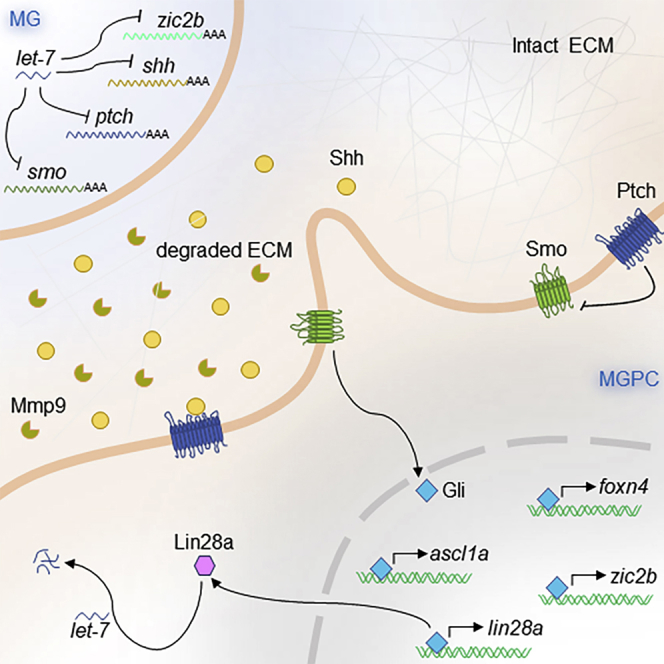
Summary
Upon injury, Müller glia cells of the zebrafish retina reprogram themselves to progenitor cells with stem cell characteristics. This necessity for retina regeneration is often compromised in mammals. We explored the significance of developmentally inevitable Sonic hedgehog signaling and found its necessity in MG reprogramming during retina regeneration. We report on stringent translational regulation of sonic hedgehog, smoothened, and patched1 by let-7 microRNA, which is regulated by Lin28a, in Müller glia (MG)-derived progenitor cells (MGPCs). We also show Shh-signaling-mediated induction of Ascl1 in mouse and zebrafish retina. Moreover, Shh-signaling-dependent regulation of matrix metalloproteinase9, in turn, regulates Shha levels and genes essential for retina regeneration, such as lin28a, zic2b, and foxn4. These observations were further confirmed through whole-retina RNA-sequencing (RNA-seq) analysis. This mechanistic gene expression network could lead to a better understanding of retina regeneration and, consequently, aid in designing strategies for therapeutic intervention in human retinal diseases.
Keywords: zebrafish, retina, regeneration, Shh, Ascl1a, Mmp9, Zic2b, Foxn4, let-7, Lin28
Graphical Abstract
Highlights
-
•
Shh signaling is essential for MG dedifferentiation during retina regeneration
-
•
Shh signaling components are regulated by let-7 microRNA in the zebrafish retina
-
•
A regulatory feedback loop between Mmp9 and Shh signaling is active in the retina
-
•
Shh signaling induced a gene-regulatory network involving mmp9, ascl1a, zic2b, and foxn4
Kaur et al. demonstrate that microRNA let-7 in injured zebrafish retina regulates the translation of shha, shhb, smo, ptch1, and zic2b mRNAs. Further, Shh signaling is necessary during retina regeneration for inducing a pro-regenerative gene expression cascade involving several genes, including ascl1a, lin28a, mmp9, foxn4, and zic2b.
Introduction
In contrast to mammals, zebrafish retina possesses remarkable regenerative capacity after an acute injury, leading to functional restoration of vision (Sherpa et al., 2008). The Müller glia (MG) cells in zebrafish retina reprogram themselves to MG-derived progenitor cells (MGPCs) that systematically differentiate into all retinal neurons, namely rods, cones, horizontal, amacrine, ganglion, bipolar cells, and MG itself (Ramachandran et al., 2010b). Although induction of MGPCs immensely contributes to the successful regeneration of zebrafish retina, the complete mechanism remains elusive. While the mechanism of retina regeneration is histologically well described, only a subset of the involved genes/proteins has been identified and characterized functionally (Goldman, 2014, Wan and Goldman, 2016). Therefore, we attempted to identify previously uncharacterized regulators of zebrafish retina regeneration using the needle-poke method of injury, which reflects the situation of mechanical damage that occurs in nature.
Even though several studies have elucidated the importance of Delta-Notch, Wnt, and Fgf signaling during retina regeneration in zebrafish, the roles of developmentally important Shh signaling remain largely underexplored (Goldman, 2014, Sun et al., 2014, Wan and Goldman, 2016). Recent studies have revealed the potential roles of Shh signaling during tissue regeneration (Ando et al., 2017, Dunaeva and Waltenberger, 2017, Thomas et al., 2018, Todd and Fischer, 2015). Therefore, we investigated the mechanistic involvement of Shh signaling during zebrafish retina regeneration. Subsequently, we hypothesized that MG dedifferentiation may depend on Shh signaling and have some similarities to the reprogramming of somatic cells by pluripotency-inducing factors (Hochedlinger and Plath, 2009, van den Hurk et al., 2016). Since we were interested in the possible involvement of Shh signaling during the early regenerative response of MG to injury, we analyzed the retina within the first few days after blockade of Shh signaling. We identified expression pattern of several important genes induced by Shh signaling and vice versa that reveal the robust regulatory network associated with retina regeneration. These include the interplay of Shh/Notch signaling components, transcription factors (namely, Ascl1a, Zic2b, Foxn4, and Insm1a), the matrix metalloproteinase Mmp9, the RNA-binding protein Lin28a, and microRNA let-7. Complete retina regeneration in zebrafish has provided valuable clues as to why their mammalian counterparts often fail (Goldman, 2014, Wan and Goldman, 2016). The findings from this study add clarity to the enigmatic process of retina regeneration lacking in mammals.
Results
Injury-Dependent Induction of Shh Signaling Is Essential for Regeneration
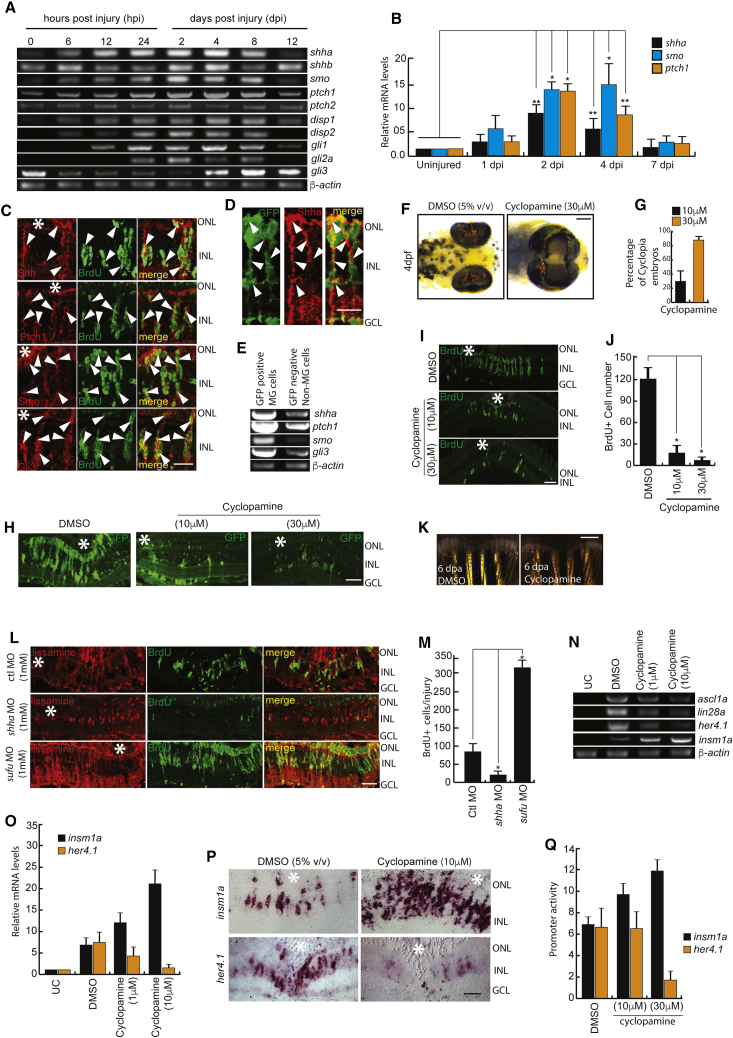
We explored the temporal expression pattern of Shh signaling component genes such as sonic hedgehog (shha, shhb), smoothened (smo), patched1 (ptch1), patched2 (ptch2), dispatched1 (disp1), dispatched2 (disp2), and glioma-associated oncogene (gli1, gli2a, and gli3) in total retina. We found that most of these genes were upregulated after retinal injury, except gli3, which showed a downregulation (Figures 1A and 1B). Moreover, the Shh signaling components Shh, Ptch1, Smo, and Gli3 showed co-localization with bromodeoxyuridine (BrdU)+ MGPCs (Figures 1C, S1A, S1B, and S7A). Western blot analysis revealed a temporal upregulation of Shh protein with a peak of expression at 4 days post-injury (dpi) (Figures S1C and S7A). The Shh protein is expressed in MG cells of wild-type (WT) injured retina marked by glutamine synthetase (GS) at 4 dpi (Figure S1D). Using tuba1a1016:GFP transgenic zebrafish (Fausett and Goldman, 2006), we showed the expression of Shh and its signaling components in proliferating MGPCs marked by GFP. Immunofluorescence (IF) studies and cell sorting revealed a relative abundance of Shh protein and its signaling components in GFP+ MGPCs compared with the rest of the cells of tuba1a1016: GFP transgenic retina at 4 dpi (Figures 1D and 1E). We confirmed the secretion of Shha and its probable autocrine action in MG using brefeldin A, a protein transport inhibitor, (Miller et al., 1992) and observed an expected increase in intracellular Shha and a decline in BrdU+ cells (Figures S1E and S1F).
