Abstract
The in vitro activity of amikacin, gentamicin, kanamycin, tobramycin, neomycin, and netilmicin against 420 Escherichia coli producing extended spectrum β-lactamases (Ec-ESBLs) and 139 Klebsiella pneumoniae producing extended spectrum β-lactamase (Kp-ESBL) collected in two multicenter studies performed in Spain in 2000 and 2006 was determined. The presence of genes encoding aminoglycoside-modifying enzymes (AMEs) and 16S ribosomal RNA (rRNA) methylases [aac(3)-Ia, aac(3)-IIa, aac(3)-IVa, aac(6′)-Ib, ant(2")-Ia, ant(4′)-IIa, aph(3′)-Ia, aph(3′)-IIa, armA, rmtB, and rmtC] was also investigated. The resistance to (one or more) aminoglycosides was significantly higher in Kp-ESBL (104/139, 74.8%) than in Ec-ESBL (171/420, 40.7%; p < 0.0001). The lowest resistance rates for both species in the two studies were observed for amikacin. The prevalence of AME genes was significantly different in Ec-ESBL (161/420, 38.3%) than in Kp-ESBL (115/139, 82.7%; p < 0.0001). The most prevalent AME genes in Ec-ESBL and Kp-ESBL were aac(6′)-Ib (16.2% and 44.6%) and aac(3)-IIa (14.7% and 43.1%), respectively. The expected phenotypic profile correlated with the found AMEs encoding genes in 59.6% Ec-ESBL and 26.1% Kp-ESBL. In Ec-ESBL, aac(6′)-Ib was usually associated in 2000 with blaSHV (26.6%), but with blaCTX-M-1 group (34.8%) in 2006, while aac(3)-IIa was coincident in 2000 with blaTEM (14.6%) and with blaCTX-M-1 group (16.3%) in 2006. Among Kp-ESBL, the aac(6′)-Ib and aac(3)-IIa genes were more frequent in strains with blaTEM (22.0% and 44.0%) in 2000 and with blaCTX-M-1 group (46.4% and 34.0%) in 2006. Resistance to aminoglycosides in Ec-ESBL and Kp-ESBL is frequent and related to production of AMEs; this limits the clinical use of aminoglycosides against these organisms.
Keywords: : aminoglycoside-modifying enzymes, Klebsiella pneumoniae, Escherichia coli, extended spectrum β-lactamases, Spain
Introduction
Enterobacteria (particularly Escherichia coli and Klebsiella pneumoniae) producing extended spectrum β-lactamases (ESBLs) represent a worldwide growing threat to public health.1,2 ESBLs include three major families, TEM, SHV, and CTX-M, and a large variety of other groups of enzymes. During the 1980s and 1990s, TEM-type and SHV-type ESBLs were dominant, but during the current century, a rapid and massive spread of organisms producing CTX-M-type enzymes has been described, and at this moment CTX-M-β-lactamases have become the most prevalent ESBLs worldwide.3 Multiple studies performed in Spain have documented the importance of ESBL production in E. coli and K. pneumoniae (and in other enterobacteria).4–7Changes along time on the relative importance of the major ESBL families were documented in two multicenter Spanish studies performed in 2000 and 2006.4–6 In the first of these two studies, TEM-type enzymes represented 18.6% and 53.8% of the ESBL identified in E. coli and K. pneumoniae, respectively, while in 2006 these figures dropped to 1.2% and 5.3%, respectively. Simultaneously, CTX-M-type enzymes increased from 50.2% to 72.0% in E. coli and from 11.5% to 66.7% in K. pneumoniae, with important variations along time of some precise enzymes, such as CTX-M-14 and CTX-M-15. The high frequency of organisms producing CTX-M-type ESBL in Spain has been confirmed later in new studies.7
ESBL-producing organisms are resistant to penicillins, cephalosporins, and monobactams and even to carbapenems when other mechanisms of resistance (i.e., porin loss, carbapenemases) are also present.8 Unfortunately, they are also frequently resistant to other families of antimicrobial agents, including fluoroquinolones and aminoglycosides.
Bacteria have developed numerous mechanisms of resistance to aminoglycosides: reduced intake into the bacterial cell,9 enzymatic inactivation by enzymes collectively known as aminoglycoside-modifying enzymes (AMEs),10 export outside the cell by active efflux pumps,11 mutation of the 16S ribosomal RNA (rRNA) or ribosomal proteins,12 or methylation of 16S rRNA by methyltransferases (a mechanism found in most aminoglycoside-producing organisms).13
In terms of frequency, the most important determinant of aminoglycoside resistance in E. coli, K. pneumoniae, and many other Gram-negative bacteria is AMEs, of which three classes are defined according to their modifying activities: acetyltransferases (AAC), nucleotidyltransferases (ANT), and phosphotransferases (APH).10 There are multiple enzymes within these families, which differ in their ability to modify aminoglycosides. In addition, one or more enzymes can be present within the same organism. For both reasons, it is very complex to infer from phenotypic data which precise AME(s) is (are) present in a considered bacterial isolate.
In addition, several 16S rRNA methylases have been identified in Enterobacteriaceae of human origin in different geographical locations, with armA, rmtB, and rmtC being the most widespread. High-level resistance (minimum inhibitory concentration [MIC] ≥128 mg/L) to most clinically relevant aminoglycosides has been described as the phenotype conferred by methylases.13
Genes encoding AMEs or methylases can be located on integrons or transposons carried by a variety of plasmids also coding for ESBLs or carbapenemases.10 Efficient mobile elements may have accelerated the simultaneous rapid spread of both ESBL and AME genes.
Because investigating carbapenem-sparing regimens for infections caused by ESBL is a medical need, there is a renewed interest in aminoglycosides as a potential alternative.14 While a large number of publications have provided detailed information on microbiological, clinical, and epidemiological aspects of β-lactam resistance in ESBL-producing enterobacteria, there are less data when considering aminoglycosides. Variations in the percentages of resistance to gentamicin (Gm), tobramycin (To), and amikacin (Ak) of Escherichia coli producing extended spectrum β-lactamase (Ec-ESBL) and K. pneumoniae (Kp-ESBL) tested in the already mentioned 2000 and 2006 Spanish studies have been previously reported, but the underlying mechanisms remain unknown.
The aim of this study was to evaluate the in vitro activity of several aminoglycosides (including clinically relevant compound not tested in the original studies) against Ec-ESBL and Kp-ESBL collected in two nationwide Spanish studies in 2000 and 2006, when a transition from TEM to CTX-M type enzymes occurred, and to screen these strains for the presence of common AME and methylase genes.
