Abstract
We present the results of studies of an unmodified version of the recombinant major barley (Hordeum vulgare) endosperm ADP-glucose pyrophoshorylase (AGPase) expressed in insect cells, which corroborate previous data that this isoform of the enzyme acts independently of the allosteric regulators 3-phosphoglycerate and inorganic phosphate. We also present a characterization of the individual subunits expressed separately in insect cells, showing that the SS AGPase is active in the presence of 3-phosphoglycerate and is inhibited by inorganic phosphate. As a step toward the elucidation of the role of the two AGPase isoforms in barley, the temporal and spatial expression profile of the four barley AGPase transcripts encoding these isoforms were studied. The results show that the steady-state level of beps and bepl, the transcripts encoding the major endosperm isoform, correlated positively with the rate of endosperm starch accumulation. In contrast, blps and blpl, the transcripts encoding the major leaf isoform, were constitutively expressed at a very low steady-state level throughout the barley plant. The implications of these findings for the evolution of plant AGPases are discussed.
ADP-Glc pyrophosphorylase (ATP: α-Glc-1-P adenylyl-transferase, EC 2.7.7.27) (AGPase) is a key enzyme in the biosynthesis of starch in plants and glycogen in bacteria. AGPase catalyzes the conversion of Glc-1-P to ADP-Glc, the glucosyl donor of starch polymers (amylose and amylopectin). The enzyme has been characterized in many different plant species, including lower plants, monocots, and dicots, and is composed of heterotetramers of two large subunits (LS) and two small subunits (SS) that are evolutionary related (Preiss, 1991). The sequence of the SS are highly conserved, whereas those of the LS are more divergent (Smith-White and Preiss, 1992). The SS from potato tuber, when expressed alone in Escherichia coli (Ballicora et al., 1995), is able to form a homotetramer with activity in the presence of high concentrations of 3-phosphoglycerate (3-PGA). This led to the suggestion that the SS plays a primary role in catalysis, whereas the LS functions to modulate the regulatory response of the SS.
In barley (Hordeum vulgare), three AGPase genes encode four transcripts throughout the plant (Kilian et al., 1994). Of these, two genes encode two different LS transcripts, bepl (barley endosperm pyrophosphorylase large) and blpl (barley leaf pyrophosphorylase large) (Villand et al., 1992; Eimert et al., 1997), first described in endosperm and leaves, respectively. In addition, one gene encodes the two SS transcripts beps (barley endosperm pyrophosphorylase small) and blps (barley leaf pyrophosphorylase small) (Villand et al., 1992; Thorbjørnsen et al., 1996b), which are identical in 90% of their length. To account for the difference of the SS transcripts in the 5′end, Thorbjørnsen et al. (1996b) proposed a model in which the unique 5′ end of the beps transcript is encoded by the first exon of the SS gene and the 5′ end of the blps transcript is encoded by the second exon, with the common part of the transcripts being coded by the remaining nine exons. Preferred expression of the two subunits in endosperm and leaves is explained by alternative promoter usage, with the promoter responsible for the major endosperm transcript being located upstream of the first exon and the leaf-preferred transcription being driven by a promoter located within the first intron of the SS gene.
In chloroplasts of photosynthetic tissues including barley leaves, the AGPase is allosterically activated by 3-PGA and inhibited by Pi (Gosh and Preiss, 1966; Sanwal et al., 1968; Plaxton and Preiss, 1987; Kleczkowski et al., 1993b). The ratio of 3-PGA/Pi being the major determinant of AGPase activity. In storage organs, however, the role of allosteric regulation of AGPase is less clear. Some non-photosynthetic tissues, such as wheat endosperm (Olive et al., 1989), pea embryo (Hylton and Smith., 1992), and bean embryo (Weber et al., 1995), contain AGPase isoforms with low sensitivity or insensitivity to 3-PGA/Pi regulation. To date, the most extensively studied unregulated storage organ AGPase is that of the barley endosperm, which in crude extracts was reported to be highly active without 3-PGA stimulation and insensitive to Pi inhibition (Kleczkowski et al., 1993a).
To eliminate proteolytic modification as a source of the barley endosperm AGPase insensitivity to allosteric regulators, the major endosperm isoform encoded by the bepl and beps transcripts was expressed in insect cells (Spodoptera frugiperda) and partially purified using a 6×His tag attached to the N terminus of the SS (Rudi et al., 1997). The results of these studies strongly support previous results showing that the major barley endosperm is insensitive to allosteric regulation. Whether the single endosperm AGPase subunits have activity as homotetramers similar to that of the potato SS (Ballicora et al., 1995) has not been determined. In this study, restoration of the N terminus of SS restored sensitivity to Pi inhibition.
Based on the expression profile of AGPase transcripts in wheat, Ainsworth et al. (1995) suggested that the SS may function as a homotetrameric enzyme during the early stages of endosperm development. Studying naturally occurring maize null mutants of the large AGPase subunit (Sh2) and the SS (Bt2) loci, Greene and Hannah (1998) concluded that, in the absence of Shrunken2, the Brittle2 subunit remains as an unactive monomer in the developing endosperm. In contrast, in the absence of BT2, the Shrunken2 protein is found in a complex of 100 kD. Interestingly, although the SS are highly conserved, they behave differently in potato, wheat, and maize.
Contrary to the plastidial location of the leaf AGPase, cell fractionation studies have indicated that the major barley and maize endosperm AGPase isoforms are predominantly located in the cytosol (Denyer et al., 1996; Thorbjørnsen et al., 1996a; Shannon et al., 1998). This is consistent with the absence of consensus transit peptides from the predicted amino acid sequences for these subunits (Thorbjørnsen et al., 1996a; Eimert et al., 1997). Based on northern-blot analysis, all four AGPase transcripts are present in the endosperm, suggesting that endosperm cells harbor the enzyme in the cytosol as well as in amyloplasts. However, the role of the two isoforms in endosperm starch synthesis is uncertain, since Thorbjørnsen et al. (1996a) reported calculations indicating that the plastidial AGPase activity in barley endosperm was sufficient to account for the total starch accumulation.
We report the results of studies of an unmodified version of the recombinant major barley endosperm AGPase expressed in insect cells, including the holoenzyme and the individual subunits. As a step toward the elucidation of the full role of the AGPase isoforms in barley, we have also studied the temporal and spatial expression of the four barley AGPase transcripts using transcript-specific probes and primers in northern-blot and reverse transcriptase (RT)-PCR analyses.
MATERIALS AND METHODS
AGPase Expression in Insect Cells
A Spodoptera frugiperda (Sf9) cell culture was purchased from Invitrogen (Carlsbad, CA). A BaculoGold transfection kit and the pAcSG-His-NT and pAcUW51 plasmids were purchased from PharMingen International (San Diego). TNM-FH medium (powder) was from Sigma Chemical (Poole, Dorset, UK). Fetal calf serum and antibiotics were from GIBCO-BRL/Life Technologies (Renfrewshire, Scotland). DNA restriction enzymes were from New England Biolabs (Hertfordshire, UK) and Promega (Madison, WI). α-d-[U-14C]Glc-1-P (specific activity 287 mCi/mmol) was purchased from Amersham Pharmacia Biotech AB (Uppsala). All other biochemicals, reagents, and coupling enzymes, unless otherwise stated, were obtained from Sigma Chemical.
Construction of Recombinant Viruses
All manipulations in molecular cloning were performed according to the method of Sambrook et al. (1989). Full-length cDNAs of bepL10 and bepsF2, encoding LS (Villand et al., 1992) and SS (Thorbjørnsen et al., 1996a), respectively, of the major barley (Hordeum vulgare) endosperm AGPase, were excised from recombinant Bluescript plasmids and subcloned into the transfer plasmid pAcUW51 (PharMingen) such that expression of the LS and SS cDNAs were under the control of the p10 and polyhedrin promoters, respectively (designated pAcUW51-AGPase). Recombinant pAcUW51 containing either the bepL10 or the bepsF2 in similar orientation was also prepared (designated pAcUW51-LS and pAcUW51-SS, respectively). Recombinant plasmid constructs were co-transfected into Spodoptera frugiperda Sf9 cells with a modified Autographa californica nuclear polyhedrosis virus DNA using the transfection kit (PharMingen), and recombinant viruses were purified with one round of viral amplification and one round of plaque assay (O'Reilly et al., 1992).