Figure 1.
Shh Signaling Is Necessary for MG Dedifferentiation in the Injured Retina
(A and B) RT-PCR (A) and qPCR (B) analysis of Shh signaling component genes in the retina at indicated time points post-injury; n = 6 biological replicates. ∗p < 0.001; ∗∗p < 0.003.
(C and D) Immunofluorescence (IF) microscopy images of Shh signaling components in wild-type BrdU+ MGPCs (C), and Shh expression in 1016 tuba1a:GFP transgenic fish at 4 dpi (D). Arrowheads mark protein expression in cells in (C) and (D).
(E) RT-PCR assay of Shh signaling component genes in GFP-positive MGPCs and the rest of the cells from 1016 tuba1a:GFP transgenic retina at 4 dpi.
(F and G) Bright-field (BF) images of 4-days post-fertilized embryos treated with 5% (v/v) DMSO and 30 μM cyclopamine (F), and quantification of the number of cyclopia embryos (G).
(H–J) IF microscopy images showing a dose-dependent decline in GFP+ and BrdU+ MGPCs in 1016 tuba1a:GFP transgenic (H) and wild-type (I) retinae, respectively, at 4 dpi upon cyclopamine treatment, which is quantified in (J).
(K) BF microscopy images of blastema during caudal fin regeneration in cyclopamine-treated wild-type zebrafish at 6 days post-amputation.
(L and M) IF microscopy images of retinal sections with shha or sufu knockdowns (L), and quantification of the number of BrdU+ cells at the injury site (M). ∗p < 0.0001; n = 4 biological replicates. Lissamine tag on MO shows red fluorescence in (L).
(N–P) RT-PCR analysis of ascl1a, lin28a, her4.1, and insm1a in uninjured control, 2.5 dpi DMSO-treated, and 2.5 dpi cyclopamine-treated retina (N); qPCR analysis of mRNA levels of insm1a and her4.1 with cyclopamine treatment (O); and BF images of corresponding mRNA in situ hybridization (ISH) of these genes in the retina at 4 dpi (P).
(Q) Single-cell-stage embryos were injected with insm1a:luciferase or her4.1:luciferase vectors along with Renilla luciferase mRNA for normalization and then treated with cyclopamine for 24 hr before lysing for quantification of insm1a and her4.1 promoter activity using a dual luciferase assay.
Scale bars represent 10 μm in (C), (D), (H), (I), (L), and (P) and 500 μm in (F) and (K). Asterisk indicates the injury site (C, H, I, L, and P). Error bars represent SD. ∗p < 0.0001 (J); ∗p < 0.001 (M). n = 6 biological replicates. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; UC, uninjured control. See also Figures S1, S2, S6, and S7.
To decipher the influence of Shh signaling on retina regeneration, we used the pharmacological agent cyclopamine (Incardona et al., 1998), a potent inhibitor of Smo (Chen et al., 2002). We found that at 30 μM concentration, 90% of zebrafish embryos exhibited cyclopia, a hallmark of impaired Shh signaling, which also impacted developmentally important genes (Figures 1F, 1G, and S1G). We then explored the impact of continuous cyclopamine exposure on MGPC induction and regeneration in WT and tuba1a1016:GFP transgenic retina at 4 dpi. Interestingly, 10 μM and 30 μM concentrations significantly inhibited MGPC induction (Figures 1H–1J, S1H, and S1I), which was not the result of enhanced apoptosis (Figure S1J). A similar reduction in fin blastema was also seen with cyclopamine treatment on the 6th day post-amputation (Figure 1K), suggesting a conserved Shh signaling mechanism across tissues during regeneration. The few residual BrdU+ MGPCs in cyclopamine-treated retina failed to form any retinal cell types (Figure S1K). Moreover, morpholino (MO)-based targeted gene knockdown of Shh signaling component genes such as shha, shhb, ptch1, ptch2, and gli2a caused progenitor reduction, and that of negative regulators sufu (suppressor of fused) (Figures 1L and 1M and S2A–S2C) and gli3 (Figures 5I, S6A, and S6B; Table S1) enhanced MGPC induction as compared with control retina at 4 dpi. These increased MGPCs when traced until 20 dpi revealed the formation of amacrine, bipolar, and MG cells, indicating their functional potential to give rise to different retinal cell types (Figures S2D and S2E). These results emphasize the importance of Shh signaling during retina regeneration.
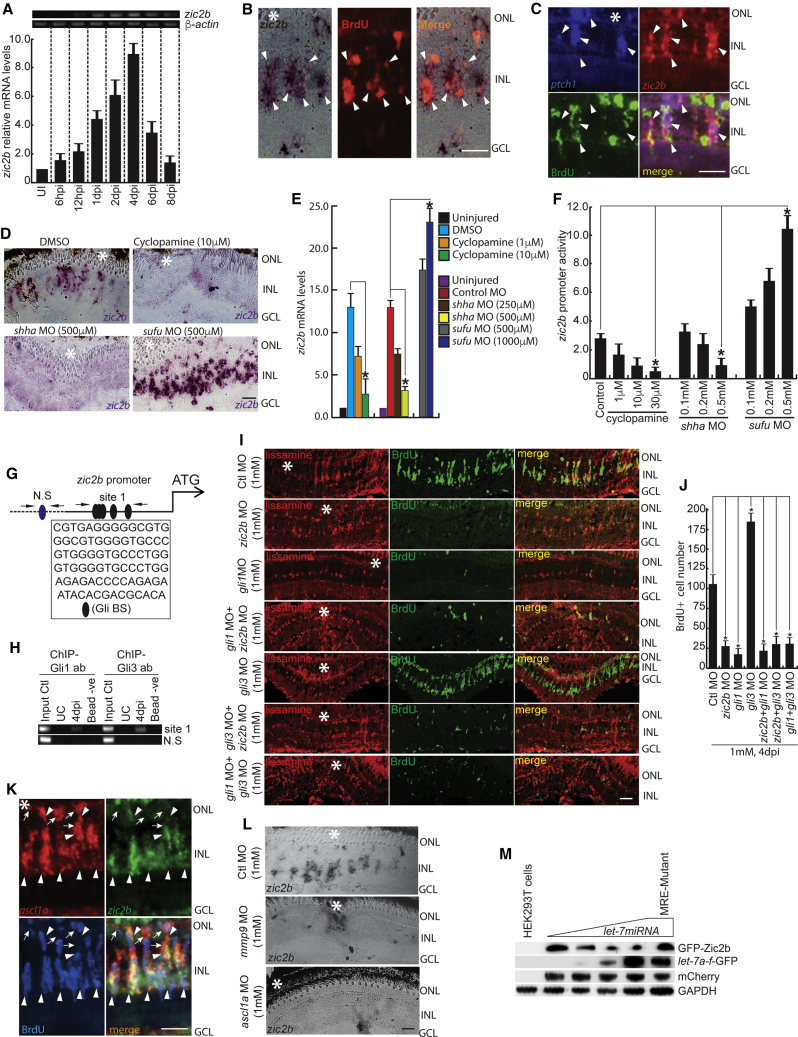
Figure 5.
The Shh-Mediated Zic2b Axis Is Necessary during Retina Regeneration
(A) RT-PCR (top) and qPCR (bottom) analysis of injury-dependent zic2b expression in the retina; n = 6 biological replicates.
(B) ISH and IF microscopy revealed co-localization of zic2b mRNA with BrdU+ MGPCs in 4 dpi retina.
(C) FISH and IF microscopy images of a 0.5-μm-thick optical section of retina showing co-localization of zic2b with ptch1 in BrdU+ MGPCs at 4 dpi.
(D and E) BF microscopy images of zic2b mRNA ISH in 4 dpi retina, with cyclopamine treatment, MO mediated shha or sufu knockdown done separately (D), which is quantified in (E).
(F) Luciferase assay in 24 hpf embryos injected with zic2b:GFP-luciferase vector with cyclopamine treatment and shha or sufu knockdowns.
(G) Schematic of the zic2b promoter with a putative Gli-BS. Arrows mark ChIP primers, N.S marks negative control devoid of Gli-BSs, and capital letters mark consensus of Gli-BSs.
(H) Retinal ChIP assay at 4 dpi showing both Gli1 and Gli3 bound to the zic2b promoter.
(I) IF microscopy images of BrdU+ cells in the regenerating retina with zic2b, gli1, and gli3 knockdowns in isolation or combination, delivered at the time of injury, compared with control MO.
(J) BrdU+ cells are quantified in the indicated knockdowns.
(K) FISH and IF microscopy images of a 0.5-μm-thick optical section of retina showing co-localization of zic2b with ascl1a in BrdU+ MGPCs at 4 dpi. Arrowheads indicate ascl1a and zic2b co-expression, whereas arrows indicate ascl1a+ but zic2b− cells.
(L) ISH microscopy retinal images of zic2b mRNA with mmp9 or ascl1a knockdown at 4 dpi.
