Abstract
Profound lung vascular permeability is a cardinal feature of acute respiratory distress syndrome (ARDS) and ventilator-induced lung injury (VILI), two syndromes known to centrally involve the nonmuscle isoform of myosin light chain kinase (nmMLCK) in vascular barrier dysregulation. Two main splice variants, nmMLCK1 and nmMLCK2, are well represented in human lung endothelial cells and encoded by MYLK, and they differ only in the presence of exon 11 in nmMLCK1, which contains critical phosphorylation sites (Y464 and Y471) that influence nmMLCK enzymatic activity, cellular translocation, and localization in response to vascular agonists. We recently demonstrated the functional role of SNPs in altering MYLK splicing, and in the present study we sought to identify the role of splicing factors in the generation of nmMLCK1 and nmMLCK2 spliced variants. Using bioinformatic in silico approaches, we identified a putative binding site for heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), a recognized splicing factor. We verified hnRNPA1 binding to MYLK by gel shift analyses and that hnRNPA1 gene and protein expression is upregulated in mouse lungs obtained from preclinical models of ARDS and VILI and in human endothelial cells exposed to 18% cyclic stretch, a model that reproduces the excessive mechanical stress observed in VILI. Using an MYLK minigene approach, we established a direct role of hnRNPA1 in MYLK splicing and in the context of 18% cyclic stretch. In summary, these data indicate an important regulatory role for hnRNPA1 in MYLK splicing, and they increase understanding of MYLK splicing in the regulation of lung vascular integrity during acute lung inflammation and excessive mechanical stress, such as that observed in ARDS and VILI.
Keywords: ventilator-induced lung injury, cyclic stretch, splicing, heterogeneous nuclear ribonucleoprotein A1, MYLK
Clinical Relevance
Critically ill patients in the ICU on mechanical ventilation experience acute respiratory distress syndrome (ARDS) and ventilator-induced lung injury (VILI). The protein product of the MYLK gene, myosin light chain kinase, is involved in the processes of vascular barrier injury and recovery from injury, key features of ARDS and VILI. MLCK has been implicated in ARDS and VILI pathogenesis. Our present study demonstrates the role of a splicing regulator in driving MYLK alternate splicing and subsequent alterations in vascular barrier integrity.
Acute respiratory distress syndrome (ARDS) is a complex disorder affecting approximately 250,000 patients each year in the United States (1). ARDS is an inflammatory disease characterized by respiratory failure resulting from profound vascular leak and lung flooding produced by inflammatory cells and by cytokines including TNF-α, IL-1β, and IL-6 (2). Current treatment is supportive therapy with mechanical ventilation, which unfortunately carries the risk of ventilator-induced lung injury (VILI), exacerbating lung inflammation via exposure of lung tissues to excessive mechanical stress, a potent inflammatory stimulus. Vascular barrier integrity is maintained by cell–cell and cell–matrix interactions and is tightly regulated by the endothelial cell (EC) cytoskeleton that generates spatially targeted contractile forces by shortening of actin and myosin filaments (3). Myosin light chain kinase (MLCK) is a central cytoskeletal bioregulatory molecule via phosphorylation of myosin light chains (4) on Ser18 and Thr19 (5). We have identified nonmuscle myosin light chain kinase (nmMLCK) as the principal isoform in ECs (6) and have demonstrated the role of nmMLCK in maintenance of the vascular barrier using cellular and murine models (7–9). Genetic variants in MYLK, the gene encoding the nmMLCK isoform, are associated with ARDS risk and mortality, likely via an influence, at least in part, on nmMLCK regulation of EC barrier integrity (3, 10–13).
We previously identified several MYLK splice variants (6) that include nmMLCK-1 and -2 and demonstrated that nmMLCK2 is the principal splice variant expressed in human lung ECs (6). The two isoforms differ only in the absence of exon 11 in nmMLCK2 that, unlike nmMLCK1, precludes nmMLCK2 regulation via phosphorylation on key tyrosine sites by c-Abl (8) and p60Src kinases (5). This absence of exon 11 and lack of post-translational modification (PTM) regulation in nmMLCK2 have detrimental consequences because nmMLCK2 exhibits proinflammatory features producing greater levels of EC barrier disruption and slower resolution of vascular integrity disruption. For example, transgenic mice with targeted overexpression of nmMLCK2 only in the lung endothelium demonstrate a profound susceptibility to inflammatory lung injury and vascular leak (9). Unlike nmMLCK1, nmMLCK2 fails to rapidly translocate to the peripheral actin cytoskeleton in response to EC barrier–enhancing agonists such as sphingosine-1-phosphate (14) and is a less active participant in vascular barrier restoration. Therefore, an understanding of splicing mechanisms generating nmMLCK splice variants nmMLCK1 and nmMLCK2 is important to developing strategies designed to limit inflammatory vascular injury.
We recently used a minigene approach to address mechanisms that potentially underlie the regulation of MYLK splicing (15) and demonstrated the functional role of SNPs in altering MYLK splicing (15). It is well known that pre-mRNA splicing occurs through the activity of the spliceosome comprised of the heterogeneous nuclear ribonucleoproteins (hnRNPs), small nuclear ribonucleoprotein particles, and other components of the splicing machinery (16). hnRNPs are a class of multifunctional proteins belonging to the heterogeneous ribonucleoprotein family involved in processing heterogeneous nuclear RNA into mRNA, with a prominent member being hnRNPA1 that is involved in alternative 5′ splice site selection (17) through a competitive mechanism with splicing factor 2/alternative splicing factor for binding (18, 19). These effects on alternative splicing are mediated primarily via direct binding of hnRNPA1 to RNA (20) at specific sites, as determined by systematic evolution of ligands by exponential enrichment (21). The two main isoforms of hnRNPA1, A1-A and A1-B, show binding affinity variation that alters splicing activity (20) because the tightly binding isoform A1-B shows a reduced activity compared with the full-length A1-A protein (22).
hnRNPA1 is essential for constitutive splicing, given its involvement in spliceosome assembly (23) and association with the essential splicing factor U2 small nuclear RNA auxiliary factor through a ternary complex with AG/uridine-rich RNA (24). hnRNPA1 functions in alternate splicing as a splicing repressor. Alternate splicing of the 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) is mediated by hnRNPA1, and overexpression of hnRNPA1 increases an HMGCR variant lacking exon 13. In addition, hnRNPA1 mediates alternate splicing of the pyruvate kinase muscle (PKM) isoform in cancer cells, leading to production of the PKM2 isoform, contributing to the switch from oxidative phosphorylation to aerobic glycolysis (25, 26), a process also involving interactions with other hnRNP family members, including hnRNPA2 and polypyrimidine tract binding protein. Therefore, we hypothesized that hnRNPA1 may be an active participant in the regulation of MYLK splicing and nmMLCK1 and nmMLCK2 generation.