Expression of Recombinant Proteins in Sf9 Cell Culture
Insect cells infected with recombinant viruses were grown at 27°C as monolayer cultures in TNM-FH complete insect medium (Sigma Chemical) supplemented with 10% (v/v) fetal calf serum, 5 μg/mL fungizone, 10 μg/mL penicillin, and 10 μg/mL streptomycin (GIBCO-BRL/Life Technologies). In a typical experiment, Sf9 cells (3.0 × 106 cells/10 mL) seeded in a 100-mm Petri dish (Corning Costar, Pleasanton, CA) were infected with at least 10 multiplicity of infection of recombinant virus and grown at 27°C for 3 d. Infected cells were harvested, collected by centrifugation at 200g for 5 min, and the pellet was washed with 1 mL of phosphate saline buffer (pH 7.4) containing 2 mm MgCl2, 10% (w/v) Suc, 2 mm DTT, 0.5 mm phenylmethylsulfonyl fluoride (PMSF), 10 μg/mL leupeptin, 10 μg/mL benamide, and 10 μg/mL pepstatin (extraction buffer). After three cycles of freezing and thawing, the cell suspension was vortexed for 15 s and centrifuged at 15,000g for 10 min. Insoluble material was washed once with 200 μL of extraction buffer. Soluble fractions were combined and stored at −20°C.
Purification of Expressed AGPase
Sf9 cells seeded in 12 100-mm Petri dishes (30 × 106 cells/dish) were infected with recombinant virus carrying both cDNAs of the barley endosperm AGPase. After 3 d, infected cells were harvested, resuspended in extraction buffer, and the crude cell extract (final volume of 12 mL) was prepared as described above. All purification steps were carried out at 4°C unless otherwise indicated. Ammonium sulfate (stock solution 50% [w/v]) was added to the crude extract to a concentration of 20% (w/v), and the precipitant was collected by centrifugation (15,000g, 15 min). The resulting pellet was resuspended in extraction buffer (2 mL), and insoluble material was further removed by centrifugation. The supernatant was diluted to 15 mL with 40 mm 3-(N-morpholino)-propanesulfonic acid (MOPS) buffer (pH 7.4) containing 2 mm MgCl2, 2 mm DTT, 0.5 mm PMSF, and 20% (w/v) Suc (buffer A), and expressed AGPase was absorbed into a high-Q column (5-mL cartridge, Econo-Pac, Bio-Rad, Hercules, CA) and eluted from the column with a linear gradient of 0 to 0.6 m NaCl prepared in the same buffer (total volume 60 mL).
Fractions containing high AGPase activity in the range of 0.2 to 0.25 m NaCl were combined and applied to a hydroxyapatite column (CHTII 1-mL cartridge, Bio-Rad), and expressed AGPase was eluted by applying a linear gradient of 0 to 200 mm phosphate prepared in buffer A (40 mL). Active fractions were pooled, equilibrated to 1 m phosphate by adding a stock solution of 2.5 m phosphate (pH 7.4) containing 20% (w/v) Suc, and incubated in batch for 1 h with 2 mL of aminopropyl-agarose. After washing with 50 mL of 1 m phosphate (pH 7.4)/20% (w/v) Suc solution, C3 resin was packed into a 10-mL disposable column (Bio-Rad), and expressed AGPase was eluted with 5 mL of buffer A. The resulting eluate was diluted at least 3-fold with buffer A and applied to the high-Q column previously equilibrated in the same buffer. Expressed AGPase was eluted with a 0 to 0.6 m NaCl linear gradient (30 mL). Fractions (2.5 mL) containing the highest enzyme activity were combined and stored in small aliquots at −80°C. Purified enzyme stored under these conditions was fully active for at least 3 months.
Preparation of Barley Endosperm Extract
Barley plants were grown in the field, and grains were harvested 15 to 20 d after anthesis and stored at −80°C (Kleczkowski et al., 1993a). Extruded starchy endosperm (from approximately 20 seeds) was ground using a mortar and pestle in 3 mL of extraction solution (40 mm MOPS buffer [pH 7.4] containing 2 mm MgCl2, 10% [w/v] Suc, 2 mm DTT, 0.5 mm PMSF, 10 μg/mL leupeptin, 10 μg/mL benzamidine, and 10 μg/mL pepstatin). The crude sample was centrifuged for 15 min at 15,000g and insoluble material was removed. The clear supernatant was immediately assayed for AGPase activity (in the pyrophosphorolysis direction) and stored in small aliquots at −80°C. All steps were carried out at 4°C unless otherwise indicated.
Protein Determination, SDS-PAGE, and Western Blotting
Protein concentration was measured using a protein assay kit (Micro BCA, Pierce Chemical, Rockford, IL) with BSA as the standard. Protein samples were separated by electrophoresis on 10% [w/v]) SDS-polyacrylamide gels (18 × 16 cm, Hoefer, San Francisco) and transferred onto Hybond-P membrane (Amersham Life Science). The apparent Mr of polypeptides on the SDS gel was determined using protein markers from Amersham Pharmacia Biotech. Following electroblotting, the membrane was treated with antibodies raised either against the synthetic peptide of the LS (Kleczkowski et al., 1993a) or against the E. coli-expressed SS (Thorbjørnsen et al., 1996a). Immunodetection was performed using the anti-rabbit secondary antibody conjugated with horseradish peroxidase and an enhanced chemiluminescence detection system (Amersham Life Science). Prior to the second immunodetection, the membrane was stripped by incubation at 50°C for 30 min in 62.5 mm Tris-HCl (pH 6.7) containing 2% (w/v) SDS and 100 mm 2-mercaptoethanol.
Stability to Proteolysis of Expressed AGPase in Insect Cell Extract
Cell lysate was incubated at 4°C after extraction, and aliquots were removed at 5-, 15-, 30-, 45-, 60-, and 120-min intervals and immediately frozen at −20°C. Protein samples were separated by SDS-PAGE and electroblotted onto Hybond-P membrane. Immunodetection was performed as described above.
Determination of Molecular Mass
Gel-exclusion chromatography was performed in triplicate at 4°C. Cell lysate (2 mL) was applied to a S300-HR column (95 × 1.5 cm) pre-equilibrated with 40 mm MOPS (pH 7.4) containing 10% (w/v) Suc, 2 mm MgCl2, 2 mm DTT, 0.5 mm PMSF, and 100 mm NaCl, and fractions of 1.4 mL were collected at 10-min intervals (0.14 mL/min). Elution of expressed AGPase was followed by assay A. Protein markers used to generate the standard curve were thyroglobulin (669 kD), apoferritin (443 kD), β-amylase (200 kD), BSA (66 kD), and carbonic anhydrase (29 kD) (Sigma Chemical).
Assay of AGPase
Enzyme activity was determined by assaying in either the pyrophosphorolysis (assay A) or the ADP-Glc synthesis (assay B) direction (Kleczkowski et al., 1993a). All assays were performed in triplicate and readings were reproducible to within 10%. Inhibition of Pi was measured by the addition of potassium phosphate solution (pH 7.5).
Assay A
Pyrophosphorolysis of ADP-Glc was measured spectrophotometrically by monitoring NADH formation at 340 nm. Diluted enzyme sample was added to a final 1 mL of 100 mm MOPS buffer (pH 7.4) containing 0.1 mg/mL BSA, 1 mm ADP-Glc, 7 mm MgCl2, 0.6 mm NAD, 1 mm sodium inorganic pyrophosphate, 2 units of Glc-6-P dehydrogenase, and 2 units of phosphoglucomutase. The reaction was initiated by the addition of ADP-Glc and was allowed to proceed for 10 min. One unit of activity was defined as the amount of enzyme required to reduce 1 μmol of NAD min−1 at 25°C.
Assay B
Diluted enzyme sample was incubated for 10 min at 25°C in 200 μL of 100 mm HEPES (pH 8.0) containing 0.5 mm [14C]Glc-1-P (1.07 × 106 cpm/μmol), 7 mm MgCl2, and 2.5 mm ATP. The reaction was initiated by the addition of [14C]Glc-1-P and was stopped by boiling for 30 s. The [14C]ADP-Glc formed was quantitated according to the method of Sanwal et al. (1968). One unit of activity was defined as the amount of enzyme catalyzing the production of 1 μmol of ADP-Glc min−1 at 25°C.
