Abstract
Cotton contains a unique group of terpenoids including desoxyhemigossypol, hemigossypol, gossypol, hemigossypolone, and the heliocides that are part of the plant's defense system against pathogenic fungi and insects. Desoxyhemigossypol is a key intermediate in the biosynthesis of these compounds. We have isolated, purified, and characterized from cotton stele tissue infected with Verticillium dahliae a methyltransferase (S-adenosyl-l-Met: desoxyhemigossypol-6-O-methyltransferase) that specifically methylates the 6-position of desoxyhemigossypol to form desoxyhemigossypol-6-methyl ether with a Km value of 4.5 μm for desoxyhemigossypol and a Kcat/Km of 5.08 × 104 s−1 (mol/L)−1. The molecular mass of the native enzyme is 81.4 kD and is dissociated into two subunits of 41.2 kD on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. The enzymatic reaction does not require Mg+2 and is inhibited 98% with 10 mm p-chloromercuribenzoate. Desoxyhemigossypol-6-methyl ether leads to the biosynthesis of methylated hemigossypol, gossypol, hemigossypolone, and the heliocides, which lowers their effectiveness as phytoalexins and insecticides.
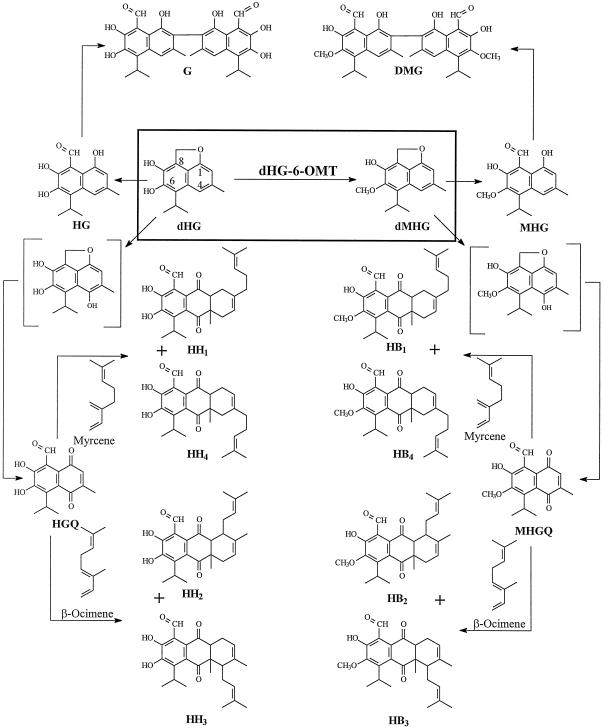
Cotton (Gossypium barbadense) is differentiated from other members of the Malvaceae family by the presence of pigment glands in the foliage and seed. The glands in the foliage contain a unique group of terpenes that include desoxyhemigossypol (dHG), hemigossypol (HG), gossypol (G), hemigossypolone (HGQ), and the heliocides H1, H2, H3, and H4 (Fig. 1). In the seed and roots gossypol is the predominant terpenoid. These compounds are important in protecting the plant from a wide range of pests. For example, gossypol, HGQ, and the heliocides, which are present in the plant's foliar glands, have been shown to be important in protecting the plant from insects such as Heliothis virescens (Hedin et al., 1992; Jenkins, 1995). So-called glandless cotton, which contains very few if any glands, is subject to attack by insects, rodents, and birds that normally are not cotton pests (Bottger et al., 1964; Lukefahr et al., 1966). Furthermore, dHG and HG are synthesized by the plant in response to invasion by pathogenic fungi such as Verticillium dahliae and Fusarium oxysporum f. sp. vasinfectum (F.o.v.) (Bell, 1967). These compounds are toxic to these pathogens (Mace et al., 1985; Zhang et al., 1993) and are therefore properly classified as phytoalexins.
Figure 1.
The proposed biosynthetic pathway for cotton terpenoids. G, Gossypol; DMG, gossypol-6,6′-dimethyl ether; HG, hemigossypol; dHG, desoxyhemigossypol; dMHG, desoxyhemigossypol-6-methyl ether; MHG, hemigossypol-6-methyl ether; HH1, 2, 3, 4, heliocides H1, 2, 3, 4; HB1, 2, 3, 4, heliocides B1, 2, 3, 4; HGQ, hemigossypolone; MHGQ, hemigossypolone-6-methyl ether.
The terpenoids indicated above are accompanied by a group of related compounds in which the hydroxyl group at C-6 is methylated. The methylated terpenoids are unique in that they are less toxic to insects such as H. virescens larvae (Stipanovic et al., 1977) and to V. dahliae and F. oxysporum f. sp. vasinfectum (Mace et al., 1985; Zhang et al., 1993). It has been previously shown that infection of cotton stele tissue with V. dahliae induced desoxyhemigossypol-6-O-methyltransferase (dHG-6-OMT) (Alchanati et al., 1994). In this paper we show that dHG-6-OMT is specific for methylating the 6-hydroxyl group of dHG and is not a general methyltransferase acting on hydroxyl groups of diphenol or dinaphthol substrates. Since dHG is an intermediate in the biosynthesis of all of the compounds shown in Figure 1, dHG-6-OMT may catalyze a key step leading to the biosynthesis of the methylated terpenoids and impacting the entire defense system of cotton toward insects and fungal pathogens.
