Abstract
A common explanation for the origins and rising prevalence of asthma is that they involve complex interactions between hereditary predispositions and environmental exposures that are incompletely understood. Yet, emerging evidence substantiates the paradigm that environmental exposures prenatally and during very early childhood induce epigenetic alterations that affect the expression of asthma genes and, thereby, asthma itself. Here, we review much of the key evidence supporting this paradigm. First, we describe evidence that the prenatal and early postnatal periods are key time windows of susceptibility to environmental exposures that may trigger asthma. Second, we explain how environmental epigenetic regulation may explain the immunopathology underlying asthma. Third, we outline specific evidence that environmental exposures induce epigenetic regulation, both from animal models and robust human epidemiological research. Finally, we review some emerging topics, including the importance of coexposures, population divergence, and how epigenetic regulation may change over time. Despite all the inherent complexity, great progress has been made toward understanding what we still consider reversible asthma risk factors. These, in time, may impact patient care.
Keywords: asthma mechanisms, environmental epigenetic regulation, perinatal environmental exposures
The prevalence of current asthma continues to rise in the United States. Recently, the U.S. Centers for Disease Control and Prevention reported that 7.8% of the population is diagnosed with current asthma (8.4% of children and 7.6% of adults) (1). Of great concern are the disparities associated with asthma rates, hospitalization, and death by race and sex, with Puerto Ricans (13.7%), African Americans (10.3%), and females (9.1%) having higher prevalence of current asthma (2). Both the prenatal and early postnatal periods have been identified as key time windows, when specific environmental exposures increase the susceptibility for later asthma. Although thoroughly reviewed in past publications (3, 4), notable findings document that prenatal exposure to stress (5) and air pollution (6) are prime examples of exposures that heighten the subsequent risk for asthma development. For the latter, greater exposure to fine particulate matter less than 2.5 μm in aerodynamic diameter, specifically during 16–25 weeks gestation, and in interaction with male sex, was associated with early childhood asthma development (6). Shortly after birth, another time period of susceptibility to asthma appears evident, as recently demonstrated by Barr and colleagues (7). They found that the estimated prevalence of self-reported asthma among Hispanics was higher when the age of immigration to the four U.S. cities under investigation (New York, NY; Chicago, IL; Miami, FL; and San Diego, CA) was within the first few years of life. This study, therefore, implicates the urban physical environment experienced at a young age, more so than during an older age or during adulthood, as impacting prevalence rates.
Epigenetic regulation after environmental exposures prenatally and during very early childhood may underlie much of these observations. Methylation, post-translational modifications of histones via acetylation, phosphorylation, ubiquitination, and the expression of inhibitory microRNAs are all regulatory mechanisms that alter gene expression without changing the coding sequence, and appear to be affected by external environmental exposures, as reviewed elsewhere (8). For example, during DNA methylation, the covalent addition of a methyl group to the 5′ position of the cytosine pyrimidine ring, often at CpG sites, may affect the metabolism of environmental compounds, degree of proinflammatory response, efficacy of pharmacological treatment, or the risk of disease development. Exposures to allergens, air pollution, and various dietary components have been observed to induce gene-specific epigenetic alterations that affect the transcription of asthma-related genes and subsequent phenotypic expression of asthma or its biomarkers (9). One demonstration of this phenomenon is that the presence of maternal asthma is a greater risk factor for offspring asthma than paternal asthma, implicating the importance of the intrauterine environment (10). In addition, uncontrolled maternal asthma has been observed recently to increase the risk of childhood early-onset persistent or transient asthma (11). Also in recent related work, maternal asthma was associated with neonatal methylation of a gene called mothers against decapentaplegic homolog 3, which was previously linked to asthma in genome-wide association studies (12), corroborating the epigenetic paradigm.
