Abstract
We have examined the relative abundance and distribution of the transcripts and protein products of a cell wall gene (Incw2) and a soluble invertase gene (Ivr1) to better understand their relative roles during maize (Zea mays L.) kernel development. In developing kernels the steady-state levels of Incw2 transcript increased dramatically from 0 to 12 d after pollination, while Ivr1 transcript, in contrast to a previous report, was undetectable. Consistent with the RNA expression data, the IVR1 protein could not be detected in kernel extracts using antisera raised to a synthetic peptide. Fractionation of the soluble form of invertase from developing kernels by isoelectric focusing and protein blots suggested that the enzyme activity was due to contamination of the cell wall invertase protein. A similar observation was made in a maize cell suspension culture in which Ivr1 RNA, but not IVR1 protein, was significantly modulated by sugars in the medium. Protein-blot analyses of the soluble enzyme activity suggested that changes in the enzyme activity are attributable to a cell wall invertase protein in the soluble fraction. Based on the collective evidence, we propose that the cell wall, but not the soluble invertase, is critical to heterotrophic sinks such as cell suspension cultures and developing kernels.
Acid invertase (β-d-fructofuranosidase, EC 3.2.1.26) catalyzes the irreversible cleavage reaction of Suc to Glc and Fru. There have been two major types of acid invertase cloned and characterized thus far in plants: a particulate, cell wall-localized form and a soluble, vacuole-localized form (Sturm and Chrispeels, 1990; Arai et al., 1992; Unger et al., 1994; Lorenz et al., 1995; Shanker et al., 1995; Xu et al., 1995; Haouazine-Takvorian et al., 1997). Using gene-specific probes, the tissue and developmental expression profiles have been demonstrated. For example, in carrot, the cell wall form of invertase (cwβF) has the most abundant steady-state mRNA levels in the first 8 weeks of growth in leaves and roots, suggesting a developmentally specific expression pattern. In contrast, transcripts of the soluble invertases (sI and sII) show both organ and temporal specificity, suggesting unique functions for each isozyme (Sturm et al., 1995). In Arabidopsis, a similar pattern of organ/tissue specificity was noted.
The two soluble invertases, Atβfruct3R and Atβfruct4, showed an organ-specific pattern of expression. Steady-state mRNA levels of Atβfruct4 were much higher in stems and flowers than in leaves and roots, while Atβfruct3R had higher transcript levels in stems, roots, and flowers and lower amounts in leaves (Haouazine-Takvorian et al., 1997). Much research has focused on assigning specific metabolic functions to invertase isozymes, taking into account their subcellular locations and tissue specificity. In one such example, a cell wall-bound form of acid invertase, INCW2, has been localized to the pedicel region of the developing kernel (Cheng et al., 1996). The maize (Zea mays L.) miniature 1 mutation and other miniature alleles, have dramatic seed phenotypes, especially the reference allele, mn1-1, with low to undetectable levels of the cell wall invertase (INCW2) and very small seeds compared with wild type. Thus, the tissue- and temporal-specific expression of an invertase is essential to normal kernel development.
The biochemical analysis of cell wall and soluble forms of invertase are usually described separately as part of intensive purification procedures (Unger et al., 1992; Michaud et al., 1993). In these instances, there is not much evidence for the presence of other contaminating invertases, with most chromatographic steps showing single elution peaks. There are, however, a few examples of tissues in which both the crude pellet (containing cell wall components) and supernatant fractions have been compared biochemically. Using wheat coleoptiles, Krishnan et al. (1985) found biochemical evidence for both forms of invertase based on pH activity profiles, Km values for Suc, the elution profile from DEAE-cellulose columns, and cytochemical localization. Three different forms were detected in elongating stem tissue from barley: a soluble, salt-extractable, cell wall-associated form and a form tightly associated with the cell wall (Karuppiah et al., 1989). The first two types were partially purified and characterized and were also found to differ significantly based on pH activity profiles, Km values for Suc, and chromatographic behavior. Further biochemical evidence for two isoforms of invertase comes from the carrot system, where the soluble and cell wall forms have different molecular masses, pI values, pH activity optima, and raffinose cleavage values (Unger et al., 1994).
Most recently, the physiological roles of both vacuolar and cell wall invertase have been investigated in carrot using antisense repression studies (Tang et al., 1999). Tap roots of plants expressing the antisense mRNA for cell wall invertase were much reduced in size and in the amount of carbohydrates, while the number of leaves was increased. In contrast, the expression of antisense mRNA for vacuolar invertase did not have such an effect on the morphology of the tap root, although it was somewhat smaller than the control. These plants also exhibited an increased number of leaves. Taken together, these data show an alteration of carbohydrate partitioning with the decrease in invertase activity and illustrate the fundamentally different physiological and developmental roles of the two forms of invertase.
In maize there are two well-characterized cell wall acid invertase genes, Incw1 and Incw2 (Taliercio et al., 1999) (GenBank nos. AF050129 and AF050128), and two other cell wall-type clones, Incw3 and Incw4 (GenBank nos. AF043346 and AF043347). Incw1 and Incw2 have shown organ specificity in their expression. RNA-blot analysis of Incw1 shows the highest levels of expression in cell-suspension cultures, etiolated shoots, roots, silks, and a low level in developing kernels. Incw2 mRNA is predominant in developing kernels but is also present in etiolated shoots (Taliercio et al., 1999). Incw2 appears to be especially important in the establishment of sink strength in kernels. Immunolocalization studies have shown cell wall invertase to be located in the pedicel region at the point where carbon must enter the developing kernel from the maternal tissue (Cheng et al., 1996). In addition, sequence analysis of Incw2 cDNA clones from the miniature seed mutant mn1-89 has shown substitution of a single amino acid relative to its parental allele Mn1, confirming our previous hypothesis that the Mn1 locus encodes the INCW2 protein (S.J. Carlson and P.S. Chourey, unpublished data).
The soluble invertase in developing kernels has been studied through enzymatic measurements of the soluble protein extracts and by the use of the reportedly non-cross-hybridizing, gene-specific probes Ivr1 and Ivr2 (Xu et al., 1996). However, prior to this report, there has been no antisera specific to any soluble form of invertase in maize. While the enzymatic measurements for soluble invertase in kernels showed the specific activity to be relatively low compared with the cell wall form (Cheng et al., 1996; Xu et al., 1996), RNA-blot analysis of the steady-state levels of Ivr1 and Ivr2 indicated that the transcript was abundant in developing kernels (Xu et al., 1996). For the first time to our knowledge, we report a comparison of the most abundant of the cell wall forms in kernels, Incw2, together with one of the soluble forms, Ivr1, in the same tissue samples, along with protein blots and biochemical characterization of IVR1 from pollen.
Some of our results appear to be inconsistent with previously published reports (Xu et al., 1996). We have found that the steady-state levels of Ivr1 are undetectable by RNA-blot analysis in developing maize kernel and cell-suspension cultures. In contrast, Ivr1 transcript is up-regulated in Suc-depleted maize cell suspensions, while the protein is undetectable with anti-IVR1 antisera. Furthermore, altered levels of invertase activity in the soluble fraction in Suc-depleted and Suc-supplemented cells correlated with corresponding changes in the levels of the cell wall invertase protein INCW1 but not with IVR1. We also present biochemical evidence to suggest that invertase activity that has previously been ascribed to the soluble form of invertase in developing kernels is indistinguishable from the cell wall form. We conclude from the collective data that the soluble fraction, i.e. the soluble invertase activity, in both kernels and Suc-depleted cell cultures is predominantly, if not entirely, due to the cell wall form of invertase.