(M) let-7 microRNA downregulated translation of the GFP construct appended with zic2b harboring microRNA responsive regions in a dose-dependent manner in HEK293T cells.
Scale bars represent 10 μm (B, C, and K) and 20 μm (D, I, and L). Asterisk indicates the injury site (B, C, D, I, K, and L). Error bars represent SD. ∗p < 0.001 (E, F, and J). n = 6 biological replicates (E and J); n = 3 (F). See also Figures S4–S7.
We also performed whole-retina RNA sequencing (RNA-seq) at 12 hr post-injury (hpi), 4 dpi, and 4 dpi with cyclopamine treatment compared with uninjured controls to get a holistic view of the blockade of Shh signaling. We found that several transcription factor genes, including ascl1a, zic2b, foxn4, and matrix metalloproteinase mmp9, are regulated with cyclopamine treatment (Table S3; Figures S1L and S1M; GEO: GSE102063).
Shh Signaling Affects Expression of Repressor Genes
We then explored the impact of compromised Shh signaling in the expression pattern of well-known regeneration-associated repressor genes such as her4.1 and insm1a (Goldman, 2014). RT-PCR and qPCR analysis in cyclopamine-treated retina revealed that the pivotal regeneration-associated genes are downregulated, with the exception of insm1a and a few Notch signaling genes (Figures 1N, 1O, and S2F). Insm1a, a known transcriptional repressor in MGPC induction and cell-cycle exit (Ramachandran et al., 2012, Zhang et al., 2009), showed upregulation, whereas levels of her4.1, one of the effectors of Notch signaling (Pasini et al., 2004, Wilson et al., 2016), showed downregulation, which was confirmed by mRNA in situ hybridization (ISH) and luciferase assays (Figures 1P and 1Q). Upregulation of insm1a and downregulation of her4.1 with blocked Shh signaling in post-injured retina led us to hypothesize the involvement of a well-known transcription factor such as Ascl1a in this regulatory loop. Insm1a, a known transcriptional repressor of ascl1a (Ramachandran et al., 2012), could influence its expression in a Shh-signaling-dependent manner. Moreover, Ascl1a could impact the expression of delta genes (Henke et al., 2009, Nelson et al., 2009), the ligand of Notch signaling, capable of inducing her4.1 expression in Notch-expressing cells (Takke et al., 1999). Thus, the Shh-signaling-dependent increase in Insm1a could cause a downregulation of ascl1a, which in turn reduces her4.1 levels in injured retina. These results suggest possible crosstalk between Shh and Notch signaling, contributing to retina regeneration.
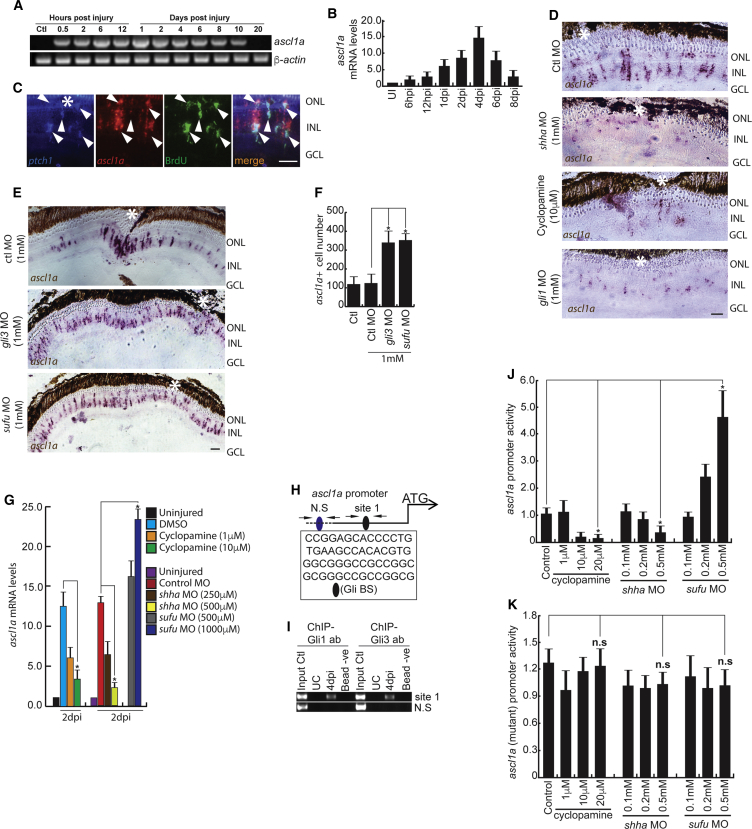
Shh Signaling Induces ascl1a during Retina Regeneration
Apart from the potential involvement of Insm1a in repressing ascl1a levels, we also speculated its direct regulation mediated through Shh signaling. This is presumably true, as the temporal expression pattern of ascl1a by RT-PCR and qPCR matched that of Shh signaling components (Figures 1A, 1B, 2A, and 2B). We found the co-expression of ptch1, a bona fide marker of active Shh signaling (Jeong and McMahon, 2005), with ascl1a mRNA in retina at 4 dpi (Figure 2C). This suggests the potential involvement of Shh signaling in ascl1a induction and vice versa. Inhibition of Shh signaling, by cyclopamine treatment or knockdown of gli1 or shha, significantly downregulated ascl1a expression (Figures 1N and S2G), which was also confirmed by mRNA ISH and qPCR in retina (Figures 2D and 2G). Conversely, knockdown of negative regulators of Shh signaling, gli3 and sufu, caused an upregulation of ascl1a (Figures 2E–2G), suggesting its possible direct regulation. This is supported by the presence of several Gli-binding sites on the ascl1a promoter, revealed by in silico analysis (Figure 2H). Further, we performed a post-injured retinal chromatin immunoprecipitation (ChIP) assay using antibodies against the Shh signaling effector proteins Gli1 and Gli3 separately to examine whether these Gli-binding sites (Gli-BSs) are functional. Interestingly, both antibodies could separately precipitate Gli-bound chromatin, supporting the direct physical interaction of Gli1/Gli3 on the ascl1a promoter (Figures 2I and S2K). Furthermore, a luciferase assay performed in zebrafish embryos confirmed the effect of stimulators and inhibitors of Shh signaling on ascl1a expression (Figure 2J). The Gli-BS mutations in the ascl1a promoter almost completely abolished the effect of inhibitors and stimulators as revealed by the luciferase assay (Table S2; Figure 2K). These results suggest that Shh signaling regulates the important gene ascl1a.
Figure 2.
Shh-Signaling-Dependent ascl1a Regulation in the Injured Retina
(A and B) RT-PCR (A) and qPCR (B) analysis of ascl1a in the post-injured retina; n = 6 biological replicates.
(C) Fluorescence ISH (FISH) and IF microscopy images of a 0.5-μm-thick optical section of retina showing co-localization of ascl1a with ptch1 in BrdU+ MGPCs at 4 dpi. Arrowheads mark co-expression of genes in BrdU+ cells.
(D–F) BF microscopy images of ascl1a mRNA ISH in retina at 4 dpi with cyclopamine treatment, shha or gli1 knockdowns (D), and gli3 or sufu knockdowns (E). The number of ascl1a+ cells from (E) is quantified in (F).
(G) qPCR analysis of ascl1a mRNA with cyclopamine treatment and shha or sufu knockdown in 2 dpi retina.
(H) Schematic of the ascl1a promoter with a putative Gli-binding site (Gli-BS) cluster. Arrows mark ChIP primers, N.S marks the negative control, and capital letters mark putative Gli-BSs.
(I) Retinal ChIP assay at 4 dpi showing both Gli1 and Gli3 bound to the ascl1a promoter.
(J) Luciferase assay in 24 hpf embryos co-injected with ascl1a:GFP-luciferase vector and sufu or shha MOs.
(K) Luciferase assay was done with mutated Gli-BS of ascl1a promoter in an experiment similar to (J).
Scale bars represent 10 μm in (C) and 20 μm in (D) and (E). Asterisk indicates the injury site (C–E). Error bars represent SD. ∗p < 0.0001 (F); ∗p < 0.01 (G); ∗p < 0.01 (J). n = 6 biological replicates (F and G); n = 3 (J). See also Figures S2, S6, and S7.