MATERIALS AND METHODS
Chemicals
S-Adenosyl-l-[methyl-3H3]Met and S-adenosyl-l-[methyl-14C]Met were purchased from Amersham (Uppsala) at specific radioactivity of 18.5 GBq/mmol (500 mCi/mmol) and 1.96 GBq/mmol (53.0 mCi/mmol), respectively. S-Adenosyl-l-[methyl-2H3]Met tri(p-toluenesulfonate) (99 atom % D, 85% chemical purity) was the product of C/D/N Isotopes. S-Adenosyl-l-Met, GSH, insoluble polyvinylpolypyrrolidone (PVP), β-NADP (NADP+), Tris, Cyt c, carbonic anhydrase, ovalbumin, albumin, alcohol dehydrogenase, β-amylase, and blue dextran were purchased from Sigma (St. Louis). 4-Methylcatechol, 2,3-dihydroxynaphthalene, caffeic acid, p-chloromercuribenzoic acid, 3-hydroxy-4-methoxyphenethylamine, 3,4-dimethoxyphenethylamine, and coniferyl alcohol were purchased from Aldrich (Milwaukee, WI). Q-Sepharose Fast Flow, 2′,5′-ADP-Sepharose 4B (adenosine-2′,5′-bisphosphate-Sepharose 4B), and Superdex 200HR 10/30 column were purchased from Pharmacia Biotech (Piscataway, NJ). Ultragel AcA34 was purchased from IBF biotechnics. Centriplus-50 concentrator was purchased from Amicon (Beverly, MA). Ready gels (10%), the Silver-Stain Plus kit, and silver stain SDS-PAGE standard (low range) were purchased from Bio-Rad (Hercules, CA). Coomassie plus protein assay reagent was purchased from Pierce Chemical (Rockford, IL).
3-Hydroxy-4-methoxyphenethylamine-Sepharose (3H4-MPEA-Sepharose) affinity column was prepared by coupling 3-hydroxy-4-methoxyphenethylamine to the Hi-Trap NHS-activated (1 mL) affinity column according to the manufacturer's procedure (Pharmacia Biotech's Hi-Trap NHS-activated 1- and 5-mL affinity columns instruction manual). 3,4-Dimethoxyphenethylamine-Sepharose (3,4DMPEA-Sepharose) affinity column was prepared by coupling 3,4-dimethoxyphenethylamine to the Hi-Trap NHS-actived (1 mL) affinity column according to the manufacturer's procedure (Pharmacia Biotech's Hi-Trap NHS-activated 1- and 5-mL affinity columns instruction manual).
Plant Material
Cotton (Gossypium barbadense cv Seabrook Sea Island 12B2) seeds were pre-germinated in paper rolls at 30°C for 48 h and then transferred to 16-ounce plastic cups. The seedlings were grown in the greenhouse to the six- to eight-true-leaf stage. The plants were then transferred to 1-gallon pots and placed in environmental growth chambers programmed to a 14-h day temperature of 28°C and a 10-h night temperature of 22°C. The plants were equilibrated in the growth chambers for 1 week prior to inoculation with conidia of Verticillium dahliae to induce dHG, dMHG, HG, MHG, and dHG-6-OMT in the stele tissue of the cotton stems.
Inoculum Preparation
V. dahliae defoliating strain V-76 was isolated from cotton plants grown in Sonora, Mexico. The fungus was grown on potato dextrose agar plates at room temperature. The agar plates were flood inoculated with 108 conidia/mL and the fungus was allowed to grow for 3 or 4 d before conidia were washed from the plates with sterile water. Conidia were diluted to a concentration of 2 to 5 × 107 cells/mL and used as inoculum.
Inoculation of the Plant
A 20-μL droplet of inoculum was placed at each of three locations equally spaced around the stem of the cotton plants ¼ inch below the cotyledons. A puncture wound was made through the droplets with a 22-gauge needle so that the inoculum was taken up by the xylem vessels. The infection of the stem tissue with V. dahliae resulted in the induction of dHG, dMHG, HG, and MHG in the stele tissue of the first internode over a 10-d period and also led to the induction of dHG-6-OMT, which peaked at 2 d following the inoculation (Alchanati et al., 1994).
Enzyme Purification
Two days after inoculating the cotton plants with V. dahliae, the plants were removed from the environmental chambers and the first internode excised. The bark was removed and 12 g of stele tissue was ground to a powder in a mortar in liquid N2. The powder was further ground in 200 mL of 50 mm Tris-HCl buffer, pH 7.85, containing 5 mm GSH and 7.5% (w/v) insoluble PVP. The homogenate was filtered through a double layer of cheesecloth and centrifuged in a refrigerated centrifuge (model J2–21, Beckman Instruments, Fullerton, CA) at 12,000 rpm for 10 min. The supernatant fraction was removed and centrifuged in an ultracentrifuge (model L8–55M, Beckman) at 100,000g for 60 min.
The soluble supernatant fraction was removed and loaded onto a 1.5- × 18.0-cm Q-Sepharose FF column which had been equilibrated with 50 mm Tris-HCl buffer, pH 7.85, containing 5 mm GSH (buffer A). The column was eluted first with 24 mL of buffer A followed by elution with 240 mL of a 0 to 1 m linear gradient of NaCl in buffer A at a flow rate of 1.5 mL/min. Three-milliliter fractions were collected and assayed for dHG-6-OMT activity and protein content. Active fractions were pooled and concentrated to 1 mL using a concentrator (Centriplus-50, Amicon). This sample was loaded onto a 1.5- × 120-cm gel-filtration column (Ultrogel AcA34, BioSepra, Marlborough, MA) that had been pre-equilibrated with 40 mm Tris-HCl buffer, pH 7.5, containing 0.2 m NaCl (buffer B). The column was eluted with the same buffer at a flow rate of 0.3 mL/min.