MATERIALS AND METHODS
Plant Material
Immature maize (Zea mays L.) kernels from the Pioneer inbred line 3165 were harvested at 4, 8, 10, 12, or 21 d after pollination (DAP). An additional inbred line, W22, was also used when indicated in the text and figures. Plants were grown in the field or the greenhouse under normal diurnal conditions and either self- or sib-pollinated. At the time of harvest, kernels were excised from the ear, taking care to include the pedicel region, and frozen in liquid nitrogen.
Other plant material used included unfertilized ovules (0 DAP) from field-grown plants, mature seedlings of the 3165 inbred line grown in the greenhouse, elongating silk and mature pollen collected from assorted genotypes in the field and greenhouse, and a maize cell-suspension culture harvested 7 d after transfer to fresh medium. All tissue was stored at −80°C until use.
For cell-suspension culture Suc-depletion experiments, 50-mL cultures were pooled and rinsed three times with an equal volume of medium without Suc. Washed cells (5 mL) were inoculated into 50-mL cultures of normal medium without Suc. Remaining cells were harvested and used for the 0-h controls. Cell cultures were grown for 48 h in the absence of Suc and then harvested or inoculated with 5 mL of 20% (w/v) Suc or mannitol solution and grown for an additional 12 h prior to harvesting.
Cloning of Ivr1 cDNA
Total RNA from pollen was prepared as previously described (Wadsworth et al., 1988). Volumes of reagents were scaled down to amounts appropriate for smaller quantities of tissue (typically 0.5 g). First-strand cDNAs were synthesized from 10 μg of total RNA using an oligo(dT) primer (5′-GCGGATCCTTTTTTTTTTTTTTTTTT) and Superscript II (BRL, Gaithersburg, MD). The reaction mix was made up according to the manufacturer's instructions and incubated at 42°C for 1 to 1.5 h. Following a 70°C incubation for 5 min, the reaction was then treated with RNase H (BRL) and incubated at 37°C for 20 to 30 min. The following primer pairs were used: (1) and (3), 5′-TTCTCACCGCCGTCGTCTCCGCCG and 3′-CTGCTGCGGGAGTGGGTCAAGTCGG;(2) and (5), 5′-GCTCTACACGGGCTCCA and 3′-CGGATGAGCGGCAAGAG; (4) and (6), 5′-CAATCCCCAGGACGGTCCTCC and 3′-GCAACGACGACTTCTCTATGA.
For PCR, 5 μL of the first-strand reaction was used per 50-μL volume. Amplification conditions were initially determined using the OptiPrime kit (Stratagene, La Jolla, CA) with either an Ivr1 genomic clone (gift from J.-L. Prioul) or maize genomic DNA as a template, and primer pair 1,3 (representing part of exon 1, all of exon 2, and a portion of exon 3; approximately 600 bp). Subsequent rounds of amplification for primer pair 1,3 were performed using OptiPrime buffer 4 in the reaction mix, along with dNTPs and Taq polymerase (both BRL), at the concentrations recommended by the manufacturer for the OptiPrime kit. Amplification conditions were as follows: 94°C for 4 min, followed by 30 cycles of 94°C for 1 min, a 60°C annealing step for 2 min, and a 72°C extension for 2 min. The program was completed with a final extension at 72°C for 10 min.
For the other primer pairs, 2,5 (representing a portion of exons 3 and 4; approximately 780 bp) and 4,6 (representing most of exon 4, and all of exons 5, 6, and 7; approximately 690 bp), OptiPrime buffer 6 (2, 5 pair) or OptiPrime buffer 9 (4, 6 pair) with a 55°C annealing temperature were used along with first-strand cDNA from pollen. Products were analyzed on 1.2% (w/v) agarose Tris-acetate EDTA gels and confirmed by hybridization prior to ligation into the vector provided with the TA cloning kit (Invitrogen, Carlsbad, CA). The manufacturer's instructions were followed for ligation and subsequent transformation steps. All clones were confirmed by hybridization and sequencing. The clones were sequenced by automatic sequencing (Applied Biosystems, Foster City, CA) initiated at one or both ends of the vector, if necessary, to complete the sequence. Therefore, both strands of each clone were not necessarily sequenced. Hereafter, the clones will be referred to as follows: primer pair 1,3 product, Ivr1-1; primer pair 2,5 product, Ivr1-2; and primer pair 4,6 product, Ivr1-3.
RNA-Blot Analyses
For all tissues other then silk, the total RNA isolation method was used (Wadsworth et al., 1988). Silk total RNA was isolated using a method employing a guanidine- and sarcosine-containing extraction buffer (Yeh et al., 1991). This method has been reported to be more efficient for high-carbohydrate-containing tissues. Green leaf/shoot tissue was from 2-week-old greenhouse-grown plants. Roots were harvested from 1-week-old greenhouse-grown plants. Elongating silk and pollen were collected from mixed genotypes grown in the field. Kernel samples were harvested from field-grown material and included the entire kernel, pedicel, and crown. Poly(A+) mRNA was isolated according to the method of Ausubel et al. (1993) using an oligo(dT)-cellulose column.
RNA was glyoxalated and separated on a 1.2% (w/v) agarose gel (Ausubel et al., 1993). RNA was transferred to Nytran membrane (Schleicher & Schull, Keene, NH) and cross-linked prior to prehybridizaton. Prehybridization buffer consisted of 50 mm PIPES, pH 6.5, 100 mm NaCl, 50 mm sodium phosphate buffer, pH 6.5, 1 mm EDTA, and 5% (w/v) ultrapure SDS. Blots were prehybridized at 65°C for 1 to 3 h. Hybridization was performed in the same buffer with 3 × 106 counts/mL of 32P-labeled probe at 65°C overnight with constant shaking. Probes were labeled using a random-priming method (BRL Random Primers DNA labeling system). Following hybridization, blots were rinsed two times for 25 min each time with a 6× SSC solution (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate) supplemented with 5 mm sodium phosphate, pH 6.5, and 5% (w/v) SDS. Two high-stringency washes of 25 min each followed, each consisting of 0.2× SSC, 5 mm EDTA, pH 8.0, 5 mm sodium phosphate, pH 6.5, and 1% (w/v) SDS. All rinses were performed at 65°C. The blots were exposed to x-ray film for 1 week with two intensifying screens or 4 d in the phosphor-imaging cassette (Molecular Dynamics, Sunnyvale, CA). Transcript size was estimated to be 2.5 to 2.7 kb based on the position of the rRNA species visualized in the ethidium-stained gel prior to blotting and by comparison with the Incw2 transcript (approximately 2.2 kb).