In the present study, we sought to understand the role of the splicing factor hnRNPA1 in alternative 5′ exon selection use of exons 10 and 11 of the MYLK gene in the generation of nmMLCK1 and nmMLCK2 spliced variants. Using our bioinformatic in silico approaches, we identified a putative binding site for hnRNPA1 on the MYLK gene, and we subsequently verified hnRNPA1 binding to MYLK by gel shift analyses and further confirmed that hnRNPA1 expression is responsive to mechanical stimuli. We demonstrated that hnRNPA1 gene and protein expression is upregulated in mouse lungs obtained from preclinical models of ARDS and VILI and in human ECs exposed to 18% cyclic stretch (CS), a model that reproduces the excessive mechanical stress observed in VILI. Using an MYLK minigene approach, we established a direct role of hnRNPA1 in MYLK splicing as well as in the context of 18% CS. In summary, these data indicate an important regulatory role for hnRNPA1 in MYLK splicing, and they increase understanding of MYLK splicing in the regulation of lung vascular integrity during acute lung inflammation and excessive mechanical stress, such as that observed in ARDS and VILI.
Methods
Analysis of MYLK for hnRNPA1 Binding
The region between exons 10 and 12 of the MYLK gene was analyzed by SpliceScan II (27), which revealed a binding site for hnRNPA1 (21). We hypothesized significant involvement of hnRNPA1 and the hnRNPA1 binding motif in regulation of MYLK alternative splicing and exon 11 inclusion ratios.
Gene Expression Profiling
Gene expression was performed using data obtained from the Gene Expression Omnibus database (GSE9368 and GSE9314) (28).
CS Design
Human pulmonary artery endothelial cells (HPAECs) (Lonza) were subjected to 5% or 18% CS using an FX-5000 Flexcell Tension Plus System (Flexcell International) (29).
RT-PCR Analysis
RNA isolated from HPAECs was reverse transcribed using SuperScript III RT (Life Technologies) and an anchored oligo(dT) (20) primer (Integrated DNA Technologies) in a 20-μl reaction at 50°C for 60 minutes and inactivated at 70°C for 15 minutes. Following RT, PCR was performed using GoTaq DNA polymerase (Promega). The primers used were MLCK Exon10F (GGCCAGAGGGATTCAGCATT) and MLCK Exon12R (ACCTCCATCACGGCAAGC) located on exon 10 and exon 12, respectively, and yielded products of 317 and 110 bp, respectively, that correspond to nmMLCK1 and nmMLCK2. Band intensities of PCR products were quantified using Image Lab software (Bio-Rad Laboratories), and ratios of exon inclusion (nmMLCK1/nmMLCK1 + nmMLCK2) and exon skipping (nmMLCK2/nmMLCK1 + nmMLCK2) were calculated.
siRNA Transfection
HPAECs were transfected with nontarget siRNA (D-001810-10-20; Dharmacon) or hnRNPA1 SMARTpool (L-008221-00-0010; Dharmacon) using siPORT amine (Ambion).
Minigene Construct and Analysis
The minigene construct was constructed as we recently reported (15). Site-directed mutagenesis was performed using the QuikChange II site-directed mutagenesis kit (Agilent Technologies). Constructs were transfected into human embryonic kidney 293 (HEK-293) cells. RNA isolation and RT were performed as described elsewhere (15). PCR and quantitation of products were performed as described above.
Cell Culture and Transfection
HEK-293 cells were maintained in Dulbecco’s modified Eagle’s medium/F-12 medium, and transfection was performed using Xfect transfection reagent (Clontech Laboratories). A total of 2.5 μg of DNA was complexed using 0.75 μl of Xfect polymer and added to HEK-293 cells in 2 ml of Dulbecco’s modified Eagle’s medium/F-12 medium. Medium was replaced after 6–8 hours.
Western Blotting
Equal amounts of protein lysates in 1× lithium dodecyl sulfate sample buffer (Life Technologies) were loaded onto 4–12% Bolt Bis-Tris Plus gels (Life Technologies), transferred onto polyvinylidene fluoride membranes, and probed using antibodies against hnRNPA1, hemagglutinin (HA)-Tag, and vinculin.
Recombinant hnRNPA1 Production
hnRNPA1 exists in two isoforms, hnRNPA1-A and hnRNPA1-B, with molecular weights of 34 and 39 kD, respectively (22). Recombinant protein was produced in HEK-293 cells and purified on anti-HA magnetic beads (Pierce Biotechnology) and eluted using HA peptide (Pierce Biotechnology).
Gel Shift Assays
Gel retardation assays were performed using the LightShift chemiluminescent RNA electrophoretic mobility shift assay kit (Pierce Biotechnology) and visualized using an MYECL imager.
Statistical Analysis
Data were analyzed using Student’s t test and ANOVA with a statistical threshold set at P < 0.05.
Results
MYLK Intron 10–11 Contains a Conserved hnRNPA1 Binding Site
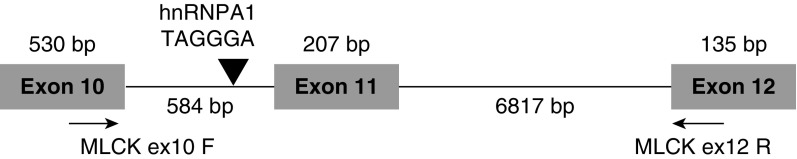
MYLK is alternatively spliced into nmMLCK1 and a predominant variant, nmMLCK2, in human ECs (6) with the inclusion of exon 11 occurring only in nmMLCK1. PTM sites within exon 11 are influenced by vascular barrier-enhancing and barrier-disruptive agents (5, 8). Therefore, we sought to identify splicing elements that potentially regulate MYLK splicing. The region between exons 10 and 12 of the MYLK gene was analyzed using SpliceScan II (27), revealing a strong splicing silencer comprised of the motif UAGGGA within intron 10–11 (Figure 1) and a binding site for hnRNPA1 (20). This splicing factor is the most abundantly expressed of the hnRNPs and has demonstrated roles in regulation of pre-mRNA processing, including modulation of alternatively skipped exon inclusion (19). We hypothesized significant involvement of hnRNPA1 and the hnRNPA1 binding motif in regulation of MYLK alternative splicing and exon 11 inclusion ratios.
Figure 1.