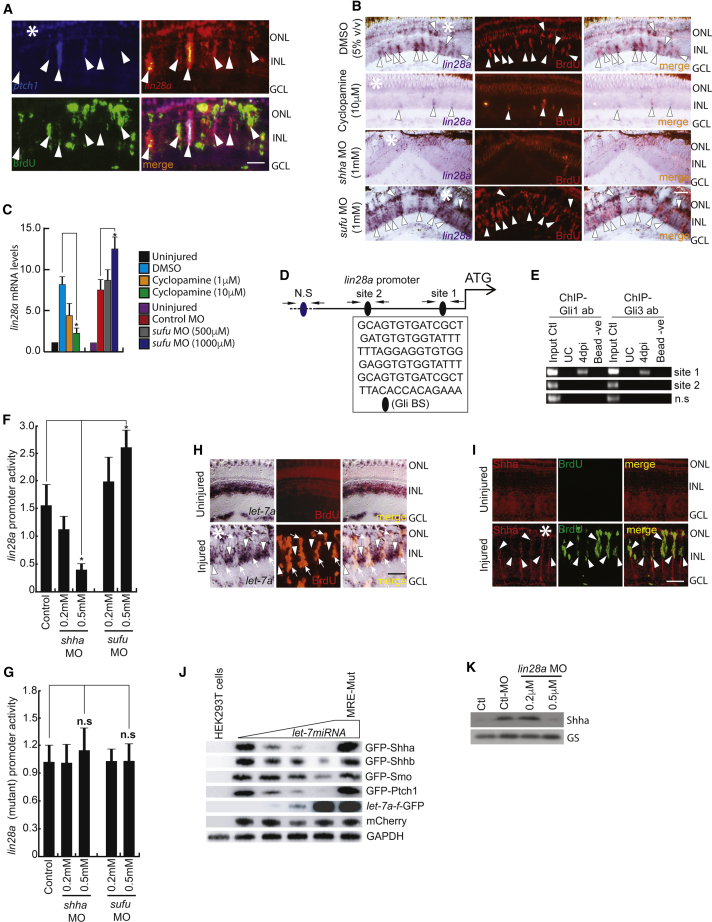
Shh Signaling/lin28a/let-7 Regulatory Loop Is Essential for MGPC Induction
We then explored whether the RNA-binding protein and pluripotency-inducing factor Lin28a, a necessary and well-known target of Ascl1a during retina regeneration, is regulated directly through Shh signaling (Ramachandran et al., 2010a). This was supported by the co-expression of ptch1 and lin28a in 4 dpi retinal sections (Figure 3A), suggesting the possible interdependency or hierarchical regulation. We further evaluated the expression pattern of lin28a that goes down with inhibited Shh signaling in retinal cross sections (Figure 3B). This was also proven by qPCR (Figure 3C). The opposite expression pattern of lin28a was found with sufu knockdown, as expected (Figures 3B and 3C). Evaluation of the lin28a promoter revealed putative Gli-BSs (Figure 3D) located as clusters, which were probed using Gli1 and Gli3 antibodies for a ChIP assay in the post-injured retina. Interestingly, both Gli1 and Gli3 bind to one of these Gli-BS clusters (Figures 3E and S2K), suggesting direct regulation of lin28a by Gli proteins. These results were further confirmed by luciferase assay performed in zebrafish embryos co-injected with lin28a:GFP-luciferase vector along with MOs against positive and negative regulators of Shh signaling (Figure 3F). The introduction of Gli-BS mutations in the lin28a promoter alleviated the impact of inhibitors and stimulators as revealed by a luciferase assay (Table S2; Figure 3G). Furthermore, let-7 microRNA, which is downregulated by Lin28a (Ramachandran et al., 2010a), was abundant in the uninjured inner nuclear layer (INL) in BrdU+ MGPCs at 4 dpi (Figure 3H). This let-7 downregulation in MGPCs is opposite to the IF pattern of Shh (Figures 3H and 3I), which suggested possible regulation of shha mRNA by let-7 microRNA. The mRNA ISH of shha and ptch1 also revealed a diffused expression pattern in both uninjured and 4 dpi retina (Figures S2H–S2J). In silico analysis predicted several let-7 microRNA-binding sites present in shha, shhb, smo, and ptch1 genes (Table S4). We cloned these four genes in-frame with GFP reporter regulated by the cytomegalovirus (CMV) promoter and transfected these constructs with increasing concentrations of let-7a and let-7f microRNA expression plasmid (Ramachandran et al., 2010a) in HEK293T cells (Figure S5F). The results showed a dose-dependent decline in GFP expression (Figure 3J), which was quantified (Figures S6C–S6F). The knockdown of lin28a led to an expected decline in Shha protein at 4 dpi (Figure 3K). These findings suggest that lin28a-mediated suppression of let-7 is required for the translational regulation of Shh signaling components in MGPCs as a part of positive feedback loop mediated through the Ascl1a-lin28a axis.
Figure 3.
Lin28a-let-7 Axis Regulates Shh Signaling Component Genes in the Injured Retina
(A) FISH and IF microscopy images of a 0.5-μm-thick optical section of retina showed co-localization of lin28a with ptch1 in BrdU+ MGPCs at 4 dpi. Arrowheads mark co-expression of genes in BrdU+ cells.
(B and C) BF microscopy images of lin28a mRNA ISH in the retina at 4 dpi with cyclopamine treatment and shha or sufu knockdown (B), which was quantified by qPCR (C). Arrowheads mark co-expression of genes in BrdU+ cells in (B).
(D and E) Schematic of the lin28a promoter with a potential Gli-BS cluster, where arrows mark ChIP primers and capital letters mark consensus sequence of Gli-BS (D). A 4 dpi retinal ChIP assay showed both Gli1 and Gli3 bound to one of the two Gli-BS clusters (E).
(F) Luciferase assay in 24 hpf embryos co-injected with lin28a:GFP-luciferase vector and sufu or shha MOs.
(G) Luciferase assay with mutated Gli-BSs of the lin28a promoter in an experiment similar to (F).
(H and I) ISH and IF microscopy of retina showing co-exclusion of let-7a microRNA (H) and co-localization of Shha protein (I) in BrdU+ MGPCs in the retina at 4 dpi. Arrowheads mark expression of let-7a in BrdU− cells and arrows mark co-exclusion of let-7a from BrdU+ cells in (H). Arrowheads mark co-expression of Shha in BrdU+ cells in (I).
(J) let-7 microRNA downregulated the translation of GFP fused with the indicated gene constructs harboring microRNA-binding regions in a dose-dependent manner in HEK293T cells.
(K) Western blot of Shha in lin28a-MO electroporated retina at 4 dpi.
Scale bars represent 10 μm (A, H, and I) and 20 μm (B). Asterisk indicates the injury site (A, B, H, and I). Error bars represent SD.∗p < 0.001 (C); ∗p < 0.001 (F). n = 6 biological replicates (C, F, and G). GS, glutamine synthetase. See also Figures S3, S6, and S7.
Mmp9 Regulates ascl1a through Shh Signaling
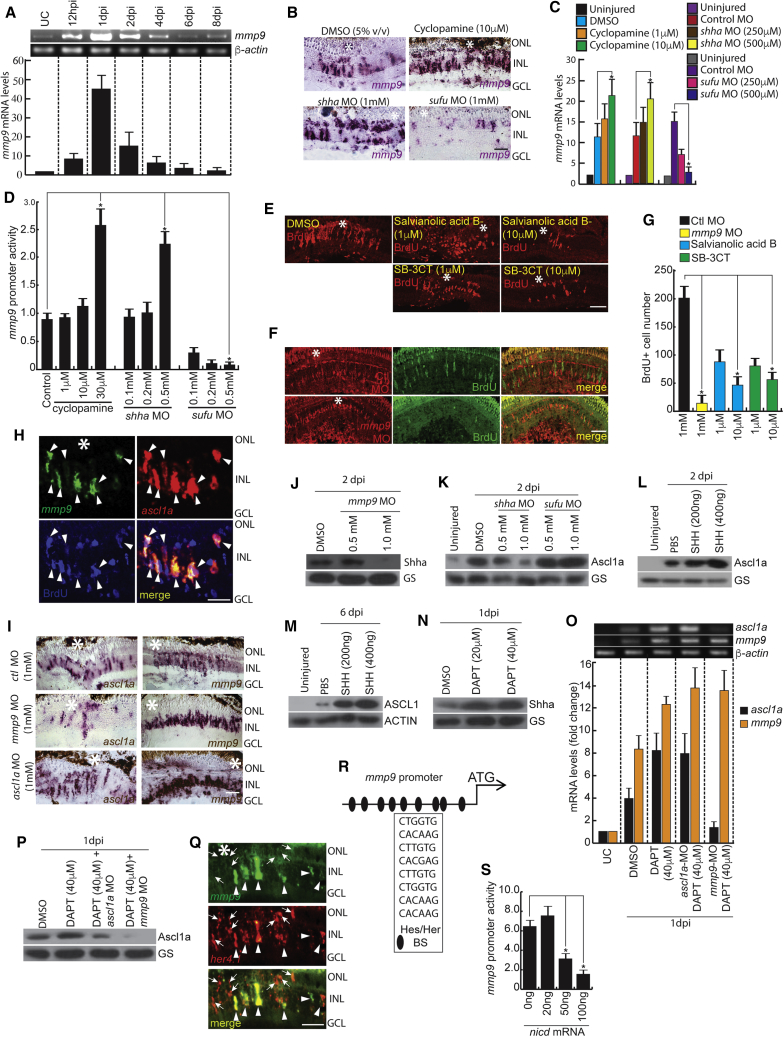
We also investigated the involvement of mmp9, a gene highly induced in regenerating MG cells, as revealed in microarray analysis (Ramachandran et al., 2012) and whole-retina RNA-seq done in the present study. Mmp9 is not only an important enzyme prerequisite for proliferative and pro-differentiative roles (Mannello et al., 2006), but also essential during fin regeneration (LeBert et al., 2015, Yoshinari et al., 2009). We found that mmp9 is rapidly induced in the injured retina, with a peak expression at 24 hpi (Figures 4A and S3A), and later (at 4 dpi), mmp9 levels were restricted to the neighboring cells of BrdU+ MGPCs (Figures S3B and S3C). Interestingly, inhibition of Shh signaling caused a significant upregulation of mmp9, and an opposite effect was seen with sufu knockdown (Figures 4B and S3D–S3F), which was confirmed by qPCR (Figure 4C) and a luciferase assay performed in zebrafish embryos injected with mmp9:GFP-luciferase vector (Figure 4D). These results suggest a negative correlation between mmp9 and active cell proliferation. However, upon inhibition of Mmp9 using pharmacological agents such as salvianolic acid B and SB-3CT, or by mmp9 targeting MO (Figures S6A, and S6B; Table S1), we found a drastic decline in BrdU+ cells in WT or GFP+ cells in tuba1016 transgenic retina (Figures 4E–4G and S3G). Interestingly, no impact was seen with mmp9 blockade after 2 dpi (Figure S3H), suggesting that its role preludes cell proliferation. To evaluate this further, we analyzed the expression pattern of an important gene, ascl1a, in mmp9-expressing cells in 4 dpi retina. We found significant co-localization of ascl1a+ cells with mmp9 expression (Figures 4H and S4A). Moreover, mmp9 knockdown caused a decline in ascl1a expression, whereas ascl1a knockdown caused an upregulation of mmp9 in 4 dpi retina (Figures 4I and S3I). Since the regulation of ascl1a is established through Shh signaling, we further explored whether Mmp9-mediated regulation of ascl1a was through Shha. Knockdown of mmp9 abolished the expression of Shha, as found with cyclopamine treatment (Figures 4J, S3J, and S7B). We also found an Shh-signaling-dependent regulation of Ascl1a protein with both shha or sufu knockdowns in 2 dpi retina (Figures 4K and S7C). Recombinant-SHH could induce Ascl1a expression and cell proliferation in zebrafish retina, similar to sufu knockdown (Figures 4L, S3K–S3M, and S7D). Interestingly, we also found a drastic increase in mRNA levels of Ascl1, Lin28a, and ASCL1 protein in injured mouse retina treated with recombinant-SHH (Figures 4M, S3N, and S7E).