Preparation of Nonparticulate Protein Extracts
Soluble protein extracts from silks, pollen, kernels, and cell suspension were prepared by grinding tissue with liquid nitrogen in a chilled mortar and pestle for approximately 15 min. After grinding, 10 volumes of chilled extraction buffer (200 mm HEPES, 5 mm MgCl2, 2 mm DTT, 1 mm PMSF, 1 mm EGTA, pH 7.5, and 10% [w/v] PVP, average Mr 40,000) was added directly to the mortar and the slurry was transferred to 15-mL tubes (Corex, Corning, NY). The extract was centrifuged for 10 min at 15,000g. The supernatant was taken to 30% saturation with solid ammonium sulfate and stirred or mixed, depending on the volume, for 30 min at 4°C. The 30% saturated solutions were then centrifuged at 15,000g for 20 min, and the resulting supernatant was desalted and the buffer exchanged to 10 mm Tris-HCl, pH 7.4, using chromatography columns (Micro Bio-Spin 6, Bio-Rad, Hercules, CA). If necessary, the equilibrated samples were then concentrated using Microcon-30 columns (Amicon, Beverly, MA). The samples prepared in this manner were used for SDS-PAGE and subsequent protein blots. For the antibody-bead titration experiments, dialysis was used for the desalting step. Protein concentrations were determined using the Bio- Rad protein assay, with bovine serum albumin (BSA) as a standard.
Invertase Activity Assays
For enzymatic activity measurements in the kernel developmental series and for the Rotofor cell (Bio-Rad) experiments, soluble and cell wall invertase fractions were prepared as previously described (Cheng et al., 1996). Acid invertase activity was measured as detailed in Tsai et al. (1970) and Miller and Chourey (1992) using Glc as a standard for the Nelson's test for reducing sugars. Fractions obtained from the Rotofor isoelectric focusing (IEF) experiments were assayed directly. Controls were performed to ensure that the ampholytes had no effect on the enzyme assay or the Nelson's test. The buffering capacity of the assay buffer (Tris acetate, pH 4.8) was not exceeded for the most basic samples, in which the measured pH of the assay reaction was less than 5.0.
Anti-IVR1 Antisera Production
A synthetic peptide to the C terminus of the predicted IVR1 sequence was synthesized at the 0.025 mmol scale, by solid phase using fluorenylmethoxycarbonyl chemistry at the Interdisciplinary Center for Biotechnology Research Protein Core Laboratory at the University of Florida (Gainesville) (peptide synthesizers from Applied Biosystems). The amino acid sequence selected was KAKSVKIWQLNSAYIR. The purity of the peptide was assessed by analytical HPLC, capillary electrophoresis, amino acid analysis, and matrix-assisted laser-desorption ionization time of flight mass spectrometry. The peptide was synthesized on a MAP (multiple antigenic peptide) Lys core with four branches (Tam, 1988). Polyclonal serum or ascites to the MAP peptide was raised in mice injected at the Interdisciplinary Center for Biotechnology Research Hybridoma Laboratory (University of Florida).
Anti-INCW1 and -INCW2 Antisera Production
INCW1 protein was expressed from a full-length clone in pET23B (Novagen, Madison, WI) using BL21 cells as a host. The overexpressed protein was excised and electroeluted from an SDS-PAGE gel. The putative INCW1, a non-glycosylated protein, was then used for injection of mice and subsequent polyclonal ascites production. The Incw1 cDNA clone has previously been described (Shanker et al., 1995). This antisera has been shown to cross-react with INCW2 (data not shown).
Antibody-Enzyme Titration
The specificity of the anti-MAP antisera was demonstrated using non-particulate protein extracts from pollen or 12-DAP kernels, as described above, and anti-mouse IgG magnetic beads (Dynal, Oslo). For the kernel sample, the 30% saturated ammonium sulfate supernatant was taken to 45% and finally 80% saturation to obtain a concentrated amount of soluble invertase activity. The M-280 beads were prepared according to the manufacturer's instructions. Three-hundred microliters of a well-mixed bead suspension was placed on the magnetic particle concentrator. The beads were washed three to four times with sterile phosphate-buffered saline (PBS), pH 7.4, and 0.1% (w/v) BSA. The prepared beads were then divided into two 150-μL aliquots. To each aliquot, an additional 50 μL of PBS was added, bringing the total volume to 200 μL. To one 200-μL aliquot, 50 μL of anti-MAP ascites was added and to the other 200-μL aliquot 50 μL of nonimmune sera was added. The two reactions were then incubated with gentle mixing for 4 h at 4°C.
During the incubation, protein samples were dialyzed against 20 mm sodium phosphate buffer, pH 6.5, using dialyzer cassettes (Slide-a-Lyzer, Pierce Chemical, Rockford, IL). After incubation, the bead/sera complexes were washed with PBS/BSA as described above. Each bead/sera mixture was resuspended in 200 μL of PBS/BSA, and then further subdivided into three 50-μL aliquots (for a total of six, three immune and three nonimmune sera bead complexes). Dialyzed protein extract (50 μL, corresponding to approximately 350 units of enzyme activity) was added to one of the bead/immune-sera reactions and its corresponding bead/nonimmune sera control reaction. This process was repeated for 25 and 10 μL (170 and 70 units of activity), with the reaction volume compensated for with addition of 0.02 m sodium-acetate buffer, pH 4.8. The six reactions were incubated overnight with gentle mixing at 4°C. The bead/sera/protein complex was collected along the side of the tube using the magnetic particle concentrator. The remaining supernatant was assayed for invertase activity in triplicate. Nonimmune sera treatments were compared with immune sera treatments to determine the percentage inhibition.
IEF in the Rotofor Cell
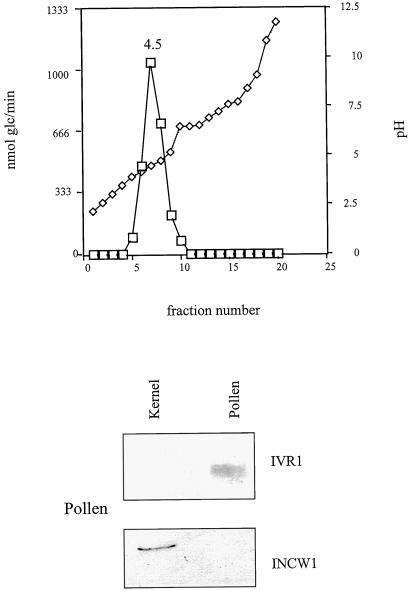
Soluble and cell wall fractions from 12-DAP kernels were prepared from 6 g of tissue as described in Cheng et al. (1996). A soluble fraction from 0.5 g of pollen was prepared as described above. The preparations were dialyzed overnight at 4°C against 1 L of 10 mm Tris-maleate buffer, pH 7.0, with one buffer change. For all of the fractions, the final sample volume was adjusted to 40 mL with 10 mm Tris-maleate buffer. Ampholytes, pH 3.0 to 10.0, were used at a 1% (v/v) concentration (Bio-Rad). Focusing and harvesting were performed according to the manufacturer's instructions. Fractions were assayed for invertase activity. Activity peaks were identified, pooled, brought to 1 m NaCl, and dialyzed against 50 mm Tris-HCl, pH 8.0 (to remove ampholytes), concentrated in Amicon units, and analyzed by SDS-PAGE followed by protein blotting. Ampholytes were not removed from the pollen samples, and as a result, the immunoreactive bands were more diffuse (Fig. 4). Control assays performed on a mock Rotofor run showed that the presence of ampholytes in the invertase reaction mixture had no background activity in the Nelson's test for reducing sugars.
Figure 4.
Rotofor fractionation of invertase activity in the supernatant fraction from pollen. The pollen protein extract was prepared as described in “Materials and Methods” and subjected to IEF in the Rotofor cell using pH 3.0 to 10.0 ampholytes. For the SDS-PAGE immunoblot, fraction 7 was concentrated and 1 μmol Glc/h was loaded. Protein (15 μg) from kernel extract was loaded as a control. Antisera dilutions and detection are described in “Materials and Methods.” The blot was first probed with anti-IVR1 antisera, stripped, and reprobed with anti-INCW1 antisera.
