Abstract
Background: Klebsiella pneumoniae (Kpn) strains are a leading cause of hospital-acquired infections, including ventilator-associated pneumonia. Resistance to antibiotics, biofilm formation, and the production of certain fimbriae play an important role in the pathogenesis.
Aim: We investigated the genetic relatedness, antibiotic resistance, virulence potential, and ability to form biofilms of Kpn strains isolated from hospital-acquired infections (n = 76). Strains were isolated at three major hospitals serving the largest metropolitan urban area in Mexico City, Mexico.
Results: Enterobacterial repetitive intergenic consensus (ERIC)–PCR demonstrated that clonal groups predominate in each hospital. Selected strains chosen from clonal groups (n = 47) were multidrug resistant (MDR, 83%), although the majority (∼70%) were susceptible to carbapenems. All strains produced robust biofilms on abiotic surfaces, and ∼90% harbored adhesin genes fimH, mrkA, and ecpA. The ultrastructure of biofilms was further studied by high-resolution confocal microscopy. The average height of Kpn biofilms on abiotic surfaces was ∼40 μm. We then assessed formation of biofilms on human lung cells, as a surrogate of lung infection. While Kpn strains formed robust biofilms on abiotic surfaces, studies on lung cells revealed attachment to human cells but scarce formation of biofilms. Gene expression studies revealed a differential temporal expression of an adhesin (ecpA) and a capsule (galF) gene when biofilms were formed on different substrates.
Conclusions: Kpn strains isolated from nosocomial infections in Mexico City are MDR, although the majority are still susceptible to carbapenems and form more robust biofilms on polystyrene in comparison to those formed on human cells.
Keywords: : K. pneumoniae, ERIC-PCR, multidrug resistance, biofilms, abiotic surfaces, gene expression
Introduction
Klebsiella pneumoniae (Kpn) is a Gram-negative bacterium ubiquitously found in different environments, including the human body, where it commonly resides in the gastrointestinal tract, skin, and nasopharynx.1,2 When favorable conditions are met, Kpn strains are opportunistic pathogens of humans and animals causing a wide range of hospital-acquired infections, particularly among immunocompromised individuals, including pneumonia, cystitis, pyelonephritis, osteomyelitis, meningitis, bacteremia, and septicemia.1
In the United States, there were ∼800,000 hospital-acquired infections in 2011 leading to U.S. ∼$6 billion in medical expenses.3 In the same study, Kpn was the third most commonly isolated pathogen.3 In developing countries, such as Mexico, hospital-acquired infections display double the prevalence observed in industrialized countries; thus, an estimated ∼21% of hospitalized patients in Mexico will develop a hospital-acquired infection. Hospital-acquired infections caused by Kpn are commonly chronic and difficult to treat due to important virulence traits, including the production of a thick capsule, antibiotic resistance, and biofilm formation.2,4
Kpn strains isolated from hospital-acquired infections often display multidrug-resistant (MDR) phenotypes caused by the presence of extended-spectrum β-lactamases or carbapenemases, making it difficult to choose appropriate antibiotics for the treatment.2 A recent article reports a Kpn strain already resistant to ceftazidime–avibactam, the first antimicrobial approved by the U.S. FDA for the treatment of carbapenem-resistant Kpn strains.5 A survey by Fernández-Canigia and Dowzicky found that Kpn strains isolated from 12 Latin American countries were resistant to cephalosporins, including ceftriaxone (97.8%), ceftazidime (81.1%), cefepime (58.8%), and also to levofloxacin (56.4%), amoxicillin/clavulanic acid (56.3%), and piperacillin/tazobactam (44.7%).6 A very similar trend of resistance to antibiotics by Kpn strains has been reported by different groups for strains isolated in Mexico.7–11 Carbapenems such as imipenem and meropenem, however, are still active against Kpn strains isolated in Latin America (>90%)6 and in Mexico.7–11 On the contrary, strains isolated from outbreaks in different hospitals in Mexico have not only demonstrated resistance to cephalosporins and quinolones, but also to carbapenems.12,13
Kpn strains have the ability to produce biofilms on abiotic surfaces, which is directly associated to hospital-acquired infections, given that structures form, and persist, on indwelling devices.14,15 Biofilm structures protect Kpn from commonly used disinfectants, antibiotics, and the attack of host immune responses near the site of contact between the human body and the indwelling devices.16,17 Kpn biofilms formed on abiotic surfaces have been associated with production of type 1 and type 3 fimbriae4,18,19 and the Escherichia coli common pili (Ecp).20 The capsule, a key virulence factor for pathogenesis, appears to be involved in early stages of biofilm formation as Kpn strains with mutations affecting the production of capsule, had a decreased biomass when compared with the wild-type strain.21,22 Regulation of biofilms has been linked, in part, to a c-di-GMP-dependent MrkH circuit, which upregulates the production of type 3 fimbriae.23,24 Supporting the existence of a tightly regulated mechanism of biofilm formation, studies have demonstrated that biomass varies when Kpn forms biofilms on different abiotic surfaces, including polystyrene, polypropylene, and catheters.23
In this study, we isolated Kpn strains from hospital-acquired infections in three major hospitals of Mexico City, including hospitals attending only children and two hospitals attending all age groups. Strains were mainly isolated from bacteremia cases (53%), followed by ventilator-associated pneumonia (VAP; 17%) and urinary tract infection (UTI; 10%). We investigated the genetic relatedness, antibiotic resistance, virulence potential, and ability to form biofilms of these strains. The clonality was evaluated by enterobacterial repetitive intergenic consensus (ERIC)-PCR. Antibiotic resistance profile and virulence traits were sought. Since biofilms produced in indwelling medical devices have been implicated as a main source of Kpn in hospital-acquired infections, the biofilm phenotype, ultrastructure of biofilms produced by selected strains, and temporal expression of genes involved in biofilm formation were also studied.
Materials and Methods
Isolation and identification of K. pneumoniae strains and bacterial culture media utilized
We collected 76 Kpn strains, in the year of 2013, from hospital-acquired infections (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/mdr) at three different hospitals in Mexico City: National Medical Center “20 de Noviembre” (NMC20N), National Medical Center “La Raza” (NMCR), and the National Institute of Pediatrics (INP). Hospital-acquired infections were determined in each hospital following international standards and criteria published by the Centers for Disease Control and Prevention (CDC).3 Strains were isolated from specimens obtained for routine testing of nosocomial pathogens at the mentioned hospitals, so neither the Institutional Review Board (IRB) approval nor informed consent were required from adult patients, or parents or legal guardians of children. Strains were identified (one strain from each specimen obtained from patients with documented nosocomial infection) at the genus and species level by a conventional method using EnteroPluri-Test system (Liofilchem, Italy) followed by a molecular identification method that amplified a 108 bp-specific fragment of the rpoB gene using primers listed in Table 1. PCR conditions were previously described.25 Reference, genome-sequenced, strain RYC492 was also utilized in this study.26 Bacterial culture media utilized throughout the study included the following: Luria-Bertani (LB) (DIBICO, Mexico), Trypticase soy agar (TSA) (Becton-Dickinson, NJ), and Mueller-Hinton agar plate (Becton-Dickinson).
Table 1.