Materials and Methods
Bacterial strains
A total of 420 Ec-ESBL and 139 Kp-ESBL clinical isolates obtained in two multicenter studies performed in 40 and 44 Spanish hospitals in 2000 and 2006, respectively, were included in this study. Overall, 168 Ec-ESBL and 70 Kp-ESBL strains were collected in 2000 and 252 Ec-ESBL and 69 Kp-ESBL in 2006. Details on prevalence, detection, and identification of ESBL genes have been described previously.4–6,15,16 To complete this study, in 123 isolates (62 Ec-ESBL and 61 Kp-ESBL) in which ESBL genes were not characterized in the original studies, these genes were identified using the same described methodology.
Antimicrobial susceptibility testing
MICs of amikacin (Ak), gentamicin (Gm), kanamycin (K), tobramycin (To) (all four agents from Sigma-Aldrich, Madrid, Spain), and neomycin (Nm) and netilmicin ([Nt], both compounds from Discovery Fine Chemicals, Wimborne, UK) against the 559 isolates were determined by broth microdilution. Clinical categories for amikacin, gentamicin, tobramycin, and netilmicin were defined using European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (2016).17 In the absence of EUCAST clinical breakpoints for kanamycin and neomycin, both E. coli and K. pneumoniae were considered resistant to those two agents taking into account the EUCAST epidemiological cutoff values (ECOFFs) for E. coli (ECOFFs >8 mg/L).
Molecular characterization of mechanisms of resistance to aminoglycosides
The presence of AME genes, commonly found in enterobacteria, was screened in all isolates by polymerase chain reaction (PCR). The rationale for this approach was to evaluate the possibility that AMEs might be present also in isolates defined as susceptible or intermediate. Previously described primers for the following genes were used: aac(3)-Ia, aac(3)-IIa, aac(3)-IVa, aac(6′)-Ib, ant(2")-Ia, ant(4′)-IIa, aph(3′)-Ia, aph(3′)-IIa, armA, rmtB, and rmtC18 (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/mdr). Twenty-three strains (21 Ec-ESBL and 2 Kp-ESBL) with an amikacin MIC of ≥16 mg/L were subjected to PCR of armA, rmtB, and rmtC methylase genes.
DNA sequencing
Amplicons representative of all AME genes obtained were sequenced using an external resource (Macrogen, Inc., Amsterdam, The Netherlands). The presence of the aac(6′)-Ib-cr variant conferring additional resistance to ciprofloxacin was inferred from the sequence of the corresponding amplicons.
Statistical analysis
The differences between groups were compared using Fisher's exact test. A p value of <0.05 was considered statistically significant.
Results
Antimicrobial susceptibility testing
The activities of the 6 tested aminoglycosides against the 559 strains are summarized in Tables 1 and 2.
Table 1.
In Vitro Susceptibility to Six Aminoglycosides in Escherichia coli Producing Extended Spectrum β-Lactamase Stratified by Year of Collection
| E. coli-ESBL collected in 2000 (n = 168) | E. coli-ESBL collected in 2006 (n = 252) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aminoglycoside | EUCAST clinical breakpointsa | Range | MIC50 (mg/L) | MIC90 (mg/L) | Intermediate | Resistant (%) | Range | MIC50 (mg/L) | MIC90 (mg/L) | Intermediate | Resistant (%) | p |
| Gentamicin | >4 | <0.125 to 128 | 1 | 128 | 10 | 45 (26.8) | <0.125 to >256 | 1 | 128 | 9 | 56 (22.2) | 0.4327 |
| Tobramycin | >4 | <0.125 to 128 | 1 | 32 | 4 | 57 (33.9) | <0.125 to >256 | 1 | 64 | 13 | 66 (26.2) | 0.2136 |
| Amikacin | >16 | 0.5 to 64 | 2 | 16 | 7 | 13 (7.7) | 0.25 to 128 | 2 | 16 | 22 | 8 (3.2) | 0.0669 |
| Kanamycinb | >8 | 1 to >256 | 4 | >256 | NA | 63 (37.5) | 0.5 to >256 | 4 | >256 | NA | 71 (28.2) | 0.1599 |
| Neomycinb | >8 | <0.125 to >256 | 0.5 | 128 | NA | 35 (20.8) | <0.125 to >256 | 1 | 4 | NA | 24 (9.5) | 0.0071 |
| Netilmicin | >4 | <0.125 to 128 | 0.5 | 128 | 2 | 50 (29.8) | <0.125 to 128 | 0.5 | 32 | 8 | 57 (22.6) | 0.2270 |
Statistically significant values are in bold type.
The clinical breakpoints and ECOFFs are done according to the EUCAST guidelines (www.escmid.org/sites).
For kanamycin and neomycin, we used EUCAST ECOFFs.
ECOFFs, epidemiological cutoff values; ESBL, extended spectrum β-lactamase; EUCAST, European Committee on Antimicrobial Susceptibility Testing; MIC, minimum inhibitory concentration; NA, not applicable.
Table 2.
In Vitro Susceptibility to Six Aminoglycosides in Klebsiella pneumoniae Producing Extended Spectrum β-Lactamase Stratified by Year of Collection
| K. pneumoniae-ESBL collected in 2000 (n = 70) | K. pneumoniae-ESBL collected in 2006 (n = 69) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aminoglycoside | EUCAST clinical breakpointsa | Range | MIC50 (mg/L) | MIC90 (mg/L) | Intermediate | Resistant (%) | Range | MIC50 (mg/L) | MIC90 (mg/L) | Intermediate | Resistant (%) | p |
| Gentamicin | >4 | <0.125 to 128 | 8 | 64 | 8 | 37 (52.9) | <0.125 to 128 | 8 | 128 | 1 | 39 (56.5) | 0.8868 |
| Tobramycin | >4 | <0.125 to 128 | 4 | 32 | 14 | 34 (48.5) | <0.125 to 128 | 8 | 32 | 6 | 45 (65.2) | 0.3254 |
| Amikacin | >16 | 0.25 to 32 | 1 | 8 | 5 | 1 (1.4) | 0.25 to 32 | 1 | 8 | 3 | 1 (1.4) | |
| Kanamycinb | >8 | 0.5 to >256 | 4 | 128 | NA | 24 (34.3) | 0.5 to >256 | 32 | >256 | NA | 37 (56.30) | 0.1681 |
| Neomycinb | >8 | <0.125 to 16 | 0.25 | 1 | NA | 1 (1.4) | <0.125 to 16 | 0.25 | 32 | NA | 12 (17.4) | 0.0055 |
| Netilmicin | >4 | <0.125 to 128 | 4 | 32 | 9 | 36 (51.4) | <0.125 to 128 | 8 | 32 | 7 | 39 (56.5) | 0.7757 |
Statistically significant values are in bold type.