Genomic organization of the myosin light chain kinase (MYLK) gene. The genomic organization of exons 10–12 of the MYLK gene and the heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) binding site on intron 10–11 are indicated. The location of the primers, myosin light chain kinase Exon10 forward (MLCKex10F) and myosin light chain kinase Exon12 reverse (MLCK ex12 R), used to distinguish the two splice variants are also shown. bp = base pair.
Expression of hnRNPA1 in Murine Lung Tissues and Human Lung ECs
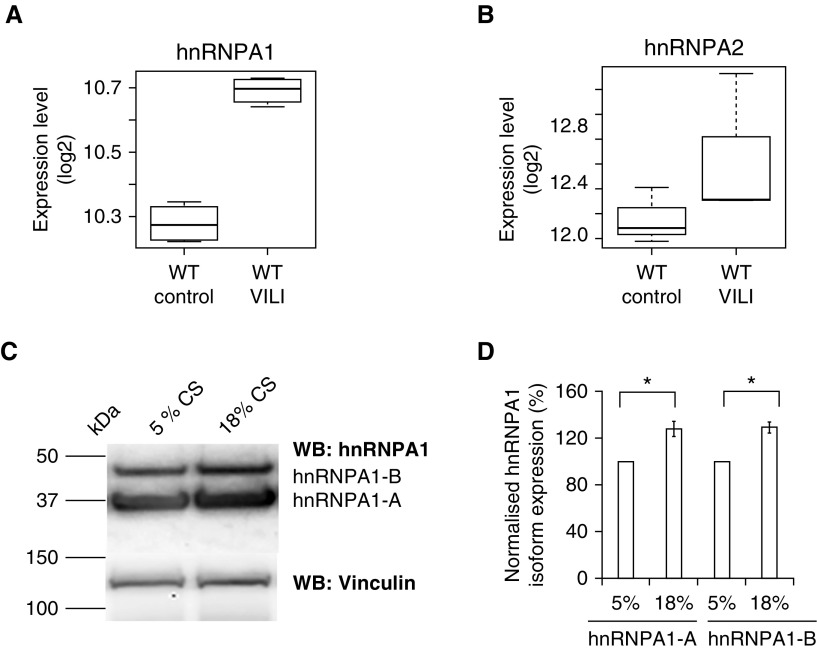
To examine hnRNPA1 regulation of MYLK in response to mechanical stress and inflammatory stimuli, we analyzed hnRNPA1 expression in lungs obtained from preclinical mouse models of VILI (12, 28). Expression of hnRNPA1 was increased in mouse lungs obtained from VILI-challenged animals (Figure 2A) compared with control animals. We also analyzed levels of the splicing factor hnRNPA2B1 (a related member of hnRNPA1) and observed no significant increase in gene expression (Figure 2B). Finally, we analyzed hnRNPA1 protein levels in human lung ECs subjected to 5% or 18% CS corresponding to normal spontaneous breathing or to high-tidal volume (Vt) mechanical ventilation. Western blotting revealed that hnRNPA1 protein levels were increased after exposure to 18% CS as compared with 5% CS (Figures 2C and 2D). These finding indicate that hnRNPA1 levels are influenced by both mechanical stress and VILI.
Figure 2.
hnRNPA1 level was increased in response to ventilator-induced lung injury (VILI) and excessive mechanical stress. (A) hnRNPA1 expression in whole mouse lungs subjected to VILI. hnRNPA1 levels were increased in VILI-challenged lungs (q < 10%) compared with lungs of the control animals. (B) hnRNPA2 expression in whole mouse lungs subjected to VILI. The expression of the related hnRNPA2 was not increased in VILI-challenged mouse lungs. (C) Western blot (WB) showing hnRNPA1 levels in human pulmonary endothelial cells in response to 5% and 18% cyclic stretch (CS; 8 h). hnRNPA1 levels (hnRNPA1-A and hnRNPA1-B) were increased in human pulmonary artery endothelial cells exposed to 18% CS compared with 5% CS. Vinculin was used as a loading control. The two isoforms hnRNPA1-A (molecular weight, 34 kD) and hnRNPA1-B (molecular weight, 38 kD) are shown. Molecular weight markers are also indicated. (D) Densitometric analysis of WBs shown in C. Quantification of the hnRNPA1 levels (hnRNPA1-A and hnRNPA1-B) that was normalized to vinculin levels. Levels of hnRNPA1 in 5% CS cells were set to 100%. Data are presented as mean ± SE (n = 3). Bars from left to right correspond to hnRNPA1-A isoform levels in 5% CS cells (set to 100%), hnRNPA1-A isoform levels in 18% CS cells (second bar from left) (*P < 0.05 compared with 5% CS [first bar from left bar]), hnRNPA1-B isoform levels in 5% CS cells (third bar from left) (set to 100%), and hnRNPA1-B isoform levels in 18% CS cells (fourth bar from left bar). *P < 0.05 compared with 5% CS (third bar from left). WT = wild type.
Mutation of the hnRNPA1 Binding Site Alters MYLK Minigene Splicing
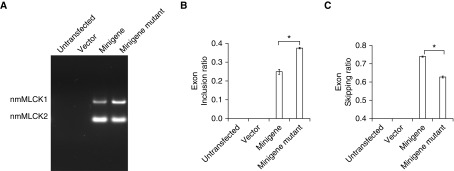
Bioinformatic analyses using SpliceScan II revealed an hnRNPA1 binding site residing between exon 10 and exon 11. We recently described and validated an MYLK minigene comprising the entire genomic region between exons 10 and 12 of MYLK and demonstrated the functional role of SNPs in MYLK splicing (15). In the present study, we used the minigene approach to determine the role of the hnRNPA1 site in MYLK splicing, and we mutated the hnRNPA1 binding site in the minigene to alter hnRNPA1 binding. HEK cells transfected with either the vector, the minigene, or the minigene lacking a functional hnRNPA1 binding site were analyzed for MYLK minigene-specific splicing products by RT-PCR using pCMV6-specific primers during RT. RT-PCR results are shown in Figure 3, and minigene-specific transcripts were obtained in cells transfected with the minigene. Exon inclusion and exon-skipping ratios that reflect the amounts of nmMLCK1 and nmMLCK2, respectively, were calculated. Our results indicate an increase in exon inclusion ratio and greater levels of nmMLCK1 generated in cells transfected with the minigene lacking a functional hnRNPA1 binding site (Figures 3A–3C) and underscore the importance of the hnRNPA1 binding site in regulation of MYLK splicing.
Figure 3.