Figure 4.
Shh-Mmp9-Ascl1a Interplay Is Necessary during MG Reprogramming
(A) RT-PCR (top) and qPCR (bottom) analysis of injury-dependent mmp9 expression in the retina; n = 6 biological replicates.
(B–D) BF microscopy images of mmp9 mRNA ISH in the retina at 4 dpi with cyclopamine treatment and shha or sufu knockdown (B), as quantified by qPCR (C), and a luciferase assay in 24 hpf embryos injected with mmp9:GFP-luciferase vector (D).
(E–G) IF microscopy images of 4 dpi retina with Mmp9 blockade using drugs (E) and MO against mmp9 (F). The number of BrdU+ MGPCs is quantified in (G).
(H) FISH and IF microscopy images of a 0.5-μm-thick optical section of retina showing co-localization of mmp9 and ascl1a in BrdU+ MGPCs at 4 dpi. Arrowheads mark co-expression of genes in BrdU+ cells.
(I) BF microscopy images of ascl1a and mmp9 mRNA ISH in ascl1a and mmp9 knockdowns in 4 dpi retina.
(J) Western blotting experiment showing Shh levels in 2 dpi retina with the mmp9 knockdown.
(K) Western blotting assay of Ascl1a in 2 dpi retina with shha or sufu knockdowns.
(L) Western blotting assay of Ascl1a in 2 dpi zebrafish retina injected with recombinant SHH protein.
(M) Western blotting assay of ASCL1 in 6 dpi mouse retina injected with recombinant SHH protein.
(N) Western blotting assay of Shha in DAPT-treated retina at 1dpi.
(O and P) RT-PCR (top) and qPCR (bottom) analysis of ascl1a and mmp9 in DAPT-treated retina, with or without ascl1a or mmp9 knockdown (O), and confirmed by western blotting assay (P).
(Q) FISH and IF microscopy images of a 0.5-μm-thick optical section of retina showed substantial co-exclusion and marginal co-localization of mmp9 with her4.1 at 4 dpi. Arrowheads mark co-expression of the gene, and arrows mark her4.1+ cells.
(R and S) Schematic of the mmp9 promoter with potential Hes/Her-BS binding sites (inside box), and luciferase assay in 24 hpf embryos co-injected with mmp9:GFP-luciferase construct and notch intracellular domain (nicd) mRNA (S).
Scale bars represent 10 μm (H and Q) and 20 μm (B, E, F, and I). Asterisk indicates the injury site (B, E, F, H, I and Q). Error bars represent SD. ∗p < 0.001 (C, D, G, and S). Biological replicates n = 6 in (C) and (G), and n = 3 in (D) and (S). See also Figures S3, S4, S6, and S7.
Inhibition of Notch signaling through N-[N-(3,5-difluorophenylacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT) treatment, which causes a decline in Her4.1 levels and enhancement of MGPCs during retina regeneration (Conner et al., 2014, Wan et al., 2012), increased mmp9, ascl1a mRNA, and Shh protein levels (Figures S4B, S4C, 4N, and S7F). We further explored whether ascl1a upregulation seen with DAPT treatment is mediated through the Mmp9/Shh axis. Interestingly, we found that in the DAPT-treated retina, ascl1a translation was nullified with mmp9 knockdown (Figures 4O, 4P, and S7G). We speculated that upregulation of mmp9 with blockade of Notch signaling is possibly due to a lack of Her4.1-mediated transcriptional repression. Expression of mmp9 and her4.1 showed co-labeling in a few and co-exclusion in the majority of retinal cells (Figure 4Q). In silico analysis of the mmp9 promoter revealed several hairy enhancer of split (Hes/Her)-binding N-boxes (Kageyama et al., 2007), suggesting its potential regulation through Notch signaling (Figure 4R). We performed a luciferase assay in zebrafish embryos co-injected with notch intracellular domain (nicd) mRNA along with mmp9:GFP-luciferase vector. nicd mRNA could cause an upregulation of Her4.1 (Nakahara et al., 2016, Wilson et al., 2016), and the luciferase assay showed dose-dependent downregulation of mmp9 promoter activity (Figure 4S), while mutations in Her4-binding sites abolished this impact (Figure S4D; Table S2). In summary, these results suggest that active Notch-signaling-mediated induction of her4.1 restricts the span of mmp9 expression at the site of injury. Further, Mmp9 coaxes MG to regenerate through Shh signaling and Ascl1a induction during retina regeneration.
Shh Signaling Regulates zic2b Expression during Regeneration
We explored a zinc-finger transcription factor, Zic2, essential for normal brain patterning during development (Elms et al., 2003), which upon mutation shows holoprosencephaly (HPE) or cyclopia (Brown et al., 2001, Teslaa et al., 2013), a phenotype similar to cyclopamine treatment. Zic2 is also known to collaborate with Gli proteins (Koyabu et al., 2001). Therefore, we investigated whether a relationship exists between Gli proteins and Zic2 during retina regeneration, because both proteins occupy the same DNA sequence of the target genes’ promoters (Vokes et al., 2007). zic2b, orthologous to the mammalian Zic2 gene, showed upregulation in the retina microarray (Ramachandran et al., 2012) and our RNA-seq analysis. zic2b is also expressed in fin blastema (Figure S4E). The temporal expression pattern of zic2b in post-injured retina showed a peak expression at 4 dpi, a time when cell proliferation is at the maximum level (Figure 5A). Pulse labeling of MGPCs with BrdU also revealed its co-localization with zic2b (Figure 5B). Co-expression of ptch1 with zic2b in BrdU+ cells suggests their interaction during regeneration (Figure 5C). The zic2b showed downregulation with blockade of Shh signaling and an upregulation with sufu knockdown (Figures 5D and 5E). These results were also confirmed by a luciferase assay done in zebrafish embryos injected with zic2b:GFP-luciferase construct along with MOs against shha and sufu and also exposed to cyclopamine (Figure 5F). Analysis of the zic2b promoter revealed a cluster of Gli-BSs (Figure 5G), and spanning chromatin was pulled down using both Gli1 and Gli3 antibodies separately (Figures 5H and S2K). Gene knockdowns of gli1, gli3, and zic2b significantly influenced MGPCs proliferation in 4 dpi retina (Figures 5I, 5J, S6A, and S6B; Table S1). The luciferase assay revealed that Shh signaling inhibitors and stimulators had a small impact on zic2b promoter activity with mutated Gli-BSs (Table S2; Figure S4F). Early or late knockdowns of gli1/zic2b caused a decline in the number of BrdU+ cells in the retina, but the opposite was seen with gli3 knockdown (Figures 5I, 5J, S4G, and S4H). zic2b showed a pan retinal expression pattern with DAPT treatment, and the same was seen with gli3/sufu knockdowns (Figures S4I–S4K). Interestingly, zic2b knockdown nullified the enhancement of MGPCs with gli3 knockdown (Figures 5I and 5J). Moreover, the induction of Gli3 seems to block the responsiveness of MGPCs to Gli1, as the late knockdowns and double knockdown of gli1 and gli3 also caused a drastic decline in cell proliferation (Figures 5I, 5J, S4G and S4H). The gli1 knockdown significantly impacted several regeneration-associated genes as the possible cause of the lack of MGPC induction (Figure S4L). These results suggest that the induction of zic2b in MGPCs largely triggers a proliferative phase mediated through Shh signaling, and it may collaborate with or outcompete Gli proteins in targeting Gli-BSs to drive MGPCs toward differentiation.
