONTRIBUTIONS
Serena Dato drafted the manuscript and generated the conception of the study, and participated in the data acquisition and interpretation of data. Mette Soerensen generated the study design and participated in the data acquisition, data management and interpretation of data. Francesco De Rango carried out the statistical analyses and participated in the interpretation of data. Giuseppina Rose participated in the interpretation of data and revision of the manuscript. Lene Christiansen participated in generating the study design, acquisition of data and interpretation of data. Giuseppe Passarino participated in the interpretation of data and revision of the manuscript. Kaare Christensen participated in the acquisition of data and interpretation of data. All authors have revised the manuscript and given their final approval.
Supporting information
ACKNOWLEDGMENTS
This study was supported by the INTERREG 4 A programme Syddanmark‐Schleswig‐K.E.R.N (by EU funds from the European Regional Development Fund), the National Institute on Aging (P01 AG08761), and the European Union's Seventh Framework Programme (FP7/2007‐2011) under Grant Agreement No. 259679, the Novo Nordisk Foundation. The Danish Aging Research Center was supported by a grant from the VELUX Foundation. SADEL S.p.A (San Teodoro, San Raffaele, Villa del Rosario, A.G.I. srl, SAVELLI HOSPITAL, Casa di Cura Madonna dello Scoglio), in the frame of the agreement “SOLUZIONI INNOVATIVE PER L'INNALZAMENTO DELLA SALUTE E DELLA SICUREZZA DELLA POPOLAZIONE” with the University of Calabria.
CONFLICT OF INTEREST
None declared.
Dato S, Soerensen M, De Rango F, et al. The genetic component of human longevity: New insights from the analysis of pathway‐based SNP‐SNP interactions. Aging Cell. 2018;17:e12755 10.1111/acel.12755
REFERENCES
- Alberts, S. C. , Archie, E. A. , Gesquiere L. R., Altmann J., Vaupel J. W., Christensen K. (2014). The male‐female health‐survival paradox: A comparative perspective on sex differences in aging and mortality In Committee on Population; Division of Behavioral and Social Sciences and Education; National Research Council ; Weinstein M., Lane M. A. (Eds.), Sociality, hierarchy, health: Comparative biodemography: A collection of papers (15 pp). Washington, DC: National Academies Press (US); 22. [PubMed] [Google Scholar]
- Bandyopadhyay, D. , Kusari, A. , Kenner, K. A. , Liu, F. , Chernoff, J. , Gustafson, T. A. , & Kusari, J. (1997). Protein‐tyrosine phosphatase 1B complexes with the insulin receptor in vivo and is tyrosine‐phosphorylated in the presence of insulin. Journal of Biological Chemistry, 272(3), 1639–1645. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. , & Yekutieli, D. (2001). The control of the false discovery rate in multiple testing under dependency. Annals of Statistics, 29, 1165–1188. [Google Scholar]
- Broer, L. , Buchman, A. S. , Deelen, J. , Evans, D. S. , Faul, J. D. , Lunetta, K. L. , … Murabito, J. M. (2015). GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 70(1), 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell, H. J. (2009). Detecting gene‐gene interactions that underlie human diseases. Nature Reviews Genetics, 10(6), 392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocco, P. , Montesanto, A. , Passarino, G. , & Rose, G. (2016). Polymorphisms falling within putative miRNA target sites in the 3'UTR region of SIRT2 and DRD2 genes are correlated with human Longevity. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 71(5), 586–592. [DOI] [PubMed] [Google Scholar]
- Curk, T. , Rot, G. , & Zupan, B. (2011). SNPsyn: Detection and exploration of SNP‐SNP interactions. Nucleic Acids Research, 39, W444–W449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dato, S. , Crocco, P. , D'Aquila, P. , de Rango, F. , Bellizzi, D. , Rose, G. , & Passarino, G. (2013). Exploring the role of genetic variability and lifestyle in oxidative stress response for healthy aging and longevity. International Journal of Molecular Sciences, 14(8), 16443–16472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dato, S. , De Rango, F. , Crocco, P. , Passarino, G. , & Rose, G. (2015). Antioxidants and quality of aging: Further evidences for a major role of TXNRD1 gene variability on physical performance at old age. Oxidative Medicine and Cellular Longevity, 2015, 926067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dato, S. , Rose, G. , Crocco, P. , Monti, D. , Garagnani, P. , Franceschi, C. , & Passarino, G. (2017). The genetics of human longevity: An intricacy of genes, environment, culture and microbiome. Mechanisms of Ageing and Development, 165(Pt B), 147–155. [DOI] [PubMed] [Google Scholar]