The clinical breakpoints and ECOFFs are done according to the EUCAST guidelines (www.escmid.org/sites).
For kanamycin and neomycin, we used EUCAST ECOFFs.
MIC50 and MIC90 values of gentamicin, tobramycin, and amikacin obtained in this study were similar (within one dilution step) to those previously obtained in the original studies of 2000 and 2006.4–6 Globally, the percentages of resistance to at least one aminoglycoside were significantly different for Ec-ESBL (171/420, 40.7%) and for Kp-ESBL (104/139, 74.8%; p < 0.0001). The compound to which lower resistance rates were observed, for both species, in the two studies of 2000 and 2006, was amikacin.
There was a trend to lower resistance to all six aminoglycosides in the Ec-ESBL isolates from 2006 in comparison with those from 2000, but this was only statistically significant for neomycin (20.8% in 2000 vs. 9.5% in 2006, p = 0.0071) (Table 1). This was not the case for Kp-ESBL, where statistically significant differences corresponded to increased resistance rates of neomycin in 2006 (1.4% in 2000 vs. 17.4% in 2006, p = 0.0055) (Table 2).
Sixteen and 13 different resistance phenotypes to aminoglycosides were observed in Ec-ESBL and Kp-ESBL, respectively. The most frequent phenotypes in Ec-ESBL were K, Nm (n = 35, 20.4%) and K, Nt, To (n = 27, 15.8%), while in Kp-ESBL, they were Gm, K, Nt, To (n = 23, 22.1%) and Gm, Nt, To (n = 18, 17.3%). The percentages of isolates resistant to two or more of the tested aminoglycosides were 37.6% (158/420) in Ec-ESBL and 59.7% (83/139) in Kp-ESBL.
Molecular characterization of mechanisms of resistance to aminoglycosides
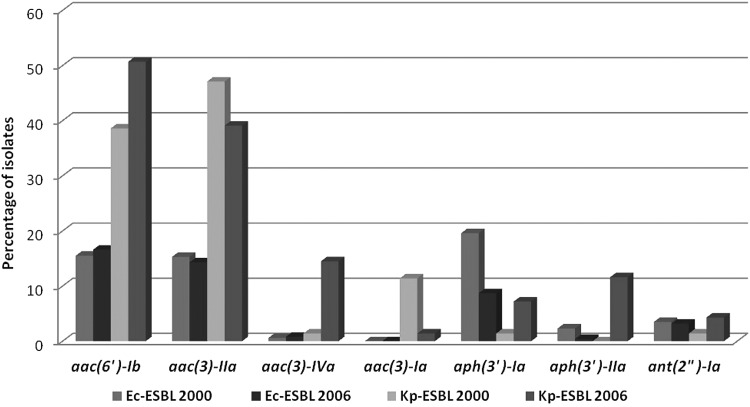
The prevalence of AME genes in the tested isolates and the correlation between expected and observed phenotypes are shown in Fig. 1 and in the Tables 3 and 4.
FIG. 1.
Prevalence of AME genes in Ec-ESBL and Kp-ESBL, stratified by year of collection. AME, aminoglycoside-modifying enzyme; Ec-ESBL, Escherichia coli producing ESBL; ESBL, extended spectrum β-lactamase; Kp-ESBL, K. pneumoniae producing ESBL.
Table 3.
Prevalence of Aminoglycoside-Modifying Enzyme Genes and Correlation with Expected and Observed Antibiograms in 420 Escherichia coli–Producing Extended Spectrum β-Lactamase
| AME gene | No. of isolates (%) | Expected resistance phenotype | Observed resistance phenotype (No. of isolates) |
|---|---|---|---|
| aac(6′)-Ib | 43 (10.2) | Ak, K, Nt, To | As expected (6) |
| K, Nt, To (26) | |||
| Ak, Gm, K, Nt, To (8) | |||
| Gm, K, Nt, To (1) | |||
| To (1) | |||
| K, To (1) | |||
| aph(3′)-Ia | 38 (9.0) | K, Nm | As expected (34) |
| Gm, K, Nm (1) | |||
| Susceptible (3) | |||
| aac(3)-IIa | 24 (5.7) | Gm, Nt, To | As expected (23) |
| Gm (1) | |||
| ant(2")-Ia | 10 (2.4) | Gm, K, To | As expected (10) |
| aac(3)-Iva | 1 (0.2) | Gm, Nt, To | Gm, K, Nt, To (1) |
| aph(3′)-IIa | 1 (0.2) | K, Nm | As expected (1) |
| aac(6′)-Ib, aac(3)-IIa | 21 (5.0) | Ak, Gm, K, Nt, To | As expected (4) |
| Ak, Gm, K, Nm, To (1) | |||
| Gm, K, Nt, To (15) | |||
| K, Nt, To (1) | |||
| aac(3)-IIa, aph(3′)-Ia | 12 (2.8) | Gm, K, Nm, Nt, To | As expected (12) |
| aac(3)-IIa, aph(3′)-IIa | 3 (0.7) | Gm, K, Nm, Nt, To | As expected (2) |
| Gm, K, Nm, To (1) | |||
| aac(6′)-Ib, ant(2")-Ia | 2 (0.5) | Ak, Gm, K, Nt, To | Gm, K, Nt, To (1) |
| Gm, K, To (1) | |||
| aph(3′)-Ia, aac(3)-IVa | 2 (0.5) | Gm, K, Nm, Nt, To | As expected (2) |
| aac(6′)-Ib, aph(3′)-IIa | 1 (0.2) | Ak, K, Nm, Nt, To | Ak, Gm, K, Nm, Nt, To (1) |
| aac(6′)-Ib, aph(3′)-Ia | 1 (0.2) | Ak, K, Nm, Nt, To | Gm, K, Nm, Nt, To (1) |
| ant(2")-Ia, aac(3)-IIa, aph(3′)-Ia | 2 (0.5) | Gm, K, Nm, Nt, To | As expected (2) |
| None | 259 (61.6) | Susceptibles (246) | |
| Gm (11) | |||
| Gm, To (1) | |||
| Ak, Gm, K, Nm, Nt, To (1) | |||
| Total No. of isolates | 420 (100) |
Ak, amikacin; AME, aminoglycoside-modifying enzyme; Gm, gentamicin; K, kanamycin; Nm, neomycin; Nt, netilmicin; To, tobramycin.
Table 4.