Kinetic Studies
Prior to kinetic studies, purified recombinant AGPase was desalted and equilibrated with the appropriate buffers: 40 mm MOPS, pH 7.4, or 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 8.0, buffers containing 2 mm MgCl2, 2 mm DTT, and 20%[w/v] Suc) using a 5-mL P-6 cartridge (Bio-Rad). Kinetic constants (Km) were determined from the double-reciprocal plots in the presence of near-saturating concentrations of other non-variable substrates as described under “Assay of AGPase.” All kinetic parameters are the means of three determinations reproducible to within 10%.
Plant Material
Barley plants were grown in a greenhouse at 15°C during the 16-h light periods and at 10°C during the 8 h of darkness. Hand-pollinated grains were harvested at the appropriate developmental stages. The barley tissues were dissected from greenhouse-grown plants, except for the root tips, which were obtained from germinating seeds kept on moistened paper towels in the dark. All samples were rapidly frozen in liquid nitrogen and stored at −80°C.
Northern Analysis
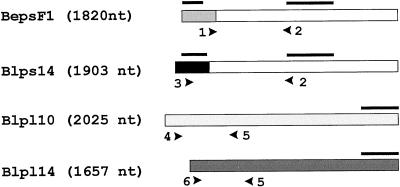
All solutions and equipment were diethyl pyrocarbonate-treated and autoclaved before use. Poly(A+)-rich RNA was extracted from barley (cv Bomi) seeds 0 to 30 d after pollination and from different barley tissues (endosperm, leaves, stem, root tip, and embryos) using unused magnetic oligo(dT) beads for each tissue sample (Dynal A/S, Oslo) (Jakobsen et al., 1990). Approximately 100 ng of poly(A+)-rich RNA from each sample was separated by 1.4% (w/v) agarose gel electrophoresis and transferred onto nylon membrane filters (Amersham Life Science) (Sambrook et al., 1989). Synthesis of 32P-DNA probes were performed using the random primer labeling kit (Rediprime, Amersham Life Science) and [32P]dCTP (Amersham Life Science). Northern filters were hybridized with 32P-labeled DNA probe (1 × 106 cpm/mL) at 65°C in the presence of 50% (v/v) formamide, 1 m NaCl, 0.1% (w/v) sodium inorganic pyrophosphate, and 0.05 m Tris-HCl (pH 7.5). Washing conditions were: 2× SSC at 25°C (2 × 5 min) and 2× SSC and 0.1% (w/v) SDS at 65°C (2 × 30 min). Filters were exposed to Amersham Hyperfilm for 1 to 3 d. Specific probes were made based on the different AGPase cDNAs. Specific probes for the SS transcripts beps and blps were made by isolating the SacI 150 and 230-nt upstream fragments of the bepsF1 and blps14 cDNAs, respectively. As a control, a fragment from the 3′ common region (bepsF1 cut with EcoRI, 600-nt) was also made. Specific probes for the bepl and blpl transcripts were the 543-nt SacI 3′ fragment of bepL10 and the 317-nt PvuII 3′ fragment of blpl14 (Fig. 3).
Figure 3.
Probes and constructs. A, Schematic outline of the barley AGPase cDNAs. Primers used in the RT-PCR are shown as arrowheads; probes used in northern-blot analysis are marked with horizontal bars. Transcript-specific primer pairs for the RT-PCR: bepsF1, 1 and 2, 516 nt; blps14, 2 and 3, 713 nt; bepl10, 4 and 5, 692 nt; and blpl14, 5 and 6, 491 nt. Transcript-specific probes for northern blot: the 150-nt upstream SacI fragment of bepsF1; the 230-nt SacI upstream fragment of blps14; the 543-nt fragment of bepl10; and the 317-nt fragment of blpl14. In addition, the 600-nt EcoRI common fragment of bepsF1/blps14 was used in northern-blot analysis (white region of bepsF1 and blps14).
cDNA Synthesis
PolyA-rich RNA was isolated as described under northern analysis. About 100 mg of plant tissue was used, except for analysis of the embryo, in which 50 mg was used. The isolated mRNA was washed twice in wash buffer (100 mm Tris-HCl [pH 8.0], 1 mm EDTA, 150 mm LiCl, 0.1% [w/v] LiDS), four times in wash buffer minus LiDS, and once in 1× RT buffer (Promega). The mRNA on the beads was resuspended in 5 μL of 1× RT buffer and used directly for first-strand cDNA synthesis. RNase inhibitor (1 μL, 40 units), 2.5 μL of dNTPs (10 mm), 6 μL of RT buffer (5×), 1 μL of avian myeloblastosis virus-RT (10 units), and distilled water to 29 μL were prewarmed to 42°C. One-fifth of the mRNA preparation was used for the cDNA synthesis (1 h at 42°C). For each tissue, one tube with no avian myeloblastosis virus-RT enzyme was prepared as a negative control. To elute mRNA, the beads were washed once with 500 μL of distilled water (room temperature) and once with 100 μL of water (65°C) for 2 min. The beads containing the first-strand cDNA were resuspended in 20 μL of distilled water, and 5 μL was used in the PCR reaction.
RT-PCR
First-strand cDNA on the beads was used as a template in the RT-PCR reaction. Transcript-specific primers were made based on the barley AGPase cDNA sequences (Fig. 3): BepsF1 forward primer (1), 5′-TGCAGATCTCAATCCCCATGCTA-3′ and reverse primer (2), 5′-CCAAATGCAGTTGCACGTTCCT-3′; Blps14 forward primer (3), 5′-GCCTCCC CTTCCAAGATCCTG-3′ and reverse primer (2); Bepl10 forward primer (4), 5′-CCGGCA GTTGCAGGTGGACT-3′ and reverse primer (5), 5′-CGRTARAGSTGATCGCCCGA-3′ (where S = CG and R = AG); Blpl14 forward primer (6), 5′-GTCTCCGTCGCGACCAC AGAG-3′ and reverse primer (5). The PCR reaction was at 94°C for 4 min, 96°C for 15 s, 57°C for 15 s, and 72°C for 1 min for 40 cycles, with an extra polymerization step at 72°C for 7 min. The primers were also used on cDNA templates. RT-PCR products were separated by 1.5% (w/v) agarose gel electrophoresis together with the PCR controls.
Sequence Analysis
All the RT-PCR products were sequenced and compared with sequences of the corresponding cDNAs encoding SS and LS. Sequencing was performed on a DNA sequencer (ABI 377, Perkin-Elmer, Foster City, CA) using cyclic sequencing with a dye terminator as recommended by the manufacturer. All PCR products were purified using a kit (QIAquick, Qiagen, Hilden, Germany) before each sequencing reaction.