Primers Used in This Study
| Primer | Sequence (5′–3′) | Target | Expected size (bp) | Reference |
|---|---|---|---|---|
| ERIC 1 | ATGTAAGCTCCTGGGGATTCAC | ERIC sequences | Variable | Versalovic et al.27 |
| ERIC 2 | AAGTAAGTGACTGGGGTGAGC | |||
| rpoF | CAACGGTGTGGTTACTGACG | rpoB | 108 | Chander et al.25 |
| rpoR | TCTACGAAGTGGCCGTTTTC | |||
| fimHF | CGCCTGGTCCTTTGCCTGCA | fimH | 817 | Cruz-Córdova et al.7 |
| fimHR | CTGCACGTTGCCGGCGGTAA | |||
| mrkAF | GTTAACGGCGGCCAGGGCAGCGA | mrkA | 382 | Cruz-Córdova et al.7 |
| mrkAR | AGGTGAAACGCGCGCCATCA | |||
| ecpAF | AATGGTTCACCGGGACATCATGTCC | ecpA | 759 | Cruz-Córdova et al.7 |
| ecpAR | AAGGATGAAATATCGCCGACATCC | |||
| qcfaF | GATGATGGAGAAAGTGAAACCAT | Ecp | 216 | This work |
| qcfaR | CGTCTTATCACCAACACCTTAAC | |||
| qgalFF | CAAAGGCAATTCCAAAGGAG | galF | 156 | This work |
| qgalFR | GCTCGTAGGAGGTGTCGAAG | |||
| qrpoDF | TAAGGAGCAAGGCTATCTGACC | rpoD | 117 | Tan et al.23 |
| qrpoDR | ACCTGAATACCCATGTCGTTG |
Cell line culture
Human lung Calu-3 cells (ATCC HTB-55) were cultured in minimal essential medium (MEM 1 × ) (Gibco by Life Technologies, NY) with 10% of heat-inactivated fetal bovine serum (FBS) and antibiotics (penicillin and streptomycin). Flasks were incubated at 37°C in a 5% CO2 atmosphere. Cells were detached with 0.25% trypsin and resuspended in 10 ml of MEM 1 × .
DNA extraction
All cultures were grown on LB agar plates at 37°C for 18 hr. Genomic DNA of each strain was obtained with the InstaGene Matrix Kit (Bio-Rad, Singapore) as per the instructions provided by the manufacturer. Once purified, genomic DNA was quantified using an AQ-07 AmpliQuant Nucleic Acid Photometer (AmpliQuant) and stored at −80°C until used.
ERIC-typing
DNA fingerprints analysis of Kpn isolates by ERIC-PCR was done as previously described27 using primers listed in Table 1. Band profiles were used to build a binary matrix which was utilized to generate a dendrogram using Jaccard similarity index and the PAST software (version 3.13).
Antimicrobial susceptibility testing
The Kirby–Bauer disc diffusion method was used to investigate susceptibility to antibiotics, according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.28 The following antibiotic discs were used (all purchased from Becton-Dickinson): gentamicin (GM) 10 μg, amikacin (AN) 30 μg, piperacillin+tazobactam (TZP) 100/10 μg, imipenem (IPM) 10 μg, meropenem (MEM) 10 μg, ertapenem (ETP) 10 μg, cefazolin (CZ) 30 μg, ceftriaxone (CRO) 30 μg, ceftazidime (CAZ) 30 μg, cefepime (FEP) 30 μg, cefoxitin (FOX) 30 μg, ciprofloxacin (CP) 5 μg, trimethoprim–sulfamethoxazole (SXT) 1.25/23.75 μg, aztreonam (ATM) 30 μg, ampicillin (AMP) 10 μg, piperacillin (PIP) 100 μg, ampicillin/sulbactam (SAM) 10/10 μg, amoxicillin+clavulanic acid (AMC) 20/10 μg, tetracycline (TE) 30 μg, and nitrofurantoin (F/M) 300 μg. Isolates were regarded as susceptible, intermediate, or resistant using breakpoints set by the CLSI. Strains were further classified as MDR if they were nonsusceptible to ≥1 agent in ≥3 antimicrobial categories and extensively drug resistant (XDR) if strains were nonsusceptible to ≥1 agent in all but ≤2 categories.29 Kpn ATCC 700603 and E. coli ATCC 25922 were used as a control.
Quantification of biofilm biomass
To measure biofilm biomass, we used the classic method with Crystal Violet (0.5% w/v) that has been widely utilized to investigate biofilms of human pathogens, including Kpn.20,30,31 Isolates were cultured on TSA plates overnight at 37°C. These cultures were utilized to make bacterial suspensions adjusted at 0.5 McFarland standard (∼1.5 × 108 CFU/ml) and ∼3 × 105 CFU/ml were inoculated into a 24-well polystyrene plate (Corning Costar, NY) containing trypticase soy broth (Becton-Dickinson) and incubated at 37°C for 24 hr. Planktonic bacteria were removed and biofilms attached to the bottom of each well were washed with sterile distilled water. Biofilms were fixed with 1 μl of glyceraldehyde 2.5% (v/v) and incubated for 3 min at room temperature. Fixed biofilms were washed three times with sterile distilled water and stained with 1 ml of Crystal Violet 0.5% for 10 min at room temperature. After two washes with distilled water, Crystal Violet was solubilized with 2 ml of ethanol–acetone (80/20, v/v) and the absorbance for each well was measured at 570 nm in a spectrophotometer Optimus 10000 xs (Spectronic 20D Genesys). The isolates were classified, based on the optical density of the assay, as high (OD570nm ≥ 0.501), moderate (OD570mn = 0.101–0.500), or low biofilm producers (OD570nm ≤ 0.100). Pseudomonas aeruginosa PAO1, a high biofilm former, was used as a positive control and E. coli K12 was used as a negative control.31,32 Each strain was tested at least five times.
Confocal microscopy analysis of K. pneumoniae biofilms
(a) Biofilms formed on abiotic surfaces. Bacterial suspensions were adjusted at 0.5 McFarland standard (∼1.5 × 108 CFU/ml) and ∼1.5 × 106 CFU were inoculated into a Chamber Slide™ (eight-well glass slide) (Lab-Tek, NY) containing LB broth and biofilms were incubated at 37°C for 24 hr.
(b) Biofilms formed on human lung Calu-3 cells. Polarized, confluent (100%), monolayers of human lung Calu-3 cells grown on an eight-well glass slide (Lab-Tek) were fixed 15 min with 2% paraformaldehyde (PFA) at room temperature, thoroughly washed with phosphate-buffered saline (PBS) to eliminate residual PFA, and MEM containing 5% of heat-inactivated FBS was added. Immobilized human lung cells were infected with ∼1.5 × 106 CFU of Kpn and biofilms were incubated for 24 hr at 37°C.
Whether produced on an abiotic surface or on human lung cells, culture medium was removed and each well was washed twice with PBS and fixed with 2% PFA for 15 min at room temperature. Then, each well was washed three times with PBS and blocked with 2% bovine serum albumin for 1 hr. Biofilms were stained with rabbit polyclonal anti-Klebsiella antibodies (1.1 μg/ml) (Thermo Fisher Scientific, IL) for 1 hr at room temperature followed by a goat anti-rabbit secondary antibody conjugated to Alexa Fluor 555 (Molecular Probes). Bacterial DNA was stained with TOPRO3 (Molecular Probes). Stained biofilms were finally washed three times with PBS, mounted with ProLong Diamond Antifade Mountant (Molecular Probes) and analyzed with an Olympus FV1000 confocal microscope. Confocal images were analyzed with ImageJ version 1.49k (National Institutes of Health) and The Imaris software (Bitplane, CT).