We also examined whether zic2b expression depends on the mmp9-shha-ascl1a signaling axis, because a substantial proportion of BrdU+ MGPCs co-expressed ascl1a and zic2b (Figure 5K). We probed for zic2b expression in 4 dpi retina electroporated with mmp9 and ascl1a MOs separately and found that zic2b levels declined drastically, as found with blockade of Shh signaling (Figures 5D and 5L). We further speculated that apart from its transcriptional control, zic2b might be regulated at translational levels. This speculation is mainly because of the presence of bona fide let-7 microRNA-binding sites in the zic2b coding region (Figure S5F). Surprisingly, we found a downregulation in the translation of GFP protein from an expression cassette appended with zic2b in HEK293T cells (Figure 5M), which was quantified (Figure S6G). These results suggest that zic2b is an essential regeneration-associated gene in zebrafish retina that is regulated through the mmp9-shha-ascl1a-lin28a-let-7 pathway.
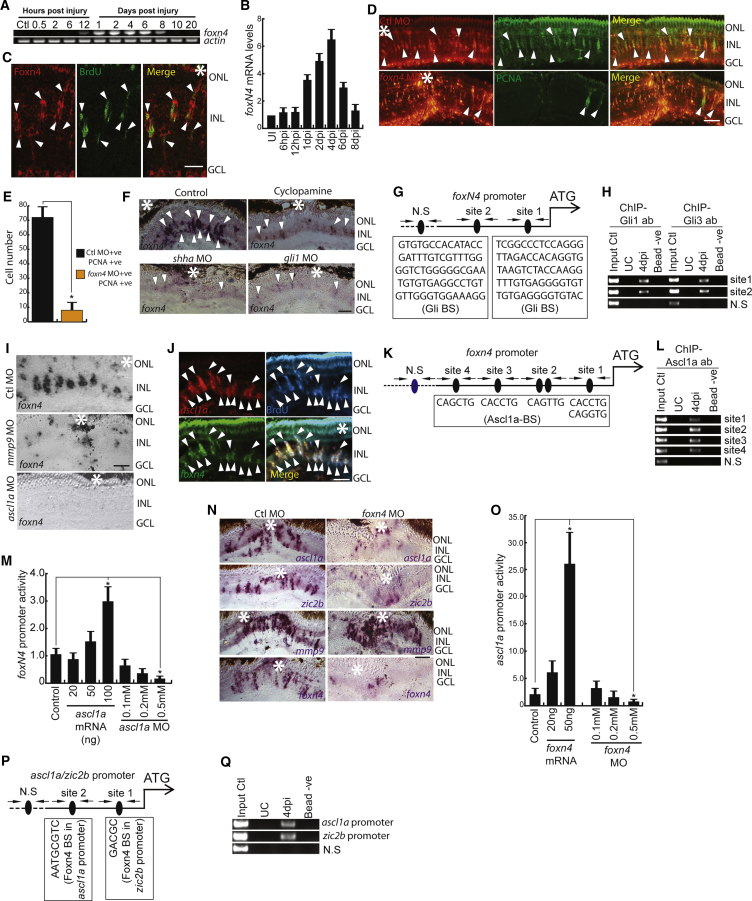
The Foxn4/Ascl1a/Shh/Zic2b Regulatory Loop Is Associated with Regeneration
Foxn4, a member of the forkhead box family of proteins and discovered in retina microarray (Ramachandran et al., 2012) and RNA-seq analyses performed in the present study, showed an upregulation, with a peak expression at 4 dpi (Figures 6A and 6B). Foxn4 expression was restricted to BrdU+ MGPCs at 4 dpi (Figure 6C). Furthermore, we explored the significance of foxn4 induction during retina regeneration. Interestingly, MO-mediated gene knockdown of foxn4 inhibited MGPC induction up to 90% (Figures 6D, 6E, and S5A).
Figure 6.
Expression Dynamics and Necessity of Foxn4 during Regeneration
(A and B) RT-PCR (A) and qPCR (B) analysis of injury-dependent foxn4 expression in the retina; n = 6 biological replicates. (C) IF microscopy of a 0.5-μm-thick optical section of retina revealing co-localization of Foxn4 with BrdU+ MGPCs in 4 dpi retina.
(D and E) IF microscopy images of the retina with foxn4 knockdown at 4 dpi (D). The number of PCNA+ MGPCs is quantified in (E).
(F) BF microscopy images of foxn4 mRNA ISH in retinal sections with cyclopamine treatment and shha or gli1 knockdowns.
(G and H) Schematic of foxn4 promoter with a putative Gli-BS cluster, where arrows mark ChIP primers, N.S marks negative control, and capital letters mark putative Gli-BSs (G). A retinal ChIP assay at 4 dpi showing both Gli1 and Gli3 bound to the foxn4 promoter (H).
(I) BF microscopy images of foxn4 mRNA ISH in retinal sections with mmp9 or ascl1a knockdowns.
(J) FISH and IF microscopy images of a 0.5-μm-thick optical section of retina showing co-localization of foxn4 and ascl1a in BrdU+ MGPCs at 4 dpi. Arrowheads mark co-expression of genes in BrdU+ cells.
(K and L) Schematic of the foxn4 promoter with a putative Ascl1a-binding site cluster, where arrows mark ChIP primers, N.S marks negative control, and capital letters mark putative Ascl1a-BS (K). A retinal ChIP assay at 4 dpi showing Ascl1a bound to the foxn4 promoter (L).
(M) Luciferase assay showing foxn4 promoter activity with overexpression or knockdown of ascl1a in 24 hpf embryos.
(N) BF microscopy images of mRNA ISH in retinal sections with foxn4 knockdown showing levels of genes (namely, ascl1a, zic2b, mmp9, and foxn4) at 4 dpi.
(O) Luciferase assay showing ascl1a promoter activity with overexpression or knockdown of foxn4 in 24 hpf embryos.
(P and Q) Schematic of ascl1a and zic2b promoter with a putative Foxn4-binding site cluster, where arrows mark ChIP primers, N.S marks negative control, and capital letters mark putative Foxn4-BS (P). A retinal ChIP assay at 4 dpi showing Foxn4 bound to both the ascl1a and zic2b promoters (Q).
Scale bars represent 10 μm (C, D, F, I, J, and N). Error bars represent SD. ∗p < 0.001 (M); ∗p < 0.04 (O). Biological replicates n = 6 in (M) and O, and n = 3 in (B). Asterisk marks injury spots in (C),(D),(F), (J) and (N). See also Figures S5–S7.
To ascertain whether foxn4 is regulated through Shh signaling or its downstream effector genes, we adopted a pharmacological inhibition or gene-knockdown approach. Blockade of Shh signaling with cyclopamine or MOs against shha or gli1 significantly abolished foxn4 expression in the retina (Figures 6F, S4L, and S5B), whereas the opposite was seen with sufu knockdown (Figures S5C and S5D). Analysis of the foxn4 promoter revealed 2 putative Gli-BS clusters (Figure 6G) that were strongly bound by Gli1 and Gli3, as revealed by a ChIP assay (Figures 6H and S2K), suggesting a direct involvement of Shh signaling in its expression. As discussed earlier, the influence of Mmp9 on expression levels of Shha led us to suspect its involvement in the regulation of foxn4. Knockdown of mmp9 in 4 dpi retina caused a significant downregulation of foxn4 (Figures 6I and S5E).
The temporal gene expression pattern and co-localization of foxn4 with MGPCs prompted us to investigate its potential parallels with ascl1a gene. Fluorescence ISH (FISH) analysis showed co-expression of ascl1a and foxn4 in BrdU+ MGPCs (Figure 6J). We then explored the possibility of a hierarchical regulation between ascl1a and foxn4 during retina regeneration, as there is already a reported role for Foxn4 in the regulation of Ascl1 expression in mouse and chick (Del Barrio et al., 2007). We found significant downregulation of foxn4 expression in retinal sections with knockdown of ascl1a (Figure 6I). foxn4 promoter analysis predicted several Ascl1a-binding E-boxes (Bertrand et al., 2002, Li et al., 2006, Ramachandran et al., 2010a, Ramachandran et al., 2011), and binding was confirmed by a ChIP assay (Figures 6K, 6L, and S5G). The transactivation of the foxn4 promoter by Ascl1a was confirmed with a luciferase assay, which was done by co-injection of ascl1a mRNA or MO against it, along with the promoter of foxn4 driving the GFP-luciferase fusion construct in zebrafish embryos (Figure 6M). The mutation of Ascl1a-BS in the foxn4 promoter had a negligible effect on its promoter activity both by ascl1a mRNA or by MO co-injections in zebrafish embryos (Figure S5H; Table S2).
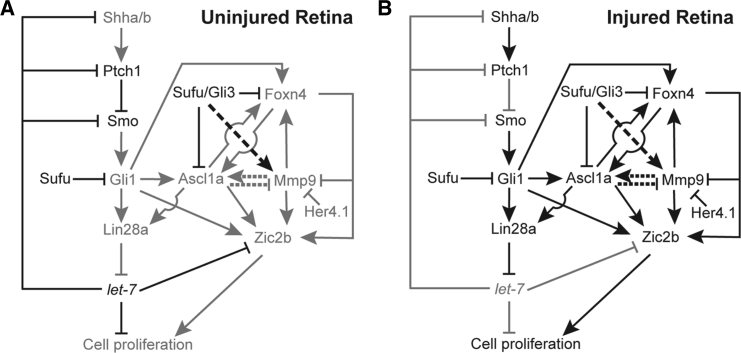
We then explored, using a knockdown approach in the retina, whether Foxn4 impacted ascl1a or other regeneration-associated genes such as zic2b and mmp9. We found that both ascl1a and zic2b were downregulated, which also explained the downregulation of foxn4 itself, whereas no appreciable change was seen in mmp9 levels (Figure 6N). A luciferase assay confirmed transactivation of the ascl1a promoter by Foxn4, which was done by co-injection of foxn4 mRNA or MO against it, along with the promoter of ascl1a driving the GFP-luciferase fusion construct in zebrafish embryos (Figure 6O). Both the ascl1a and zic2b promoters harbor 2 potential Foxn4-binding sites (Luo et al., 2012) (Figure 6P), and this was confirmed by a ChIP assay, which was done using an antibody targeting Foxn4 (Figure 6Q). Mutated Foxn4-BS on the ascl1a promoter caused an almost complete alleviation of upregulated luciferase activity, as seen by its overexpression (Figures 6O and S5I; Table S2). These results suggest that foxn4 expression is dependent on Shh signaling directly as well as through other genes such as ascl1a, which in turn regulates another regeneration-associated gene such as zic2b in a feedback loop. The findings from this study are summarized in a model (Figures 7A and 7B).