RESULTS
The Unmodified Recombinant Version of the Major Endosperm AGPase Holoenzyme from Insect Is Fully Active in the Absence of 3-PGA and Is Not Inhibited by Pi
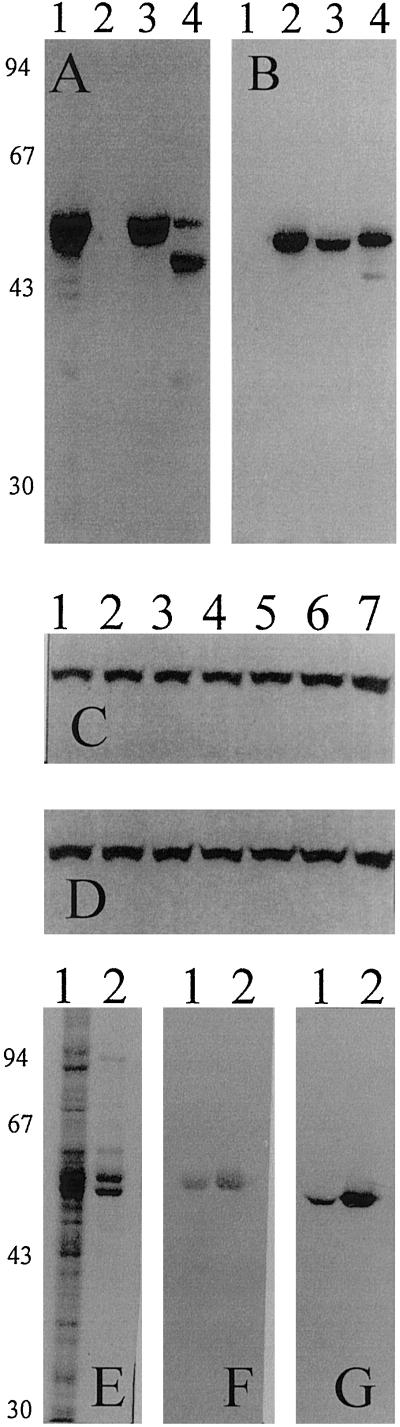
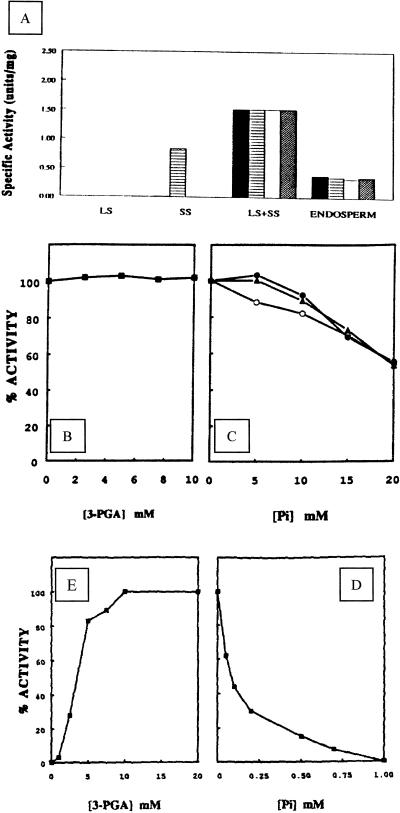
Full-length cDNAs carrying the complete coding sequence and a 5′-untranslated sequence (Villand et al., 1992; Thorbjørnsen et al., 1996a) encoding an unmodified LS and the SS of the major barley endosperm AGPase were expressed separately and together in insect cells using the baculovirus system (Fig. 1). In the crude extract of lysate from these cells, high catalytic activity of the holoenzyme was not affected either by 3-PGA or by 20 mm Pi when assayed in the pyrophosphorolysis direction (Fig. 2A). The expressed AGPase is a hetrotetramer of approximately 240 kD as determined by gel filtration, and consists of two different subunits of 58 and 51 kD (Fig. 1). These values are in agreement with the computed masses deduced from the cDNAs, as well as the previous estimate of 60 kD for LS (Kleczkowski et al., 1993a).
Figure 1.
Heterologous expression of the major barley endosperm AGPase in insect cells. A and B, Comparison of subunits of insect-cell-expressed and barley endosperm AGPases. Proteins from insect cell lysates containing only the expressed LS (lane 1, 10 μg), SS (lane 2, 10 μg), or both subunits (lane 3, 10 μg), and from barley endosperm extract (lane 4, 30 μg) were denatured, separated by 10% (w/v) SDS-PAGE, and transferred onto nylon membrane. Immunoblot was probed consecutively with antibodies recognizing the LS (A) and the SS (B). Molecular masses of the marker proteins are indicated on the left in kD. C and D, Stability at 4°C of expressed AGPase subunits in insect cell extract containing protease inhibitors. Insect cell extract containing the expressed AGPase (lane 1) was incubated at 4°C after extraction and aliquots were removed after 5 min (lane 2), 15 min (lane 3), 30 min (lane 4), 45 min (lane 5), 60 min (lane 6), and 120 min (lane 7), respectively. Proteins from different stages of incubation were denatured, separated on 10% (w/v) SDS-PAGE, and transferred onto nylon membrane that was then probed consecutively with antibodies recognizing the LS (C) and the SS (D). E to G, SDS-PAGE and immunoblots of the purified recombinant AGPase. Insect cell extract (lane 1, 20 μg) and purified recombinant AGPase (lane 2, 5 μg) were denatured, separated on 10% (w/v) SDS-PAGE, and either visualized with Coomassie Brilliant Blue R (E) or transferred onto nylon membrane that was then probed with antibodies recognizing the LS (F) and the SS (G). The molecular masses of marker proteins are indicated on the left in kD.
Figure 2.
Catalytic activities of the holoenzyme and its single subunits in insect cells. A, Insect cell extracts containing only expressed LS, SS, or both subunits were assayed in the pyrophosphorolysis direction (█) or in the presence of 10 mm 3-PGA (▤), 20 mm Pi (□), or both 10 mm 3-PGA and 20 mm Pi (▨). Enzyme activity from extruded endosperm was assayed immediately after extraction. B and C, Effect of 3-PGA and Pi on catalytic activity of the purified AGPase. Enzyme activity of the purified recombinant AGPase was determined in the ADP-Glc synthesis direction in the presence of 3-PGA at different concentrations (B), or with elevated levels of Pi (C) together with 3-PGA in the assay medium: 0 mm (○), 5 mm (●), and 10 mm (▴). D and E, Effect of 3-PGA and Pi on enzyme activity of SS expressed in insect cells. D, Insect cell extract containing only the expressed SS was assayed in the pyrophosphorolysis direction with various concentrations of 3-PGA. E, Cell extract was assayed in the presence of 10 mm 3-PGA together with various concentration of Pi.
Two major polypeptide bands of 58 and 48 kD were detected from barley endosperm extracts (Fig. 1A, lane 4) by polyclonal antibodies raised against synthetic peptides based on the sequence of the LS (Kleczkowski et al., 1993a). In this gel, the lower band is the product of protein degradation of the 58-kD band, since prolonged incubation of endosperm extracts at 4°C led to disappearance of the upper band, and an increase in the intensity of the 48-kD band and other low-molecular-mass bands (data not shown). Both the expressed LS and the undegraded LS from barley endosperm have similar mobility on SDS-PAGE (Fig. 1A, lanes 1 and 3 versus 4). Similarly, polyclonal antibodies raised against the E. coli-expressed SS (Thorbjørnsen et al., 1996b) recognized the recombinant SS as well as a major polypeptide from barley endosperm extract, both with similar mobility on SDS-PAGE (Fig. 1B, lanes 2 and 3 versus 4). In the present investigation, the extraction of proteins from infected insect cells was carried out in the presence of protein inhibitors, and there was no indication of proteolysis (Fig. 1, C and D). Like the native enzyme, the recombinant enzyme was susceptible to denaturation when heated to 60°C for 5 min, resulting in an almost complete loss of catalytic activity (data not shown). No enzyme activity was detectable in noninfected cells.
We observed that the catalytic activity assayed immediately after extraction was not significantly enhanced nor inhibited by 10 mm 3-PGA or 20 mm Pi. After storage at −80°C and subsequent incubation at 4°C, the LS of the endosperm AGPase was found to be extensively degraded (Fig. 1A, lane 4). In these preparations, enzyme activity in the crude extract was altered by the addition of 3-PGA and Pi in the assay medium (data not shown).
To further corroborate our previous studies of the His-tagged recombinant AGPase (Rudi et al., 1997), we partially purified the present unmodified version of the enzyme from insect cells. In the experiments reported here, we routinely obtained 5 to 7 units of AGPase/10 mL of insect cell culture. After five purification steps, the AGPase was purified 44-fold, to a specific activity of 56 units/mg as determined by enzyme assay in the pyrophosphorolysis direction in the absence of the activator 3-PGA (Table I). Attempts to obtain the NH2-terminal sequence of the two subunits have so far been unsuccessful. Like the enzyme in crude extracts (Fig. 2A), the purified recombinant AGPase was not affected by 3-PGA or Pi when assayed in the pyrophosphorolysis direction (data not shown). When assayed in the ADP-Glc synthesis direction, the purified recombinant AGPase was also insensitive to 3-PGA (Fig. 2B), but was inhibited at unphysiological concentrations of Pi with reduction of catalytic activity of 46% at 20 mm (Fig. 2C).
Table I.