Molecular detection of adhesin-encoding genes
Conventional PCR was used to detect genes encoding for type 1, type 3, or Ecp adhesins, that is, genes fimH, mrkA, or ecpA, respectively. PCR primers are listed in Table 1. The reaction was carried out with 5 μl of 10 × PCR buffer, 1.8 μl of MgCl2 (50 mM), 7.5 μl dNTPs (2 mM), 1 μl of primer F (100 μM), 1 μl of primer R (100 μM), 0.5 μl of Taq polymerase (5 U/μL), 2 μl of DNA (80–100 ng), and enough molecular-grade water to adjust to a total of 50 μl. Reactions were performed under the following conditions: initial denaturing at 92°C for 5 min and 35 cycles of 92°C for 60 sec, 56°C for 45 sec, and 72°C for 45 sec, with a final extension at 72°C for 5 min.
Phenotypic detection of production of siderophores
The Chrome Azurol S (CAS) assay was used to determine the production of siderophores. Approximately 1.5 × 105 CFU of each strain was inoculated into CAS medium and incubated at 37°C for 24 hr. The presence of a yellow or orange halo around the colony in the media was indicative of siderophores production.33 Aeromonas hydrophila ATCC 7966T and Aeromonas caviae Sch3 were used as a control for siderophores production.34
Gene expression studies
We performed gene expression assays using biofilms harvested from six-well plates cultured on an abiotic surface (polystyrene, i.e., plastic), immobilized Calu-3 cells (using 2% PFA), or on live Calu-3 cells. Plates were infected with ∼3.5 × 107 CFU of Kpn RYC492. Planktonic cells and biofilms were obtained at 2, 4, 8, and 24 hr postincubation at 37°C in 5% of CO2 atmosphere. RNA was purified from planktonic cells and biofilms using the RNeasy® Mini Kit (Qiagen, MD) according to the manufacturer's instructions. To eliminate DNA contamination, RNA preps were treated with TURBO DNA-free™ (Ambion, Life Technologies) and 50 ng of purified, DNA-free, RNA was reverse transcribed into cDNA with iScript™ Reverse Transcription system (Bio-Rad). qPCRs with iQ™ SYBR® Green Supermix (Bio-Rad) were used to measure the relative expression of cfa (ecpA homologous gene in Kpn) and galF with primers listed in Table 1. Recommended efficiency of reactions (>90% to <110%)35 using these primers was confirmed. Reactions were carried out with 300 nM primers and 2 μl of cDNA in a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) under the following conditions: 1 cycle at 95°C for 3 min and 40 cycles of 95°C for 15 sec, 57°C for 30 sec, and 72°C for 30 sec. Melting curves were generated by a cycle of 95°C for 1 min, 55°C for 1 min, and 80 cycles starting at 55°C with 0.5°C increments.36,37 Average CT values were normalized to the rpoD gene, and the fold differences were calculated using the comparative CT method (2−ΔΔCT)38 with the Bio-Rad CFX manager software, version 3.0 (Bio-Rad).
Results
Genetic relatedness of K. pneumoniae strains isolated from nosocomial infections across three major hospitals in Mexico City
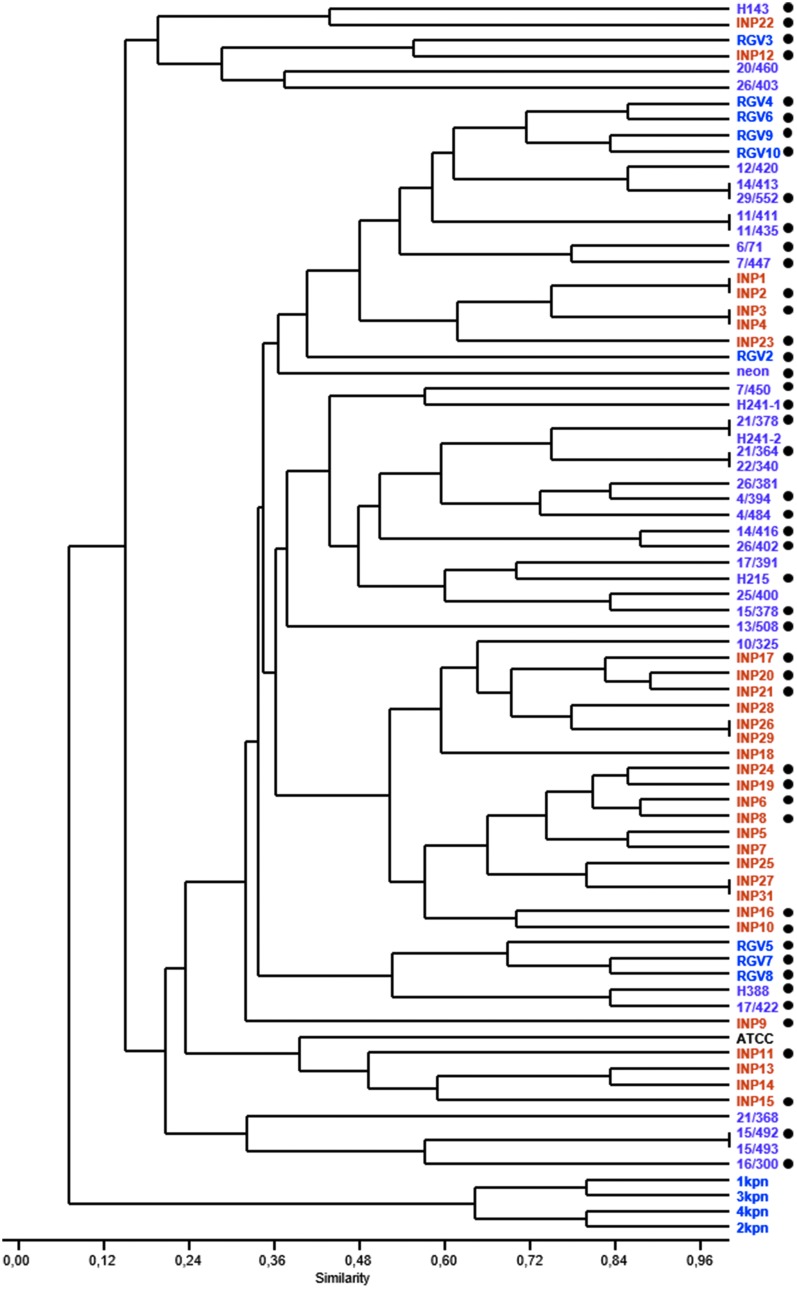
Kpn strains (n = 76) were isolated in three different hospitals, mainly from hospital-acquired infections, including bacteremia (n = 40), VAP (n = 13), UTI (n = 8), wound infection (WI; n = 7), meningitis (n = 4), upper respiratory infection (n = 3), and diarrhea (n = 1) (Supplementary Table S1). Genetic relatedness was investigated by ERIC-PCR analysis and demonstrated 47 unique patterns (Fig. 1 and Supplementary Fig. S1). Strains were positioned in two clusters. The smallest cluster included four Kpn strains isolated from the same hospital (NMCR), three of which were isolated from UTI and one isolated from VAP. The second cluster contained Kpn strains isolated across the three hospitals. Regardless of where the hospital-acquired infection strains were isolated from (e.g., bacteremia, UTI, etc.), genetic relatedness was observed in strains isolated from the same hospital (Fig. 1). For example, Kpn strains isolated at the INP from hospital-acquired infections in children formed a clonal group within the second cluster, separated from strains infecting adults isolated from NMCLR.