Experimental models in mice have provided direct evidence of prenatal environmental epigenetic regulation. In one study, our group examined CpG methylation in promoter regions of the T-helper genes, interleukin (IL)-4, and interferon-γ, in grand-offspring mice (F2) after prenatal sensitization and gestational exposure of female mice (F0) to inhaled Aspergillus fumigatus allergen (13). Methylation at several IL-4 and interferon-γ CpG sites in lung tissue was lower in grand offspring of mice dosed with A. fumigatus during gestation. In another model, prenatal allergen sensitization followed by suboptimal sensitization (considered to make offspring “at risk” for asthma-like phenotype) induced distinct patterns of DNA methylation and transcription in dendritic cells. Most of the genes with differential methylation in offspring mice also showed transcriptional changes after the first encounter with the allergen, the majority of the genes identified as involved in asthma or allergic inflammation (14). Another group studied the effect of prenatal exposure to the gram-negative, farm-derived isolate, Acinetobacter lwoffii F78, to model the mechanisms of asthma and allergy prevention and immune modulation among children raised on farms (15). The effect of this exposure on histone 4 acetylation (associated with transcriptionally active chromatin) and its mitigation of T-helper cytokine activity in CD4+ T-cells, after ovalbumin sensitization of offspring, was assessed. Ovalbumin sensitization and challenge of the offspring of prenatally sham-exposed mothers demonstrated lower levels of histone 4 acetylation at the counter-regulatory interferon-γ promoter when compared with levels measured among sham-exposed offspring. This effect was associated with altered interferon-γ expression, and reversed among ovalbumin-sensitized and -challenged offspring of mothers exposed to A. lwoffii F78. Prenatal A. lwoffii also reduced histone 4 acetylation levels in sensitized/challenged A. lwoffii–exposed offspring at the proallergic IL-4 (but not IL-5 or conserved noncoding sequence 1) promoter. Notably, prenatal A. lwoffii did not induce any changes in the CpG methylation at these sites (16). The authors interpreted these results to indicate that A. lwoffii modulated the allergic immune response through the transcriptional control of histone 4 acetylation. Jahreis and colleagues (17) observed, in the LINA (Lifestyle and Environmental Factors and their Influence on Newborns Allergy Risk) mother–child cohort, an increased risk of asthma in children of mothers having an elevated level of mono-n-butyl phthalate, a major, butyl benzyl phthalate metabolite. A coordinating mouse model demonstrated that prenatal and perinatal exposure to butyl benzyl phthalate exacerbated the airway hyperreactivity associated with elevated ovalbumin-specific immunoglobin E and proallergic T-helper cell type 2 cytokines, IL-5 and IL-13. Isolation of differentially methylated and differentially expressed genes revealed three hypermethylated CD4+ T-cell genes, including fatty acid desaturase 1, fanconi anemia complementation group A, and zinc finger protein 1. Specifically highlighted was the GATA binding protein 3–regulating zinc finger protein 1 gene, the decreased expression and increased enhancer methylation of which was observed in the prenatally phthalate-exposed children of the LINA cohort.
Robust human studies also suggest that environmental exposures induce epigenetic outcomes relevant to asthma. These include ex vivo studies, such as one that isolated airway epithelial cells from adults with asthma versus those without asthma and cultured them in the presence of IL-13 as a cellular model of the proallergic external environment. After discovering an IL-13 epigenetic signature in culture, validation experiments were performed on the airway epithelial cells isolated from subjects with and without asthma. Epigenetic findings clustered into one distinct module that correlated with asthma severity and lung function, and another distinct module that correlated with eosinophilia, suggesting environmental epigenetic regulation of distinct asthma endotypes (18). Also supportive of the importance of the childhood environment was recent work from a study of monozygotic twins, a cohort that allows natural controls for the genetic background and, presumably, the intrauterine, and to some extent, early postnatal environment. Murphy and colleagues (19) examined numerous differences in genome-wide DNA methylation patterns by the presence or absence of childhood asthma at age 10 years, when the external environments have started to diverge. The data also suggested that some of these differences may persist through age 18 years, in contrast to measures among healthy twins or those with remitted disease; however, statistical corrections for multiple comparisons were less supportive. In addition, environmental influences on epigenetic regulation were suggested from observations derived from 6- to 12-year-old children participating in the Inner-City Asthma Consortium. Presumably, these children with asthma have airway disease that is affected by urban environmental exposures when compared with health control subjects. A subset of their genes discovered on genome-wide analyses exhibited high correlations between DNA methylation and expression levels that were replicated in a second independent urban cohort (20).