Purification of barley endosperm AGPase expressed in insect cells (from 120-mL culture)
| Step | Protein | Activity | Specific Activity | Yield | Purification |
|---|---|---|---|---|---|
| mg | unitsa | units/mg | % | −fold | |
| Crude extract | 70.2 | 88.83 | 1.27 | 100 | 1.0 |
| (NH4)2SO4 | 18 | 96.46 | 5.36 | 109 | 4.2 |
| High-Q | 4.75 | 76.37 | 16.08 | 86 | 12.7 |
| CHTII | 2.55 | 56.03 | 21.97 | 63 | 17.3 |
| C3 | 1.08 | 48.23 | 44.66 | 54 | 35.2 |
| High-Q | 0.5 | 28.14 | 56.28 | 32 | 44.3 |
Activity was measured in the pyrophosphorolysis direction (assay A).
Micromoles of Glc-1-P formed/min at 25°C.
Comparison of the kinetic properties of the present recombinant AGPase with those of the native enzyme also indicated comparable values (Table II). The addition of 3-PGA to the assay medium at concentrations of 0.5 and 10 mm slightly protected the enzyme from this inhibition, but did not seem to have any appreciable effect on either inducing or overcoming the inhibition (Fig. 2B). These data strongly support previous observations in our laboratory about the native barley endosperm enzyme (Kleczkowski et al., 1993a), as well as data on the recombinant version of the enzyme with an appended His-tag (Rudi et al., 1997). These data showed that 3-PGA and Pi are not involved in the regulation of the catalytic activity major AGPase from barley endosperm either in the pyrophosphorolysis or in the ADP-Glc synthesis direction.
Table II.
Kinetic properties of expressed AGPase
| Substrate |
Km
|
|
|---|---|---|
| Native AGPasea | Expressed AGPase | |
| mm | ||
| ADP-Glc | 0.13 | 0.14 |
| PPi | 0.027 | 0.035 |
| Glc-1-P | 0.12 | 0.13 |
| ATP | 0.31 | 0.25 |
Kinetic values obtained from the partially purified AGPase (Kleczkowski et al., 1993a).
The SS of the Major Barley Endosperm AGPase Is Activated by 3-PGA and Inhibited by Pi
To determine whether the separate subunits of the recombinant AGPase possess activity on their own, LS and SS were expressed individually in insect cells. In these experiments, no activity was detectable in cells expressing the LS (Fig. 2A). In cells expressing SS, a low activity was detectable (0.0018 unit/mg). However, the addition of 10 mm 3-PGA in the assay reaction led to an increase of approximately 460-fold in the SS enzyme activity, the specific activity being 0.83 unit/mg (Fig. 2, A and D). The activity resulting from the addition of 10 mm 3-PGA is highly susceptible to Pi inhibition (I0.5 of 0.085 mm), with complete loss of activity occurring at 1 mm Pi (Fig. 2D). The barley enzyme is the second recombinant small AGPase subunit shown to have activity on its own, and, similar to the potato SS (Ballicora et al., 1995), responded to allosteric activation.
The Steady-State Level of the Major Endosperm AGPase Transcripts Correlates Positively with the Rate of Endosperm Starch Deposition
In an attempt to implicate the unregulated AGPase isoform in endosperm starch metabolism, we investigated the temporal pattern of expression of the four AGPase transcripts in developing barley grains using transcript-specific probes and primers. Northern-blot analysis was carried out using poly(A+)-rich RNA from different developmental stages. In these analyses, the unique 5′ends of beps and blps were used as probes for the SS transcripts (Fig. 3A). As a control, a probe was also included that represents the common region of the two cDNAs. For the LS transcripts, 3′ fragments of the bepl10 and blpl14 cDNAs were used as probes. For an overview of probes and transcript sizes, see Figure 3A.
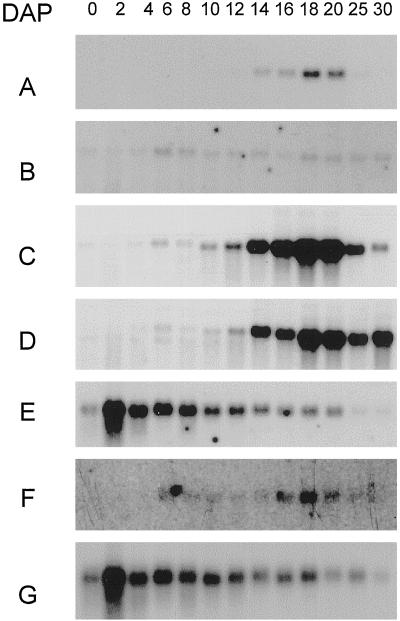
Northern-blot analysis demonstrated that the steady-state level of the two major endosperm AGPase transcripts, beps and bepl, peaks midway through seed development at 19 d after pollination (DAP) (Fig. 4, A and D). Comparing the profile of expression of these two transcripts, both have the same pattern with low levels before 6 DAP, peaking around 18 DAP, and decreasing after 20 DAP. As expected, using the probe representing the common region of the two SS transcripts, a similar pattern of expression was observed (Fig. 4C).
Figure 4.
Northern-blot analysis of beps, bepl, blps, blpl, and histone H3 transcripts in developing barley grains. Each lane was loaded with poly(A+)-rich RNA (100 ng/lane) extracted from intact ovaries and grains harvested 0 to 30 DAP (days after pollination). The probes (single-stranded) used were bepsF1 (A), blps14 (B), bepsF1/blpsl4 (C, 3′ common region), bepl10 (D), blpl14 (F), and histone H3 (E and G). The same blot was used in A to E. A new blot was used in F and G. The histone probe (Doan et al., 1996) was used to monitor gel loading.
In contrast to the steady-state levels of beps and the bepl transcripts, which are high during the most active period of starch deposition in the endosperm (Fig. 4, A and D), the blps and blpl transcripts were constitutively expressed throughout grain development at a much lower level (Fig. 4, B and F). Both transcripts were present in unfertilized ovules, demonstrating that the transcripts are also present in maternal tissues. From these data we conclude that the beps and bepl transcripts, which encode the unregulated AGPase isoform purified from insect cells, are expressed in a temporal pattern that correlates positively with the rate of endosperm starch deposition.
The Major Endosperm AGPase Isoform Transcripts Are Also Expressed in Vegetative Tissues, Including Leaves
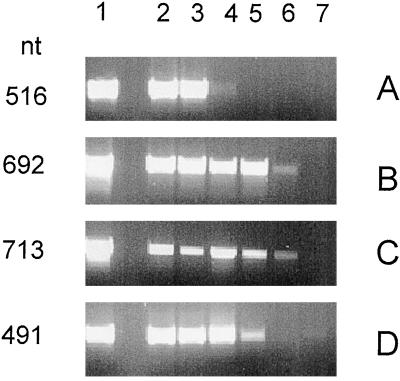
As seen in the northern-blot analysis of AGPase transcripts in developing grains (Fig. 4), the level of the major leaf transcripts was very low, bordering on the detection limit, even when using poly(A+)-rich RNA. Since preliminary analysis with poly(A+)-rich RNA extracted from other tissues also indicated low transcript levels (data not shown), RT-PCR analysis was performed with transcript-specific primer pairs (see Fig. 3A). To test the specificity of the primers, each primer pair was used in reactions with bepsF1, bepl10, blps14, and blpl14 cDNAs as template DNA, giving the expected product lengths of 516, 692, 713, and 491 nt, respectively (Fig. 5, lane 1). As a control for DNA contamination in the RT-PCR reaction, samples were run in parallel without RT, giving a negative result in all cases (not shown).
Figure 5.
RT-PCR analysis using specific primer pairs for the barley AGPase transcripts beps (A), bepl (B), blps (C), and blpl (D). Lane 1 (A–D) shows PCR product from each primer pair with control cDNAs as templates. Poly(A+)-rich RNA used in the analysis was extracted from 100 mg of endosperm (14 and 18 DAP) (lanes 2 and 3, respectively), leaf (lane 4), root-tip (lane 5), stem (lane 6), and embryo (from 50 mg of tissue) (20 DAP) (lane 7). The gel was loaded with 10 μL from a total reaction volume of 50 μL (except for embryos, for which 20 μL was used).
To further eliminate the possibility of contaminating products from the genomic DNA template, the primer pair for the beps transcript was designed to span the first two introns of the small AGPase gene, giving an unlikely product of approximately 3.3 kb. Likewise, for the blps transcript, the primer pair was expected to amplify a product of approximately 1.8 kb with genomic DNA as a template. Products of these sizes were not observed either with the cloned AGPase gene (Thorbjørnsen et al., 1996b) as a template or with barley genomic DNA in control RT-PCR reactions (data not shown). Using total genomic DNA as a template with the primer pairs for the bepl and blpl transcripts, no amplification products were detectable (data not shown), indicating the presence of introns in these genes as well.