Mutation of the putative hnRNPA1 site alters MYLK splicing. (A) Representative gel showing RT-PCR analysis of the MYLK minigene. HEK-293 cells were transfected with constructs as indicated, and RT-PCR was performed. First lane from left = untransfected cells; second lane from left = vector-transfected cells; third lane from left = MYLK minigene with wild-type hnRNPA1 site; fourth lane from left = MYLK minigene-ΔhnRNPA1 site. (B) Densitometric analysis of the RT-PCR products in A showing exon inclusion ratio (nonmuscle isoform of myosin light chain kinase 1 [nmMLCK1]/[nmMLCK1 + nmMLCK2]). Bars from left to right correspond to untransfected cells, vector-transfected cells, minigene, and minigene with the hnRNPA1 binding site mutated, respectively. Data are presented as mean ± SE (n = 3). *P < 0.05 comparing minigene and minigene with hnRNPA1 binding site mutated. Mutation of the hnRNPA1 binding site elicited an increase in the exon inclusion ratio with greater levels of nmMLCK1. (C) Densitometric analysis of the RT-PCR products in A showing exon-skipping ratio (nmMLCK2/[nmMLCK1 + nmMLCK2]). Bars from left to right represent untransfected, vector-transfected cells, MYLK minigene, and MYLK minigene with the hnRNPA1 binding site mutated, respectively. Data are presented as mean ± SE (n = 3). *P < 0.05 comparing MYLK minigene and MYLK minigene with hnRNPA1 binding site mutated. Loss of the hnRNPA1 binding site reduced nmMLCK2 expression (compare the two right bars, minigene versus minigene mutant).
Both hnRNPA1 Splice Isoforms Bind to the Putative MYLK hnRNPA1 Binding Site
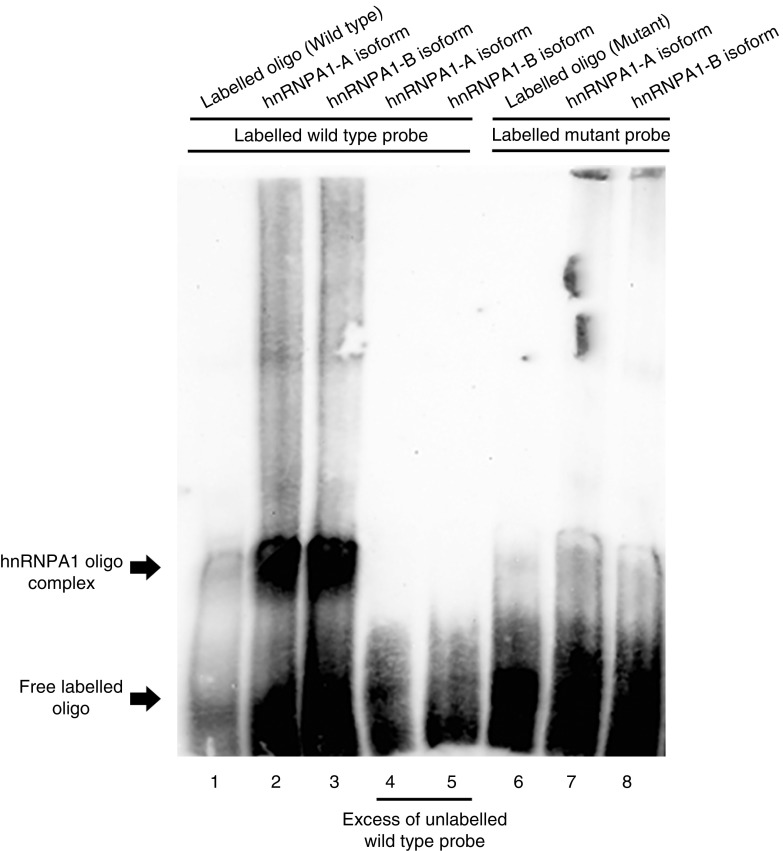
Two hnRNPA1 isoforms (A and B) exist that differ in the presence of 52 amino acids (372 vs. 320 amino acids) (20). We identified the presence of a functional MYLK hnRNPA1 binding site and sought to determine if hnRNPA1 effects on splicing were the result of direct MYLK binding. Gel shift assays were performed, and an RNA oligonucleotide harboring the wild-type hnRNPA1 binding site, as well as an oligonucleotide with a mutated putative hnRNPA1 binding site, was synthesized. Interactions of these hnRNPA1 isoform oligonucleotides, obtained via expression in HEK-293 cells followed by purification on anti-HA magnetic beads, were analyzed on 5% gels with chemiluminescence imaging. Our results indicate that both hnRNPA1 isoforms are capable of direct binding to the labeled oligonucleotide (Figure 4, lanes 2 and 3) with complete abolishment by an excess of unlabeled oligonucleotide (Figure 4, lanes 4 and 5). No bands were seen in the absence of the hnRNPA1 (Figure 4, lane 1) and in lanes in which a putative binding site had been mutated (Figure 4, lanes 7 and 8), indicating that hnRNPA1 binds directly and specifically to the binding site on MYLK intron 10–11.
Figure 4.
hnRNPA1 directly binds to the putative binding site on intron 10–11 of the MYLK gene. Representative gel showing hnRNPA1 binding to a labeled RNA oligo of MYLK intron 10–11 that contains either a wild-type hnRNPA1 or the site mutated. Lane 1 and lane 6 represent labeled wild-type and mutated oligonucleotides, respectively (without hnRNPA1 isoform). Addition of hnRNPA1-A (lane 2) and hnRNPA1-B (lane 3) isoforms in the presence of labeled wild-type oligo produces a specific band (indicated by the arrow). The specific bands can be competed using excess of wild-type unlabeled oligo in the presence of hnRNPA1-A (lane 4) and hnRNPA1-B (lane 5). Mutation of the hnRNPA1 binding site on the oligo caused a loss of binding of the hnRNPA1-A (lane 7) and hnRNPA1-B (lane 8) isoforms.