Other cohort studies have measured exposures in the environment more directly and related their levels to altered methylation levels. Our group at the Columbia Center for Children’s Environmental Health (New York, NY) examined prenatal exposure to the combustion products of polycyclic aromatic hydrocarbons, and found that these measures were associated with altered methylation in cord blood at several sites of the interferon-γ promoter, although, these differences were not associated with later parental report of asthma (21). Prenatal exposure to the pollutant NO2, measured by land-use regression models, similarly was associated with a number of altered methylation sites found using epigenome-wide meta-analysis from specimens derived from four studies participating in the Pregnancy and Childhood Epigenetics consortium. Moreover, cigarette smoking, particularly during pregnancy, has been linked frequently in cohort studies with altered methylation levels in epigenome-wide analyses in cord blood. These include studies that discovered specific genes, aryl-hydrocarbon receptor repressor, FERM (4.1 ezrin radixin moesin) domain containing 4A, chromosome 11 open reading frame 52, and c-Jun N-terminal kinase 2, and specific areas of enrichment, such as CpG island shores, enhancers, and DNAase hypersensitivity sites within these genes (22–24). The links to asthma, via altered DNA methylation, have been more elusive, but investigators at Columbia and University of Cincinnati (Cincinnati, OH) found parallel links between prenatal polycyclic aromatic hydrocarbon exposure and altered methylation of the acyl-coenzyme A synthetase long-chain family member 3 gene, and altered methylation of acyl-coenzyme A synthetase long-chain family member 3 with parental report of childhood asthma in the Columbia Center for Children’s Environmental Health cohort (25). Exposure to phthalates, also previously implicated with asthma risk in the Columbia Center for Children’s Environmental Health cohort (26), when assessed using the urinary metabolite as the biomarker, was associated with altered methylation in tumor necrosis factor-α. The group validated this finding in a second cohort, and then linked the altered methylation to asthma cases in the Childhood Environment and Allergic diseases and Isle of Wight cohorts (27). Asthma gene–specific altered methylation was also found after prenatal exposure to farm living, previously associated with protection from the development of allergic immune responses in the Protection against Allergy: Study in Rural Environment group. Orosomucoid-like sphingolipid biosynthesis regulator (known as ORMDL) 1, an important ORMDL3 paralog sharing similar metabolic functions, and signal transducer and activator of transcription 6, which is important to immunoglobin E regulation, were demethylated, and RAD50 double strand break repair protein (known as RAD50) and IL13 were hypermethylated, in children of farmers compared with children of nonfarmers (15). Finally, differential methylation by season of birth was discovered and separately validated as associated with altered methylation in several genes, such as vitamin K epoxide reductase complex subunit 1, protein tyrosine phosphatase receptor type N2 (autumn), and brain-specific serine/threonine-protein kinase 2, Toll-interacting protein (summer), and pyruvate dehydrogenase kinase 1 (spring) (28). Toll-interacting protein is a negative regulator of Toll-like receptor signaling (29), and its downregulation has been observed to result in decreased airflow in individuals with asthma (30). Informative human longitudinal cohorts also include LINA, which demonstrated that high measures of maternal stress during pregnancy were associated with genome-wide differential DNA methylation and wheeze in children (5).