Prevalence of Aminoglycoside-Modifying Enzyme Genes and Correlation with Expected and Observed Antibiograms in 139 Klebsiella pneumoniae Producing Extended Spectrum β-Lactamase
| AME gene | No. of isolates (%) | Expected resistance phenotype | Observed resistance phenotype (No. of isolates) EUCAST |
|---|---|---|---|
| aac(6′)-Ib | 28 (20.1) | Ak, K, Nt, To | As expected (0) |
| K, Nt, To (14) | |||
| To (3) | |||
| K, To (3) | |||
| K, Nt (2) | |||
| Gm, K, Nt, To (1) | |||
| K (1) | |||
| Susceptible (4) | |||
| aac(3)-Ia | 9 (6.5) | Gm | As expected (2) |
| Susceptible (7) | |||
| aph(3′)-Ia | 2 (1.4) | K, Nm | As expected (2) |
| aac(3)-IIa | 31 (22.3) | Gm, Nt, To | As expected (13) |
| Gm, To (2) | |||
| Gm, Nt (2) | |||
| Gm (14) | |||
| aac(3)-IVa | 2 (1.4) | Gm, Nt, To | As expected (2) |
| aac(6′)-Ib, aac(3)-IIa | 28 (20.1) | Ak, Gm, K, Nt, To | As expected (1) |
| Gm, K, Nt, To (21) | |||
| Gm, K, Nm, Nt, To (1) | |||
| Gm, Nt, To (3) | |||
| Gm, To (1) | |||
| Gm, Nt (1) | |||
| aph(3′)-IIa, aac(3")-IVa | 7 (5.0) | Gm, K, Nm, Nt, To | As expected (7) |
| aac(6′)-Ib, aph(3′)-Ia | 2 (1.4) | Ak, K, Nm, Nt, To | Gm, K, Nt, To (1) |
| K (1) | |||
| aac(6′)-Ib, aph(3′)-IIa | 1 (0.7) | Ak, K, Nm, Nt, To | K, Nm, Nt, To (1) |
| aac(6′)-Ib, ant(2")-Ia | 1 (0.7) | Ak, Gm, K, Nt, To | K, Nt, To (1) |
| ant(2")-Ia, aac(3)-IVa | 1 (0.7) | Gm, K, Nt, To | As expected (1) |
| aph(3′)-Ia, aac(3)-IVa | 1 (0.7) | Gm, K, Nm, Nt, To | As expected (1) |
| aac(6′)-Ib, ant(2")-Ia, aph(3′)-Ia | 1 (0.7) | Ak, Gm, K, Nm, Nt, To | Gm, K, Nm, Nt, T (1) |
| aac(6′)-Ib, aac(3)-IIa, ant(2")-Ia | 1 (0.7) | Ak, Gm, K, Nt, To | As expected (1) |
| None | 24 (17.26) | Susceptible | As expected (24) |
| Total No. of isolates | 139 (100) |
Among the 420 Ec-ESBL, 161 strains (38.3%) harbored at least one of the investigated AME genes. The most prevalent was aac(6′)-Ib (68 strains, 16.2%), aac(3)-IIa (62 strains, 14.7%), and aph3-Ia (55 strains, 13.1%). Significant differences between strains collected in 2000 versus 2006 were found only for the aph(3)-Ia gene (19.6% vs. 8.7%, respectively; p = 0.006). One hundred and seventeen (72.6%) of the strains with AMEs presented one of the investigated genes and 42 (26.1%) harbored two AME genes. The combination of aac(6′)-Ib and aac(3)-IIa was the most common one (21 isolates, 5.0%), followed by aac(3)-IIa plus aph(3′)-Ia (12 isolates, 2.8%). Two isolates harbored the ant(2")-Ia, aac(3)-IIa, and aph(3′)-Ia (Table 3).
In 246 out of 259 (95.0%) Ec-ESBL not resistant to at least 1 aminoglycoside, none of the investigated AME genes were present.
One hundred and fifteen out of 139 (82.7%) Kp-ESBL strains harbored AME genes, including 72 (62.6%) isolates with 1 of the evaluated genes, 41 (35.6%) isolates with 2 genes, and 2 isolates (1.7%) with 3 genes. The most common genes in this species were aac(6′)-Ib (62 strains, 44.6%), aac(3)-IIa (60 strains, 43.1%), and aac(3)-IVa (11 strains, 7.9%). The combination of aac(6′)-Ib and aac(3)-IIa was observed in 28 (20.1%) isolates and that of aac(3)-IVa plus aph(3′)-IIa in 7 (5.0%) isolates. Significant differences were found in the prevalence of AME genes in strains collected in 2000 and in 2006: aac(3)-IVa 1.4% versus 14.5% (p = 0.010), aac(3)-Ia 11.4% versus 1.4% (p = 0.035), and aph(3′)-IIa 0% versus 11.6% (p = 0.006).
All 24 (17.3%) Kp-ESBL isolates lacking any of the investigated genes were susceptible to all the aminoglycosides tested. Seven aminoglycoside-susceptible Kp-ESBL contained the aac(3)-Ia gene and four contained the aac(6′)-Ib gene.
Overall, the prevalence of AME genes was significantly different in Ec-ESBL (38.3%) and in Kp-ESBL (82.7%; p < 0.0001), especially when considering the genes aac(3)-IVa (1.9% vs. 9.6%; p = 0.0104), aac(3)-Ia (0% vs.7.8%, p = 0.0005), and aph(3′)-Ia (34.1% vs.5.2%, p < 0.0001). None of the 559 isolates contained the ant(4")-IIa gene.
There was a complete concordance between resistance phenotypes and genes detected by PCR in 96 (59.6%) Ec-ESBL and 30 (26.1%) Kp-ESBL. Detailed information for different enzymes is given below.
The aac(6′)-Ib gene was detected in 68 E. coli (42.2%) and 62 K. pneumoniae (53.9%), alone or in combination with other AME genes. The expected resistance phenotype for this enzyme (amikacin, kanamycin, tobramycin, and netilmicin) was only detected in 10/68 (14.7%) Ec-ESBL and 2/62 (3.2%) Kp-ESBL. This was in many cases due to unexpected results for amikacin: among 26 isolates of Ec-ESBL with only aac(6′)-Ib, the MIC of amikacin was 16 mg/L (intermediate) for17 isolates and 8 mg/L (susceptible) for 9 isolates. Similarly, for 28 Kp-ESBL with only aac(6′)-Ib, MICs of amikacin corresponded to the intermediate or susceptible categories in 4 and 24 isolates, respectively. In addition, aac(6′)-Ib was detected in 20 of 21 (95.2%) amikacin-resistant Ec-ESBL. Four K. pneumoniae susceptible to all tested aminoglycosides also contained this resistance determinant.