FIG. 1.
Genetic relatedness of Klebsiella pneumoniae strains isolated from cases of hospital-acquired infection in Mexico City. K. pneumoniae strains isolated from National Medical Center “20 de Noviembre” (NMC20N, purple), National Medical Center “La Raza” (NMCLR, blue), National Institute of Pediatrics (INP, orange), were analyzed by ERIC-PCR. A matrix that considered the presence, or absence, of PCR products was entered into the PAST software, and a dendrogram was generated. The Jaccard similarity index is shown. A black dot at right indicates K. pneumoniae strains representative from all divergent strains and hospital or isolation that were chosen for further analysis. ERIC, enterobacterial repetitive intergenic consensus.
Antibiotic resistance among K. pneumoniae strains
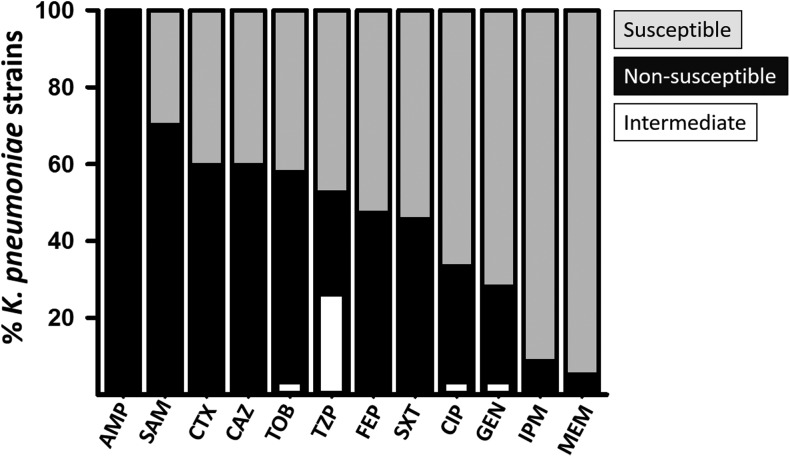
Since antibiotic resistance of Kpn strains isolated from hospital-acquired infections has been increasingly reported,6,14 antibiotic susceptibility was evaluated in 47 representative Kpn strains belonging to most clonal groups identified in Fig. 1. Our studies revealed that >60% of strains were resistant to ampicillin (AMP), piperacillin (PIP), cefazolin (CFZ), piperacillin/tazobactam (TZP), ceftriaxone (CRO), nitrofurantoin (NIT), ampicillin/sulbactam (SAM), amoxicillin/clavulanic acid (AMC), and ceftazidime (CAZ), whereas <30% of strains were resistant to imipenem (IPM), ertapenem (ETP), and cefoxitin (FOX) (Fig. 2 and Supplementary Fig. S2). Based on antibiotic resistance profiles, strains were grouped using the Jaccard similarity index and most strains (n = 38, 80%) were classified as MDR, whereas one Kpn strain showed resistance to all antibiotics (XDR) tested in this study (Supplementary Fig. S2). Only 17% of strains (n = 8) had a wild-type phenotype, that is, they were resistant to less than three categories of antibiotics. Therefore, in this study, we found that ∼83% of Kpn strains tested displayed resistance to multiple antibiotics.
FIG. 2.
Antibiotic susceptibility of K. pneumoniae strains. Selected K. pneumoniae strains (n = 47) were tested for susceptibility to antibiotics as specified in the Materials and Methods section and classified as resistant, intermediate resistance (both grouped within the nonsusceptible group), and susceptible based on criteria established by the CLSI. Antibiotics used were the following: ampicillin (AMP), ampicillin/sulbactam (SAM), cefotaxime (CTX), ceftazidime (CAZ), tobramycin (TOB), piperacillin+tazobactam (TZP), cefepime (FEP), trimethoprim-sulfamethoxazole (SXT), ciprofloxacin (CIP), gentamicin (GEN), imipenem (IPM), and meropenem (MEM). CLSI, Clinical and Laboratory Standards Institute.
K. pneumoniae biofilms
Kpn strains were isolated from hospital-acquired infections, therefore strains may possess virulence factors allowing them to survive in the hospital environment and cause infection. Given that biofilm-related infections account for ∼50% of all nosocomial infections,3 we investigated the ability of strains to form biofilms. Our studies demonstrate that all strains were able to produce biofilms on abiotic surfaces, that is, polystyrene. Most of them produced high biofilm biomass, in comparison to the control strain, and therefore were classified as high biofilm producers (72% [n = 34]) or moderate producers (28% [n = 13]) (Table 2).
Table 2.
Biofilm Phenotype, Detection of Gene Encoding for Adhesins, cas Genes, and Siderophore Production in Klebsiella pneumoniae Strains
| Biofilm phenotype | |||
|---|---|---|---|
| Gene encoding fimbriae or adhesion | High producer (n = 34) n (%) | Medium producer (n = 13) n (%) | Total (n = 47) n (%) |
| fimH | 30 (88.2) | 12 (92.3) | 42 (89.3) |
| mrkA | 31 (91.7) | 13 (100) | 44 (93.6) |
| ecpA | 33 (97.1) | 13 (100) | 46 (97.9) |
| None | 1 (2.1) | 0 (0) | 1 (2.1) |
| cas genes | 3 (6.4) | 1 (2.1) | 4 (8.5) |
| Production of siderophores | 27 (79.4) | 7 (53.8) | 34 (72.3) |
Kpn's ability to form biofilms has been attributed to the production of three main fimbriae named type 1, type 3, and the Ecp.2,20 To assess whether the isolated Kpn strains carry genes involved in the production of these fimbriae, genes fimH, mrkA, and ecpA were screened by PCR as a marker for type 1, type 3, or Ecp, respectively. Genes were carried by >89% of strains (Table 2). Using the χ2 test, there was no statistically significant difference in the carriage of any of the above genes when high biofilm producers were compared with moderate biofilm producers (not shown).
Ultrastructure of biofilms formed by K. pneumoniae strains on abiotic surfaces
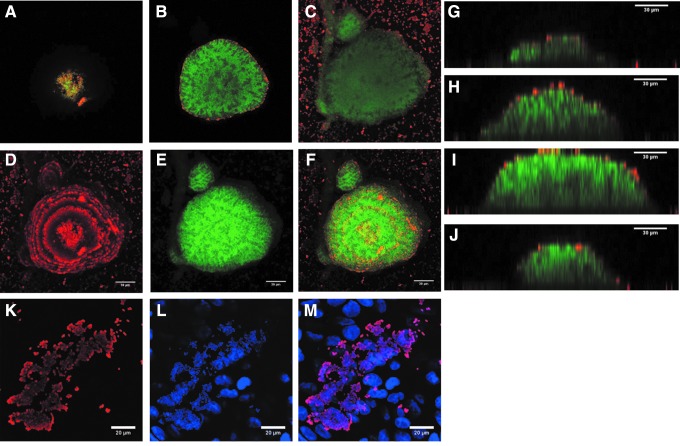
Despite Kpn strains being recognized for their ability to form biofilms, the ultrastructure of Kpn biofilms has not been thoroughly investigated. We selected strains producing high biofilm biomass as detected by Crystal Violet, including reference strain Kpn RYC492. As shown in Fig. 3, biofilms made by strain RYC492 produced structures that initiated concentrically and then matured toward the top, resembling domes. Biofilm structures were built up from specific points of initiation, that is, not all attached bacteria produced biofilm structures (Fig. 3). A few, nonbiofilm-forming, Kpn bacteria adhered to the polystyrene substratum and were detected with anticapsule antibodies; therefore, they were producing capsule. In contrast, capsule staining was only observed on cells surrounding the biofilm structure, but not in those present in the core. Similar observations were made when biofilms of the other three Kpn strains were visualized by confocal microscopy (Fig. 4, left panels). The size of Kpn biofilms ranged from ∼20 μm to as high as 60 μm (Figs. 3 and 4).