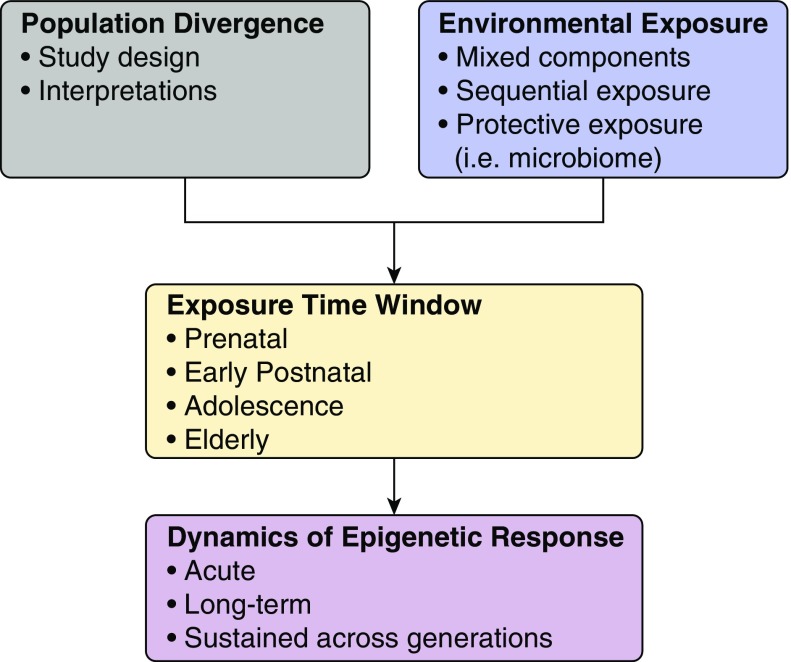
So, what topics are emerging in this rapidly evolving field? As this field advances, the relatively simple model that tests one exposure at one time point for epigenetic events has evolved to more granular research that examines coexposures, and the ensuing dynamics of the epigenetic responses (Figure 1). This was evident in research that re-examined the previously observed priming of diesel exposure on allergic sensitization (31). Growing literature continues to suggest that early-childhood exposures to multiple toxicants may impact the risk for asthma (32). Yet studies on their epigenetic regulation of combined exposures are more limited. In one, Rider and colleagues (33) conducted a double-blind, randomized, cross-over–controlled study of 2 hours of filtered air or diesel exhaust (300 μg particulate matter less than 2.5 μm in diameter per m3) exposure, followed by saline-controlled segmental bronchial allergen challenge, on 18 adult volunteers with atopy. Interestingly, they reported significant effects of diesel and allergen coexposure on both microRNA and messenger RNA expression without evidence for an interaction between them. This compares with recent work by Clifford and colleagues (34) among 17 adult volunteers. Here, in a randomized, cross-over–controlled exposure study design, diesel exhaust (also 300 μg particulate matter less than 2.5 μm in diameter per m3) or filtered air was administered for 2 hours, followed 1 hour later by allergen extract (vs. saline control), at a concentration 10-fold lower than the minimal dose producing a positive wheal, instilled in a lower-lobe bronchial segment. Four weeks later, the process was repeated with opposing exposures. At 48 hours after both, bronchial epithelial cells were collected from brushings during bronchoalveolar lavage and used to measure DNA methylation. They found that an initial allergen exposure primed DNA methylation of a set of CpG sites that differed from those first exposed to diesel. Furthermore, although only a relatively small number of CpG sites exhibited altered methylation 48 hours after initial exposure, upon repeat exposure 4 weeks later, there were large alterations in DNA methylation manifested. The authors interpreted these findings to indicate that, although the lung was highly sensitive to the effects of short-term exposure, the consequences required a second toxic exposure to manifest substantially. Clearly, controlled and cross-over exposures will be important in sorting out the effects of mixed exposures on epigenetic regulation, and remain a significant current research gap, in large part due to barriers inherent in pediatric research.
Figure 1.
Emerging concepts in environmental epigenetic regulation in asthma. Emerging concepts include assessing a greater understanding of the importance of individual versus mixtures and sequential environmental exposures, both toxic and protective; their time course may affect the timeline of epigenetic regulation. Although the perinatal time-window of susceptibility has emerged as key, others, such as adolescence and elderly periods, are under investigation. Studies may need to consider epigenetic patterns in the setting of divergence of whole populations over time, and examine the dynamics of the epigenetic responses, including over generations of individuals.