The results of the RT-PCR analysis using first-strand cDNA template from different barley tissues are shown in Figure 5, the PCR products obtained from endosperm (14 and 18 DAP), mature leaves, root-tip, stem, and 20 DAP embryos. In all reactions, the RT-PCR products had the same length as in the control reactions with cDNA templates. As expected, the analysis confirmed the presence of both major endosperm transcripts, beps and bepl, in endosperm, with the two bands having similar intensity (Fig. 5, A and B, lanes 2 and 3). In addition, both transcripts were also detectable in leaves, although the beps transcript was very weak (Fig. 5, A and B, lane 4). Using bepl-specific primers, products of the expected size was also detected in root tips (same intensity as in leaves) and in stems (Fig. 5B, lanes 5 and 6, respectively). No product was obtained using the beps primers with first-strand cDNA from either root tips or stems (Fig. 5A, lanes 5 and 6, respectively). The major endosperm transcripts beps and bepl were not detectable in embryos (Fig. 5, A and B, lane 7).
RT-PCR analysis confirmed the results of northern-blot analysis for the major leaf AGPase transcripts blps and blpl in endosperm (Fig. 5, C and D, lanes 2 and 3). As expected, blps and blpl primer pairs gave bands of the expected size using first-strand cDNA from leaves as a template (Fig. 5, C and D, lane 4). In addition, blps and blpl transcripts were detectable in root tips and stems (Fig. 5, C and D, lanes 5 and 6). Of the transcripts, only the blpl primer pair gave a detectable, although weak, band using template cDNA from embryos (Fig. 5D, lane 7). The staining intensity of the blpl product from endosperm and leaves was the same, whereas in root tips the band was weaker. In stems and embryos the band was very weak. For the blps primer pair, the bands in endosperm, leaves, and root tips had the same intensity as those from reactions with template DNA, whereas the signal from stems was very weak.
Based on these data, we conclude that the major barley endosperm transcripts beps and bepl are not restricted to the endosperm, but are expressed in leaves as well, although at a much lower steady-state level than in endosperm. From the control experiments we are confident that the PCR products seen on the agarose gel in Figure 5, A to D, reflect the pattern of expression of the four AGPase transcripts and not genomic DNA contamination. We found no indication of the existence of additional AGPase transcripts that had previously remained undetected.
DISCUSSION
The high activity of the major barley endosperm AGPase in the absence of 3-PGA, as reported here from studies of an unmodified recombinant version of the enzyme, from previous studies of a recombinant enzyme with an attached His-tag to the amino terminus of the SS (Rudi et al., 1997), and from observations on the native enzyme (Kleczkowski et al., (1993a), is in contrast to most other AGPases, which have low activity without 3-PGA stimulation. One possible explanation for this unexpected feature of the barley endosperm enzyme is proteolytic modification leading to 3-PGA and Pi insensitivity (Plaxton and Preiss, 1987; Sivak and Preiss, 1995). This point is of particular relevance, since restoration of the N terminus of the potato SS restored the sensitivity of the holoenzyme to Pi inhibition (Ballicora et al., 1995). Although proteolysis occurs vigorously in endosperm extracts, effectively preventing the purification of an intact form of the enzyme, proteolysis does not occur at any appreciable level in extracts from insect cells after the addition of inhibitors of Ser proteases (Fig. 1). In addition, the fact that SS alone possesses activity sensitive to 3-PGA/Pi makes it unlikely that the AGPase expressed in insect cells is modified in such a way that it can no longer respond to these allosteric regulators.
The activity of the recombinant AGPase determined in the ADP-Glc synthesis direction was not affected by 3-PGA concentrations as high as 10 mm, and was only inhibited approximately 50% by unphysiological concentrations of Pi such as 20 mm (Fig. 2, B and C). Unlike the barley leaf enzyme, a varied ratio of 3-PGA and Pi in the reaction medium did not affect the catalytical properties of the enzyme. In comparison, the catalytic activity of the recombinant potato AGPase from E. coli cells was activated 43-fold with 1 mm 3-PGA in the ADP-Glc synthesis direction (Iglesias et al., 1993). Pi had little effect on the catalytic activity of the potato enzyme, causing only 20% reduction at 4 mm in the absence of 3-PGA. Maximum Pi inhibition was observed at 2 mm when assayed together with various concentrations of 3-PGA in the ADP-Glc synthesis direction (Iglesisas et al., 1993).
Based on the data for the recombinant enzyme, we conclude that the properties described here, i.e. full activity in the absence of 3-PGA and no inhibitory effect of Pi, is an intrinsic property of the major barley endosperm AGPase. Similar to the potato SS expressed in E. coli cells (Iglesias et al., 1993; Ballicora et al., 1995), the catalytic activity of the recombinant SS barley endosperm AGPase from insect cells was activated by 3-PGA and inhibited by Pi (Fig. 2). As is commonly observed for plant AGPase holoenzymes, the unstimulated enzyme displayed very little activity, with 3-PGA leading to a 460-fold increase (Fig. 2A). The independence of allosteric regulators suggests that the major barley endosperm AGPase isoform has potential to increase the starch content in plants, the convenience of an unregulated enzyme for this purpose being demonstrated by the use of the modified glgC16 E. coli AGPase to increase potato tuber starch (Stark et al., 1992).
Based on northern-blot and PCR experiments using gene-specific probes and primers, the steady-state level of the beps and bepl transcripts increased around the time of onset of the linear growth phase of the endosperm, being high during the time of maximum endosperm starch deposition. In contrast, the transcripts for the major leaf endosperm form, blps and blpl, appear to be constitutively expressed in the grain at a much lower steady-state level. A positive correlation between the rate of starch deposition and AGPase transcript levels has previously been reported for maize, rice, and wheat endosperm, although gene-specific probes were not used in these studies (Perez et al., 1975; Reeves et al., 1986; Anderson et al., 1991; Prioul et al., 1994; Ainsworth et al., 1995) or in cotyledons from fava bean (Weber et al., 1995) or pea (Burgess et al., 1997).
The same correlation has also been seen in potato leaves, but only for the LS RNA (Müller-Röber et al., 1992). Based on this correlation, and on the assumption that the two AGPase subunits combine in stoichiometric quantities to form the holoenzyme, we infer that the beps and bepl transcripts encode the AGPase isoform involved in the deposition of the bulk of endosperm starch. Since this enzyme is insensitive to allosteric regulation, transcript levels should reflect protein levels, and hence enzyme activity, assuming equal transcript stability and translational efficacy throughout the grain-filling period. However, other functions for this AGPase isoform cannot be completely ruled out. Thorbjørnsen et al. (1996a) pointed out that, according to their estimates, the major leaf AGPase isoform that they assumed to be present in the endosperm amyloplast fraction, is sufficient to account for total endosperm starch accumulation. However, based on the profile of expression of the major leaf AGPase in endosperm, we infer that the AGPase isoform encoded by the blps and blpl transcripts plays a minor role in endosperm starch deposition.
Studying the pattern of AGPase transcript accumulation in developing wheat endosperm, Ainsworth et al. (1995) published data from a northern-blot experiment in which SS but not LS AGPase transcripts were detectable prior to 10 DAP. Since low AGPase activity was measured before 10 DAP, the authors suggested that the SS was capable of forming an active homotetramer in the young endosperm. Although the SS of the major AGPase isoform from barley endosperm is capable of forming an active enzyme complex, no difference in the timing of SS and LS transcription was detectable in the present analysis of developing barley grains. Therefore, a role for a barley AGPase SS homotetramer, if it exists, remains to be determined.