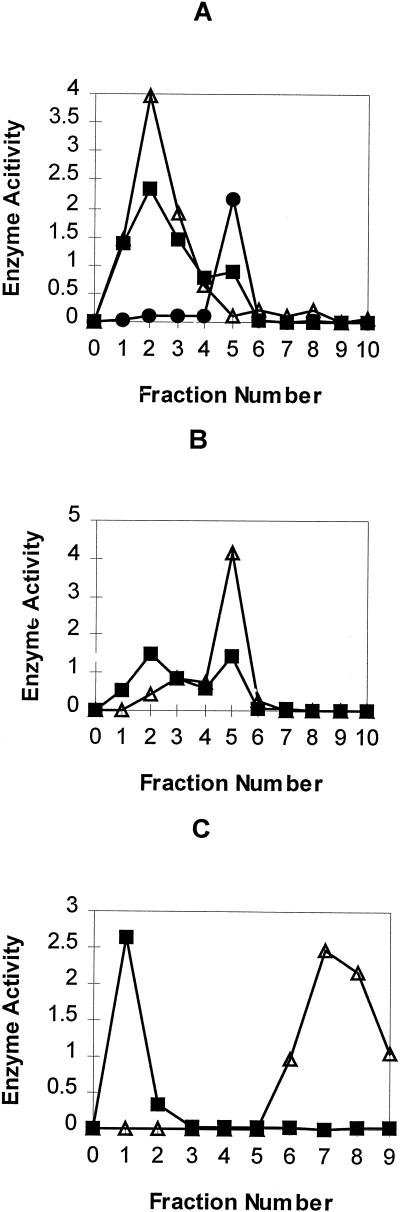
Fractions of 3.0 mL were collected and assayed for dHG-6-OMT activity and protein. Active fractions were pooled, desalted, and concentrated to 0.5 mL using a concentrator and loaded onto a 3H4MPEA-Sepharose affinity column that had been pre-equilibrated with buffer A (Fig. 2A). The column was first eluted with 8.0 mL of buffer A, then with 2.0 mL of 0.1, 0.2, 0.3, 0.4, 0.5, and 1.0 m NaCl in buffer A. Fractions of 2.0 mL were collected and assayed for dHG-6-OMT activity. Fraction 1 to 3 were pooled, concentrated to 0.5 mL using a concentrator and loaded onto a 3,4 DMPEA-Sepharose affinity column pre-equilibrated with buffer A. The column was eluted with 8.0 mL of buffer A, followed by 2.0 mL of 0.1, 0.2, 0.3, 0.4, and 1.0 m NaCl in buffer A (Fig. 2B). The dHG-6-OMT active fraction was loaded onto a 2′,5′-ADP-Sepharose 4B column. The column was eluted with 8.0 mL of buffer C, followed by 2.0 mL of 0.25 m NaCl in buffer C and 8.0 mL of 1 m NaCl in buffer C (Fig. 2C). The fraction containing the highest dHG-6-OMT activity was stored at −20°C after the addition of 0.45 mL of glycerol.
Figure 2.
Purification of dHG-6-OMT on affinity columns: A, 3-Hydroxy-4-methoxyphenethylamine-Sepharose; B, 3,4-dimethoxyphenethylamine-Sepharose; C, 2′,5′-ADP-Sepharose 4B. Enzyme activity: dHG-6-OMT (▪, pkat/mL); o-diphenol-OMT (●, pkat/mL); cinnamyl alcohol dehydrogenase (▵, pkat/mL × 10−2).
Preparation of dHG
dMHG was isolated from cotton stems infected with V. dahliae by the procedure of Stipanovic et al. (1975). dHG is highly labile and was prepared by demethylation of dMHG by the procedure of Stipanovic et al. (1992). The crystallized dHG had a Tm of 145°C to 147°C crystallization from ether-hexane solution.
The dHG-6-OMT Assay
The assay mixture contained 50 μL of enzyme preparation, 60 μL of 0.1 m Tris-HCl buffer, pH 7.5, containing 5 mm GSH, 40 μL of 188 μm dHG dissolved in ethanol, and 0.1 m Tris-HCl buffer, pH 7.5, containing 5 mm GSH in a 1:9 ratio, 10 μL of S-adenosyl-l-[methyl-3H3]Met ([methyl-3H3]SAM) (16.2 nmol, 1.25 μCi of radioactivity) for a total volume of 160 μL. The reaction mixture was incubated for 1 h at 30°C and the reaction was stopped by extracting the aqueous phase with 2.0 mL of hexane:EtOAc (3:1, v/v). A 200-μL aliquot of the hexane:EtOAc extract was assayed for radioactivity in a liquid scintillation spectrometer (Beckman).
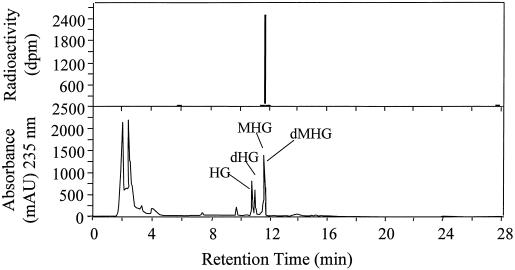
The hexane:EtOAc phase was concentrated under N2 and the radioactive enzymatic product(s) in these extracts was separated on a 250- × 4-mm MOS-Hypersil-1 C-8 column (5 μm) (Scientific Glass Engineering, Austin, TX) at a column temperature of 40°C and a flow rate of 1.25 mL/min using a HPLC equipped with a diode array detector (model 1090, Hewlett-Packard, Miami). A linear methanol:water gradient containing 0.07% (w/v) phosphoric acid was used for the column chromatography. The initial methanol: water ratio was 2:8, progressing to 7:3 over 7 min, to 8:2 over the next 5 min, to 9:1 over the next 7 min, and to 100% methanol over the last 4 min. The eluate was monitored at 235 nm. The column eluate was collected at 15-s intervals and an aliquot assayed for radioactivity. MHG and dMHG, and the radioactivity eluted at 11.66 min. The identity of the radiolabeled compound was ascertained using 2H-labeled SAM (see below).