SDS-PAGE and Protein Blots
SDS-PAGE gels were run according to the Bio-Rad Mini-Protean II manual. In all cases, a 7.5% (w/v) acrylamide Tris-HCl gel was used, following the method of Laemmli (1970). Protein blotting to nitrocellulose membrane was performed in the Bio-Rad Mini Trans-Blot apparatus following the manufacturer's instructions. As an additional blotting control, prestained standards were loaded along with the samples (Bio-Rad, Kaleidoscope Markers). For IVR1 size estimation, biotinylated molecular mass markers were used according to the manufacturer's instructions (Bio-Rad).
Antibody detection was performed with enhanced chemiluminescent substrate (Pierce Super Signal Substrate) following the manufacturer's protocol. Anti-MAP antisera (IVR1 synthetic peptide) was used at a 1:200 dilution, anti-SS1 polyclonal antisera at 1:1,000, and anti-INCW1 at 1:1,000. Anti-rabbit HRP-labeled secondary antisera was used for the detection of SS1 and anti-mouse HRP-labeled secondary antisera was used in all other cases. All secondary antisera were used at a 1:12,500 dilution. Some protein blots were stripped and reprobed by washing the exposed blots four times for 5 min each wash in PBS-T (PBS-Tween). The washed membranes were incubated in 62.5 mm Tris-HCl, pH 6.8, 2% (w/v) SDS, and 100 mm 2-mercaptoethanol for 30 min at 50°C. The membranes were then washed six times for 5 min each wash in PBST, blocked, and reprobed.
RESULTS
Cloning of Maize Ivr1-Encoding cDNAs
Three overlapping Ivr1 cDNA clones, ranging in length from approximately 600 to 800 bp each, were obtained from reverse transcriptase (RT)-PCR products. The primers and their positions used to generate cDNAs for this cloning are described in “Materials and Methods.” These clones, Ivr1-1, Ivr1-2, and Ivr1-3, constitute almost all of the exonic sequences derived from the published Ivr1 genomic sequence (GenBank accession no. U16123), with approximately 100 bp at the 5′-end excluded. These clones have been deposited as a contiguous sequence in GenBank (accession no. AF171874). Pollen total RNA was used as the template for first-strand synthesis. Total RNA from other tissues, such as 4- to 5- and 12-DAP kernels and cell-suspension culture also gave products in the RT-PCR reactions (data not shown), but the pollen products were the most abundant and were used for subsequent TA cloning.
The Ivr1 partial genomic clone (1 kb) was also used to screen a maize cDNA library prepared from poly(A+) mRNA isolated from the basal third of 13-DAP maize kernels. This library previously had been used for the cloning of the cell wall invertase Incw2. Under moderate stringency, 55°C, and 6×/1× SSC washes, no positive plaques were detected among the 105 plaque-forming units screened. All three RT-PCR clones, Ivr1-1, Ivr1-2, and Ivr1-3, were sequenced entirely in at least one direction, and their identities as Ivr1 were confirmed using the BLAST homology search. An alignment of the RT-PCR clones with the published Ivr1 cDNA sequence shows these clones to be practically identical. The minor sequence variation that does occur may be due to artifacts of Taq polymerase, sequencing errors, or genotypic differences.
Expression of Ivr1 mRNA in Maize
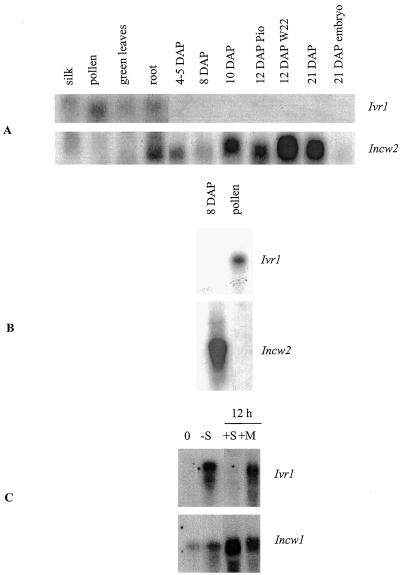
As seen in Figure 1, Ivr1 transcript appears in pollen, silks, green leaves from 2-week-old seedlings, and roots from 1-week-old green seedlings. The 2.5- to 2.7-kb transcript appears to be in greater abundance in pollen (Fig. 1, A and B) than the other tissues, although these results were not quantified. This size range is the same as is reported for Ivr2 (Pelleschi et al., 1999). Ivr1 transcript is not detectable in any of the developmental series of kernel samples surveyed using probes generated from Ivr1-1, Ivr1-2, or Ivr1-3. The same results were obtained using 32P-labeled probes and either traditional x-ray film exposure with intensifying screens or phosphor-imaging detection techniques for the RNA blots. Ivr1 transcript was also not detected in a poly(A+) mRNA sample from the 8-DAP stage (Fig. 1B), while the pollen sample on this same blot showed hybridization. Following hybridization with Ivr1-2, these same blots were re-hybridized with Incw2 (Fig. 1, A and B). Abundant Incw2 transcript can be seen, thus confirming the quality of the RNA samples.
Figure 1.
RNA gel blot with total (A) or poly(A+) RNA (B) from various tissues as shown. Amounts loaded are as follows: A, 10 μg of total RNA; B, 3 μg of poly(A+) RNA from kernel and 12 μg of total RNA from pollen; C, 20 μg of total RNA from suspension-cultured cells. The blots were initially hybridized with an Ivr1 (exons 3–D4 region) 32P-labeled probe and subsequently hybridized with a probe consisting of the entire coding region of Incw2 or Incw1. −S, 48 h Suc-depeletion; +S and +M, 12-h additions of Suc or mannitol, respectively.
Because invertase gene expression is modulated by sugars (Xu et al., 1996; Ehness et al., 1997), we utilized a maize cell suspension culture to determine if Ivr1 is also regulated by sugars in the medium, as is the case with the Incw1 gene in the same cell culture (Cheng et al., 1999). Indeed, cell cultures grown in Suc-free medium after culture in normal Suc-supplemented medium for 7 d, show a substantial increase in the steady-state levels of Ivr1 transcript (Fig. 1C). Cells treated with mannitol continued to show Ivr1 expression, suggesting that the response is specific to Suc and not the result of osmotic stress.
Expression of IVR1 Protein in Maize
To study IVR1 expression at the protein level, a synthetic MAP-peptide was generated and used to generate polyclonal antisera in mice. The carboxyl-terminal region of the protein was selected for the peptide as it is more likely to be exposed in the native protein. This region shows no sequence homology to the other known maize invertases, including IVR2, in multiple alignments of protein sequences. Therefore, antisera raised to this region of the protein must be specific to IVR1.