The aac(6′)-Ib-cr variant, conferring additional resistance to ciprofloxacin, was found in 39 of the 68 (57.3%) Ec-ESBL and in 28 of the 62 (41.9%) Kp-ESBL, all of them collected in 2006. Two Kp-ESBL simultaneously harbored aac(6′)-Ib and aac(6′)-Ib-cr.
The aac(3)-IIa gene was detected in 62 Ec-ESBL (14.7%) and 60 Kp-ESBL (43.1%) and was frequently associated with aac(6′)-Ib. Twenty-three out of 24 (95.8%) Ec-ESBL with aac(3)-IIa alone had a resistance phenotype (gentamicin, tobramycin, and netilmicin) consistent with that described for the encoded enzyme and for them MICs of gentamicin (64–128 mg/L) were higher than those of tobramycin and netilmicin (8–32 mg/L). This correlation between the presence of the gene and the expected phenotype was noted in only 13 of 31 (41.9%) Kp-ESBL, and again in this species, the isolates presented higher MICs of gentamicin than of tobramycin or netilmicin (Table 4).
In Ec-ESBL the third most frequent AME gene was aph(3′)-Ia, detected in 55 (13.1%) isolates. Of the 38 isolates positive only for aph(3′)-Ia gene, 34 (89.5%) had the expected pattern of resistance to kanamycin and neomycin; this gene was also detected in three aminoglycoside susceptible (Table 3). In contrast, this gene was detected in only six (4.3%) Kp-ESBL, all of them resistant to kanamycin and neomycin. Data on other less frequently found genes are shown in Tables 3 and 4.
Although in this study none of the isolates showed high-level resistance (MIC ≥128 mg/L) to all tested aminoglycosides, 23 strains with an amikacin MIC of ≥16 mg/L were screened by PCR for armA, rmtB, and rmtC methylase genes and all of them were negative for these genes.
Correlation between AME genes detected and ESBL types
Ec-ESBL producing enzymes of the CTX-M-1 group were significantly more frequently resistant to an aminoglycoside than susceptible to all six tested compounds, while the reverse was observed for strains producing ESBLs of the CTX-M-9 group. In the case of Kp-ESBL, resistance to at least one compound was more frequent than susceptibility to all six tested drugs in strains producing TEM or CTX-M group-1 ESBLs (Supplementary Table S2). These differences are likely due to different plasmids or plasmid gene content in the indicated group of isolates.
The different AME genes obtained in Ec-ESBL and Kp-ESBL related to ESBL types are shown in Tables 5 and 6. Detailed information on the concrete ESBL genes identified in those isolates is presented in Supplementary Table S3.
Table 5.
Correlation Between Extended Spectrum β-Lactamase Families and Aminoglycoside-Modifying Enzyme Genes Detected in E. coli Producing Extended Spectrum β-Lactamase
| Types of ESBLa(No. of isolates) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ec-ESBL with AMEs from the 2000 study (n = 75) | Ec-ESBL with AMEs from the 2006 study (n = 86) | ||||||||||
| AME genes detectedb(No. of isolates in 2000; 2006) | CTXM-G9 (21) | CTXM-G1 (1) | TEM (17) | SHV (30) | CTXM-G2+SHV (1) | CTXM-G9+SHV (1) | Not identified (4) | CTXM-G9 (32) | CTXM-G1 (35) | CTXM-G9+SHV (4) | SHV (15) |
| aac(6′)-Ib (26; 42) | 1 | 1 | 20 | 1 | 1 | 2 | 7 | 30 | 2 | 3 | |
| aac(3)-IIa (26; 36) | 8 | 11 | 6 | 1 | 11 | 14 | 1 | 9 | |||
| aac(3)-IVa (1; 2) | 1 | 2 | |||||||||
| aph(3′)-Ia (33; 22) | 14 | 1 | 13 | 4 | 1 | 14 | 3 | 1 | 4 | ||
| aph(3′)-IIa (4; 1) | 4 | 1 | |||||||||
| ant(2")-Ia (6; 8) | 1 | 1 | 2 | 2 | 4 | 1 | 2 | 1 | |||
CTXM-G: production of an ESBL of the CTX-M-group indicated by the corresponding figure.
Some strains with two ESBLs and two or three AME genes.
Ec-ESBL, E. coli producing ESBL.
Table 6.
Correlation Between Extended Spectrum β-Lactamase Families and Aminoglycoside-Modifying Enzyme Genes Detected in K. pneumoniae Producing Extended Spectrum β-Lactamase
| Types of ESBLa(No. of isolates) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Kp-ESBL with AMEs from the 2000 study (n = 59) | Kp-ESBL with AMEs from the 2006 study (n = 56) | |||||||
| AME genes detectedb(No. of isolates in 2000; 2006) | TEM (36) | SHV (18) | No identified (6) | CTXM-G1 (38) | CTXM-G9 (5) | TEM (6) | SHV (13) | No identified (1) |
| aac(6′)-Ib (27; 35) | 13 | 9 | 5 | 26 | 3 | 3 | 5 | |
| aac(3)-IIa (33; 27) | 26 | 5 | 3 | 19 | 1 | 2 | 4 | 1 |
| aac(3)-IVa (1; 10) | 1 | 7 | 2 | 2 | 3 | |||
| aac(3)-Ia (8; 1) | 8 | 1 | ||||||
| aph(3′)-IIa (0; 8) | 7 | 3 | ||||||
| aph(3′)-Ia (1; 5) | 1 | 3 | 1 | 1 | 3 | |||
| ant(2")-Ia (1; 3) | 1 | 1 | 1 | 1 | ||||
CTXM-G: production of an ESBL of the CTX-M-group indicated by the corresponding figure.
Some strains with two ESBLs and two or three AME genes.
Kp-ESBL, K. pneumoniae producing ESBL.
In Ec-ESBL with one or more AME genes collected in 2000, the most prevalent ESBLs were the SHV family (42.6%) while in 2006 were the CTX-M-9 or -1 groups (41.8% and 40.7%, respectively).
As shown in the Table 5, the aac(6′)-Ib gene was usually associated in 2000 with SHV-type ESBL (20 isolates, 26.6%) but with CTX-M-1 group enzymes in 2006 (30 isolates, 34.8%). In contrast, the aac(3)-IIa gene was correlated in the collection from 2000 with TEM enzymes (11 isolates, 14.6%) and with CTX-M-1 group ESBLs in 2006 (14 strains, 16.3%). Finally, the aph(3′)-Ia gene was associated with ESBLs of the CTX-M-9 group or TEM family in 2000 (14 and 13 isolates, 18.6% and 17.3%) but with CTX-M-9 group in 2006 (15 strains, 17.4%).