The same procedure used to assay the dHG-6-OMT activity was used to assay for o-diphenol-OMT (o-DP-OMT) activity using 4-methylcatechol as substrate, and 2,3-dihydroxynaphthalene was substituted for dHG in the assay of o-dinaphthol-OMT (o-DN-OMT) activity.
Cinnamyl Alcohol Dehydrogenase (CAD) Assay
The enzymatic oxidation of coniferyl alcohol was measured by the increase in A400 due to the formation of coniferaldehyde according to the procedure of Wyrambick and Grisebach (1975). The assay mixture contained 50 μL of enzyme, 0.1 mm coniferyl alcohol, 0.2 mm NADP+, and 200 mm Tris-HCl buffer, pH 8.8, in a total volume of 1 mL.
Identification of the dHG-6-OMT Enzymatic Product
To establish the identity of the product from the dHG-6-OMT catalyzed methylation of dHG with SAM the reaction was carried out with [2H]SAM, the reaction product isolated and analyzed by GC-MS. The reaction mixture contained 1 mL of partially purified dHG-6-OMT isolated from the Ultrogel chromatography step containing in 0.1 m Tris-HCl buffer, pH 7.5, and 5 mm GSH, 0.4 mL of 188 μm dHG (75.2 nmol) dissolved in ethanol and 0.1 m Tris-HCl buffer, pH 7.5, in a ratio of 1:9 (v/v), and 0.2 mL of 446 μm (89.3 nmol) S-adenosyl-l-[methyl-2H3]Met (total volume 1.6 mL). The reaction was incubated for 2 h at 30°C and the reaction was stopped with the addition of 2.0 mL of hexane:EtOAc (3:1, v/v). The aqueous phase was vigorously extracted with this solvent. The organic phase was separated and the extraction repeated twice. The organic phases were combined and evaporated to dryness at reduced pressure at room temperature. The residue was dissolved in 100 μL of ethyl acetate and transferred to a 0.3-mL microfuge tube. The solvent was evaporated to dryness with N2 and the residue dissolved in 50 μL of methanol:water (9:1, v/v).
This 50-μL sample was separated on a 250- × 4.6-mm MOS-Hypersil-1 C-8 column (5 μm) in a single injection at a column temperature of 40°C and a flow rate of 1.25 mL/min using an HPLC equipped with a diode array detector. The linear gradient of methanol:water described in the previous section was used to chromatograph the 2H product. The fraction eluting between 11.5 and 11.8 min with a UV spectrum identical to dMHG was collected and evaporated to dryness under reduced pressure at room temperature. The residue was dissolved in 50 μL of ethyl acetate and transferred to a 0.3-mL microfuge tube. The extract was reduced to 15 μL with N2 and a 1 μL-aliquot analyzed by GC-MS.
The 2H product was analyzed using a mass spectrometer (model 5989B, Hewlett-Packard) coupled to a gas chromatograph (model 5890II, Hewlett-Packard) utilizing a 25-m column (BP-1, Scientific Glass Engineering) with an i.d. of 0.22 mm and film thickness of 0.25 μm. The 1-μL sample was injected through a splitless injector with the purge off for 1 min. The carrier gas was He with a flow rate of 1 mL/min. The temperatures were set as follows: source, 280°C; mass analyzer, 100°C; injector, 210°C; transfer line, 280°C. The following temperature program was followed: initial temperature 60°C, hold 8 min; to 180°C at 10°C/min, hold 1 min; to 280°C at 15°C/min, hold 5 min. The mass spectrum of the 2H reaction product showed ions at m/z (%): 262 (17), 261 (100), 259 (35), 247 (15), 246 (91), 243 (10), 242 (37), 228 (24), 225 (11), 212 (11), 153 (11), 152 (10), 142 (11), 141 (20), 130 (12), 129 (15), 128 (26), 127 (12), 115 (37), 114 (10), 113 (10), 99 (11), 98 (10), 91 (15), 77 (16), 76 (19), 63 (10). The mass spectrum of dMHG isolated from cotton stele tissue infected with V. dahliae showed ions at m/z (%): 259 (14), 258 (79), 256 (12), 244 (17), 243 (100), 241 (20), 229 (15), 228 (42), 227 (9), 213 (13), 211 (9), 153 (8), 152 (8), 141 (10), 129 (15), 128 (17), 115 (15), 114 (9).
Estimation of Protein
The protein concentration in the different extracts was determined by the method of Bradford (1977).
SDS-PAGE
SDS-PAGE of the purified protein was carried out according to the procedure of Laemmli (1970) in 10% (w/v) separation gel and 5% (w/v) stacking gel. Proteins were stained on the gels using the silver-staining kit.
Molecular Mass Determination of Native dHG-6-OMT
The molecular mass of the native dHG-6-OMT was determined by gel filtration chromatography. The fractions eluted from the gel-filtration column (fractions 38–42, 15.8 mL) were pooled and concentrated to 2.0 mL using a concentrator. A 200-μL aliquot was injected onto a Superdex 200HR 10/30 column installed on an HPLC system (Waters, Milford, MA). The column had been previously equilibrated with 40 mm Tris-HCl buffer, pH 7.5, containing 0.2 m NaCl and eluted with the same buffer at a flow rate of 0.5 mL/min. Fractions of 0.5 mL were collected and assayed for dHG-6-OMT activity. The dHG-6-OMT had a retention time of 28.05 min, which was used to determine the molecular mass of the enzyme.