The specificity of this antisera was demonstrated using a magnetic bead-antibody-antigen complex. Using protein extracts from pollen, three different amounts of total invertase activity, ranging from 100 to 400 units were incubated with equal amounts of anti-mouse IgG Dynabeads coupled to either nonimmune ascites or anti-MAP sera. Using the nonimmune ascites as the control, the percent inhibition for each reaction was calculated. The pollen extract experiments consistently gave linear results in a dose-dependent fashion. The incubations with the highest amounts of initial invertase activity (380 nmol Glc/h) were the least inhibited (28%), and the incubations with the least amount of invertase activity (94 nmol Glc/h) were the most inhibited (92%) (Table I). Therefore, the linear relationship between anti-MAP antisera and invertase activity demonstrates the antigen-antibody specificity. Unlike the pollen extract, similar bead-antisera incubations with the kernel extract failed to show antigen-antibody specificity (Table I). Instead of a dose-dependent relationship between the antisera and enzyme activity, no inhibition was observed.
Table I.
Inhibition of invertase activity from maize extracts with antisera/bead complexes
| Total Activity
|
Inhibition | |||
|---|---|---|---|---|
| Initial | Remaining
|
|||
| Anti-MAP | Non-immune | |||
| nmol Glc/h | % | |||
| Pollen | 376 | 270 | 380 | 28 |
| 188 | 130 | 190 | 32 | |
| 94 | 5 | 58 | 92 | |
| Endosperm | 400 | 360 | 300 | 0 |
| 100 | 135 | 105 | 0 | |
The results of titration experiments using pollen or kernel extracts incubated with anti-IVR1 MAP antisera/bead or nonimmune sera/bead complexes. All enzymatic measurements were in triplicate, with sd values less than 10%.
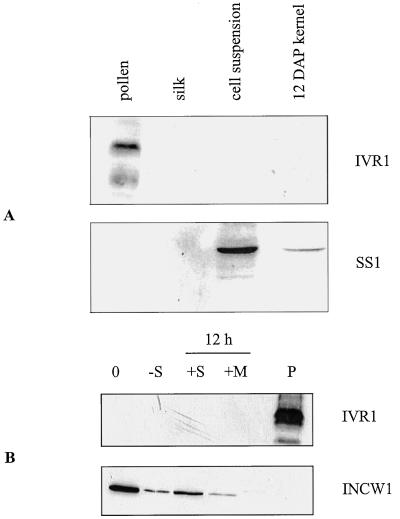
Figure 2A is a protein blot of silk, suspension culture, pollen, and kernel concentrated extracts. Of the samples analyzed (which also included leaf and root samples, not shown), only pollen had an immunoreactive band of 62 ± 4 kD (Fig. 2). This immunoreactive species has a lower limit of detectability of 6 μg of total pollen protein or 75 nmol Glc/h (total activity). Occasionally a lower-molecular-mass band was also detected, perhaps a breakdown product, as has been previously reported for soluble invertases (Unger et al., 1992). As a control, nonimmune ascites from mouse was used, with no immunoreactive bands appearing in any of the samples (data not shown). This nonimmune sera originated from the same SP20 myeloma cell line that was used for the MAP-peptide injections. As an additional control, protein blots were stripped of the anti-MAP antisera and re-probed with anti-SS1 polyclonal antisera (Fig. 2). This antisera detected SS1 in both the kernel and cell suspension samples; however, no signal was detected in the pollen or silk lanes. The absence of SS1 protein in pollen corroborates with the lack of SuSy enzyme activity in this tissue (Bryce and Nelson, 1979).
Figure 2.
Protein gel-blot analysis of selected maize tissues. A, Desalted, 30% saturated ammonium sulfate extracts were loaded in the following amounts: pollen, 20 μg; silk, 10 μg; suspension-cultured cells, 20 μg; and 12-DAP kernel, 15 μg. The blot was first probed with anti-MAP antisera (IVR1 synthetic peptide) and then stripped and reprobed with anti-SS1 antisera, as detailed in “Materials and Methods.” B, Soluble (crude supernatant) protein extracts from suspension-cultured cells. 0 h, 7-d-old, washed cells; −S, 48 h Suc-depeletion; +S, +M, 12-h additions of Suc or mannitol; P, pollen. All samples were loaded with 20 μg of total protein.
Protein-blot analysis of Suc-depleted cell suspension culture shows no new detectable IVR1 protein, despite an up-regulation of Ivr1 RNA. The results were the same for SDS-PAGE gels loaded with equal amounts of total activity (150 nmol Glc/h, data not shown) or with equal amounts of total protein loaded. A control blot with the same samples incubated with INCW1 antisera, showed a decrease in the amount of INCW1 protein in the absence of Suc and an increase with the addition of Suc but not mannitol in the supernatant fraction (Fig. 2B). Enzymatic activity measurements parallel these observations (Table II).
Table II.
Invertase activity from various sugar treatments of maize cell-suspension cultures
| Treatment | Fraction | nmol Glc μg−1/h−1 | Percentage of Initial |
|---|---|---|---|
| 0 h | Supernatant | 25 | 100 |
| Pellet | 64 | 100 | |
| 48 h − Suc | Supernatant | 6.7 | 27 |
| Pellet | 12 | 18 | |
| 12 h + Suc | Supernatant | 11 | 44 |
| Pellet | 23 | 36 | |
| 12 h + mannitol | Supernatant | 5.5 | 22 |
| Pellet | 10 | 16 |
The results of Suc starvation and subsequent addition of Suc or mannitol to suspension-cultured cells. Seven-day-old cultures (0 h) were washed with Suc-free medium and grown without Suc for 48 h prior to the addition of 2% Suc or mannitol for 12 h. All enzymatic measurements were in triplicate, with sd values less than 10%.
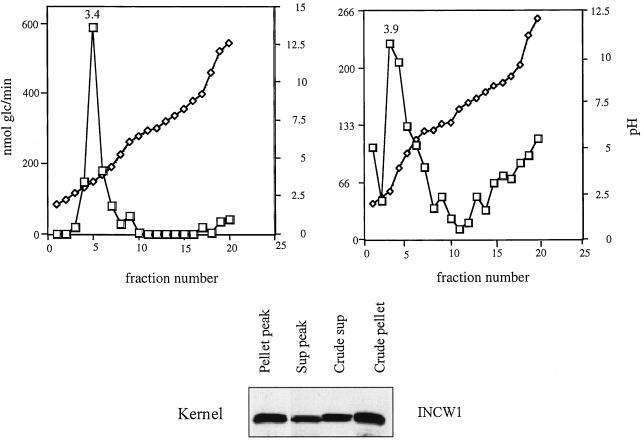
To further investigate the nature of the soluble invertase activity, IEF was used on kernel extracts to characterize the biochemical properties of the crude pellet and crude supernatant fractions. Additionally, a 30% saturated ammonium sulfate preparation prepared from pollen crude supernatant was also studied. We used the Bio-Rad Rotofor cell, which allows for IEF in solution. This non-denaturing method allows for further enzymatic assays of the recovered fractions. Using this method, the NaCl-extracted pellet from the crude kernel preparation was subjected to IEF. This preparation consistently gave activity peaks in the pH 3.0 to 4.0 region and a minor peak at pH 9.0 (Fig. 3). An immunoblot of the fractions pooled from the peak at pH 3.4 has a band detectable with INCW1 polyclonal antisera (Fig. 3). In addition, a single band was detected with INCW1 polyclonal antisera in concentrated samples of the crude supernatant and crude pellet (Fig. 3). For the supernatant, or soluble fraction from 12-DAP kernels, a single peak of invertase activity was detected, corresponding to pH 3.9 (Fig. 3). The pooled fractions from this peak had a single band on a protein blot detected with INCW1 polyclonal antisera (Fig. 3) but not anti-IVR1 antisera (not shown). It is important that the predicted pI value for INCW2 is 9.4 and that of IVR1 is 4.98. No immunoreactive species could be detected in the basic samples from either the pellet or the supernatant IEF preparations.