Figure 7.
Schematic Representation of the Gene Regulatory Network during Retina Regeneration
(A and B) Genetic interrelationships in uninjured (A) and injured (B) retina. Faded arrows and gene names show absence and bold shows presence. See also Figures S1–S7.
Discussion
In this study, we explored the significance and potential regulators of Shh signaling during zebrafish retina regeneration. Our findings unravel mechanisms through which Shh signaling contributes to retina regeneration. We propose that Shh-dependent induction of Ascl1a and Lin28a contributes to Müller glia dedifferentiation through let-7 microRNA-mediated translational downregulation of shha, shhb, smo, ptch1, and zic2b from respective mRNAs. Such stringent translational regulation probably accounts for the lack of an immature regenerative response despite the marginal expression of Shh signaling components such as shha, shhb, smo, and ptch in the uninjured retina. Cyclopamine-mediated repression of MGPCs might result from a decline in the regeneration-specific genes ascl1a and lin28a. This situation could be further exacerbated by upregulation of the repressor insm1a and the lack of the Delta-Notch signaling effector her4.1. These observations suggest the ability of Shh signaling to impinge upon various other signaling pathways important for regeneration.
Our results also show that Shh signaling impacted regeneration not only through transcription factors but also through negative regulation of enzymes such as Mmp9. Moreover, Mmp9-dependent expression of Shha causes the induction of Ascl1a as a prelude to MG dedifferentiation and MGPC induction. The increased expression of Mmp9 in a regeneration-compromised scenario like cyclopamine treatment (shha or ascl1a knockdown retina) suggests the existence of a feedback loop between Mmp9 and Shh signaling. The abundance of Mmp9 is probably due to the lack of Shha protein to give a feedback response for a decrease in its expression in MG to induce MGPCs. This observation is also supported by the sufu knockdown-mediated decline in mmp9 expression. Co-labeling of ascl1a and mmp9, which was seen in a good number of cells, may appear paradoxical, but they all need not be Shh-positive or BrdU+. Only a subset of ascl1a-positive cells is ptch1 positive and can have active Shh signaling and downregulated mmp9. The remainder of the ascl1a positive cells can have upregulated mmp9 due to the lack of Shh signaling. Moreover, the Mmp9 expression is necessary for normal cycling of MGPCs during regeneration, and the repression of mmp9 by Her4.1 could enable its expression restricted to the injury site at a later time. We anticipate a much wider role for the Shha-Mmp9-Ascl1a-Lin28a-let-7 regulatory loop during retinal regeneration.
The induction of repressor Gli3 might cause the exit of MGPCs from the cell cycle to restrict the impact of a transcriptional activator, Gli1. This is evident from the knockdown results of gli1 and gli3 either in isolation or in combination. The gli1 knockdown indicated a decline in the number of MGPCs, whereas gli3 inhibition caused an expansion of MGPCs. Interestingly, double knockdown of gli1 and gli3 resulted in significant decline in MGPCs, suggesting that the Gli3 is necessary to quit the cell cycle as a prelude to differentiation. Similar results were seen with zic2b knockdown or cyclopamine treatment. This could be due to the impact of Shh signaling on the expression of downstream genes through Zic2b, although both Gli and Zic2b may compete or collaborate with the same binding sites on DNA. As zic2b mRNA shows a translational regulation through let-7 microRNA, one could speculate that the role of Zic2b protein is restricted to Ascl1a- or Lin28a-expressing MGPCs.
The forkhead box gene family member foxn4 is unique in its expression pattern during zebrafish development, with multiple isoforms in the thymus, skin, and brain (Danilova et al., 2004). We show the brain-specific isoform of foxn4 is rapidly induced by Shh signaling, which orchestrates a series of gene expression events in response to retinal injury. Gli-BSs on the foxn4 promoter is functional and probably explains the lack of its expression in the cyclopamine-treated retina. The regeneration-associated transcription factor Ascl1a significantly contributes to the induction of foxn4, suggesting dual control of its expression. Moreover, Foxn4 deficiency caused a significant reduction in MGPC number, probably through its effect on other regeneration-associated genes, which form a regulatory loop. To support this view, the proof that FoxN4 binds to promoters of ascl1a and zic2b at its consensus-binding sites (obtained from ChIP) makes it one of the central pillars of regeneration.
Taken together, our study sheds light on the mechanisms of MGPC induction in zebrafish retina in response to injury in an Shh-signaling-dependent manner and the significance of its downstream effector genes such as ascl1a, lin28a, zic2b, foxn4, and mmp9. These findings also suggest ways to coax mammalian MG dedifferentiation that may enable us to find ample solutions to intervene therapeutically for an efficient regenerative response.
Experimental Procedures
Further details and an outline of resources used in this work can be found in Supplemental Experimental Procedures.
Animals and Retinal Injury
Zebrafish were maintained at 26–28°C on a 14 hr/10 hr light/dark cycle for all experiments unless otherwise specified. The retinal injury was performed using a 30G needle as described previously (Fausett and Goldman, 2006). The C57BL/6 mice used in this study were maintained on a 12 hr/12 hr light/dark cycle with continuous access to food and water.
RNA-Seq Analysis
The RNA-seq analysis of the total RNA of the retina at different time points post-injury and with cyclopamine treatment was performed as described previously (Brooks et al., 2012).
Statistical Analysis
Observed data were analyzed for statistical significance by comparisons done using a two-tailed unpaired Student’s t test to analyze data from all experiments. Error bars represent SD in all histograms.
Acknowledgments
This work was supported by the Department of Biotechnology (DBT) (S.K.), the Indian Council of Medical Research (S.G.), IISER Mohali (S.M., M.A.K., and M.C.), the Wellcome Trust/DBT India Alliance (Intermediate Fellowship IA/I/12/2/500630, to R.R.), extramural research funding from DBT India (102/IFD/SAN/3975/2015-2016 and 102/IFD/SAN/2255/2017-2018, to R.R.), and intramural funding support from IISER Mohali. The authors would like to express their gratitude to Daniel Goldman (Molecular and Behavioral Neuroscience Institute, University of Michigan) for sharing promoter clones of 1016tuba1a, lin28a, ascl1a, and insm1a genes. The authors also thank Dr. Kuljeet Singh Sandhu and Mr. Arashdeep Singh (IISER Mohali) for their help predicting Gli-BSs, Dr. Mahak Sharma’s laboratory (IISER Mohali) for cell transfection experiments, and Dr. Adrene Freeda D’cruz and Dr. Rayman Matharu (IISER Mohali) for critical reading of the manuscript.
Author Contributions
R.R. conceived the study and designed experiments. S.K. performed the majority of experiments. S.G. and M.C. contributed to western blotting assays. M.A.K. conducted the fin experiments, S.M. performed RNA-seq Venn diagrams, and A.J.K. helped with cell sorting. R.R., S.K., and S.M. analyzed the experimental data. R.R. wrote the manuscript with critical input from S.K., S.G., M.C., and S.M.
Declaration of Interests
The authors declare no competing interests.
Published: May 1, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and five tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.04.002.
Data and Software Availability
The accession number for the RNA-seq data reported in this paper is GEO: GSE102063.
Supplemental Information
The table shows a list of transcription factors, transcription co-factors and chromatin remodeling factors along with their ENSEMBL IDs, obtained from RNA-seq data that are regulated during various time points, and Shh signaling inhibition with cyclopamine, post-retinal injury.
All the primers used in this study are listed with their code names indicating the genes in the first column and the 5′ to 3′ sequence, in the next column.