FIG. 3.
Ultrastructure of K. pneumoniae biofilms. K. pneumoniae strain RYC492 was incubated for 24 hr on abiotic surfaces (A–J) or on immobilized human lung cells (K–M). Bacteria were stained with an anti-K. pneumoniae antibody followed by an Alexa 555 conjugated secondary antibody and bacterial DNA was stained with TOPRO3, which was pseudocolored in green (A–J) or blue (L, M) for better visualization. Confocal microscopy XY images showing optical sections (A) top, (B) middle, and (C) bottom. XY projections of (D) capsule stain, (E) DNA stain, or (F) merged images. (G–J) Optical sections of YZ images collected across the biofilm structure. XY projection of (K) capsule stain, (L) DNA stain, or (M) merged images.
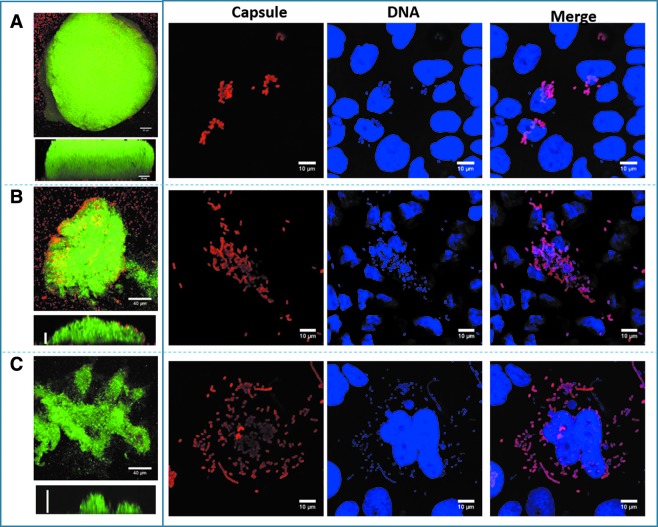
FIG. 4.
K. pneumoniae forms robust biofilms on abiotic surfaces, but flat structures on lung cells. K. pneumoniae strain INP15 (A), RGV2 (B), or 4/484 (C) was incubated for 24 hr on an abiotic surface (left panels) and on immobilized human lung cells (right panel). Bacteria were stained with an anti-K. pneumoniae antibody followed by an Alexa 555 conjugated secondary antibody and DNA was stained with TOPRO3, which was pseudocolored in green (left panels) or blue (right panels) for better visualization. Left panels show an XY projection and underneath a YZ optical middle section from each strain. Right panels show capsule stain, DNA stain, and merged.
Comparison of K. pneumoniae biofilms formed on plastic versus those formed on human cells
Nosocomial, biofilm-forming, Kpn strains ultimately produce human disease by colonizing human epithelia (i.e., the lungs). Strains adhere to in vitro cultures of human cells using fimbriae and the Ecp pili.20 We, therefore, investigated the biofilm phenotype on polarized cultures of human lung Calu-3 cells. The same four strains utilized to inoculate abiotic surfaces, that is, polystyrene plates, were tested. Since Kpn might detach in vitro cultured cells within 24 hr of incubation, our experiments immobilized Calu-3 cell cultures to assure the cell substrate remained attached for Kpn to form biofilms.
In contrast to those large biofilm structures formed by Kpn strains on abiotic surfaces, biofilms on human lung Calu-3 cells were barely detected 24 hr postinoculation (Fig. 4, compare left vs. right panels). Therefore, Kpn forms robust biofilms on abiotic surfaces, but these structures are not produced on human cells within 24 hr.
Gene expression of K. pneumoniae planktonic cells and biofilms
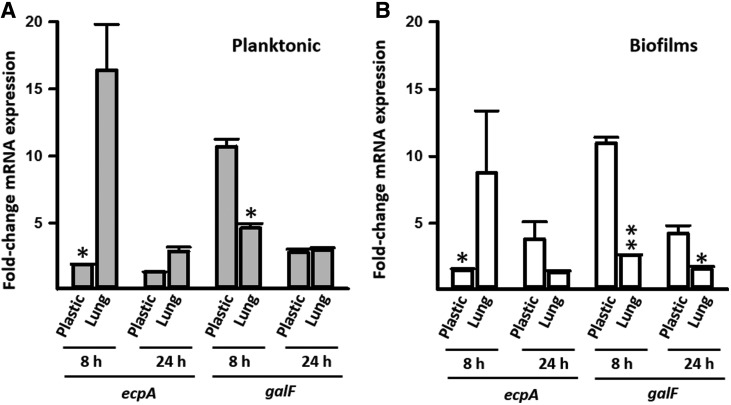
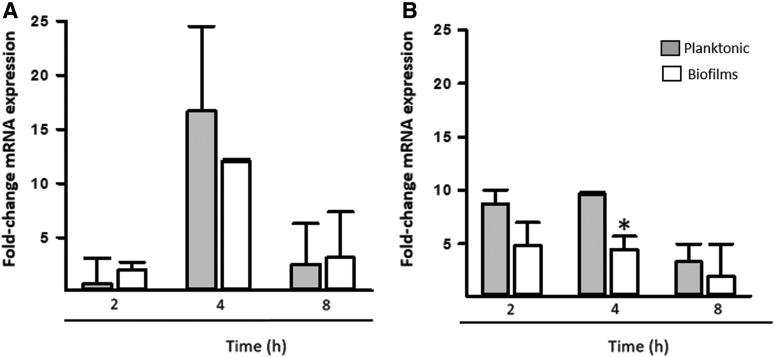
To gain insights into a potential regulatory mechanism controlling the formation of robust biofilms on abiotic surfaces, we evaluated gene expression of planktonic cells and biofilms. We selected the capsule gene, galF, and the gene encoding the Ecp fimbriae (ecpA). Overall transcription of the galF gene was significantly upregulated when on abiotic surfaces compared with immobilized lung cells, whereas transcripts of the ecpA gene were significantly upregulated when Kpn was inoculated onto immobilized lung cells. Upregulated expression of these genes was generally more prominent 8 hr postinoculation than after 24 hr of incubation (Fig. 5). We then studied transcription in living cultures of lung cells. Consistent with Fig. 5 experiments, in this model, the ecpA gene was highly upregulated in comparison to galF (Fig. 6). Upregulated transcription of ecpA at 4 hr postinoculation was similar in planktonic cells in comparison to transcription in biofilms (Fig. 6A), whereas galF transcription was significantly higher in planktonic cells 4 hr postinoculation (Fig. 6B).
FIG. 5.