Figure 3.
Rotofor fractionation of invertase activity in supernatant and pellet fractions from 12-DAP kernels. Protein extracts were subjected to IEF in solution, using a Rotofor cell and pH 3.0 to 10.0 ampholytes. The activity peaks from soluble (Sup) and pellet fractions were pooled and concentrated as described in “Materials and Methods.” For the SDS-PAGE immunoblot, equal amounts of total activity were loaded at 12 μmol/h. Numbers above activity peaks correspond to the measured pH for that fraction. Antisera dilutions and detection are described in “Materials and Methods.”
The pollen supernatant (or soluble) preparation gave a single strong activity peak at pH 4.5 (Fig. 4). This is much closer to the predicted pI value of 4.98 for IVR1, and also suggests that the pollen supernatant consists of predominantly one form of invertase. The cell wall-bound fraction from pollen was not examined, as the extremely particulate nature of the salt-extracted pollen pellet made it unsuitable for the Rotofor cell. Previous RNA blots of pollen failed to show any Incw1 or Incw2 transcript, and protein blots of pollen salt-extracted pellet also showed no immunoreactive bands with INCW1 or INCW2 antisera (not shown), even though this fraction has measurable invertase activity (Fig. 5). It seems likely that some of the soluble form of invertase may adhere to the sticky pollen pellet fraction. Alternatively, a pollen cell wall invertase may be sufficiently different from Incw1 or Incw2 as to be undetectable with their respective probes.
Figure 5.
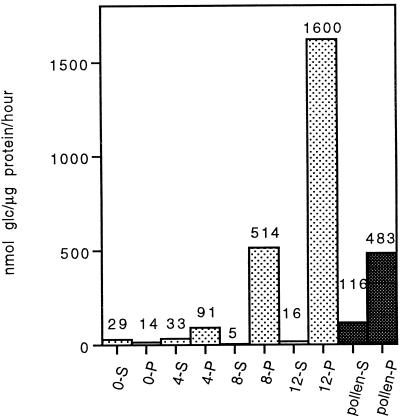
Histogram of invertase specific activities measured in a kernel developmental series from 0 DAP to 12 DAP. Whole kernels were used to prepare supernatant (S) and pellet (P) fractions from each stage. Specific activity values are the means of three separate measurements, with sd values less than 10%. Samples from mature pollen are included for comparison.
DISCUSSION
We have shown that the cell wall form of invertase is the predominant form of invertase in developing kernel and undifferentiated cell suspension culture from maize. Using gene-specific probes for a cell wall (Incw2) and a soluble form (Ivr1), we show that Ivr1 transcript is undetectable in developing kernels, while steady-state levels of Incw2 transcript increase in the 0- to 12-DAP developmental period. Furthermore, we present evidence that the IVR1 protein is undetectable in both the developing kernels and cell suspension, even though the transcript is up-regulated in the latter tissue by Suc depletion. Currently, we propose and present evidence to support the idea that much of the enzyme activity attributable to soluble invertase was in fact due to the presence of cell wall invertase in the crude supernatant fraction. Thus, the previous observations regarding soluble invertase activity were artifactual due to an imperfect method of subcellular fractionation and a lack of suitable antisera for detecting soluble invertase protein.
The lack of a detectable Ivr1 transcript in developing kernels is inconsistent with previously published data that describes Ivr1 RNA of an undetermined size at the 6- to 12-DAP stages (Xu et al., 1996). While the presence of small amounts of Ivr1 transcript in the kernel cannot be entirely ruled out based on RT-PCR amplifications, as mentioned in the “Materials and Methods,” the steady-state levels of Incw2 mRNA must be vastly greater, as Ivr1 transcript cannot be detected at all under the same conditions. The blots presented here were hybridized with the clone Ivr1-2, covering the exon 3 and 4 regions of Ivr1, the same region covered in the genomic probe of Xu et al. (1996). Care was taken to look at kernel developmental stages previously reported to have the greatest amount of transcript, in particular the 8-DAP stage, which is predominantly the maternal nucellar tissue. As an additional precaution, entire kernels were used for RNA isolation, rather than the crown or base portion, so as not to skew the results. We examined two inbred stocks, W22 and Pioneer 3165, both of which exhibit the same pattern shown in Figure 1.
In other plant systems, soluble invertase transcripts show a tissue and temporal specificity, and are not always expressed at the same time or place as cell wall invertase. For example, in Arabidopsis the gene for a vacuolar invertase, Atβfruct3, is predominant in very young tissues such as cotyledons and first leaves, while the cell wall form, Atβfruct1, is not detectable (Tymowska-Lalanne and Kreis, 1998). Similarly, in carrot, a soluble invertase gene, sII, is the only invertase detectable in developing roots from 10 to 20 weeks post-germination (Sturm et al., 1995).
The expression of IVR1 protein in the maize tissues examined appears to be limited to pollen. Evidence from both antibody-titration experiments and protein blots of SDS-PAGE gels suggests that the anti-MAP antiserum specifically recognizes an invertase in pollen protein extracts, but not extracts from kernels (Table I; Fig. 2A). Additionally, IVR1 is not detectable in protein extracts from Suc-depleted suspension-cultured cells (Fig. 2B). These results, in combination with the RNA data (Fig. 1C), strongly suggest that in Suc-depleted cell cultures, Ivr1 is transcribed but does not lead to a detectable IVR1 protein. A similar up-regulation of Ivr1 transcript was reported by Xu et al. (1996) in sugar-starved excised root tips. Although we cannot rule out the possibility that the polyclonal anti-IVR1 antisera does not cross-react with the enzyme responsible for soluble invertase activity from cell culture, it is striking that an INCW1 polypeptide is seen in the soluble fractions (Fig. 2B). A similar example has been seen in the same cell lines with Incw1 transcript. In this instance, two transcripts are detected: a larger transcript, Incw-L, and a smaller transcript, Incw-S. The larger transcript is not detectably translated (Cheng et al., 1999). There are numerous examples showing a lack of concordance between the levels of mRNA and corresponding protein (Vayda and Webster, 1998).
In addition to the enzymatic titration data, evidence from protein blots using the antisera raised to the synthetic peptide also suggest that the antisera detects an invertase. The 62-kD estimated size is well within the predicted range for this protein, based on deduced amino acid sequence (72 kD deduced from the cDNA and 65 kD deduced from a putative cleavage site at the N terminus, predicted by the SignalP V1.1 program) and other soluble type invertases. Examples of SDS-PAGE estimates of other purified soluble invertases include Lilium longiflorum flower buds with isoforms at 78, 54, 52, and 24 kD (Ranwala et al., 1998), carrot dry seeds and seedlings with a 68-kD polypeptide and proteolytic fragments at 43 and 25 kD (Unger et al., 1992), elongating stem tissue from barley at 60 kD (Karuppiah et al., 1989), Jerusalem artichoke shoot extracts at 58 kD (Goupil et al., 1988), and wheat coleoptiles at 53 kD (Krishnan et al., 1985). Somewhat smaller vacuolar forms of invertase have also been reported, including a 35-kD polypeptide from potato tuber (Isla et al., 1998) and a 42-kD protein from sweet pepper (Michaud et al., 1993).