Another area of complexity gaining greater presence in the published literature is the impact of population divergence on patterns of DNA methylation and presumably other epigenetic indicators. For example, Fraser and colleagues (35) measured DNA methylation near the transcription start sites of over 14,000 genes in 180 cell lines derived from one West African and one Northern European population. Studying 30 mother/father/offspring trios from each group, they found population-specific patterns of DNA methylation in over one-third of all genes. Upon further analyses of trios, they also found that the heritability of CpG methylation differed by population. The impact of these findings is large, because it uncovers additional variability at the level of DNA methylation that should be considered, perhaps, in all clinical studies of genetic-by-environment-by-epigenetic relationships.
Finally, environmental exposures are constantly changing in the short term, and there are genuine concerns about our changing climate, and of known asthma environmental triggers, such as allergens and air pollutants, in the long term (36, 37). The epigenetic consequences of the latter can only be speculated from the limited studies available on the former. These include cell studies that used time-lapse microscopy to discern targeted chromatin regulator recruitment and tracked its acute effects on reporter gene activity and cell silencing. Wide variations in silencing events were observed among cells, and even between sister cells. In addition, the rate of silencing depended largely on the chromatin regulator involved (38). Human cohort studies that measured epigenetic responses over time also are scant. However, the Protection against Allergy: Study in Rural Environment group conducted pooled analysis of German discovery samples and Austrian and Swiss samples from a replication data set at two time points. Early-childhood asthma was associated with a change in blood methylation between birth and age 4.5 years in the RAD50 double strand break repair gene and CpG island 5′ of the transcription start site of IL-13 (15). In addition, in the intronic region of the orosomucoid-like sphingolipid biosynthesis regulator 3 gene, previously linked to asthma in a genome-wide association study (39), a significant decrease in methylation over time was observed in children with asthma exposed prenatally to farm living in contrast to an increase in methylation in children with asthma not exposed prenatally to farm living and to children without asthma (15). Human cohort studies that measure responses over changing environmental exposures are also scarce. Our group conducted one such randomized intervention study with a cohort of children and adolescents aged 5–17 years. We investigated the effect of mouse allergen–targeted integrated pest management and education (versus education only) on asthma morbidity. Although no significant difference was observed in asthma morbidity between the two groups after intervention, a decrease in buccal DNA, Forkhead box P3 promoter methylation was observed in the mouse-specific integrated pest management group (40).
Conclusions
So what are the lessons learned regarding our investigation of root causes of asthma? Despite all the inherent complexity, several themes do emerge. For one, the prenatal and early postnatal time periods appear to be vulnerable time windows of susceptibility to epigenetic regulation (Table 1). Other periods, such as adolescence and during older adulthood, warrant further investigations. Second, epigenetic regulation, in both controlled laboratory experiments and rigorous cohort studies, is linked to asthma-related outcomes. Factors, such as mixed exposures, dynamic timeline of epigenetic responses to changing environmental conditions, and population divergence, should not be underestimated in study designs or in the interpretation of their results. Still, great progress has been made toward understanding what we still consider reversible asthma risk factors. These, in time, may impact patient care.
Table 1.