RESULTS
Purification of dHG-6-OMT
The purification achieved at each step in the isolation procedure of dHG-6-OMT is presented in Table I. The enzyme was purified 851-fold with a final specific activity of 2,926 pkat mg−1 protein. In addition to the induced dHG-6-OMT activity found in the crude extract of cotton stele tissue infected with V. dahliae compared with extracts of stele tissue inoculated with water (Alchanati et al., 1994), the crude extract of the cotton stele tissue contained induced o-DP-OMT, o-DN-OMT, and CAD activities that co-eluted with the dHG-6-OMT activity through the first two steps of purification. For example, the crude soluble supernatant was first fractionated on a Q-Sepharose FF column and the dHG-6-OMT activity eluted in fractions 83 to 87 (3-mL fractions). These fractions were contaminated with o-DP-OMT, o-DN-OMT, and CAD. Fractions 83 to 87 were concentrated and subjected to purification by gel filtration.
Table I.
Purification of dHG-6-OMT from cotton stele tissue infected with V. dahlia
| Purification Stepa | Volume | Protein | Total Activity | Specific Activity | Purification | Recovery |
|---|---|---|---|---|---|---|
| mL | mg | pkat | pkat mg−1 protein | -fold | % | |
| Crude extract | 304 | 86.8 | 298.7 | 3.44 | 1 | 100 |
| Q-Sepharose, FF | 30 | 3.504 | 73.7 | 21.0 | 6.1 | 25 |
| Ultragel AcA34 | 31.5 | 0.797 | 46.4 | 58.2 | 16.9 | 16 |
| 3H4MPEA-Sepharose | 16 | 0.402 | 23.8 | 59.4 | 17.3 | 8 |
| 3,4DMPEA-Sepharose | 4 | 0.0095 | 5.75 | 608 | 177 | 1.9 |
| 2′,5′-ADP-Sepharose | 3 | 0.0014 | 3.95 | 2,926 | 851 | 1.3 |
3H4MPEA-Sepharose: 3-Hydroxy-4-methoxyphenethylamine-Sepharose; 3,4DMPEA-Sepharose: 3,4-dimethoxyphenethylamine-Sepharose; 2′,5′-ADP-Sepharose: adenosine 2′,5′-diphosphate-Sepharose 4B.
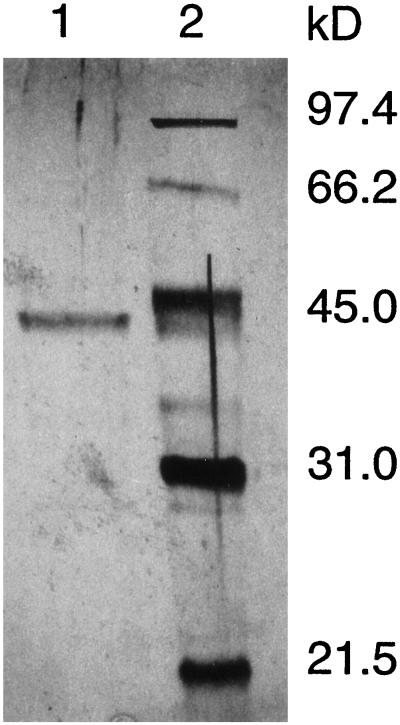
The dHG-6-OMT active fractions were found to be in tubes 38 to 42 (3-mL fractions collected). These fractions were contaminated with o-DP-OMT, o-DN-OMT, and CAD. These contaminants could only be removed by utilizing affinity chromatography. This required the use of three affinity columns (Fig. 2). As shown in Figure 2A, chromatography of the partially purified dHG-6-OMT on the 3H4MPEA-Sepharose affinity column separated the dHG-6-OMT and CAD activities from the o-DP-OMT and o-DN-OMT activities. The o-DP-OMT and o-DN-OMT active fraction showed a major band at 44.2 kD on an SDS-PAGE silver-stained gel. Chromatography of the fractions containing the dHG-6-OMT and CAD on a 3,4-DMPEA-Sepharose affinity column (Fig. 2B) separated several proteins from the dHG-6-OMT and CAD. SDS-PAGE of fraction 5 showed two major bands on the silver-stained gel at 42.1 and 43.7 kD. Chromatography of fraction 5 on 2′,5′-ADP-Sepharose 4B affinity column (Fig. 2C) separated dHG-6-OMT from CAD. The protein heterogeneity of fraction 1 containing the dHG-6-OMT activity as judged by the silver-staining pattern after SDS-PAGE is shown in Figure 3. The purified dHG-6-OMT fraction contained a single band at 42.1 kD together with trace bands between 66.2 and 97.4 kD and between 21.5 and 31.0 kD.
Figure 3.
SDS-PAGE of the dHG-6-OMT active fraction from 2′,5′-ADP-Sepharose 4B affinity column (lane 1). Low-range silver-stain SDS-PAGE standards (Bio-Rad) (lane 2) are also shown.
Molecular Mass Determination of the Native dHG-6-OMT
The retention times of partially purified dHG-6-OMT and authentic proteins were determined on a gel-filtration column. The molecular mass of the dHG-6-OMT was found to be 81.4 kD (data not shown). We judge that the single band of the purified dHG-6-OMT on SDS-PAGE gels (Fig. 3) is a subunit of the native enzyme with a molecular mass of 42.1 kD and the native enzyme is composed of two subunits.
The dHG-6-OMT Activity with Different Cosubstrates
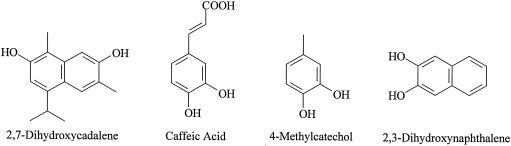
The methylation activity of purified dHG-6-OMT with different cosubstrates is shown in Table II using [methyl-3H]SAM as a cosubstrate with each hydroxylated compound. Optimum methylation activity was achieved with the presence of dHG in the complete reaction mixture. Omitting Mg2+ from the complete reaction mixture had little effect on the enzymatic activity. There was essentially no methylation activity in reaction mixtures substituting boiled enzyme for the native dHG-6-OMT. Considering the boiled enzyme as a blank, the enzyme did not significantly methylate (<2%) HG, 2,7-dihydroxycadalene, caffeic acid, 4-methylcatechol, or 2,3-dihydroxynaphthalene (Fig. 4) when they were used as substrates. An HPLC analysis (Fig. 5) of the reaction product(s) from the complete reaction mixture when dHG was used as the cosubstrate gave as the sole 3H-reaction product a peak that had the same retention time as that of dMHG.