The biochemical characteristics of the two fractions from kernels were further analyzed using IEF. The similar IEF fractionation pattern of the cell wall and soluble preparations and the presence of immunoreactive INCW proteins strongly suggest that the predominant invertase activity in the supernatant or soluble fraction is contamination from the cell wall forms. The activity peaks identified by the cell wall antisera are not in agreement with predicted or measured pI values of cell wall-bound acid invertases from maize (INCW2, 9.4) or two other plants (9.9 for suspension-cultured carrot cells; Unger et al., 1994), and 9.5 for Agrobacterium tumefaciens-transformed tobacco cells (Weil and Rausch, 1994); nevertheless, the enzyme activity peaks corresponded to bands of the correct size on a protein blot. Additionally, very few plant invertase pI values have been reported. The previously reported pI values were estimated using different methods, with narrow pH gradients. Granular IEF was used in the case of the tobacco cells and flatbed IEF in the carrot example.
The Rotofor cell is unique in that it separates proteins based on pI in solution at very low buffer concentrations (10 mm or less) in the absence of salt. It is possible that using this method and a broad range of ampholytes (pH 3.0–10.0) allows for different interactions and conformations of the proteins, and thus very different observed pI values. It is also possible that other post-translational modifications of these glycoproteins contribute to the measured acidic pI. For example, in yeast, invertase has multiple states of phosphorylation of the mannoprotein chains attached to the peptide, resulting in much polymorphism by IEF (Frevert and Ballou, 1982). While it is not known if this sort of modification occurs in this instance, plant invertases are well-known as glycoproteins and the role of post-translational modifications of the cell wall bound invertases of maize remains to be investigated. Charge heterogeneity has been observed in cell wall and cytoplasmic (soluble) isozymes of radish invertase (Faye et al., 1986). Even though these isozymes showed single bands on SDS-PAGE immunoblots, IEF experiments showed considerable heterogeneity, with some forms of cell wall invertase having pI values less than 7.
Traditionally, the invertase activity remaining in the crude supernatant after cell disruption has been considered to be due to the soluble type of invertase, a different form than the cell wall type, which has been shown to be in the crude pellet after high-salt extraction. The relative amounts of invertase specific activity measured in the crude supernatant from kernels is always very low, as shown here and as reported by others (Bryce and Nelson, 1979). Figure 5 shows a developmental profile of both soluble and cell wall invertase activity. Typically, the specific activity of the supernatant (or soluble) fraction is one-tenth that of the pellet (or cell wall-bound) fraction (Miller and Chourey, 1992; Cheng et al., 1996; Xu et al., 1996).
We previously reported a coordinate control in the enzyme activities of soluble and cell wall invertase in developing endosperm (Miller and Chourey, 1992; Cheng et al., 1996) and maize suspension-cultured cells (Cheng and Chourey, 1998) based solely on enzymatic data. Based on the data in this study, it is now possible to attribute such regulation to low levels of cell wall invertase protein in the soluble fraction. Table II gives further support to such an argument. Suc-depleted suspension-cultured cells showed a reduction in invertase activity measured in both the supernatant and pellet fractions and a corresponding increase in activity in both fractions with the addition of Suc. However, protein blots of the supernatant fractions show a good correlation with the presence of INCW1 but not IVR1 (Fig. 2B). Similar observations have previously been made in sugar beet callus and seedling protein preparations (Masuda et al., 1988). A comparison of activity profiles, Km values for Suc, and elution profiles on DEAE-cellulose chromatography of both soluble and cell wall protein preparations led the authors to conclude that extracellular invertases were probably identical to the invertases found in the soluble fraction.
IVR1 does appear to be of major importance to the mature pollen grain. In this tissue it is present under conditions where cell wall forms of invertase are undetectable. Using antisera raised against a synthetic peptide to the carboxy terminus of IVR1, the presence of the enzyme in pollen was confirmed using both protein blots and enzyme-antisera titration experiments. The molecular mass of the denatured protein was estimated from pollen to be 62 ± 4 kD, with a pI of 4.5. Protein blots of other tissues where Ivr1 transcript had been detected had no visible immunoreactive bands. This does not rule out the presence of IVR1 protein in these tissues, but may reflect significantly lower levels of the enzyme, or possibly further post-translational modifications that make the mature form of the enzyme undetectable with the anti-MAP antisera. Unger et al. (1994) note that the sequences of soluble invertases may include short C-terminal vacuolar-targeting extensions. From the data presented here it seems logical that the soluble and cell wall-bound forms of invertase in maize have spatial specificities of expression, with the soluble form, IVR1, being predominant in certain sink tissues such as floral organs and roots, and the cell wall-bound form, INCW2, being prevalent in rapidly dividing sink tissues such as endosperm.
ACKNOWLEDGMENTS
We thank Drs. Ed Echeverria, Earl Taliercio, and Thomas Rausch for critical reading of the manuscript. In addition, we thank Dr. Earl Taliercio for technical assistance with analysis of RNA from cell-suspension cultures and for the production of INCW1 antisera. We gratefully acknowledge the services of the DNA Sequencing, Hybridoma and Protein Chemistry Core laboratories of the Interdisciplinary Center for Biotechnology Research at the University of Florida. We also thank Mr. Mark Ross for assistance with figure preparation. This was a cooperative investigation of the U.S. Department of Agriculture-Agricultural Research Service and the Institute of Food and Agricultural Science, University of Florida.
NOTE ADDED IN PROOF
We recently cloned a portion of Ivr2, corresponding to the published sequence (GenBank accession no. U31451). Hybridization of RNA blots with this clone show abundant expression of Ivr2 in root, pollen, and silk with low levels of expression in 4- to 5-DAP and 8-DAP kernels. These young kernels consist primarily of maternal, nucellar tissue. At 12 DAP, no expression is seen in Mn1 kernels, but trace levels of Ivr2 RNA are detected in mn1-1 kernels that retain a much greater proportion of nucellar tissue than the wild type. Based on these data, it appears that within the kernel Ivr2 expression is limited to maternal tissues rather than developing endosperm.
Footnotes
This work was supported in part by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (grant no. 98–35301–6135). This paper is Florida Agricultural Experiment Station Journal Series no. R–06927.