Recent experimental versus epidemiological evidence of perinatal exposures and epigenetic regulation
| Exposure | Outcome | Reference |
|---|---|---|
| Epidemiological studies | ||
| Prenatal stress | Subsequent wheeze and genome-wide differential DNA methylation patterns | 5 |
| Maternal asthma | Neonatal SMAD3 methylation | 12 |
| Asthma discordant twins | Genome-wide differential DNA methylation patterns | 19 |
| Inner city | Correlated changes in DNA methylation and expression in select genes | 20 |
| Prenatal cigarette | Epigenome-wide and gene-specific altered methylation (C11orf52 and JNK2) | 22–24 |
| Prenatal PAH | Altered ACSL3 methylation and subsequent asthma | 25 |
| Prenatal phthalates | Altered TNF-α methylation and subsequent asthma | 27 |
| Prenatal farm living | Altered ORMDL1, STAT6, RAD50, and IL-13 methylation | 15 |
| Season of birth | Genome-wide differential DNA methylation patterns | 28 |
| Experimental studies | ||
| Prenatal Aspergillus fumigatus | Altered lung IL-4 and IFN-γ methylation in grandoffspring | 13 |
| “At risk” neonatal mice allergen challenge | Differential methylation and transcription | 14 |
| Prenatal Acinetobacter lwoffii F78 | Increased H4ac at INF-γ and reduced H4ac at IL-4 promoters | 16 |
| IL-13 | Epigenetic signature associated with asthma severity or eosinophilia | 18 |
| Diesel exhaust and subsequent allergen challenge | Altered microRNA and messenger RNA expression | 33 |
| Diesel exhaust and subsequent allergen challenge | Altered DNA methylation in bronchial epithelial cells | 34 |
Definition of abbreviations: ACSL3 = acyl-coenzyme A synthetase long-chain family member; C11orf52 = chromosome 11 open reading frame 52; H4ac = histone 4 acetylation; IL = interleukin; IFN = interferon; JNK2 = c-Jun N-terminal kinase 2; ORMDL1 = orosomucoid-like sphingolipid biosynthesis regulator 1; PAH = polycyclic aromatic hydrocarbon; RAD50 = RAD50 double strand break repair protein; SMAD3 = mothers against decapentaplegic homolog 3 gene; STAT6 = signal transducer and activator of transcription 6; TNF = tumor necrosis factor.
Supplementary Material
Footnotes
Supported by National Institutes of Health grants 4R01ES13163 (R.L.M.) and 1R01HL118612A1 (R.L.M.).
Author Contributions: R.L.M. and J.L. co-authored the manuscript.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Smoke S. Secondhand smoke (SHS) facts. Atlanta: Centers for Disease Control and Prevention; 2015. [Google Scholar]
- 2.Moorman J, Akinbami L, Bailey C, Zahran HS, King MD, Johnson CA, et al. National surveillance of asthma: United States 2001–2010Vital and Health Statistics201231–57. [PubMed] [Google Scholar]
- 3.Miller RL, Peden DB. Environmental effects on immune responses in patients with atopy and asthma. J Allergy Clin Immunol. 2014;134:1001–1008. doi: 10.1016/j.jaci.2014.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaffin JM, Kanchongkittiphon W, Phipatanakul W. Perinatal and early childhood environmental factors influencing allergic asthma immunopathogenesis. Int Immunopharmacol. 2014;22:21–30. doi: 10.1016/j.intimp.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trump S, Bieg M, Gu Z, Thürmann L, Bauer T, Bauer M, et al. Prenatal maternal stress and wheeze in children: novel insights into epigenetic regulation. Sci Rep. 2016;6:28616. doi: 10.1038/srep28616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu HH, Chiu YH, Coull BA, Kloog I, Schwartz J, Lee A, et al. Prenatal particulate air pollution and asthma onset in urban children. Identifying sensitive windows and sex differences. Am J Respir Crit Care Med. 2015;192:1052–1059. doi: 10.1164/rccm.201504-0658OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barr R, Avilés-Santa L, Davis S, Aldrich T, Gonzalez F, et al. Pulmonary disease and age at immigration among Hispanics: results from the Hispanic Community Health Study/Study of Latinos. Am J Respir Crit Care Med. 2016;193:386–395. doi: 10.1164/rccm.201506-1211OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Planell-Saguer M, Lovinsky-Desir S, Miller RL. Epigenetic regulation: the interface between prenatal and early-life exposure and asthma susceptibility. Environ Mol Mutagen. 2014;55:231–243. doi: 10.1002/em.21836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller R. Environmental medical epigenetics: a review of epigenetically-induced medical risks generated from exposures in our air, food, and personal products. In: Tollesfsbol T, editor. Medical epigenetics. New York: Elsevier; 2016. pp. 103–125. [Google Scholar]