hnRNPA1 Promotes Skipping of MYLK Exon 11
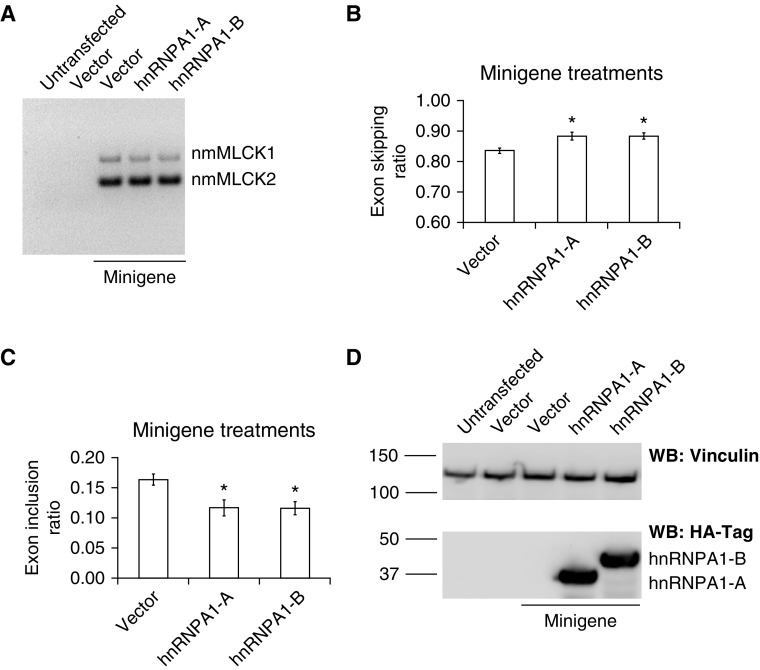
To directly validate the effect of hnRNPA1 on MYLK splicing, we transfected HEK-293 cells with the minigene alone or with either of the two hnRNPA1 isoforms and analyzed exon inclusion and skipping ratios by RT-PCR (Figure 5A). Our results indicate that hnRNPA1 overexpression promotes exon skipping (Figure 5B, middle [hnRNPA1-A] and right [hnRNPA1-B] bars) and inhibited exon inclusion (Figure 5C, middle [hnRNPA1-A] and right [hnRNPA1-B] bars), indicating increased levels of nmMLCK2 and reduced levels of nmMLCK1, respectively. To confirm overexpression of hnRNPA1, we analyzed protein levels by Western blotting using antibodies specific to hnRNPA1 (see Figure E1 in the data supplement) or directed against the HA epitope (Figure 5D), which confirms overexpression of recombinant hnRNPA1 isoforms, as shown in Figure 5D (last two lanes at right). Taken together, our results indicate that hnRNPA1 is functionally involved in regulation of MYLK splicing.
Figure 5.
hnRNPA1 promotes exon 11 skipping in an MYLK minigene system. (A) Representative gel showing RT-PCR analysis of the MYLK minigene. HEK-293 cells were transfected with constructs as indicated. Lanes from left to right show untransfected, vector-transfected cells, MYLK minigene, MYLK minigene + hnRNPA1-A, and MYLK minigene + hnRNPA1-B, respectively. Addition of hnRNPA1 promotes exon skipping. RT-PCR was performed, resulting in two specific products, nmMLCK1 and nmMLCK2, as indicated. (B and C) Densitometric analysis of the RT-PCR products in A showing (B) exon-skipping ratios (nmMLCK2/nmMLCK1 + nmMLCK2) and (C) exon inclusion ratios (nmMLCK1/nmMLCK1 + nmMLCK2). Bars correspond to MYLK minigene 2 (left), MYLK minigene + hnRNPA1-A (middle), and MYLK minigene + hnRNPA1-B (right). The data are presented as mean ± SE (n = 3). The data indicate that hnRNPA1 promotes exon skipping. *P < 0.05 comparing hnRNPA1-A or hnRNPA1-B with vector. (D) Western blot analysis using hemagglutinin-Tag (HA-Tag)-specific antibodies showing hnRNPA1 overexpression. Lanes from left to right represent untransfected cells, vector-transfected cells, MYLK minigene, MYLK minigene + hnRNPA1-A, and MYLK minigene + hnRNPA1-B, respectively. Vinculin was used as a loading control. The two isoforms hnRNPA1-A (molecular weight, 34 kD) and hnRNPA1-B (molecular weight, 38 kD) are shown. Molecular weight markers are also indicated.
hnRNPA1 and CS
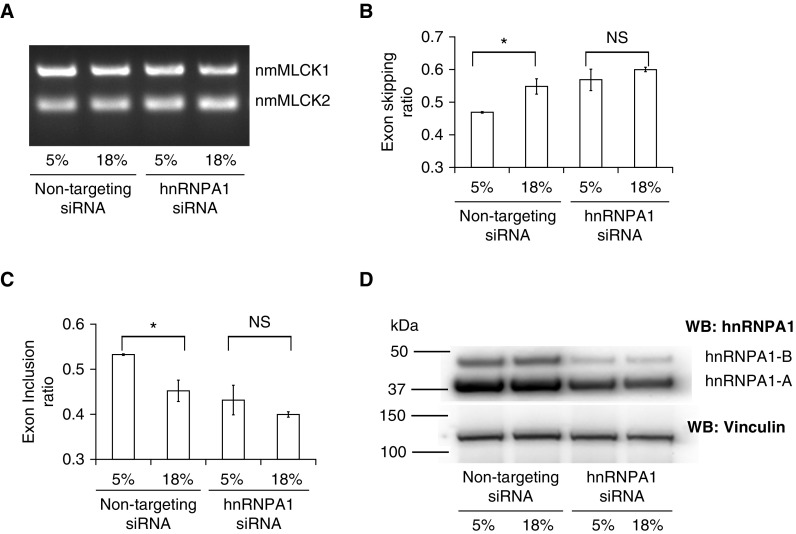
We have recently demonstrated that ECs subjected to 18% CS showed an increase in nmMLCK2 expression compared with cells exposed to 5% CS (15). Because in the present study we demonstrated that hnRNPA1 is increased in response to CS, we sought to investigate the role of hnRNPA1 in MYLK splicing and nmMLCK2 expression, and we silenced hnRNPA1 in ECs using hnRNPA1-specific siRNAs. Both hnRNPA1-silenced cells and nonspecific siRNA-transfected cells were subjected to 5% and 18% CS as described, and MYLK splicing was determined by RT-PCR (Figure 6A). The results indicated that in nontargeting siRNA-transfected cells, 18% CS caused increased nmMLCK2 expression revealed by an increase in exon-skipping ratio and a decrease in exon inclusion ratio (compared with 5% CS) (Figures 6B and C, first two bars at left). However, in hnRNPA1-silenced cells, there was no significant difference in nmMLCK2 expression after exposure to either 5% or 18% CS (Figure 6A and B, third and fourth lanes/bars from left). The efficacy of hnRNPA1 knockdown was validated using Western blotting (Figure 6D, last two lanes at right), which showed a reduction in hnRNPA1 protein expression. These results demonstrate a positive role of hnRNPA1 in MYLK splicing, promoting increased exon skipping and resulting in greater nmMLCK2 expression.