Transcription of cfa and galF gene of K. pneumoniae planktonic cells and biofilms. K. pneumoniae RYC492 was inoculated on polystyrene plates (plastic), or on immobilized human Calu-3 cells (lung) followed by incubation at 37°C for 8 or 24 hr. (A) Planktonic cells or (B) biofilms were harvested, RNA extracted, and used as a template in qRT-PCRs targeting the ecpA or galF gene. Average Ct values were normalized to the rpoD gene, and the fold differences were calculated using the comparative CT method (2−ΔΔCT).38 Graphs show fold changes in mRNA expression of 8 and 24-hr planktonic cells and biofilms, relative to the inoculum. Error bars represent the standard errors of the mean calculated using data from at least two independent experiments. Gene expression on plastic was compared with that on immobilized lung cells using two-tailed Student t test, *p ≤ 0. 05, **p < 0.006.
FIG. 6.
Transcription of cfa and galF gene of K. pneumoniae inoculated on living cultures of human lung cells. A monolayer of human Calu-3 cells was infected with K. pneumoniae strain RYC492 and incubated for 2, 4, and 8 hr. Planktonic cells (gray boxes) and biofilms (white boxes) were harvested, RNA extracted, and used as the template in qRT-PCRs targeting the (A) ecpA or (B) galF gene. Average Ct values were normalized to the rpoD gene, and the fold differences were calculated using the comparative CT method (2−ΔΔCT).38 Graphs show fold changes in mRNA expression of biofilms and planktonic cells relative to inoculum. Error bars represent the standard errors of the mean calculated using data from at least two independent experiments. Two-tailed Student t test compared gene expression in biofilms versus planktonic cells, at each time point, *p ≤ 0.05.
Production of siderophores
Another important virulence factor of Kpn is the capability to acquire iron from the host. At least 12 distinct iron uptake systems can be identified in Kpn NTUH-K2044, including siderophores. We detected siderophore production phenotypically by CAS assay in 34 out of 47 strains (72.3%) (Table 2). The nature of these siderophores was not investigated further.
Discussion
We demonstrated in this study that Kpn strains isolated from hospital-acquired infections, from Mexico City hospitals, were resistant to first-line antibiotics, although the majority was still susceptible to carbapenems. Genetic studies found that Kpn strains isolated from each hospital were genetically related, indicating a common source of contamination. As expected, given that strains were isolated from nosocomial infections, biofilms were formed by all Kpn strains with the majority (74%) classified as high biofilm producers on abiotic surfaces. Kpn biofilms growing on abiotic surfaces, which mimic conditions found in hospital environments, grew as high as ∼60 μm.
Biofilm structures were not formed when strains colonized human lung Calu-3 cells during the same period, that is, 24 hr, and under the same culture conditions. Limited formation of biofilms within the first 16 hr postinoculation has also been described on human bronchial epithelial cells cultivated in a flow chamber.39 Altogether, these pieces of evidence provide further explanation for the high burden of hospital-acquired infections caused by Kpn, despite the low prevalence of community-acquired infections attributed to these bacteria. Kpn biofilms produced on abiotic surfaces (average ∼40 μm) were larger or comparable to biofilms produced by other human pathogens, including Streptococcus pneumoniae (∼10 μm), and Vibrio cholerae (∼30 μm), respectively.40,41
In 2011, Kpn strains were the third most important cause of hospital-acquired infections in the United States, accounting for 9.9% of all cases; just below two other important nosocomial pathogens: Clostridium difficile (12.1%) and Staphylococcus aureus (10.7%).3 In Mexico, the prevalence of hospital-acquired infections produced by Kpn was similar to that reported in the United States, with 8.5% of cases attributed to Kpn (www.epidemiologia.salud.gob.mx).
Kpn can be involved in a variety of biofilm-related hospital-acquired infections, including pneumonia, surgical-site infections, bacteremia, and UTI.3 Accordingly, most strains utilized in this study were isolated from biofilm-related diseases and were MDR (83%). Similar antibiotic resistance findings have been published by studies conducted in different settings in Mexico.7–11,42,43 The situation in other Latin American countries appears to be similar. For example, a recent survey by Fernández-Canigia and Dowzicky found that Kpn strains isolated from 12 Latin American countries were resistant to cephalosporins, including ceftriaxone (97.8%), ceftazidime (81.1%), cefepime (58.8%), and to levofloxacin (56.4%), amoxicillin/clavulanic acid (56.3%), and piperacillin/tazobactam (44.7%).6
Resistance to most commonly prescribed antibiotics and new generations of cephalosporins is worrisome. Carbapenems such as imipenem and meropenem, however, are still active against Kpn strains isolated in Latin America (>90%)6 and in Mexico (this study and Refs.7–11,42,43). It is expected, however, that antibiotic resistance rates will be maintained at steady levels now that a new law, which requires certified prescriptions for purchasing antibiotics, has been recently implemented in Mexico.44
Besides antibiotic resistance, strains evaluated in this study produced biofilms, with 72% being classified as high biofilm producers. A study by Cruz-Córdoba et al. assessed biofilms of Kpn strains isolated from nosocomial infections in children and demonstrated a similar prevalence.7 Other studies have reported ∼80% of prevalence for strains forming biofilms.17,20 The variance in prevalence observed in other studies may be due to differences in the methodology utilized or selection bias. For example, while the current study, and that one by Cruz-Córdoba et al., only included nosocomial isolates, those published by other groups evaluated strains isolated from different sources, including community-acquired infections.17,20 We hypothesize that strains isolated from nosocomial infection may have an increased ability to form biofilms.
Kpn produces different fimbriae implicated in the production of biofilms. These fimbriae include type 1 and type 3, encoded by the genes fimH and mrkA, respectively.2 Another adhesin known as the E. coli common pilus, or Ecp (encoded by ecpA), has recently been shown to be involved in adhesion of Kpn to host cells and biofilm formation.20 This survey demonstrated that ∼89%, ∼94%, and ∼98% of hospital-acquired Kpn isolates carried the fimH, mrkA, and ecpA genes, respectively. Prevalence of these genes is in agreement with other epidemiological studies. In our survey, however, we did not target the fimA gene, which has been reported to produce another type 1 fimbriae. As expected, given that most strains carried the three genes, no associations could be detected between the presence of individual genes and the biofilm phenotype (not shown).
Biofilms were more robust when Kpn strains grew on abiotic surfaces in comparison to those attached bacteria observed on cultures of human lung cells. It has also been demonstrated that Kpn strains attached better to human lung cells than to intestinal cells,7 further supporting that colonization of the substrate is a tightly regulated process. Moreover, biomass was different when Kpn formed biofilms on different abiotic surfaces, including polystyrene, polypropylene, and catheters.23 Formation of biofilms was regulated by a c-di-GMP-dependent MrkH circuit, which upregulated the production of type 3 fimbriae.23,24
Mutagenesis analysis has suggested that the capsule is required for the formation of Kpn biofilms.21,22 Mutants deficient in capsule production were unable to attach to either glass or polyvinyl chloride substrates, and microscopy studies also revealed that the capsule was required for appropriate initial coverage of the substrate.21 The demonstrated need for the capsule is consistent with the upregulated expression of the capsule gene, galF, observed in gene expression studies presented in this work. Both planktonic Kpn and biofilms upregulated transcription of galF at early stages of biofilm formation on abiotic surfaces, whereas only a modest increase in transcription was observed when Kpn was inoculated onto cultures of human lung cells. In contrast, the ecpA gene was only upregulated when Kpn infected human lung cells, but not during formation of biofilms on polystyrene. These observations support the main role of Ecp for colonization of human HeLa cells, recently demonstrated by Alcantar-Curiel et al. The authors demonstrated that Ecp is produced by all strains when colonizing human cells, but its production when forming biofilms was less clear, that is, only 8% of strains produced the fimbriae on abiotic surfaces.20
The specific mechanism by which Kpn strains regulate production of robust biofilms on abiotic surfaces, versus those “flat structures” of attached bacteria formed on polarized cultures of human lung cells, warrants further investigation. Several lines of evidences suggest the existence of an efficient regulatory mechanism controlling formation of robust biofilms on abiotic substrates, different to that one allowing bacteria to colonize human cells. To gain further insights into this regulatory mechanism, more sophisticated studies are underway in our laboratories.