- 10.Lim RH, Kobzik L, Dahl M. Risk for asthma in offspring of asthmatic mothers versus fathers: a meta-analysis. PLoS One. 2010;5:e10134. doi: 10.1371/journal.pone.0010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Agerbo E, Schlünssen V, Wright RJ, Li J, Munk-Olsen T.Maternal asthma severity and control during pregnancy and risk of offspring asthma J Allergy Clin Immunol [online ahead of print] 27 Jun 2017DOI: 10.1016/j.jaci.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 12.DeVries A, Wlasiuk G, Miller SJ, Bosco A, Stern DA, Lohman IC, et al. Epigenome-wide analysis links SMAD3 methylation at birth to asthma in children of asthmatic mothers. J Allergy Clin Immunol. 2017;140:534–542. doi: 10.1016/j.jaci.2016.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niedzwiecki M, Zhu H, Corson L, Grunig G, Factor PH, Chu S, et al. Prenatal exposure to allergen, DNA methylation, and allergy in grandoffspring mice. Allergy. 2012;67:904–910. doi: 10.1111/j.1398-9995.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikhaylova L, Zhang Y, Kobzik L, Fedulov AV. Link between epigenomic alterations and genome-wide aberrant transcriptional response to allergen in dendritic cells conveying maternal asthma risk. PLoS One. 2013;8:e70387. doi: 10.1371/journal.pone.0070387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel S, Busato F, Genuneit J, Pekkanen J, Dalphin JC, Riedler J, et al. PASTURE Study Group. Farm exposure and time trends in early childhood may influence DNA methylation in genes related to asthma and allergy. Allergy. 2013;68:355–364. doi: 10.1111/all.12097. [DOI] [PubMed] [Google Scholar]
- 16.Brand S, Teich R, Dicke T, Harb H, Yildirim AÖ, Tost J, et al. Epigenetic regulation in murine offspring as a novel mechanism for transmaternal asthma protection induced by microbes. J Allergy Clin Immunol. 2011;128:618–625.e1–7. doi: 10.1016/j.jaci.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 17.Jahreis S, Trump S, Bauer M, Bauer T, Thürmann L, Feltens R, et al. Maternal phthalate exposure promotes allergic airway inflammation over 2 generations through epigenetic modifications. J Allergy Clin Immunol. 2018;141:741–753. doi: 10.1016/j.jaci.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 18.Nicodemus-Johnson J, Naughton KA, Sudi J, Hogarth K, Naurekas ET, Nicolae DL, et al. Genome-wide methylation study identifies an IL-13–induced epigenetic signature in asthmatic airways. Am J Respir Crit Care Med. 2016;193:376–385. doi: 10.1164/rccm.201506-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy TM, Wong CC, Arseneault L, Burrage J, Macdonald R, Hannon E, et al. Methylomic markers of persistent childhood asthma: a longitudinal study of asthma-discordant monozygotic twins. Clin Epigenetics. 2015;7:130. doi: 10.1186/s13148-015-0163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang IV, Pedersen BS, Liu A, O’Connor GT, Teach SJ, Kattan M, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 2015;136:69–80. doi: 10.1016/j.jaci.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang WY, Levin L, Talaska G, Cheung YY, Herbstman J, Tang D, et al. Maternal exposure to polycyclic aromatic hydrocarbons and 5′-CpG methylation of interferon-γ in cord white blood cells. Environ Health Perspect. 