LITERATURE CITED
- Arai M, Mori H, Imaseki H. Cloning and sequence of cDNAs for an intracellular acid invertase from etiolated hypocotyls of mung bean and expression of the gene during growth of seedlings. Plant Cell Physiol. 1992;33:245–252. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1993. [Google Scholar]
- Bryce WH, Nelson OE. Starch-synthesizing enzymes in the endosperm and pollen of maize. Plant Physiol. 1979;63:312–317. doi: 10.1104/pp.63.2.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W-H, Chourey PS. Two novel modes of regulation of invertases by sugars in suspension-cultured cells of maize (abstract no. 753) Plant Physiol. 1998;117:S-155. [Google Scholar]
- Cheng W-H, Taliercio EW, Chourey PS. The Miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell. 1996;8:971–983. doi: 10.1105/tpc.8.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W-H, Taliercio EW, Chourey PS. Sugars modulate an unusual mode of control of the cell-wall invertase gene (Incw1) through its 3′ untranslated region in a cell suspension culture of maize. Proc Natl Acad Sci USA. 1999;96:10512–10517. doi: 10.1073/pnas.96.18.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehness R, Ecker M, Godt DE, Roitsch T. Glucose and stress independently regulate source and sink metabolism and defense mechanisms via signal transduction pathways involving protein phosphorylation. Plant Cell. 1997;9:1825–1841. doi: 10.1105/tpc.9.10.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye L, Mouatassim B, Gorbel A. Cell wall and cytosolic isozymes of radish β-fructosidase have different N-linked oligosaccharides. Plant Physiol. 1986;80:27–33. doi: 10.1104/pp.80.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frevert J, Ballou CE. Yeast invertase polymorphism is correlated with variable states of oligosaccharide chain phosphorylation. Proc Natl Acad Sci USA. 1982;79:6147–6150. doi: 10.1073/pnas.79.20.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goupil P, Croisille Y, Croisille F, Ledoigt G. Jerusalem artichoke invertases: immunocharacterization of a soluble form and its putative precursor. Plant Sci. 1988;54:45–54. [Google Scholar]
- Haouazine-Takvorian N, Tymowska-Lalanne Z, Takvorian A, Tregear J, Lejeune B, Lecharny A, Kreis M. Characterization of two members of the Arabidopsis thaliana gene family, Atβfruct3 and Atβfruct4, coding for vacuolar invertases. Gene. 1997;197:239–251. doi: 10.1016/s0378-1119(97)00268-0. [DOI] [PubMed] [Google Scholar]
- Isla MI, Vattuone MA, Sampietro AR. Hydrolysis of sucrose within isolated vacuoles from Solanum tuberosum L. tubers. Planta. 1998;205:601–605. doi: 10.1007/s004250050362. [DOI] [PubMed] [Google Scholar]
- Karuppiah N, Vadlamudi B, Kaufman PB. Purification and characterization of soluble (cytosolic) and bound (cell wall) isoforms of invertases in barley (Hordeum vulgare) elongating stem tissue. Plant Physiol. 1989;91:993–998. doi: 10.1104/pp.91.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HB, Blanchette JT, Okita TW. Wheat invertases: characterization of cell wall-bound and soluble forms. Plant Physiol. 1985;78:241–245. doi: 10.1104/pp.78.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lorenz K, Lienhard S, Sturm A. Structural organization and differential expression of carrot β-fructofuranosidase genes: identification of a gene coding for a flower bud-specific isozyme. Plant Mol Biol. 1995;28:189–194. doi: 10.1007/BF00042049. [DOI] [PubMed] [Google Scholar]
- Masuda H, Takahashi T, Sugawara S. Acid and alkaline invertases in suspension cultures of sugar beet cells. Plant Physiol. 1988;86:312–317. doi: 10.1104/pp.86.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud D, Seye A, Driouich A, Yelle S, Faye L. Purification and partial characterization of an acid β-fructosidase from sweet-pepper (Capsicum annuum L.) fruit. Planta. 1993;191:308–315. [Google Scholar]
- Miller ME, Chourey PS. The maize invertase-deficient miniature-1 seed mutation is associated with aberrant pedicel and endosperm development. Plant Cell. 1992;4:297–305. doi: 10.1105/tpc.4.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelleschi S, Guy S, Kim J-Y, Pointe C, Mahé A, Barthes L, Leonardi A, Prioul J-L. Ivr2, a candidate gene for a QTL of vacuolar invertase activity in maize leaves: gene-specific expression under water stress. Plant Mol Biol. 1999;39:373–390. doi: 10.1023/a:1006116310463. [DOI] [PubMed] [Google Scholar]
- Ranwala AP, Baird WV, Miller WB. Organ-specific localization and molecular properties of three soluble invertases from Lilium longiflorum flower buds. Physiol Plant. 1998;103:551–559. [Google Scholar]
- Shanker S, Salazar RW, Taliercio EW, Chourey PS. Cloning and characterization of full-length cDNA encoding cell-wall invertase from maize. Plant Physiol. 1995;108:873–874. doi: 10.1104/pp.108.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Chrispeels M. cDNA cloning of carrot extracellular β-fructosidase and its expression in response to wounding and infection. Plant Cell. 1990;2:1107–1119. doi: 10.1105/tpc.2.11.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Sebkova V, Lorenz K, Hardegger M, Lienhard S, Unger C. Development- and organ-specific expression of the genes for sucrose synthase and three isoenzymes of acid β-fructofuranosidase in carrot. Planta. 1995;195:601–610. [Google Scholar]
- Taliercio EW, Kim J-Y, Mahé A, Shanker S, Choi J, Cheng W-H, Prioul J-L, Chourey PS. Isolation, characterization and expression analyses of two cell wall invertase genes in maize. J Plant Physiol. 1999;155:197–204. [Google Scholar]
- Tam JP. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc Natl Acad Sci USA. 1988;85:5409–5413. doi: 10.1073/pnas.85.15.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G-Q, Lüscher M, Sturm A. Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell. 1999;11:177–189. doi: 10.1105/tpc.11.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CY, Salamini F, Nelson OE. Enzymes of carbohydrate metabolism in the developing endosperm of maize. Plant Physiol. 1970;46:299–306. doi: 10.1104/pp.46.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymowska-Lalanne Z, Kreis M. Expression of the Arabidopsis thaliana invertase gene family. Planta. 1998;207:259–265. doi: 10.1007/s004250050481. [DOI] [PubMed] [Google Scholar]
- Unger C, Hardegger M, Lienhard S, Sturm A. cDNA cloning of carrot (Daucus carota) soluble acid β-fructofuranosidases and comparison with the cell wall isoenzyme. Plant Physiol. 1994;104:1351–1357. doi: 10.1104/pp.104.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger C, Hofsteenge J, Sturm A. Purification and characterization of a soluble β-fructofuanosidase from Daucus carota. Eur J Biochem. 1992;204:915–921. doi: 10.1111/j.1432-1033.1992.tb16712.x. [DOI] [PubMed] [Google Scholar]
- Vayda ME, Webster C. Translational regulation during periods of environmental stress. In: Bailey-Serres J, Gallie DR, editors. A Look Beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. Rockville, MD: American Society of Plant Physiologists; 1998. pp. 102–114. [Google Scholar]
- Wadsworth GJ, Redinbaugh MG, Scandalios JG. A procedure for the small-scale isolation of plant RNA suitable for RNA-blot analysis. Anal Biochem. 1988;172:279–283. doi: 10.1016/0003-2697(88)90443-5. [DOI] [PubMed] [Google Scholar]
- Weil M, Rausch T. Acid invertase in Nicotiana tabacum crown-gall cells: molecular properties of the cell-wall isoform. Planta. 1994;193:430–437. doi: 10.1007/BF00201824. [DOI] [PubMed] [Google Scholar]
- Xu J, Avigne WT, McCarty DR, Koch KE. A similar dichotomy of sugar modulation and developmental expression affects both paths of sucrose metabolism: evidence from a maize invertase gene family. Plant Cell. 1996;8:1209–1220. doi: 10.1105/tpc.8.7.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Pemberton GH, Almira EC, McCarty DR, Koch KE. The Ivr1 gene for invertase in Zea mays L. Plant Physiol. 1995;108:1293–1294. doi: 10.1104/pp.108.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-W, Juang R-H, Su J-C. Focus, BRL Product Literature. Vol. 13. Gaithersburg, MD: BRL; 1991. A rapid and efficient method for RNA isolation from plants with high carbohydrate content; pp. 102–103. [Google Scholar]