Figure 6.
hnRNPA1 knockdown prevents the 18% cyclic stretch–induced increases in exon skipping in endothelial cells. (A) Representative gel showing RT-PCR analysis of nmMLCK1 and nmMLCK2 in human pulmonary artery endothelial cells (HPAECs). HPAECs were transfected with siRNA as indicated, and RT-PCR was performed. The two lanes at left represent HPAECs exposed to 5% and 18% cyclic stretch, respectively, transfected with nontargeting siRNA. The two lanes at right represent HPAECs exposed to 5% and 18% cyclic stretch, respectively, transfected with hnRNPA1-specific siRNA. (B and C) Densitometric analysis of the RT-PCR products in A showing ratios of (B) exon skipping (nmMLCK2/nmMLCK1 + nmMLCK2) and (C) exon inclusion (nmMLCK1/nmMLCK1 + nmMLCK2). Left bars represent HPAECs exposed to 5% and 18% cyclic stretch, respectively, transfected with nontargeting siRNA. Right bars represent HPAECs exposed to 5% and 18% cyclic stretch, respectively, transfected with hnRNPA1-specific siRNA. The data are presented as mean ± SE (n = 3). ANOVA was used to compare the means. The data indicate that 18% cyclic stretch promotes nmMLCK2 expression, as indicated by increased exon skipping (comparison of first two bars at left). *P < 0.05 comparing the two left bars, 5% and 18% transfected with nontargeting siRNA. NS = not significant. (D) Western blot analysis using hnRNPA1-specific antibodies showing hnRNPA1 knockdown. Two lanes at left represent HPAECs exposed to 5% and 18% cyclic stretch, respectively, transfected with nontargeting siRNA. Two lanes at right represent HPAECs exposed to 5% and 18% cyclic stretch, respectively, transfected with hnRNPA1-specific siRNA. Vinculin was used as a loading control. The two hnRNPA1 isoforms are indicated (hnRNPA1-A, 34 kD; hnRNPA1-B, 38 kD).
hnRNPA1 Regulates Transendothelial Electrical Resistance
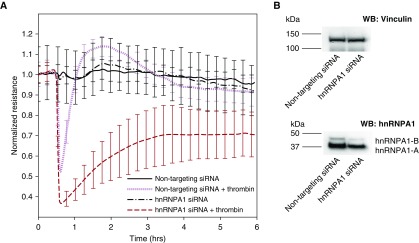
We have previously determined that silencing nmMLCK isoforms 1 and 2 alters transendothelial resistance (TER) in ECs in response to thrombin (15). Because we demonstrated a role of hnRNPA1 in regulating nmMLCK isoform splicing in the present study, we sought to determine the role of hnRNPA1 in affecting vascular barrier responses in human lung ECs. Consequently, we silenced hnRNPA1 in ECs subsequently challenged with thrombin (0.5 U/ml) and determined changes in electrical resistance, a measure of vascular barrier response. hnRNPA1-silenced cells demonstrated greater changes in TER responses than nontargeting siRNA-treated cells treated with thrombin (Figure 7A). The efficacy of silencing was validated using Western blot analysis (Figure 7B). These results indicate that hnRNPA1 regulates vascular barrier function in ECs.
Figure 7.
hnRNPA1 knockdown regulates vascular permeability in human lung endothelial cells. (A) Transendothelial resistance responses in endothelial cells upon thrombin stimulation. hnRNPA1 siRNA-treated cells showed an increased disruption upon thrombin treatment compared with nontargeting siRNA-treated cells. The values are means of three replicates. (B) Western blotting showing the efficiency of hnRNPA1 silencing. Vinculin was used as a loading control.
Discussion
In the present study, we have demonstrated a functional role for the splicing factor hnRNPA1 in the context of pathological 18% CS, generating the proinflammatory MYLK splice variant nmMLCK2. Understanding the splicing mechanisms regulating nmMLCKL2 generation is critical because regulatory PTMs that occur in response to vascular barrier agonists involve key residues (Y464, Y471) residing in exon 11 present only in nmMLCK1. The role of MYLK in generation of spatially specific contractile forces is underscored in nonmuscle isoform-specific MYLK knockout (30), nmMLCK-silenced mice (12), and nmMLCK2-overexpressing mice (9), highlighting the critical role of MYLK in vascular barrier regulation.
The MYLK gene is alternatively spliced into several isoforms, and the predominant form in lung endothelium is nmMLCK2, characterized by the absence of exon 11 (6). The two major isoforms nmMLCK1 and nmMLCK2 have similar enzymatic activities; however, only MLCK1 kinase activity is increased in response to phosphorylation by the p60src kinase (5), indicating that differential regulation of nmMLCK kinase activity occurs via exon 11 phosphorylation. Additionally, Y464 and Y471 are phosphorylated by activated c-Abl kinase in response to vascular barrier–promoting stimuli (8), post-translational events that we have shown to be associated with promotion of nmMLCK translocation to the cellular periphery and participation in vascular barrier regulation (31). However, the absence of exon 11 in nmMLCK2 precludes regulation of its activity and promoting an inflammatory phenotype because mice overexpressing nmMLCK2 show an increased injury in response to LPS and VILI (9). Taken together, these data suggest that splicing events promoting nmMLCK2 generation retard barrier recovery and promote vascular permeability.
We have demonstrated that hnRNPA1 expression levels are increased in response to VILI in mouse lungs and that excessive mechanical stress produced by CS. hnRNPA1 can function both as a transcription factor (32) and as a splicing factor (25, 26, 33–35). As a splicing factor, hnRNPA1 competes with the serine/arginine-rich family of proteins at overlapping exonic splicing enhancer and exonic splicing silencer sites (36) or at nonoverlapping sites where cooperative hnRNPA1 binding prevents serine/arginine-rich protein binding (37) or at intronic splicing silencer sites (38), ultimately promoting exon skipping.