In conclusion, Kpn strains isolated from hospital-acquired infections from different hospitals located in Mexico City were resistant to most commonly prescribed antibiotics, but a high proportion of isolates remained susceptible to carbapenems. All strains produced biofilms and those structures formed by selected strains were robust on abiotic surfaces when compared with those attached bacteria observed on human lung cells.
Supplementary Material
Acknowledgments
The authors thank Dr. Rosa González-Vázquez from Laboratorio de Hematología Especial, Instituto Mexicano del Seguro Social CMN-La Raza, Hospital de Especialidades, México D.F., México, MSc. Cristina Majalca from Laboratorio de Pruebas Especiales, Centro Médico Nacional (CMN) 20 de Noviembre Instituto de Seguridad y Servicios Sociales de los Trabajadores del Estado (ISSSTE), Mexico City, Mexico for providing strains. Prof. Rosalba Lagos and Dr. Andrés Marcoleta from the Department of Biology, University of Chile, for providing K. pneumoniae strain RYC492 and Dr. Wendy Garret from Harvard Medical School for providing us with purified gDNA from K. pneumoniae isolates. Authors also thank Faidad Khan, and Virginia Stringer from Emory University as well as Adrian Rodríguez from Laboratorio de Bacteriología Médica of Instituto Politécnico Nacional for their assistance in some laboratory methods. Special thanks to David Watson from Emory University for reading and his suggestions to our article. G.C.-E. and J.A.I. received support from Estímulos al Desempeño en Investigación and Comisión y Fomento de Actividades Académicas (Instituto Politécnico Nacional) and Sistema Nacional de Investigadores (SNI, CONACyT). This study was funded by the research Secretaría de Investigación y Posgrado del Instituto Politécnico Nacional (SIP 20160609 and 20161245) and by a grant from the National Institutes of Health (NIH; R21AI112768-01A1 to J.E.V.). The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health. Confocal studies were in part supported by funds from the Integrated Cellular Imaging (ICI) pediatric core and the Emory+Children's Pediatric Research Center to JEV. The SIP-IPN was not involved in the development of the study design, the collection, analysis, and interpretation of data, in the writing of the report, nor in the decision to submit the article for publication. M.L.O.H. had a scholarship from CONACyT and BEIFI.
Disclosure Statement
No competing financial interests exist.
References
- 1.Davis G.S., and Price L.B., 2016. Recent research examining links among Klebsiella pneumoniae from food, food animals, and human extraintestinal infections. Curr. Environ. Health. Rep. 3:128–135 [DOI] [PubMed] [Google Scholar]
- 2.Li B., Zhao Y., Liu C., Chen Z., and Zhou D., 2014. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 9:1071–1081 [DOI] [PubMed] [Google Scholar]
- 3.Magill S.S., Edwards J.R., Bamberg W., Beldavs Z.G., Dumyati G., Kainer M.A., Lynfield R., Maloney M., McAllister-Hollod L., Nadle J., Ray S.M., Thompson D.L., Wilson L.E., and Fridkin S.K. Emerging Infections Program Healthcare-Associated Iinfections and Antimicrobial Use Prevalence Survey Team, 2014. Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med. 370:1198–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy C.N., and Clegg S., 2012. Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation. Future Microbiol. 7:991–1002 [DOI] [PubMed] [Google Scholar]
- 5.Camargo J.F., Simkins J., Beduschi T., Tekin A., Aragon L., Perez-Cardona A., Prado C.E., Morris M.I., Abbo L.M., and Canton R., 2015. Successful treatment of carbapenemase-producing pandrug-resistant Klebsiella pneumoniae bacteremia. Antimicrob. Agents Chemother. 59:5903–5908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Canigia L., and Dowzicky M.J., 2012. Susceptibility of important Gram-negative pathogens to tigecycline and other antibiotics in Latin America between 2004 and 2010. Ann. Clin. Microbiol. Antimicrob. 11:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz-Córdova A., Esteban-Kenel V., Espinosa-Mazariego K., Ochoa S.A., Moreno Espinosa S., de la Garza Elhain A., Fernández Rendón E., López Villegas E.O., and Xicohtencatl-Cortes J., 2014. Pathogenic determinants of clinical Klebsiella pneumoniae strains associated with their persistence in the hospital environment. Bol. Méd. Hosp. Infant. Méx. 71:15–24 [Google Scholar]
- 8.Ares M.A., Alcantar-Curiel M.D., Jimenez-Galicia C., Rios-Sarabia N., Pacheco S., and De la Cruz M.A., 2013. Antibiotic resistance of gram-negative bacilli isolated from pediatric patients with nosocomial bloodstream infections in a Mexican tertiary care hospital. Chemotherapy 59:361–368 [DOI] [PubMed] [Google Scholar]
- 9.Morfin-Otero R., Mendoza-Olazaran S., Silva-Sanchez J., Rodriguez-Noriega E., Laca-Diaz J., Tinoco-Carrillo P., Petersen L., Lopez P., Reyna-Flores F., Alcantar-Curiel D., Garza-Ramos U., and Garza-Gonzalez E., 2013. Characterization of Enterobacteriaceae isolates obtained from a tertiary care hospital in Mexico, which produces extended-spectrum beta-lactamase. Microb. Drug Resist. 19:378–383 [DOI] [PubMed] [Google Scholar]
- 10.Silva-Sanchez J., Garza-Ramos J.U., Reyna-Flores F., Sanchez-Perez A., Rojas-Moreno T., Andrade-Almaraz V., Pastrana J., Castro-Romero J.I., Vinuesa P., Barrios H., and Cervantes C., 2011. Extended-spectrum beta-lactamase-producing enterobacteriaceae causing nosocomial infections in Mexico. A retrospective and multicenter study. Arch. Med. Res. 42:156–162 [DOI] [PubMed] [Google Scholar]
- 11.Alcantar-Curiel D., Tinoco J.C., Gayosso C., Carlos A., Daza C., Perez-Prado M.C., Salcido L., Santos J.I., and Alpuche-Aranda C.M., 2004. Nosocomial bacteremia and urinary tract infections caused by extended-spectrum beta-lactamase-producing Klebsiella pneumoniae with plasmids carrying both SHV-5 and TLA-1 genes. Clin. Infect. Dis. 38:1067–1074 [DOI] [PubMed] [Google Scholar]
- 12.Torres-Gonzalez P., Bobadilla-Del Valle M., Tovar-Calderon E., Leal-Vega F., Hernandez-Cruz A., Martinez-Gamboa A., Niembro-Ortega M.D., Sifuentes-Osornio J., and Ponce-de-Leon A., 2015. Outbreak caused by Enterobacteriaceae harboring NDM-1 metallo-beta-lactamase carried in an IncFII plasmid in a tertiary care hospital in Mexico City. Antimicrob. Agents Chemother. 59:7080–7083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garza-Ramos U., Barrios H., Reyna-Flores F., Sanchez-Perez A., Tamayo-Legorreta E., Ibarra-Pacheco A., Salazar-Salinas J., Nunez-Ceballos R., and Silva-Sanchez J., 2014. Characteristics of KPC-2-producing Klebsiella pneumoniae (ST258) clinical isolates from outbreaks in 2 Mexican medical centers. Diagn. Microbiol. Infect. Dis. 79:483–485 [DOI] [PubMed] [Google Scholar]