Among Kp-ESBL, CTX-M-type enzymes were only present in strains collected in 2006 (43 isolates, 76.8%) while TEM-type was most prevalent in Kp-ESBL collected in 2000 (36 isolates, 61.0%). In this species, the most frequent AME genes were aac(6′)-Ib and aac(3)-IIa, which in 2000 were more often present in strains with TEM ESBL, while in 2006 they were associated to ESBL of the CTX-M-1 group (Table 6).
Discussion
The percentages of aminoglycoside resistance in this study among Ec-ESBL (24% to gentamicin, 29.3% to tobramycin, and 5% to amikacin) are slightly higher than those obtained for 100 ESBL-E. coli collected in 2004 from a single center in Spain, with rates of resistance of 19% to gentamicin, 17% to tobramycin, and 0% to amikacin.19 In contrast, the overall incidence of aminoglycoside resistance that we have found, particularly to gentamicin and tobramycin, is lower than the observed in many other reports evaluating Ec-ESBL or Kp-ESBL in other countries. Our results contrast with those obtained by Schwaber et al. in 2005 in Israel who found 84% and 11% Kp-ESBL resistant to gentamicin and amikacin, respectively, and 54% and 14% Ec-ESBL resistant to these aminoglycosides.20 Similarly, among 60 ESBL-E. coli collected in Norway, a total of 45%, 57%, and 8.3% of them displayed resistance to gentamicin, tobramycin, and amikacin, respectively.21 In another study in Norway, 63 Ec-ESBL showed rates of resistance of 73%, 94%, and 6% to gentamicin, tobramycin, and amikacin, respectively.22 High resistance rates to amikacin have also been obtained in Kp-ESBLs in Iran (51%), but this was related with the presence of methylase genes.23 It is important to underline the significant geographical variability in resistance, and therefore, aminoglycosides should not be discarded as a potential useful alternative for ESBL without considering the local resistance rates.
The most common AME gene detected in this study was aac(6′)-Ib followed by aac(3)-IIa and aph(3′)-Ia in Ec-ESBL or aac(3)-IVa in Kp-ESBL. In 152 E. coli and 115 K. pneumoniae isolates with MIC ≥2 mg/L to third-generation cephalosporins collected in 7 hospitals in Australia between 2008 and 2009, the most frequent genes were aac(3)-IIa (46.0% and 74.0%), aac(6′)-Ib (3.3% and 21.7%), and ant(2")-Ia (2.0% and 3.4%).24 Our results agree with those from Norwegian researches, who observed that in Ec-ESBL the most prevalent AME gene was aac(6′)-Ib followed by aac(3)-IIa.21 In another study by our group with E. coli resistant to amoxicillin/clavulanic acid collected in seven Spanish hospitals the most common AME gene detected was also aac(6′)-Ib, followed by aph(3′)-Ia, ant(2")-Ia, and aac(3)-IIa.18
We have observed disagreements between resistance phenotypes and corresponding AME genotypes in 65 (40.4%) E. coli and 85 (73.9%) K. pneumoniae isolates. Ginn et al.24 also observed genotypic–phenotypic discrepancies with aminoglycosides in 7.4% of the strains they studied.
Inferring aminoglycoside resistance mechanisms from phenotypic data is particularly complex because the enzymatic modification of an aminoglycoside can be mediated by different enzymes, a single enzyme can modify different drugs of this family and a single isolate can express different AMEs, and very likely regulation of AME genes is not completely understood. In addition, detection of an AME gene does not imply that the organism is resistant to the compounds expected to be modified by the corresponding enzyme.
The discrepancies we have observed have been particularly relevant in relation to the aac(6′)-Ib gene and amikacin, as only 9.9% of the isolates containing just aac(6′)-Ib (and no other AME genes) expressed phenotypic resistance to this aminoglycoside. Similar findings have been previously reported.21,22,25 This could be due to different possibilities: altered gene expression26 or inefficient acetylation, insufficient to completely inactivate the aminoglycoside.27 Amikacin (4 × the MIC) has been shown to be bactericidal against carbapenemase-producing K. pneumoniae strains harboring the aac(6′)-Ib gene, but at serum concentrations that can be achieved in humans, most strains showed regrowth,25 which can be considered an indication that AAC(6′)-Ib actually causes intermediate resistance to amikacin. In addition, the EUCAST recommends that if a member of the Enterobacteriaceae tests as tobramycin intermediate or resistant and gentamicin and amikacin susceptible, its amikacin susceptibility status should be revised to “intermediate,”28 because production of the AAC(6′)-I enzyme may not translate into phenotypic resistance despite modification of amikacin.
The results we have obtained in this study and those obtained by other authors suggest that it would be convenient to reevaluate this expert rule and to obtain additional information on the mechanisms of regulation and expression of aac(6′)-Ib.
It is notable that one E. coli with low-level resistance to all six antibiotics we tested (MIC at the limit of the breakpoints) lacked any of the tested AME genes. It is possible that additional undetected enzymes, decreased permeability, and/or active efflux are involved in this uncommon resistance phenotype. Whole genome sequencing of the organism would likely provide relevant information.
The highest frequency of some AME genes could be partly due to their presence in successful plasmids containing also other resistance genes, which may allow co-selection by different antibiotics. In our study we have detected that 14 out of 36 (38.9%) E. coli producing CTX-M-15 also had the aac(6′)-Ib-cr and aac(3)-IIa genes, in agreement with others studies that have also found this association in similar proportions.23,24
One of the objectives of this study was to analyze whether the important changes observed in the types of ESBL in E. coli and K. pneumoniae occurring in Spain between 2000 and 2006, with a rapid decrease in TEM type and an increase in CTX-M-type ESBLs, were accompanied by similar changes in the prevalence of AME genes. In general terms, this was not the case (Tables 5 and 6), with the observation that the aac(6′)-Ib-cr variant was more frequent in 2006. Additional studies using whole genome sequencing are in progress to gain information on plasmid content in our isolates and on the genes these plasmids contain.
Conclusions
ESBL-producing strains of E. coli and K. pneumoniae isolated in Spain in two multicenter studies in 2000 and 2006 were frequently resistant to gentamicin and tobramycin, but not to amikacin, which was very active in vitro against both Ec-ESBL and Kp-ESBL (95% and 98.5% susceptible isolates, respectively). Resistance to aminoglycosides was predominantly caused by the AME genes aac(6′)-Ib and aac(3)-IIa, which were usually associated with ESBL of the TEM family in isolates collected in 2000 and with those of the CTX-M-1 group in strains of 2006. The strains exhibited a remarkable AME diversity; we have identified 14 AME patterns, which correlated with different levels of aminoglycoside resistance.