We have identified an hnRNPA1 binding site located on intron 10–11 and demonstrated hnRNPA1 binding to this MYLK site using gel shift assays. We have demonstrated a functional role of hnRNPA1 in MYLK splicing, and it is highly possible that other splicing factors not identified in our study are involved in MYLK splicing. Using a minigene system (15), we have shown that mutation of the site causes a reduction in nmMLCK2 levels and is a result of direct binding of hnRNPA1 revealed by gel shift analysis. Similar splicing of PKM occurs through an hnRNPA1 concentration–dependent mechanism (25). In cancer cells, increased hnRNPA1 expression binds to exon 9 of PKM, promoting exclusion of exon 9. However, at lower concentrations, binding occurs at downstream intronic sites, promoting inclusion of exon 9 (25, 26). We have observed elevated levels of hnRNPA1 in response to excessive CS, and these results suggest a concentration-dependent effect of hnRNPA1 on MYLK splicing whereby elevated levels of hnRNPA1 result in binding of its cognate site on intron 10–11 of MYLK, promoting exon skipping and greater nmMLCK2 generated, a form of nmMLCK that produces greater vascular leak and inflammation. Upon hnRNPA1 silencing, exon skipping occurs at an increased level that may result from compensatory expression of other splicing factors including other members belonging to the hnRNP family of proteins or other splicing repressors. However, silencing of hnRNPA1 prevents increase of nmMLCK2 under 18% CS. In summary, the data indicate an important role of hnRNPA1 in MYLK splicing and promoting nmMLCK2 expression.
We validated the role of hnRNPA1 in determining the vascular barrier integrity in response to thrombin by silencing hnRNPA1 in ECs and determined that loss of hnRNPA1 caused an increase in vascular barrier disruption. This finding indicated that compensatory mechanisms through expression of other hnRNP family members or splicing repressors can promote exon skipping and generation of nmMLCK2, as we have observed. Alternatively hnRNPA1 is involved in splicing or regulation of other genes involved in regulation of vascular barrier integrity. We have previously demonstrated that silencing nmMLCK1/2 or nmMLCK2 altered vascular barrier responses in ECs (15). Taken together, our TER experiments further support the role of hnRNPA1 in vascular barrier regulation through nmMLCK splicing regulation.
The regulation of hnRNPA1 is not well characterized. Cancer cells show elevated levels of c-myc and knockdown of c-myc using shRNA reduced levels of hnRNPA1, hnRNPA2, and other splicing factors in NIH3T3 cells (33). However, only marginal decreases in levels of the splicing factor were seen in HeLa cells, indicating the involvement of other splicing factors (33). Other factors, including MYCN, directly activate both hnRNPA1 and polypyrimidine tract binding protein and directly influence PKM2 expression through splicing (39). We have observed elevated myc expression in lungs obtained from VILI-exposed mice (data not shown). It is also possible that increased hnRNPA1 levels may result from reduced protein turnover because proteasomal inhibition increases hnRNPA1 levels in human ECs correlated with differential gene expression, suggesting that hnRNPA1 is likely regulated through the proteasomal pathway (40).
Splicing by hnRNPA1 is involved in spinal muscular atrophy caused by loss/mutation of the survival motor neuron-1 (SMN1) gene. SMN2 is identical to SMN1 but has a single nucleotide difference that negatively affects splicing, and overexpression of hnRNPA1 inhibits exon 7 inclusion (41). Overexpression of hnRNPA1 decreases expression of an HMGCR splice variant lacking exon 13 that is important in response to statins, a class of cholesterol-lowering drugs (42). A single base change (c.1007G>A) within the neurofibromatosis type 1 (NF1) gene generates a pathogenic mutation that creates a binding site for hnRNPA1 and exon 9 exclusion (43). hnRNPA1 also plays an important role in generation of androgen receptor splice variants (AR-V7). A strong positive correlation was observed between hnRNPA1 and AR-V7, and knockdown of hnRNPA1 caused concomitant reduction in AR-V7 levels and increased sensitivity to enzalutamide. Together, these results strongly indicate that dysregulation of hnRNPA1 potentially alters disease phenotypes via splicing regulation.
We report a direct and important regulatory role of hnRNPA1 in MYLK splicing as well as in the context of 18% CS, resulting in greater expression of the proinflammatory nmMLCK2 variant. These findings increase understanding of MYLK splicing in the regulation of lung vascular integrity during acute lung inflammation and excessive mechanical stress, such as observed in ARDS and VILI.
Footnotes
Supported by National Institutes of Health grants R01 HL91889 and P01 HL126609.
Author Contributions: J.B.M.: performed and analyzed the experiments and was the primary writer; A.Y.T. and S.M.D.: contributed to writing the manuscript and performing bioinformatic analyses; T.Z. and T.W.: contributed to writing and analysis; and J.G.N.G.: was the project leader and contributed to experimental design and writing the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2017-0141OC on October 27, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Butt Y, Kurdowska A, Allen TC. Acute lung injury: a clinical and molecular review. 2016;140:345–350. doi: 10.5858/arpa.2015-0519-RA. [DOI] [PubMed] [Google Scholar]
- 2.Suter PM, Suter S, Girardin E, Roux-Lombard P, Grau GE, Dayer JM. High bronchoalveolar levels of tumor necrosis factor and its inhibitors, interleukin-1, interferon, and elastase, in patients with adult respiratory distress syndrome after trauma, shock, or sepsis. 1992;145:1016–1022. doi: 10.1164/ajrccm/145.5.1016. [DOI] [PubMed] [Google Scholar]
- 3.Dudek SM, Garcia JG. Cytoskeletal regulation of pulmonary vascular permeability. 2001;91:1487–1500. doi: 10.1152/jappl.2001.91.4.1487. [DOI] [PubMed] [Google Scholar]
- 4.Garcia JG, Pavalko FM, Patterson CE. Vascular endothelial cell activation and permeability responses to thrombin. 1995;6:609–626. doi: 10.1097/00001721-199510000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Birukov KG, Csortos C, Marzilli L, Dudek S, Ma SF, Bresnick AR, et al. Differential regulation of alternatively spliced endothelial cell myosin light chain kinase isoforms by p60(Src) 2001;276:8567–8573. doi: 10.1074/jbc.M005270200. [DOI] [PubMed] [Google Scholar]
- 6.Lazar V, Garcia JG. A single human myosin light chain kinase gene (MLCK; MYLK) 1999;57:256–267. doi: 10.1006/geno.1999.5774. [DOI] [PubMed] [Google Scholar]
- 7.Dudek SM, Birukov KG, Zhan X, Garcia JG. Novel interaction of cortactin with endothelial cell myosin light chain kinase. 2002;298:511–519. doi: 10.1016/s0006-291x(02)02492-0. [DOI] [PubMed] [Google Scholar]