- 14.Vuotto C., Longo F., Balice M.P., Donelli G., and Varaldo P.E., 2014. Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens 3:743–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juma NA, and Forsythe S.J., 2015. Microbial biofilm development on neonatal enteral feeding tubes. Adv. Exp. Med. Biol. 830:113–121 [DOI] [PubMed] [Google Scholar]
- 16.Broberg C.A., Palacios M., and Miller V.L., 2014. Klebsiella: a long way to go towards understanding this enigmatic jet-setter. F1000prime Rep. 6:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Fertas-Aissani R., Messai Y., Alouache S., and Bakour R., 2013. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol. Biol. 61:209–216 [DOI] [PubMed] [Google Scholar]
- 18.Stahlhut S.G., Chattopadhyay S., Kisiela D.I., Hvidtfeldt K., Clegg S., Struve C., Sokurenko E.V., and Krogfelt K.A., 2013. Structural and population characterization of MrkD, the adhesive subunit of type 3 fimbriae. J. Bacteriol. 195:5602–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Struve C., Bojer M., and Krogfelt K.A., 2008. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 76:4055–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcantar-Curiel M.D., Blackburn D., Saldana Z., Gayosso-Vazquez C., Iovine N.M., De la Cruz M.A., and Giron J.A., 2013. Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence 4:129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balestrino D., Ghigo J.M., Charbonnel N., Haagensen J.A., and Forestier C., 2008. The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ. Microbiol. 10:685–701 [DOI] [PubMed] [Google Scholar]
- 22.Boddicker J.D., Anderson R.A., Jagnow J., and Clegg S., 2006. Signature-tagged mutagenesis of Klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infect. Immun. 74:4590–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan J.W., Wilksch J.J., Hocking D.M., Wang N., Srikhanta Y.N., Tauschek M., Lithgow T., Robins-Browne R.M., Yang J., and Strugnell R.A., 2015. Positive autoregulation of mrkHI by the cyclic di-GMP-dependent MrkH protein in the biofilm regulatory circuit of Klebsiella pneumoniae. J. Bacteriol. 197:1659–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilksch J.J., Yang J., Clements A., Gabbe J.L., Short K.R., Cao H H., Cavaliere R., James C.E., Whitchurch C.B., Schembri M.A., Chuah M.L., Liang Z.X., Wijburg O.L., Jenney A.W., Lithgow T., and Strugnell R.A., 2011. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7:e1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chander Y., Ramakrishnan M., Jindal N., Hanson K., and Goyal S.M., 2011. Differentiation of Klebsiella pneumoniae and K. oxytoca by multiplex polymerase chain reaction. Int. J. Appl. Res. Vet. Med. 9:138 [Google Scholar]
- 26.Marcoleta A., Gutierrez-Cortez S., Maturana D., Monasterio O., and Lagos R., 2013. Whole-genome sequence of the microcin E492-producing strain Klebsiella pneumoniae RYC492. Genome Announc. 1:e00178-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Versalovic J., Koeuth T., and Lupski J.R., 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute. 2013. Performance Standards for Antimicrobial Susceptibility Testing—Approved Standard M100-S23. 23rd ed. CLSI, Wayne, PA [Google Scholar]
- 29.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., Paterson D.L., Rice L.B., Stelling J., Struelens M.J., Vatopoulos A., Weber J.T., and Monnet D.L., 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18:268–281 [DOI] [PubMed] [Google Scholar]
- 30.Vidal J.E., Ludewick H.P., Kunkel R.M., Zahner D., and Klugman K.P., 2011. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect. Immun. 79:4050–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christensen G.D., Simpson W.A., Younger J.J., Baddour L.M., Barrett F.F., Melton D.M., and Beachey E.H., 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Toole G.A. 2011. Microtiter dish biofilm formation assay. J Vis Exp. 47:2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwyn B., and Neilands J.B., 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47–56 [DOI] [PubMed] [Google Scholar]
- 34.Funahashi T., Tanabe T., Miyamoto K., Tsujibo H., Maki J., and Yamamoto S., 2013. Characterization of a gene encoding the outer membrane receptor for ferric enterobactin in Aeromonas hydrophila ATCC 7966(T). Biosci. Biotechnol. Biochem. 77:353–360 [DOI] [PubMed] [Google Scholar]
- 35.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., and Wittwer C.T., 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 36.Vidal J.E., Shak J.R., and Canizalez-Roman A., 2015. The CpAL quorum sensing system regulates production of hemolysins CPA and PFO to build Clostridium perfringens biofilms. Infect. Immun. 83:2430–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shak J.R., Ludewick H.P., Howery K.E., Sakai F., Yi H., Harvey R.M., Paton J.C., Klugman K.P., and J.E., 2013. Novel role for the Streptococcus pneumoniae toxin pneumolysin in the assembly of biofilms. MBio 4:e00655-00613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak K.J., and Schmittgen T.D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 39.Jagnow J., and Clegg S., 2003. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology. (Reading, England). 149:2397–2405 [DOI] [PubMed] [Google Scholar]
- 40.Drescher K., Dunkel J., Nadell C.D., van Teeffelen S., Grnja I., Wingreen N.S., Stone H.A., and Bassler B.L., 2016. Architectural transitions in Vibrio cholerae biofilms at single-cell resolution. Proc. Natl. Acad. Sci. USA. 113:E2066–E2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidal J.E., Howery K.E., Ludewick H.P., Nava P., and Klugman K.P., 2013. Quorum-sensing systems LuxS/autoinducer 2 and Com regulate Streptococcus pneumoniae biofilms in a bioreactor with living cultures of human respiratory cells. Infect. Immun. 81:1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miranda G., Castro N., Leanos B., Valenzuela A., Garza-Ramos U., Rojas T., Solorzano F., Chihu L., and Silva J., 2004. Clonal and horizontal dissemination of Klebsiella pneumoniae expressing SHV-5 extended-spectrum beta-lactamase in a Mexican pediatric hospital. J. Clin. Microbiol. 42:30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosqueda-Gomez J.L., Montano-Loza A., Rolon A.L., Cervantes C., Bobadilla-del-Valle J.M., Silva-Sanchez J., Garza-Ramos U., Villasis-Keever A., Galindo-Fraga A., Palacios G.M., Ponce-de-Leon A., and Sifuentes-Osornio J., 2008. Molecular epidemiology and risk factors of bloodstream infections caused by extended-spectrum beta-lactamase-producing Klebsiella pneumoniae A case-control study. Int. J. Infect. Dis. 12:653–659 [DOI] [PubMed] [Google Scholar]
- 44.Dreser A., Vazquez-Velez E., Trevino S., and Wirtz V.J., 2012. Regulation of antibiotic sales in Mexico: an analysis of printed media coverage and stakeholder participation. BMC Public Health 12:1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.