In addition to endosperm, the beps and bepl transcripts were also detectable in leaves in northern-blot and RT-PCR experiments (Figs. 4 and 5). These data are in contrast to the results presented by Villand et al. (1992), Thorbjørnsen et al. (1996b), and Eimert et al. (1997), who failed to detect the beps and bepl transcript in leaves. Most likely, the discrepancy between their results and ours are technical, with the transcript levels in leaves being relatively low. In pea, one of the AGPase transcripts, agpS2, which is present at high steady-state levels during the period of intensive starch accumulation in the major seed starch storage organ, is expressed in leaves (Burgess et al., 1997). In contrast, the corresponding bean transcript, agpc, was only detected in cotyledons. For barley roots, the data presented in this paper and those reported by Villand et al. (1992) showing that the bepl transcript is present in roots, are different from those of Eimert et al. (1997), who were unable to detect the transcript in this organ. As in leaves, part of the explanation for this discrepancy may be technical. Additionally, the growing conditions for the plants or seeds from which the roots were harvested may influence bepl steady-state levels. Thus, in their experiment, Eimert et al. (1997) used roots from greenhouse-grown plants, while we in the current study and Villand et al. (1992) used roots grown in the dark on paper towels. This conclusion is strengthened by the fact that comparison of roots grown under both conditions in our laboratory showed that only root tips from seeds germinated in the dark on paper towels gave a positive signal for bepl in the RT-PCR analysis (data not presented).
The role, if any, of the major endosperm AGPase isoform in vegetative tissues remains unclear. The general picture emerging from studies of other higher plant AGPases is that the same SS gene is often expressed in both photosynthetic and non-photosynthetic tissues, whereas the LS is expressed by separate genes in photosynthetic and non-photosynthetic tissues (Müller-Röber et al., 1990; Weber et al., 1995; Burgess et al., 1997). This suggests the possibility that increased variability of AGPase enzymes may be created by different combinations of SS and LS. As discussed above, based on the similarity of the steady-state levels of the transcripts for the two isoforms in endosperm, this is not likely to be the case in barley. Also, this would meet with additional difficulty if the two isoforms were confined to different cellular compartments (the cytosol and the plastids). In leaf tissues, however, the level of expression is more similar and the possibility of hybrid AGPase formation cannot be ruled out. Studies of the properties of such hybrid AGPases are currently under way in our laboratory using the baculovirus-insect cell system. As pointed out by Kleczkowski (1996), a cytosolic AGPase in endosperm cells would give the possibility of linking Suc breakdown and ADP-Glc formation. Whether such a link would be favorable in vegetative tissue is unclear.
Finally, the possibility has to be considered that the beps and bepl transcripts are present in vegetative tissues due to so-called promoter leakage, and that the enzyme is not active in such tissues. Obviously, to fully understand the role of the two barley AGPase isoforms in different tissues, mutant studies are needed in which the major barley isoform (insensitive to 3-PGA/Pi regulation) and the major barley leaf isoform (sensitive to 3-PGA/Pi regulation) has been inactivated. With recent improvements of barley transformation technology (Wan and Lemaux, 1994), antisense experiments addressing these questions should be feasible in the near future. Alternatively, similar studies could be carried out in maize, where reverse genetics is well established using the Mu-transposable element (Bensen et al., 1995).
The existence of an APGase isoform that is independent of 3-PGA adds another chapter to the fascinating evolutionary history of this important enzyme. The higher plant AGPase is believed to have evolved from homotetrameric forms similar to those seen in enteric bacteria today, where the enzyme is activated by Fru 1,6-bisphosphate and inhibited by Pi. A next step in AGPase evolution appears to have been an adoption to different allosteric activators (3-PGA and Pi), the effectors of the cyanobacterial AGPase, although the enzyme is active as a homotetramer. The heterotetrameric forms of the enzyme are believed to have evolved through duplication and divergence of the ancestral AGPase gene, facilitating the evolvement of an interaction between a main (SS) catalytic subunit and a regulatory (LS) subunit. Interestingly, the SS of higher plants such as potato (Ballicora et al., 1995) and barley (this study) has not lost its activity as a homotetramer, an activity that, similar to the ancestral enzyme, is responsive to 3-PGA and Pi.
Our studies of the major barley endosperm AGPase suggest that the interaction between the two subunits of higher plant AGPase is more diverse than previously believed. Thus, in addition to the well-characterized activation resulting from interactions of the LS and the SS of, for example, several leaf and the potato tuber enzymes, the interaction between the LS and the SS of the major barley endosperm enzyme leads to an AGPase activity that is independent of allosteric regulators. In addition to the barley endosperm enzyme, other AGPases from non-photosynthetic tissues have also been reported to be insensitive or only weakly affected by the effectors, including those from pea embryos (Hylton and Smith, 1992) and wheat grains (Olive et al., 1989). Further studies are needed to verify the dependence of these enzymes on 3-PGA activation.
What is the nature of the interaction between the two types of subunits of the major barley endosperm enzyme leading to the 3-PGA-independent activity? Most likely, this effect is mainly attributable to LS, with the two SS of the barley AGPase isoforms differing only in the amino terminus, whereas the sequence of the two barley LS are more diverse. In an attempt to identify the AGPase binding-sites for 3-PGA, Preiss and co-workers carried out binding studies using the activator analog pyridoxal-5-phosphate (Morell et al., 1988; Ball and Preiss, 1994). Of the three putative regulatory sites on the LS, the highly conserved Lys residue in site 3 of regulated AGPases is substituted by a Met in the LS of the major endosperm AGPase (Rudi et al., 1997).
The phylogenetic distance tree of the LS generated from all available higher plant AGPases indicates that this change occurred relatively recently, which, assuming that this residue is involved in 3-PGA regulation, may explain the large variation in AGPase characteristics between higher plants. One explanation for the variability of higher plant AGPases is possibly the high selection pressure for increased grain quality by human nutrition needs. Whether the subcellular location in the cytosol, as indicated for the barley and corn enzymes, is a property of unregulated AGPases remains to be determined. In barley, it is tempting to speculate that the unique 5′ end of the SS transcript evolved by the addition of the endosperm-preferred promoter of the SS gene studied here (plus the first exon) led to a peptide lacking the ability to translocate to amyloplast rather than causing 3-PGA insensitivity. We intend to use the insect cell system to determine whether the Met residue of LS site 3 or other unrelated parts of the LS is responsible for the difference between the barley endosperm and leaf AGPase by forming recombinant hybrid enzymes consisting of various combinations of the subunits of the two isoforms.
ACKNOWLEDGMENTS
We thank Hege Munck for support with molecular analysis, Drs. Leszek Kleczkowski and Per Villand for helpful suggestions during the initial construction of the baculovirus expression vector, and Peter Sekkelsten and Berit Morken for technical support and careful nursing of barley materials. Drs. Knut Rudi and Robert Wilson are acknowledged for helpful comments on the manuscript.
Footnotes
This work has been funded by the European Union project “Genetic Tailoring of Novel Starch Polymers-CT95–0568” and the Biotechnology Program of the Norwegian Research Council.