- 8.Dudek SM, Chiang ET, Camp SM, Guo Y, Zhao J, Brown ME, et al. Abl tyrosine kinase phosphorylates nonmuscle myosin light chain kinase to regulate endothelial barrier function. 2010;21:4042–4056. doi: 10.1091/mbc.E09-10-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moitra J, Evenoski C, Sammani S, Wadgaonkar R, Turner JR, Ma SF, et al. A transgenic mouse with vascular endothelial over-expression of the non-muscle myosin light chain kinase-2 isoform is susceptible to inflammatory lung injury: role of sexual dimorphism and age. 2008;151:141–153. doi: 10.1016/j.trsl.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birukov KG, Birukova AA, Dudek SM, Verin AD, Crow MT, Zhan X, et al. Shear stress-mediated cytoskeletal remodeling and cortactin translocation in pulmonary endothelial cells. 2002;26:453–464. doi: 10.1165/ajrcmb.26.4.4725. [DOI] [PubMed] [Google Scholar]
- 11.Garcia JG, Davis HW, Patterson CE. Regulation of endothelial cell gap formation and barrier dysfunction: role of myosin light chain phosphorylation. 1995;163:510–522. doi: 10.1002/jcp.1041630311. [DOI] [PubMed] [Google Scholar]
- 12.Mirzapoiazova T, Moitra J, Moreno-Vinasco L, Sammani S, Turner JR, Chiang ET, et al. Non-muscle myosin light chain kinase isoform is a viable molecular target in acute inflammatory lung injury. 2011;44:40–52. doi: 10.1165/rcmb.2009-0197OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T, Brown ME, Kelly GT, Camp SM, Mascarenhas JB, Sun X, et al. Myosin light chain kinase (MYLK) coding polymorphisms modulate human lung endothelial cell barrier responses via altered tyrosine phosphorylation, spatial localization and lamellipodial protrusions 2018822045894018764171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia JG, Liu F, Verin AD, Birukova A, Dechert MA, Gerthoffer WT, et al. Sphingosine 1-phosphate promotes endothelial cell barrier integrity by Edg-dependent cytoskeletal rearrangement. 2001;108:689–701. doi: 10.1172/JCI12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mascarenhas JB, Tchourbanov AY, Fan H, Danilov SM, Wang T, Garcia JG. Mechanical stress and single nucleotide variants regulate alternative splicing of the MYLK gene. 2017;56:29–37. doi: 10.1165/rcmb.2016-0053OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 17.Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 18.Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 19.Ge H, Zuo P, Manley JL. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 20.Jean-Philippe J, Paz S, Caputi M. hnRNP A1: the Swiss army knife of gene expression. 2013;14:18999–19024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayeda A, Munroe SH, Cáceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavanez JP, Madl T, Kooshapur H, Sattler M, Valcárcel J. hnRNP A1 proofreads 3′ splice site recognition by U2AF. 2012;45:314–329. doi: 10.1016/j.molcel.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 25.Chen M, David CJ, Manley JL. Concentration-dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. 2012;19:346–354. doi: 10.1038/nsmb.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M, Zhang J, Manley JL. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. 2010;70:8977–8980. doi: 10.1158/0008-5472.CAN-10-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Churbanov A, Vorechovský I, Hicks C. A method of predicting changes in human gene splicing induced by genetic variants in context of cis-acting elements. 2010;11:22. doi: 10.1186/1471-2105-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong SB, Huang Y, Moreno-Vinasco L, Sammani S, Moitra J, Barnard JW, et al. Essential role of pre-B-cell colony enhancing factor in ventilator-induced lung injury. 2008;178:605–617. doi: 10.1164/rccm.200712-1822OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X, Elangovan VR, Mapes B, Camp SM, Sammani S, Saadat L, et al. The NAMPT promoter is regulated by mechanical stress, signal transducer and activator of transcription 5, and acute respiratory distress syndrome-associated genetic variants. 2014;51:660–667. doi: 10.1165/rcmb.2014-0117OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wainwright MS, Rossi J, Schavocky J, Crawford S, Steinhorn D, Velentza AV, et al. Protein kinase involved in lung injury susceptibility: evidence from enzyme isoform genetic knockout and in vivo inhibitor treatment. 2003;100:6233–6238. doi: 10.1073/pnas.1031595100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown M, Adyshev D, Bindokas V, Moitra J, Garcia JG, Dudek SM. Quantitative distribution and colocalization of non-muscle myosin light chain kinase isoforms and cortactin in human lung endothelium. 2010;80:75–88. doi: 10.1016/j.mvr.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y, Lin L, Yu X, Wen G, Pu X, Zhao H, et al. Functional involvements of heterogeneous nuclear ribonucleoprotein A1 in smooth muscle differentiation from stem cells in vitro and in vivo. 2013;31:906–917. doi: 10.1002/stem.1324. [DOI] [PubMed] [Google Scholar]
- 33.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. 2007;16:3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- 35.Nadiminty N, Tummala R, Liu C, Lou W, Evans CP, Gao AC. NF-κB2/p52:c-Myc:hnRNPA1 pathway regulates expression of androgen receptor splice variants and enzalutamide sensitivity in prostate cancer. 2015;14:1884–1895. doi: 10.1158/1535-7163.MCT-14-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 37.Okunola HL, Krainer AR. Cooperative-binding and splicing-repressive properties of hnRNP A1. 2009;29:5620–5631. doi: 10.1128/MCB.01678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo R, Li Y, Ning J, Sun D, Lin L, Liu X. HnRNP A1/A2 and SF2/ASF regulate alternative splicing of interferon regulatory factor-3 and affect immunomodulatory functions in human non-small cell lung cancer cells. 2013;8:e62729. doi: 10.1371/journal.pone.0062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang S, Wei JS, Li SQ, Badgett TC, Song YK, Agarwal S, et al. MYCN controls an alternative RNA splicing program in high-risk metastatic neuroblastoma. 2016;371:214–224. doi: 10.1016/j.canlet.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bieler S, Hammer E, Gesell-Salazar M, Völker U, Stangl K, Meiners S. Low dose proteasome inhibition affects alternative splicing. 2012;11:3947–3954. doi: 10.1021/pr300435c. [DOI] [PubMed] [Google Scholar]
- 41.Wee CD, Havens MA, Jodelka FM, Hastings ML. Targeting SR proteins improves SMN expression in spinal muscular atrophy cells. 2014;9:e115205. doi: 10.1371/journal.pone.0115205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu CY, Theusch E, Lo K, Mangravite LM, Naidoo D, Kutilova M, et al. HNRNPA1 regulates HMGCR alternative splicing and modulates cellular cholesterol metabolism. 2014;23:319–332. doi: 10.1093/hmg/ddt422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernández-Imaz E, Martín Y, de Conti L, Melean G, Valero A, Baralle M, et al. Functional analysis of mutations in exon 9 of nf1 reveals the presence of several elements regulating splicing. 2015;10:e0141735. doi: 10.1371/journal.pone.0141735. [DOI] [PMC free article] [PubMed] [Google Scholar]