2012;120:1195–1200. doi: 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breton CV, Siegmund KD, Joubert BR, Wang X, Qui W, Carey V, et al. Asthma BRIDGE consortium. Prenatal tobacco smoke exposure is associated with childhood DNA CpG methylation. PLoS One. 2014;9:e99716. doi: 10.1371/journal.pone.0099716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98:680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauer T, Trump S, Ishaque N, Thürmann L, Gu L, Bauer M, et al. Environment-induced epigenetic reprogramming in genomic regulatory elements in smoking mothers and their children. Mol Syst Biol. 2016;12:861. doi: 10.15252/msb.20156520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perera F, Tang WY, Herbstman J, Tang D, Levin L, Miller R, Ho SM. Relation of DNA methylation of 5′-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS One. 2009;4:e4488. doi: 10.1371/journal.pone.0004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whyatt RM, Perzanowski MS, Just AC, Rundle AG, Donohue KM, Calafat AM, et al. Asthma in inner-city children at 5–11 years of age and prenatal exposure to phthalates: the Columbia Center for Children’s Environmental Health Cohort. Environ Health Perspect. 2014;122:1141–1146. doi: 10.1289/ehp.1307670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang I-J, Karmaus WJ, Chen S-L, Holloway JW, Ewart S. Effects of phthalate exposure on asthma may be mediated through alterations in DNA methylation. Clin Epigenetics. 2015;7:27. doi: 10.1186/s13148-015-0060-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lockett GA, Soto-Ramírez N, Ray MA, Everson TM, Xu CJ, Patil VK, et al. Association of season of birth with DNA methylation and allergic disease. Allergy. 2016;71:1314–1324. doi: 10.1111/all.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritter M, Mennerich D, Weith A, Seither P. Characterization of Toll-like receptors in primary lung epithelial cells: strong impact of the TLR3 ligand poly(I:C) on the regulation of Toll-like receptors, adaptor proteins and inflammatory response. J Inflamm (Lond) 2005;2:16. doi: 10.1186/1476-9255-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang C, Jiang D, Francisco D, Berman R, Wu Q, Ledford JG, et al. Tollip SNP rs5743899 modulates human airway epithelial responses to rhinovirus infection. Clin Exp Allergy. 2016;46:1549–1563. doi: 10.1111/cea.12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diaz-Sanchez D, Tsien A, Fleming J, Saxon A. Combined diesel exhaust particulate and ragweed allergen challenge markedly enhances human in vivo nasal ragweed-specific IgE and skews cytokine production to a T helper cell 2–type pattern. J Immunol. 1997;158:2406–2413. [PubMed] [Google Scholar]
- 32.Wang IJ, Tung TH, Tang CS, Zhao ZH. Allergens, air pollutants, and childhood allergic diseases. Int J Hyg Environ Health. 2016;219:66–71. doi: 10.1016/j.ijheh.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Rider CF, Yamamoto M, Günther OP, Hirota JA, Singh A, Tebbutt SJ, et al. Controlled diesel exhaust and allergen coexposure modulates microRNA and gene expression in humans: effects on inflammatory lung markers. J Allergy Clin Immunol. 2016;138:1690–1700. doi: 10.1016/j.jaci.2016.02.038. [DOI] [PubMed] [Google Scholar]
- 34.Clifford RL, Jones MJ, MacIsaac JL, McEwen LM, Goodman SJ, Mostafavi S, et al. Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. J Allergy Clin Immunol. 2017;139:112–121. doi: 10.1016/j.jaci.2016.03.046. [DOI] [PubMed] [Google Scholar]
- 35.Fraser HB, Lam LL, Neumann SM, Kobor MS. Population-specificity of human DNA methylation. Genome Biol. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bayram H, Bauer AK, Abdalati W, Carlsten C, Pinkerton KE, Thurston GD, et al. Environment, global climate change, and cardiopulmonary health. Am J Respir Crit Care Med. 2017;195:718–724. doi: 10.1164/rccm.201604-0687PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlsten C, Rider CF. Traffic-related air pollution and allergic disease: an update in the context of global urbanization. Curr Opin Allergy Clin Immunol. 2017;17:85–89. doi: 10.1097/ACI.0000000000000351. [DOI] [PubMed] [Google Scholar]
- 38.Bintu L, Yong J, Antebi YE, McCue K, Kazuki Y, Uno N, et al. Dynamics of epigenetic regulation at the single-cell level. Science. 2016;351:720–724. doi: 10.1126/science.aab2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 40.Miller RL, Zhang H, Jezioro J, De Planell Saguer M, Lovinsky-Desir S, Liu X, Perzanowski M, et al. Reduced mouse allergen is associated with epigenetic changes in regulatory genes, but not mouse sensitization, in asthmatic children. Environ Res. 2017;156:619–624. doi: 10.1016/j.envres.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.