Table II.
The activity of purified dHG-6-OMT with different cosubstrates
| Reactiona | Enzyme Activity |
|---|---|
| at/mg | |
| Complete | 2.67 |
| +Mg2+ (5 mm) | 2.49 |
| −dHG | 0.08 |
| −dHG, + hemigossypol | 0.06 |
| −Enzyme, + boiled enzyme | 0.06 |
| −dHG, + 2.7-dihydroxycadalene | 0.06 |
| −dHG, + caffeic acid | 0.09 |
| −dHG, + 4-methylcatechol | 0.09 |
| −dHG, + 2,3-dihydroxynaphthalene | 0.11 |
Complete reaction mixture contained 50 μL of purified enzyme preparation, 60 μL of 0.1 m Tris-HCI buffer, pH 7.5, containing 5 mm GSH, 40 μL of 188 μm dHG dissolved in ethanol, and 0.1 m Tris-HCI buffer, pH 7.5, in a 1:9 ratio, and 10 μL of [methyl- 3H]-SAM (16.2 nmol and 1.25 μCi of radioactivity) for a total volume of 160 μL. Numbers in parentheses refer to the final concentration. Substrates hemigossypol, 2,7-dihydroxycadalene, caffeic acid, 4-methylcatechol, and 2,3-dihydroxynaphthalene were added in the same amounts as for dHG.
Figure 4.
Structures of dHG-6-OMT substrate analogs used in this study.
Figure 5.
HPLC chromatograms of radiolabeled enzymatic product from incorporation of 14C-methyl group from SAM into dMHG in the complete reaction mixture (top) and the extract from V. dahliae inoculated cotton stele tissue (bottom).
Identification of the Product of the Reaction Catalyzed by dHG-6-OMT
The 2H reaction product from the complete reaction mixture containing buffer, dHG-6-OMT, [methyl-2H3]SAM, and dHG was isolated and chromatographed on HPLC columns by the procedures described in “Materials and Methods.” The 2H product was analyzed by GC-MS. The MS spectrum had prominent ions at m/z 261, 246, and 228. The ion at m/z 261 accounts for the parent ion [dMHG (d3)]. The loss of a methyl group from the isopropyl side chain produces an ion at m/z 246. The peak at m/z 228 results from the loss of CD3 from the methoxy group at position 6 from the ion at m/z 246. The fragmentation pattern for the naturally occurring dMHG from the cotton stele tissue gives major ions at m/z 258, 243, and 228. These analyses are consistent with the product of the reaction catalyzed by dHG-6-OMT as desoxyhemigossypol-6-methyl (d3) ether. In these reaction mixtures there was no MHG (d3) isolated.
The dHG-6-OMT Activity with Different Inhibitors and Metal Ions
The effect of different metal ions, chelating agent and thiol-blocking agents on the activity of the dHG-6-OMT was examined. The enzyme activity was not affected by the addition of EDTA, and as indicated from the data in Table II, there was no evidence that the enzyme required Mg2+. The enzyme was strongly inhibited (98%) by the addition of 10 mm p-CMB and was inhibited 63% by the addition of 10 mm iodoacetamide. The enzyme was inactivated by 5 mm concentrations of heavy metals such as cobalt (96%), copper (98%), manganese (82%), and zinc (98%). A similar effect was observed with the phenolic-OMT from Phanerochaete chrysosporium (Coulter et al., 1993).
Substrate-saturation kinetic data were obtained with the dHG-6-OMT preparations purified to near homogeneity, and were typical Michaelis-Menton type. A Km of 4.55 μm and a Kcat/Km of 5.08 × 104 s−1 (mol/L)−1 were determined for dHG and a Km of 81.4 μm and a Kcat/Km of 1.83 × 103 s−1 (mol/L)−1 were determined for SAM. The enzyme showed strong affinity toward dHG. The Kcat or turnover number with dHG is 0.231/s.
DISCUSSION
In this investigation it was demonstrated that the crude extracts of the cotton stele tissue infected with V. dahliae contained SAM-dependent O-methyltransferase activities of 3.44, 15.93, 20.71, and 27.56 pkat mg−1 protein with the diverse cosubstrates dHG, 4-methylcatechol, 2,3-dihydroxynaphthalene, and ethyl 3,4-dihydroxyhydrocinnamate, respectively. The rate of the enzymatic methylation of the cosubstrates was lowest for dHG. Since it has been reported that V. dahliae induces lignin biosynthesis in cotton hypocotyl tissue (Smit and Dubery, 1997), it was conceivable that methylation of dHG by the cotton stele extract was the result of utilizing o-diphenol or o-dinaphthol cosubstrates. However, the dHG-6-OMT purified in this present work showed no methylating activity (Table II), with a diverse array of o-diphenol and o-dinaphthol cosubstrates such as: HG, caffeic acid, 4-methylcatechol, 2,3-dihydroxynaphthalene, and ethyl 3,4-dihydroxyhydrocinnamate (Fig. 4).