For 40.4% E. coli and 73.9% K. pneumoniae, the aminoglycoside resistance phenotype was an inadequate predictor of the AME genotype [particularly when considering the aac(6′)-Ib gene], an indication of the possible contribution of multiple concurrent resistance mechanisms, the insufficient knowledge on regulation and expression of AME genes, and the need of reconsidering currently available breakpoints for aminoglycoside mechanisms.
Concomitant aminoglycoside resistance in ESBL-producing Enterobacteriaceae is of public health interest as aminoglycosides are a neglected therapeutical alternative, but also because inappropriate aminoglycoside use might promote the spread of ESBL genes.29
Supplementary Material
Contributor Information
Collaborators: for the Spanish Network for the Research in Infectious Diseases (REIPI) and the Spanish Group for Nosocomial Infections (GEIH), L. Michaus, C. Martínez Peinado, A. Yagüe, A. Torreblanca, A. Fleites, J.F. Ordás, J.J. Moreno, E. Garduño, J. Gil, A. Oliver, M.A. Domínguez, F. Marco, O. del Valle, F. Navarro, G. Prats, F. Corcoy, E. Ojeda, P. Marín, C. Fernández, L. Martínez, R. Carranza, F. Rodríguez, C. García Tejero, F. Artiles, I. Álamo, B. Palop, M. De la Rosa, J. Gutiérrez, M. Gomáriz, I. Cuesta, M. Cartelle, M. Rodríguez, I. Fernández, E. Ugalde, J.J. Picazo, F. Chaves, R. Cantón, E. Cercenado, L. Folgueira, A. Delgado Iribarren, C. Guerrero, L. Torroba, J.J. García Irure, B. Fernández, M. García, F. Lueiro, I. Otero, E. García Sánchez, J. Elías, M. Treviño, J.R. Hernández, M. Ruiz, M.A. Díaz, A. Moreno, M. Lara, C. Aspiroz, L. Torres, E. García Leoni, D. Navarro, M. Gobernado, A. Tenorio, C. Ezpeleta, J. Castillo, and J. García Moya
Acknowledgments
This study was supported by Ministerio de Ciencia e Innovacion, Instituto de Salud Carlos III, cofinanced by European Development Regional Fund “A way to achieve Europe” ERDF Spanish Network for Research in Infectious Diseases (REIPI RD12/0015) and the Fondo de Investigación Sanitaria (grants PI11/1117 and PI14/01911).
The GEIH study group includes L. Michaus (Vitoria, Álava), C. Martínez Peinado (Villajoyosa, Alicante), A. Yagüe (Orihuela, Alicante), A. Torreblanca (Cangas del Narcea, Asturias), A. Fleites (Oviedo, Asturias), J.F. Ordás (Cangas de Nancea, Asturias), J.J. Moreno (Mérida, Badajoz), E. Garduño (Badajoz), J. Gil (Palma de Mallorca, Baleares), A. Oliver (Palma de Mallorca, Baleares), M.A. Domínguez (Barcelona), F. Marco (Barcelona), O. del Valle (Barcelona), F. Navarro (Barcelona), G. Prats (Barcelona), F. Corcoy (San Pedro de Rivas, Barcelona), E. Ojeda (Burgos), P. Marín (Cádiz), C. Fernández (Santander, Cantabria), L. Martínez (Santander, Cantabria), R. Carranza (Alcázar de S. Juan, Ciudad Real), F. Rodríguez (Córdoba), C. García Tejero (Figueras, Gerona), F. Artiles (Gran Canaria), I. Álamo (Gran Canaria), B. Palop (Granada), M. De la Rosa (Granada), J. Gutiérrez (Granada), M. Gomáriz (San Sebastián, Guipúzcoa), I. Cuesta (Jaén), M. Cartelle (La Coruña), M. Rodríguez (Ferrol, La Coruña), I. Fernández (León), E. Ugalde (Logroño), J. J. Picazo (Madrid), F. Chaves (Madrid), R. Cantón (Madrid), E. Cercenado (Madrid), L. Folgueira (Madrid), A. Delgado Iribarren (Alcorcón, Madrid), C. Guerrero (Murcia), L. Torroba (Pamplona, Navarra), J.J. García Irure (Pamplona, Navarra), B. Fernández (Orense), M. García (Pontevedra), F. Lueiro (Pontevedra), I. Otero (Vigo, Pontevedra), E. García Sánchez (Salamanca), J. Elías (Salamanca), M. Treviño (Santiago de Compostela), J.R. Hernández (Seville), M. Ruiz (Seville), M.A. Díaz (Seville), A. Moreno (Tenerife), M. Lara (Tenerife), C. Aspiroz (Teruel), L. Torres (Teruel), E. García Leoni (Toledo), D. Navarro (Valencia), M. Gobernado (Valencia), A. Tenorio (Valladolid), C. Ezpeleta (Bilbao, Vizcaya), J. Castillo (Zaragoza), and J. García Moya (Zaragoza).
Disclosure Statement
No competing financial interests exist.