- De Luca, M. , Crocco, P. , De Rango, F. , Passarino, G. , & Rose, G. (2016). Association of the Laminin, Alpha 5 (LAMA5) rs4925386 with height and longevity in an elderly population from Southern Italy. Mechanisms of Ageing and Development, 155, 55–59. [DOI] [PubMed] [Google Scholar]
- Debrabant, B. , Soerensen, M. , Flachsbart, F. , Dato, S. , Mengel‐From, J. , Stevnsner, T. , … Christiansen, L. (2014). Human longevity and variation in DNA damage response and repair: Study of the contribution of sub‐processes using competitive gene‐set analysis. European Journal of Human Genetics, 22(9), 1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen, J. , Beekman, M. , Uh, H. W. , Broer, L. , Ayers, K. L. , Tan, Q. , … Slagboom, P. E. (2014). Genome‐wide association meta‐analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Human Molecular Genetics, 23(16), 4420–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen, J. , Uh, H. W. , Monajemi, R. , van Heemst, D. , Thijssen, P. E. , Böhringer, S. , … Beekman, M. (2013). Gene set analysis of GWAS data for human longevity highlights the relevance of the insulin/IGF‐1 signaling and telomere maintenance pathways. Age (Dordr), 35(1), 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista, J. , Manning, A. K. , Florez, J. C. , & Groop, L. (2016). The (in)famous GWAS P‐value threshold revisited and updated for low‐frequency variants. European Journal of Human Genetics, 24, 1202–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z. , & Levine, A. J. (2010). The regulation of energy metabolism and the IGF‐1/mTOR pathways by the p53 protein. Trends in Cell Biology, 20, 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke, J. , Lorenz, K. , & Cohen, B. (2009). Genetic interactions between transcription factors cause natural variation in yeast. Science, 323, 498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Rodríguez, A. , Más‐Gutierrez, J. A. , Mirasierra, M. , Fernandez‐Pérez, A. , Lee, Y. J. , Ko, H. J. , … Valverde, A. M. (2012). Essential role of protein tyrosine phosphatase 1B in obesity‐induced inflammation and peripheral insulin resistance during aging. Aging Cell, 11(2), 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskind, A. M. , McGue, M. , Holm, N. V. , Sørensen, T. I. , Harvald, B. , & Vaupel, J. W. (1996). The heritability of human longevity: A population‐based study of 2872 Danish twin pairs born 1870‐1900. Human Genetics, 97(3), 319–323. [DOI] [PubMed] [Google Scholar]
- Johnson, S. C. , Dong, X. , Vijg, J. , & Suh, Y. (2015). Genetic evidence for common pathways in human age‐related diseases. Aging Cell, 14, 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , & Li, C. (2008). Assessing departure from Hardy‐Weinberg equilibrium in the presence of disease association. Genetic Epidemiology, 32, 589–599. [DOI] [PubMed] [Google Scholar]
- Lin, R. , Zhang, Y. , Yan, D. , Liao, X. , Gong, G. , Hu, J. , … Cai, W. (2016). Association of common variants in TOMM40/APOE/APOC1 region with human longevity in a Chinese population. Journal of Human Genetics, 61(4), 323–328. [DOI] [PubMed] [Google Scholar]
- Moore, J. H. (2003). The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Human Heredity, 56, 73–82. [DOI] [PubMed] [Google Scholar]
- Moore, J. H. (2004). Computational analysis of gene‐gene interactions using multifactor dimensionality reduction. Expert Review of Molecular Diagnostics, 4, 795–803. [DOI] [PubMed] [Google Scholar]
- Napolioni, V. , Carpi, F. M. , Giannì, P. , Sacco, R. , Di Blasio, L. , Mignini, F. , … Persico, A. M. (2011b). Age‐ and gender‐specific epistasis between ADA and TNF‐α influences human life‐expectancy. Cytokine, 56(2), 481–488. [DOI] [PubMed] [Google Scholar]
- Napolioni, V. , Giannì, P. , Carpi, F. M. , Predazzi, I. M. , & Lucarini, N. (2011a). APOE haplotypes are associated with human longevity in a Central Italy population: Evidence for epistasis with HP 1/2 polymorphism. Clinica Chimica Acta, 412(19–20), 1821–1824. [DOI] [PubMed] [Google Scholar]
- Neel, B. G. , Tonks, N. K. (2016). Protein tyrosine phosphatases in cancer. 10.1007/978-1-4939-3649-6: Springer Editors. [DOI] [Google Scholar]
- Newman, A. B. , Walter, S. , Lunetta, K. L. , Garcia, M. E. , Slagboom, P. E. , Christensen, K. , … Murabito, J. M. (2010). A meta‐analysis of four genome‐wide association studies of survival to age 90 years or older: The Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 65(5), 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybo, H. , Gaist, D. , Jeune, B. , Bathum, L. , McGue, M. , Vaupel, J. W. , & Christensen, K. (2001). The Danish 1905 cohort: A genetic‐epidemiological nationwide survey. Journal of Aging and Health, 13(1), 32–46. [DOI] [PubMed] [Google Scholar]