The specific utilization of dHG as a cosubstrate by the purified dHG-6-OMT, demonstrates that the dHG-6-OMT induced by V. dahliae in cotton stele tissue requires a uniquely substituted o-dinaphthol substrate such as the dihydroxy substituted naphthofuran found in dHG. The dHG-6-OMT in cotton is different from the lignan synthesis OMTs found in tobacco. These latter OMTs utilize 4-methylcatachol and caffeic acid (Collendavelloo et al., 1981). It is also interesting that dHG-6-OMT does not methylate the cotton phytoalexin 2,7-dihydroxycadalene to produce 2-hydroxy-7-methoxycadalene (Fig. 4). These latter compounds are produced in cotton leaves in response to infection by Xanthomonas campestris (Essenberg et al., 1990). This indicates that cotton probably produces a unique 2,7-dihydroxycadalene-OMT.
The procedures outlined in this paper have resulted in purifying dHG-6-OMT from cotton stele tissue to near homogeneity. The results are consistent with the conclusion that the molecular mass of the native dHG-6-OMT is 81.4 kD and consists of two subunits with molecular masses of 42.1 kD. These molecular masses correspond closely to the native molecular masses of 66 and 78 to 80 kD and subunit molecular masses of 43 and 39 kD for hydroxymaackiain-OMT from pea (Preisig et al., 1989) and hydroxyindole-OMT from mammalian tissue (Lovenberg, 1982); respectively. However, many OMTs from plants have been reported to consist of single subunits with molecular masses of about 40 kD (Edwards and Dixon, 1991a, 1991b). The purified dHG-6-OMT does not require Mg2+ for the methylation reaction and the reaction was unaffected by the addition of EDTA.
These observations agree with the reports that many small molecular mass OMTs from plants do not require Mg2+ for catalytic activity (Kuhnl et al., 1989; Edward and Dixon, 1991a, 1991b; Sato et al., 1993). The activity of dHG-6-OMT was inhibited by iodoacetamide and p-CMB similarly to other OMTs from plants (Khouri et al., 1986; Edwards and Dixon, 1991a, 1991b; Sato et al., 1993), indicating the necessity for a -SH group for enzymatic activity. The Km values of dHG-6-OMT for dHG and SAM were 4.6 and 81.4 μm, respectively. These kinetic values compare well with those of flavonoid-ring-OMTs (DeLuca and Ibrahim, 1985; Khouri et al., 1986; Frenzel and Zenk, 1990) and to hydroxymaackiain-3-OMT (Preisig et al., 1989). Many small molecular mass OMTs from plants had higher Km values in the range of 20 to 100 μm for non-SAM cosubstrates (DeCarolis and Ibrahim, 1989; Pakusch et al., 1989; Edwards and Dixon, 1991a; Sato et al., 1993).
Alchanati et al. (1994) demonstrated that infection of cotton stele tissue with V. dahliae induced the formation of dHG, HG, dMHG, and MHG. The dHG-6-OMT activity was increased from 0 level of activity at time 0 of inoculation to a peak level of activity of 13.2 nmol h−1 g−1 stele tissue in 48 h. Thereafter, the dHG-6-OMT activity slowly declined over 8 d. These analyses, together with the demonstration in this paper that the purified dHG-6-OMT showed very strong cosubstrate specificity toward the o-dihydroxynaphthofuran ring of dHG, indicates that the probable function of this O-methyltransferase in cotton stele tissue is to methylate dHG. Since the purified enzyme does not utilize HG as a cosubstrate, the induction of MHG in the cotton stele tissue probably arises by the conversion of dMHG to MHG in a reaction analogous to the conversion of dHG to HG. We have now shown that dHG is the unique substrate that links the terpenoids with a free phenol group at C-6 to their methylated counterparts and purified and characterized the enzyme that controls this step. This provides the basis for future work on developing sense or antisense constructs to block synthesis of this enzyme. Because of the generally reduced effectiveness of the methylated terpenoids in protecting cotton from insects and pathogens, down-regulation of the gene that controls methylation of dHG may lead to a more resistant plant.
Footnotes
This work was supported in part by Texas A&M Agricultural Experiment Station, Cotton Incorporated, and the U.S. Department of Agriculture.
LITERATURE CITED
- Alchanati I, Benedict CR, Stipanovic RD. The enzymatic conversion of desoxyhemigossypol to desoxy methyl hemigossypol in cotton stems: dHG-O-methyltransferase. In: Jividen G, Benedict RC, editors. Proceedings of the Biochemistry of Cotton Workshop. Raleigh, NC: Cotton Incorporated; 1994. pp. 35–39. [Google Scholar]
- Bell AA. Formation of gossypol in infected or chemically irritated tissues of Gossypium spp. Phytopathology. 1967;57:759–764. [Google Scholar]
- Bell AA. Mechanisms of disease resistance in Gossypium species and variation in Verticillium dahliae. In: Constable GA, Forrester NW, editors. Challenging the Future, Proceedings of the World Cotton Research Conference-1. Melbourne, Australia: Commonwealth Scientific and Industrial Research Organization; 1995. pp. 225–235. [Google Scholar]
- Bottger GT, Sheehan ET, Lukefahr MJ. Relationship of gossypol content of cotton plants to insects resistance. J Econ Entomol. 1964;57:283–285. [Google Scholar]
- Bradford NM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1977;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Collendavelloo J, Legrand M, Geoffroy P, Barthelemy J, Fritig B. Purification and properties of the three o-diphenol-O-methyltransferases of tobacco leaves. Phytochemistry. 1981;20:611–616. [Google Scholar]