References
- 1.Cantón R., Novais A., Valverde A., Machado E., Peixe L., Baquero F., and Coque T.M. 2008. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14:144–153 [DOI] [PubMed] [Google Scholar]
- 2.Chong Y., Ito Y., and Kamimura T. 2011. Genetic evolution and clinical impact in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Genet. Evol. 11:1499–1504 [DOI] [PubMed] [Google Scholar]
- 3.Adler A., Katz D.E., and Marchaim D. 2016. The continuing plague of extended-spectrum β-lactamase-producing Enterobacteriaceae infections. Infect. Dis. Clin. North Am. 30:347–375 [DOI] [PubMed] [Google Scholar]
- 4.Hernández J.R., Martinez-Martinez L., Cantón R., Coque T.M., and Pascual A. 2005. Nationwide study of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases in Spain. Antimicrob. Agents Chemother. 49:2122–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Díaz M.A., Hernández-Bello J.R., Rodríguez-Baño J., Martínez-Martínez L., Calvo J., Blanco J., and Pascual A.; Spanish Group for Nosocomial Infections(GEIH). 2010. Diversity of Escherichia coli strains producing extended-spectrum β-lactamases in Spain: second nationwide study. J. Clin. Microbiol. 48:2840–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz de Alegría C., Rodríguez-Baño J., Cano M.E., Hernández-Bello J.R., Calvo J., Román E., Díaz M.A., Pascual A., and Martínez-Martínez L.; Spanish Group for Nosocomial Infections(GEIH). 2011. Klebsiella pneumoniae strains producing extended-spectrum β-lactamases in Spain: microbiological and clinical features. J. Clin. Microbiol. 49:1134–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merino I., Shaw E., Horcajada J.P., Cercenado E., Mirelis B., Pallare M.A., et al. 2016. CTX-M-15-H 30Rx-ST131 subclone is one of the main causes of healthcare-associated ESBL-producing Escherichia coli bacteraemia of urinary origin in Spain. J. Antimicrob. Chemother. 71:2125–2130 [DOI] [PubMed] [Google Scholar]
- 8.Bush K. 2010. Alarming beta-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 13:558–564 [DOI] [PubMed] [Google Scholar]
- 9.Över U., Gür D., Ünal S., and Miller G.H. 2001. The changing nature of aminoglycoside resistance mechanisms and prevalence of newly recognized resistance mechanisms in Turkey. Clin. Microbiol. Infect. 7:470–478 [DOI] [PubMed] [Google Scholar]
- 10.Ramirez M.S., and Tolmasky M.E. 2010. Aminoglycoside modifying enzymes. Drug Resist. Updat. 13:151–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poole K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10:12–26 [DOI] [PubMed] [Google Scholar]
- 12.Magnet S., and Blanchard J.S. 2005. Molecular insights into aminoglycoside action and resistance molecular insights into aminoglycoside action and resistance. Chem. Rev. 105:477–498 [DOI] [PubMed] [Google Scholar]
- 13.Wachino J.I., and Arakawa Y. 2012. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist. Updat. 15:133–148 [DOI] [PubMed] [Google Scholar]
- 14.Rodríguez-Baño J. 2015. The times they are a-changin’: carbapenems for extended-spectrum-β-lactamase-producing bacteria. Antimicrob. Agents Chemother. 59:5095–5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernández J.R., Pascual Á., Cantón R., and Martínez-Martínez L.; Grupo de Estudio de Infección Hospitalaria(GEIH). 2003. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Spanish hospitals (GEIH-BLEE Project 2000). Enferm. Infecc. Microbiol. Clin. 21:77–82 [PubMed] [Google Scholar]
- 16.Díaz M.A., Hernández J.R., Martínez-Martínez L., Rodríguez-Baño J., and Pascual A.; Grupo de Estudio de Infección Hospitalaria(GEIH). 2009. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Spanish hospitals: 2nd multicenter study (GEIH-BLEE project, 2006). Enferm. Infecc. Microbiol. Clin. 27:503–510 [DOI] [PubMed] [Google Scholar]
- 17.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 6.0, 2016. Available at www.eucast.org (accessed December29, 2016)
- 18.Fernández-Martínez M., Miró E., Ortega A., Bou G., González-López J.J., Oliver A., Pascual A., Cercenado E., Oteo J., Martínez-Martínez L., and Navarro F.; Spanish Network for the Research in Infectious Diseases(REIPI). 2015. Molecular identification of aminoglycoside-modifying enzymes in clinical isolates of Escherichia coli resistant to amoxicillin/clavulanic acid isolated in Spain. Int. J. Antimicrob. Agents 46:157–163 [DOI] [PubMed] [Google Scholar]
- 19.Ruiz del Castillo B., Vinué L., Román E.J., Guerra B., Carattoli A., Torres C., and Martínez-Martínez L. 2013. Molecular characterization of multiresistant Escherichia coli producing or not extended-spectrum β-lactamases. BMC Microbiol. 13:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwaber M.J., Navon-Venezia S., Schwartz D., and Carmeli Y. 2005. High levels of antimicrobial coresistance among extended-spectrum-β -lactamase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 49:2137–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haldorsen B.C., Simonsen G.S., Sundsfjord A., and Samuelsen; Norwegian Study Group on Aminoglycoside Resistance O. 2014. Increased prevalence of aminoglycoside resistance in clinical isolates of Escherichia coli and Klebsiella spp. in Norway is associated with the acquisition of AAC(3)-II and AAC(6′)-Ib. Diagn. Microbiol. Infect. Dis. 78:66–69 [DOI] [PubMed] [Google Scholar]
- 22.Lindemann P.C., Risberg K., Wiker H.G., and Mylvaganam H. 2012. Aminoglycoside resistance in clinical Escherichia coli and Klebsiella pneumoniae isolates from Western Norway. APMIS 120:495–502 [DOI] [PubMed] [Google Scholar]
- 23.Peerayeh S.N., Rostami E., Siadat S.D., and Derakhshan S. 2014. High rate of aminoglycoside resistance in CTX-M-15 producing Klebsiella pneumoniae isolates in Tehran, Iran. Lab. Med. 45:231–237 [DOI] [PubMed] [Google Scholar]
- 24.Ginn A.N., Zong Z., Wiklendt A.M., Thomas L.C., Merlino J., Gottlieb T., van Hal S., Harkness J., Macleod C., Bell S.M., Leroi M.J., Partridge S.R., and Iredell J.R. 2013. Limited diversity in the gene pool allows prediction of third-generation cephalosporin and aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae. Int. J. Antimicrob. Agents 42:19–26 [DOI] [PubMed] [Google Scholar]
- 25.Bremmer D.N., Clancy C.J., Press E.G., Almaghrabi R., Chen L., Doi Y., Nguyen M.H., and Shields R.K. 2014. KPC-producing Klebsiella pneumoniae strains that harbor AAC(6′)-Ib exhibit intermediate resistance to amikacin. Antimicrob. Agents Chemother. 58:7597–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen D., and Murchie A.I.H. 2014. An aminoglycoside sensing riboswitch controls the expression of aminoglycoside resistance acetyltransferase and adenyltransferases. Biochim. Biophys. Acta 1839:951–958 [DOI] [PubMed] [Google Scholar]
- 27.Green K.D., Chen W., and Garneau-Tsodikova S. 2011. Effects of altering aminoglycoside structures on bacterial resistance enzyme activities. Antimicrob. Agents Chemother. 55:3207–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leclercq R., Cantón R., Brown D.F., Giske C.G., Heisig P., MacGowan A.P., Mouton J.W., Nordmann P., Rodloff A.C., Rossolini G.M., Soussy C.J., Steinbakk M., Winstanley T.G., and Kahlmeter G. 2013. EUCAST expert rules in antimicrobial susceptibility testing. Clin. Microbiol. Infect. 19:141–160 [DOI] [PubMed] [Google Scholar]
- 29.Cantón R., and Ruiz-Garbajosa P. 2011. Co-resistance: an opportunity for the bacteria and resistance genes. Curr. Opin. Pharmacol. 11:477–485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.