- Pedersen, C. B. , Gotzsche, H. , Moller, J. O. , & Mortensen, P. B. (2006). The Danish Civil Registration System. A cohort of eight million persons. Danish Medical Bulletin, 53, 441–449. [PubMed] [Google Scholar]
- Purcell, S. , Neale, B. , Todd‐Brown, K. , Thomas, L. , Ferreira, M. A. R. , Bender, D. , … Sham, P. C. (2007). PLINK: A toolset for whole‐genome association and population‐based linkage analysis. American Journal of Human Genetics, 81(3), 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, H. C. , & Schumacher, B. (2012). The p53 network: Cellular and systemic DNA damage responses in aging and cancer. Trends in Genetics, 28(3), 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M. D. , Hahn, L. W. , Roodi, N. , Bailey, L. R. , Dupont, W. D. , Parl, F. F. , & Moore, J. H. (2001). Multifactor‐dimensionality reduction reveals high‐order interactions among estrogen‐metabolism genes in sporadic breast cancer. American Journal of Human Genetics, 69, 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, G. , Crocco, P. , De Rango, F. , Corsonello, A. , Lattanzio, F. , De Luca, M. , & Passarino, G. (2015). Metabolism and successful aging: Polymorphic variation of syndecan‐4 (SDC4) gene associate with longevity and lipid profile in healthy elderly Italian subjects. Mechanisms of Ageing and Development, 150, 27–33. [DOI] [PubMed] [Google Scholar]
- Schaid, D. J. , Batzler, A. J. , Jenkins, G. D. , & Hildebrandt, M. A. (2006). Exact tests of Hardy‐Weinberg equilibrium and homogeneity of disequilibrium across strata. American Journal of Human Genetics, 79, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani, P. , Nussbaum, L. , Andersen, S. L. , Black, M. J. , & Perls, T. T. (2016). Increasing Sibling Relative Risk of Survival to Older and Older Ages and the Importance of Precise Definitions of “Aging”, “Life Span”, and “Longevity”. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 71(3), 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani, P. , Solovieff, N. , Dewan, A. T. , Walsh, K. M. , Puca, A. , Hartley, S. W. , … Perls, T. T. (2012). Genetic signatures of exceptional longevity in humans. PLoS ONE, 7(1), e29848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skytthe, A. , Christiansen, L. , Kyvik, K. O. , Bødker, F. L. , Hvidberg, L. , Petersen, I. , … Christensen, K. (2013). The Danish Twin Registry: Linking surveys, national registers, and biological information. Twin Research and Human Genetics, 16, 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So, H. C. , & Sham, P. C. (2011). Robust association tests under different genetic models, allowing for binary or quantitative traits and covariates. Behavior Genetics, 41(5), 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen, M. , Dato, S. , Christensen, K. , McGue, M. , Stevnsner, T. , Bohr, V. A. , & Christiansen, L. (2010). Replication of an association of variation in the FOXO3A gene with human longevity using both case‐control and longitudinal data. Aging Cell, 9(6), 1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen, M. , Dato, S. , Tan, Q. , Thinggaard, M. , Kleindorp, R. , Beekman, M. , … Christiansen, L. (2012). Human longevity and variation in GH/IGF‐1/insulin signaling, DNA damage signaling and repair and pro/antioxidant pathway genes: Cross sectional and longitudinal studies. Experimental Gerontology, 47(5), 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, L. , Liu, G. , Wang, H. , Tian, Y. , Zhou, Z. , Han, L. , & Yan, L. (2015). Research on single nucleotide polymorphisms interaction detection from network perspective. PLoS ONE, 10(3), e0119146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Q. , Soerensen, M. , Kruse, T. A. , Christensen, K. , & Christiansen, L. (2013). A novel permutation test for case‐only analysis identifies epistatic effects on human longevity in the FOXO gene family. Aging Cell, 12(4), 690–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T. (2005). A Package for Survival Analysis in S. version 2.38. Retrieved from https://CRAN.R-project.org/package=survival.
- Therneau, T. M. , & Grambsch, P. M. (2000). Modeling survival data: Extending the cox model. New York: Springer. [Google Scholar]
- Zeng, Y. , Cheng, L. , Chen, H. , Cao, H. , Hauser, E. R. , Liu, Y. , … Vaupel, J. W. (2010). Effects of FOXO genotypes on longevity: A biodemographic analysis. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 65(12), 1285–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y. , Nie, C. , Min, J. , Liu, X. , Li, M. , Chen, H. , … Vaupel, J. W. (2016). Novel loci and pathways significantly associated with longevity. Scientific Reports, 6, 21243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