References
- Ando K., Shibata E., Hans S., Brand M., Kawakami A. Osteoblast production by reserved progenitor cells in zebrafish bone regeneration and maintenance. Dev. Cell. 2017;43:643–650. doi: 10.1016/j.devcel.2017.10.015. [DOI] [PubMed] [Google Scholar]
- Bertrand N., Castro D.S., Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Brooks M.J., Rajasimha H.K., Swaroop A. Retinal transcriptome profiling by directional next-generation sequencing using 100 ng of total RNA. Methods Mol. Biol. 2012;884:319–334. doi: 10.1007/978-1-61779-848-1_23. [DOI] [PubMed] [Google Scholar]
- Brown L.Y., Odent S., David V., Blayau M., Dubourg C., Apacik C., Delgado M.A., Hall B.D., Reynolds J.F., Sommer A. Holoprosencephaly due to mutations in ZIC2: alanine tract expansion mutations may be caused by parental somatic recombination. Hum. Mol. Genet. 2001;10:791–796. doi: 10.1093/hmg/10.8.791. [DOI] [PubMed] [Google Scholar]
- Chen J.K., Taipale J., Cooper M.K., Beachy P.A. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner C., Ackerman K.M., Lahne M., Hobgood J.S., Hyde D.R. Repressing notch signaling and expressing TNFα are sufficient to mimic retinal regeneration by inducing Müller glial proliferation to generate committed progenitor cells. J. Neurosci. 2014;34:14403–14419. doi: 10.1523/JNEUROSCI.0498-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N., Visel A., Willett C.E., Steiner L.A. Expression of the winged helix/forkhead gene, foxn4, during zebrafish development. Brain Res. Dev. Brain Res. 2004;153:115–119. doi: 10.1016/j.devbrainres.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Del Barrio M.G., Taveira-Marques R., Muroyama Y., Yuk D.I., Li S., Wines-Samuelson M., Shen J., Smith H.K., Xiang M., Rowitch D., Richardson W.D. A regulatory network involving Foxn4, Mash1 and delta-like 4/Notch1 generates V2a and V2b spinal interneurons from a common progenitor pool. Development. 2007;134:3427–3436. doi: 10.1242/dev.005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaeva M., Waltenberger J. Hh signaling in regeneration of the ischemic heart. Cell. Mol. Life Sci. 2017;74:3481–3490. doi: 10.1007/s00018-017-2534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elms P., Siggers P., Napper D., Greenfield A., Arkell R. Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev. Biol. 2003;264:391–406. doi: 10.1016/j.ydbio.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Fausett B.V., Goldman D. A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina. J. Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D. Müller glial cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 2014;15:431–442. doi: 10.1038/nrn3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke R.M., Meredith D.M., Borromeo M.D., Savage T.K., Johnson J.E. Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev. Biol. 2009;328:529–540. doi: 10.1016/j.ydbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K., Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona J.P., Gaffield W., Kapur R.P., Roelink H. The teratogenic Veratrum alkaloid cyclopamine inhibits sonic hedgehog signal transduction. Development. 1998;125:3553–3562. doi: 10.1242/dev.125.18.3553. [DOI] [PubMed] [Google Scholar]
- Jeong J., McMahon A.P. Growth and pattern of the mammalian neural tube are governed by partially overlapping feedback activities of the hedgehog antagonists patched 1 and Hhip1. Development. 2005;132:143–154. doi: 10.1242/dev.01566. [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- Koyabu Y., Nakata K., Mizugishi K., Aruga J., Mikoshiba K. Physical and functional interactions between Zic and Gli proteins. J. Biol. Chem. 2001;276:6889–6892. doi: 10.1074/jbc.C000773200. [DOI] [PubMed] [Google Scholar]
- LeBert D.C., Squirrell J.M., Rindy J., Broadbridge E., Lui Y., Zakrzewska A., Eliceiri K.W., Meijer A.H., Huttenlocher A. Matrix metalloproteinase 9 modulates collagen matrices and wound repair. Development. 2015;142:2136–2146. doi: 10.1242/dev.121160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu Q., Qiu M., Pan Y., Li Y., Shi T. Identification and analysis of the mouse basic/Helix-Loop-Helix transcription factor family. Biochem. Biophys. Res. Commun. 2006;350:648–656. doi: 10.1016/j.bbrc.2006.09.114. [DOI] [PubMed] [Google Scholar]
- Luo H., Jin K., Xie Z., Qiu F., Li S., Zou M., Cai L., Hozumi K., Shima D.T., Xiang M. Forkhead box N4 (Foxn4) activates Dll4-Notch signaling to suppress photoreceptor cell fates of early retinal progenitors. Proc. Natl. Acad. Sci. USA. 2012;109:E553–E562. doi: 10.1073/pnas.1115767109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannello F., Tonti G.A., Bagnara G.P., Papa S. Role and function of matrix metalloproteinases in the differentiation and biological characterization of mesenchymal stem cells. Stem Cells. 2006;24:475–481. doi: 10.1634/stemcells.2005-0333. [DOI] [PubMed] [Google Scholar]
- Miller S.G., Carnell L., Moore H.H. Post-Golgi membrane traffic: brefeldin A inhibits export from distal Golgi compartments to the cell surface but not recycling. J. Cell Biol. 1992;118:267–283. doi: 10.1083/jcb.118.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara Y., Muto A., Hirabayashi R., Sakuma T., Yamamoto T., Kume S., Kikuchi Y. Temporal effects of Notch signaling and potential cooperation with multiple downstream effectors on adenohypophysis cell specification in zebrafish. Genes Cells. 2016;21:492–504. doi: 10.1111/gtc.12358. [DOI] [PubMed] [Google Scholar]
- Nelson B.R., Hartman B.H., Ray C.A., Hayashi T., Bermingham-McDonogh O., Reh T.A. Acheate-scute like 1 (Ascl1) is required for normal delta-like (Dll) gene expression and notch signaling during retinal development. Dev. Dyn. 2009;238:2163–2178. doi: 10.1002/dvdy.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini A., Jiang Y.J., Wilkinson D.G. Two zebrafish Notch-dependent hairy/Enhancer-of-split-related genes, her6 and her4, are required to maintain the coordination of cyclic gene expression in the presomitic mesoderm. Development. 2004;131:1529–1541. doi: 10.1242/dev.01031. [DOI] [PubMed] [Google Scholar]
- Ramachandran R., Fausett B.V., Goldman D. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R., Reifler A., Parent J.M., Goldman D. Conditional gene expression and lineage tracing of tuba1a expressing cells during zebrafish development and retina regeneration. J. Comp. Neurol. 2010;518:4196–4212. doi: 10.1002/cne.22448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R., Zhao X.F., Goldman D. Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proc. Natl. Acad. Sci. USA. 2011;108:15858–15863. doi: 10.1073/pnas.1107220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R., Zhao X.F., Goldman D. Insm1a-mediated gene repression is essential for the formation and differentiation of Müller glia-derived progenitors in the injured retina. Nat. Cell Biol. 2012;14:1013–1023. doi: 10.1038/ncb2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherpa T., Fimbel S.M., Mallory D.E., Maaswinkel H., Spritzer S.D., Sand J.A., Li L., Hyde D.R., Stenkamp D.L. Ganglion cell regeneration following whole-retina destruction in zebrafish. Dev. Neurobiol. 2008;68:166–181. doi: 10.1002/dneu.20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Li P., Carr A.L., Gorsuch R., Yarka C., Li J., Bartlett M., Pfister D., Hyde D.R., Li L. Transcription of the SCL/TAL1 interrupting Locus (Stil) is required for cell proliferation in adult Zebrafish Retinas. J. Biol. Chem. 2014;289:6934–6940. doi: 10.1074/jbc.M113.506295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takke C., Dornseifer P., v Weizsäcker E., Campos-Ortega J.A. her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development. 1999;126:1811–1821. doi: 10.1242/dev.126.9.1811. [DOI] [PubMed] [Google Scholar]
- Teslaa J.J., Keller A.N., Nyholm M.K., Grinblat Y. Zebrafish Zic2a and Zic2b regulate neural crest and craniofacial development. Dev. Biol. 2013;380:73–86. doi: 10.1016/j.ydbio.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.L., Morgan G.W., Dolinski K.M., Thummel R. Characterization of the pleiotropic roles of Sonic Hedgehog during retinal regeneration in adult zebrafish. Exp. Eye Res. 2018;166:106–115. doi: 10.1016/j.exer.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd L., Fischer A.J. Hedgehog signaling stimulates the formation of proliferating Müller glia-derived progenitor cells in the chick retina. Development. 2015;142:2610–2622. doi: 10.1242/dev.121616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk M., Kenis G., Bardy C., van den Hove D.L., Gage F.H., Steinbusch H.W., Rutten B.P. Transcriptional and epigenetic mechanisms of cellular reprogramming to induced pluripotency. Epigenomics. 2016;8:1131–1149. doi: 10.2217/epi-2016-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes S.A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W.J., Davidson E.H., Wong W.H., McMahon A.P. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- Wan J., Goldman D. Retina regeneration in zebrafish. Curr. Opin. Genet. Dev. 2016;40:41–47. doi: 10.1016/j.gde.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Ramachandran R., Goldman D. HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev. Cell. 2012;22:334–347. doi: 10.1016/j.devcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.G., Wen W., Pillai-Kastoori L., Morris A.C. Tracking the fate of her4 expressing cells in the regenerating retina using her4:Kaede zebrafish. Exp. Eye Res. 2016;145:75–87. doi: 10.1016/j.exer.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari N., Ishida T., Kudo A., Kawakami A. Gene expression and functional analysis of zebrafish larval fin fold regeneration. Dev. Biol. 2009;325:71–81. doi: 10.1016/j.ydbio.2008.09.028. [DOI] [PubMed] [Google Scholar]
- Zhang T., Liu W.D., Saunee N.A., Breslin M.B., Lan M.S. Zinc finger transcription factor INSM1 interrupts cyclin D1 and CDK4 binding and induces cell cycle arrest. J. Biol. Chem. 2009;284:5574–5581. doi: 10.1074/jbc.M808843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table shows a list of transcription factors, transcription co-factors and chromatin remodeling factors along with their ENSEMBL IDs, obtained from RNA-seq data that are regulated during various time points, and Shh signaling inhibition with cyclopamine, post-retinal injury.
All the primers used in this study are listed with their code names indicating the genes in the first column and the 5′ to 3′ sequence, in the next column.