LITERATURE CITED
- Ainsworth C, Hosein F, Tarvis M, Weir F, Burrell M, Devos KM, Gale MD. Adenosine diphosphate glucose pyrophosphorylase genes in wheat: differential expression and gene mapping. Planta. 1995;197:1–10. doi: 10.1007/BF00239933. [DOI] [PubMed] [Google Scholar]
- Anderson JM, Larsen R, Laudencia D, Kim WT, Morrow D, Okita TW, Preiss J. Molecular characterization of the gene encoding a rice endosperm-specific ADP-glucose pyrophosphorylase subunit and its developmental pattern of transcription. Gene. 1991;97:199–205. doi: 10.1016/0378-1119(91)90052-d. [DOI] [PubMed] [Google Scholar]
- Ball S, Preiss J. Allosteric sites of the large sub-unit of the ADP-glucose pyrophosphorylase. J Biol Chem. 1994;269:24706–24711. [PubMed] [Google Scholar]
- Ballicora MA, Laughlin MJ, Fu YB, Okita TW, Barry GF, Preiss J. Adenosine 5′-diphosphate-glucose pyrophosphorylase from potato tuber: significance of the N terminus of the SS for catalytic properties and heat stability. Plant Physiol. 1995;109:245–251. doi: 10.1104/pp.109.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP. Cloning and characterization of the maize An1 gene. Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess D, Penton A, Dunsmuir P, Dooner H. Molecular cloning and characterization of ADP-glucose pyrophosphorylase cDNA clones from pea cotyledons. Plant Mol Biol. 1997;33:431–444. doi: 10.1023/a:1005752311130. [DOI] [PubMed] [Google Scholar]
- Denyer K, Dunlap F, Thorbjørnsen T, Keeling P, Smith AM. The major form of ADPglucose pyrophosphorylase activity in maize (Zea mays L.) endosperm is cytosolic. Plant Physiol. 1996;112:779–785. doi: 10.1104/pp.112.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan DNP, Linnestad C, Olsen O-A. Isolation of molecular markers from the barley endosperm coenocyte and the surrounding nucellus cell layers. Plant Mol Biol. 1996;31:877–886. doi: 10.1007/BF00019474. [DOI] [PubMed] [Google Scholar]
- Eimert K, Luo C, Villand P, Thorbjørnsen T, Kleczkowski LA. Molecular cloning and expression of the LS of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves. Gene. 1997;189:79–82. doi: 10.1016/s0378-1119(96)00837-2. [DOI] [PubMed] [Google Scholar]
- Gosh HP, Preiss J. Adenosine diphosphate glucose pyrophosphorylase: a regulatory enzyme in the biosynthesis of starch in spinach chloroplasts. J Biol Chem. 1966;241:4491–4505. [PubMed] [Google Scholar]
- Greene TW, Hannah LC. Maize endosperm ADP-glucose pyrophosphorylase SHRUNKEN2 and BRITTLE2 subunit interactions. Plant Cell. 1998;10:1295–1306. doi: 10.1105/tpc.10.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylton C, Smith AM. The rb mutation of peas causes structural and regulatory changes in ADP-glucose pyrophosphorylase from developing embryos. Plant Physiol. 1992;99:1626–1634. doi: 10.1104/pp.99.4.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias AA, Barry GF, Meyer C, Bloksberg L, Nakata PA, Green T, Laughlin MJ, Okita TW, Kishore GM, Preiss J. Expression of the potato tuber ADP-glucose pyrophosphoryalse in E. coli. J Biol Chem. 1993;268:1081–1086. [PubMed] [Google Scholar]
- Jakobsen KS, Breivold E, Hornes E. Purification of mRNA directly from crude plant tissues in 15 min using oligo dT microspheres. Nucleic Acids Res. 1990;18:3669. doi: 10.1093/nar/18.12.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian A, Kleinhofs A, Villand P, Thorbjørnsen T, Olsen O-A, Kleckowski LA. Mapping of the ADP-glucose pyrophosphorylase genes in barley. Theor Appl Genet. 1994;87:869–871. doi: 10.1007/BF00221140. [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA. Back to the drawing board: redefining starch synthesis in cereals. Trends Plant Sci. 1996;1:363–402. [Google Scholar]
- Kleczkowski LA, Villand P, Luthi E, Olsen O-A, Preiss J. Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate regulation. Plant Physiol. 1993a;101:179–186. doi: 10.1104/pp.101.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski LA, Villand P, Preiss J, Olsen O-A. Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves. J Biol Chem. 1993b;268:6228–6233. [PubMed] [Google Scholar]
- Morell M, Bloom M, Preiss J. Affinity labeling of the allosteric activator site(s) of spinach leaf. J Biol Chem. 1988;263:633–637. [PubMed] [Google Scholar]
- Müller-Röber B, Sonnewald U, Willmitzer L. Inhibition of ADPglucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage proteins genes. EMBO J. 1992;11:1229–1238. doi: 10.1002/j.1460-2075.1992.tb05167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Röber BT, Kossmann J, Hannah LC, Willmitzer L, Sonnewald U. One of two different ADP-glucose pyrophosphorylase genes responds strongly to elevated levels of sucrose. Mol Gen Genet. 1990;224:136–146. doi: 10.1007/BF00259460. [DOI] [PubMed] [Google Scholar]
- O'Reilly DR, Miler LK, Luckow VA. Baculovirus Expression Vectors: A Laboratory Manual. W.H. New York: Freeman; 1992. [Google Scholar]
- Olive M, Ellis RJ, Schuch WW. Isolation and nucleotide sequences of cDNA clones encoding ADP-glucose pyrophosphorylase peptides from wheat leaf and endosperm. Plant Mol Biol. 1989;12:525–538. doi: 10.1007/BF00036967. [DOI] [PubMed] [Google Scholar]
- Perez CM, Perdon AA, Resurreccion AP, Villareal RM, Juliano BO. Enzymes of carbohydrate metabolism in developing rice grain. Plant Physiol. 1975;56:579–586. doi: 10.1104/pp.56.5.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Preiss J. Purification and properties of nondeproteolytic degraded ADPglucose pyrophosphorylase from maize endosperm. Plant Physiol. 1987;83:105–112. doi: 10.1104/pp.83.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J. Biology and molecular biology of starch synthesis and its regulation. Oxf Surv Cell Mol Biol. 1991;7:59–114. [Google Scholar]
- Prioul J-L, Jeanette E, Reyss A, Grègory N, Giroux M, Hannah LC, Causse M. Expression of ADP-glucose pyrophosphorylase in maize (Zea mays L.) grain and source leaf during grain filling. Plant Physiol. 1994;104:179–187. doi: 10.1104/pp.104.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves CD, Khrisnan HB, Okita TW. Gene expression in developing wheat endosperm: accumulation of gliadin and ADPglucose pyrophosphorylase messenger RNAs and polypeptides. Plant Physiol. 1986;82:34–40. doi: 10.1104/pp.82.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudi H, Doan DNP, Olsen O-A. A (His)6-tagged recombinant barley (Hordeum vulgare L.) endosperm ADP-glucose pyrophosphorylase expressed in the baculovirus-insect cell system is insensitive to allosteric regulation by 3-phospho glycerate and inorganic phosphate. FEBS Lett. 1997;419:124–130. doi: 10.1016/s0014-5793(97)01448-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanwal GG, Greenberg E, Hardie J, Cameron E, Preiss J. Regulation of starch biosynthesis in plant leaves: activation and inhibition of ADPglucose pyrophosphorylase. Plant Physiol. 1968;43:417–427. doi: 10.1104/pp.43.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC, Pien TM, Cao H, Liu KC. Brittle-1, an adenylate translocator, facilitates transfer of extraplastidial synthesized ADP-glucose into amyloplasts of maize endosperms. Plant Physiol. 1998;117:1235–1252. doi: 10.1104/pp.117.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivak MN, Preiss J. Starch synthesis in seeds. In: Kigel J, Galili D, editors. Seed Development and Germination. New York: Marcel Decker; 1995. pp. 139–168. [Google Scholar]
- Smith-White BJ, Preiss J. Comparison of proteins of ADP-glucose pyrophosphorylase from diverse sources. J Mol Evol. 1992;34:449–464. doi: 10.1007/BF00162999. [DOI] [PubMed] [Google Scholar]
- Stark DM, Timmerman KP, Barry GF, Preiss J, Kishore GM. Regulation of the amount of starch in plant tissues by ADP glucose pyrophosphorylase. Science. 1992;258:287–292. doi: 10.1126/science.258.5080.287. [DOI] [PubMed] [Google Scholar]
- Thorbjørnsen T, Villand P, Denyer K, Olsen O-A, Smith AM. Distinct isoforms of ADPglucose pyrophosphorylase occur inside and outside the amyloplasts in barley endosperm. Plant J. 1996a;10:243–250. [Google Scholar]
- Thorbjørnsen T, Villand P, Kleczkowski LA, Olsen O-A. A single gene encodes two different transcripts for the ADP-glucose pyrophosphorylase SS from barley (Hordeum vulgare) Biochem J. 1996b;313:149–154. doi: 10.1042/bj3130149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villand P, Aalen R, Olsen O-A, Lüthi E, Lønneborg A, Kleckowski LA. PCR amplification and sequences of cDNA clones for the small and LS of ADP-glucose pyrophosphorylase from barley tissues. Plant Mol Biol. 1992;19:381–189. doi: 10.1007/BF00023385. [DOI] [PubMed] [Google Scholar]
- Wan Y, Lemaux PG. Generation of large numbers of independently transformed fertile barley plants. Plant Physiol. 1994;104:37–48. doi: 10.1104/pp.104.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Stitt M, Heldt HW. Cell-type specific, coordinate expression of two ADP-glucose pyrophosphorylase genes in relation to starch biosynthesis during seed development of Vicia faba L. Planta. 1995;195:352–361. doi: 10.1007/BF00202592. [DOI] [PubMed] [Google Scholar]