- Coulter C, Kennedy JT, McRoberts WC, Harper DB. Purification and properties of an S-adenosylmethionine: 2,4-disubstituted phenol O-methyltransferase from Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:706–711. doi: 10.1128/aem.59.3.706-711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCarolis E, Ibrahim RK. Purification and kinetics of phenylpropanoid O-methyltransferase activities from Brassica oleracea. Biochem Cell Biol. 1989;67:763–769. [Google Scholar]
- De Luca V, Ibrahim RK. Enzymatic synthesis of polymethylated flavonols in Chrysosplenium americanum. II. Substrate interaction and product inhibition studies of flavonol 3-, 6-, and 4′-O-methyltransferases. Arch Biochem Biophys. 1985;238:606–618. doi: 10.1016/0003-9861(85)90206-1. [DOI] [PubMed] [Google Scholar]
- Edwards R, Dixon R. Isoflavone O-methyltransferase activities in elicitor-treated cell suspension cultures of Medicago Sativa. Phytochemistry. 1991a;30:2597–2606. [Google Scholar]
- Edwards R, Dixon R. Purification and characterization of S-adenosyl-l-methionine: caffeic acid 3-O-methyltransferase from suspension cultures of alfalfa (Medicago sativa L.) Arch Biochem Biophys. 1991b;287:372–379. doi: 10.1016/0003-9861(91)90492-2. [DOI] [PubMed] [Google Scholar]
- Essenberg M, Grover PB, Cover EC. Accumulation of antibacterial sesquiterpenoids in bacterially inoculated Gossypium leaves and cotyledons. Phytochemistry. 1990;29:3107–3113. [Google Scholar]
- Frenzel T, Zenk MH. S-Adenosyl-l-methionine: 3′hydroxy-N-methyl-(S)-coclaurine-4′-O-methyltransferase, a regio- and stereoselective enzyme of the (S)-reticuline pathway. Phytochemistry. 1990;29:3505–3511. [Google Scholar]
- Hedin PA, Parrot WL, Jenkins JN. Relationship of glands, cotton square terpenoid aldehydes and other allelochemicals to larvae growth of Heliothis virescens (Lepidoptera: Noctui) J Econ Entomol. 1992;85:359–364. [Google Scholar]
- Jenkins JN. Host resistance to insects in cotton. In: Constable GA, Forrester NW, editors. Challenging the Future, Proceedings of the World Cotton Research Conference-1. Melbourne, Australia: Commonwealth Scientific and Industrial Research Organization; 1995. pp. 359–372. [Google Scholar]
- Khouri HE, Ishikura N, Ibrahim RK. Fast protein liquid chromatographic purification and some properties of a partially O-methylated flavonol glucoside 2′-/5′-O-methyltransferase. Phytochemistry. 1986;25:2475–2479. [Google Scholar]
- Kuhnl T, Koch U, Heller W, Wellmann E. Elicitor induced S-adenosyl-l-methionine: caffeoyl-CoA 3-O-methyltransferase from carrot cell suspension cultures. Plant Sci. 1989;60:21–25. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lovenberg W. Methylation of small molecules: an overview. In: Usdin E, Borchardt RT, Creveling CR, editors. Biochemistry of S-Adenosylmethionine and Related Compounds. Macmillan Press, Great Britain. 1982. pp. 427–436. [Google Scholar]
- Lukefahr MJ, Noble LW, Houghtaling JE. Growth and infestation of bollworms and other insects on glanded and glandless strains of cotton. J Econ Entomol. 1966;59:817–820. [Google Scholar]
- Mace ME, Stipanovic RD, Bell AA. Toxicity and role of terpenoid phytoalexins in Verticillium wilt resistance in cotton. Physiol Plant Pathol. 1985;26:209–218. [Google Scholar]
- Pakusch AE, Kneusel RE, Matern U. S-Adenosyl-l-methionine: trans-caffeoyl-CoA 3-O-methyltransferase from elicitor-treated parsley cell suspension cultures. Arch Biochem Biophys. 1989;271:488–494. doi: 10.1016/0003-9861(89)90299-3. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Geoffroy P, Fritig B, Legrand M. Molecular cloning and expression of a new class of ortho-diphenol-O-methyltransferases induced in tobacco (Nicotiana tabacum L.) leaves by infection or elicitor treatment. Plant Physiol. 1993;103:509–517. doi: 10.1104/pp.103.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisig CL, Mathews DE, VanEtten HD. Purification and characterization of S-adenosyl-l-methionine: 6a-hydroxymaack-iain 3-O-methyltransferase from Pisum sativum. Plant Physiol. 1989;91:559–566. doi: 10.1104/pp.91.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F, Takeshita N, Fitchem JH, Fujiwara H, Yamada Y. S-Adenosyl-l-methionine: scoulerine-9-O-methyltransferase from cultured Coptis japonica cells. Phytochemistry. 1993;32:659–664. [Google Scholar]
- Smit F, Dubery IA. Cell wall reinforcement in cotton hypocotyls in response to a Verticillium dahliae elicitor. Phytochemistry. 1997;44:811–814. [Google Scholar]
- Stipanovic RD, Bell AA, Lukefahr MJ. Natural insecticides from cotton (Gossypium) Am Chem Soc Symp Ser. 1977;62:197–214. [Google Scholar]
- Stipanovic RD, Bell AA, Mace ME, Howell CR. Antimicrobial terpenoids of Gossypium: 6-methoxygossypol and 6,6′-dimethoxygossypol. Phytochemistry. 1975;14:1077–1081. [Google Scholar]
- Stipanovic RD, Mace ME, Bell AA, Beier RC (1992) The role of free radicals in the decomposition of the phytoalexin desoxyhemigossypol. J Chem Soc, Perkin Trans I 3189–3192
- Wyrambick D, Grisebach H. Purification and properties of isoenzymes of cinnamyl-alcohol dehydrogenase from soybean-cell-suspension cultures. Eur J Biochem. 1975;59:9–15. doi: 10.1111/j.1432-1033.1975.tb02418.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Mace ME, Stipanovic RD, Bell AA. Production and fungitoxicity of the terpenoid phytoalexins in cotton inoculated with Fusarium oxysporum f. sp. vasinfectum. J Phytopathol. 1993;139:247–252. [Google Scholar]