Summary
Aging is associated with tissue-level changes in cellular composition that are correlated with increased susceptibility to disease. Aging human mammary tissue shows skewed progenitor cell potency, resulting in diminished tumor-suppressive cell types and the accumulation of defective epithelial progenitors. Quantitative characterization of these age-emergent human cell subpopulations is lacking, impeding our understanding of the relationship between age and cancer susceptibility. We conducted single-cell resolution proteomic phenotyping of healthy breast epithelia from 57 women, aged 16–91 years, using mass cytometry. Remarkable heterogeneity was quantified within the two mammary epithelial lineages. Population partitioning identified a subset of aberrant basal-like luminal cells that accumulate with age and originate from age-altered progenitors. Quantification of age-emergent phenotypes enabled robust classification of breast tissues by age in healthy women. This high-resolution mapping highlighted specific epithelial subpopulations that change with age in a manner consistent with increased susceptibility to breast cancer.
Keywords: human mammary epithelia, aging, mass cytometry, single-cell analysis, heterogeneity, breast cancer
Graphical Abstract
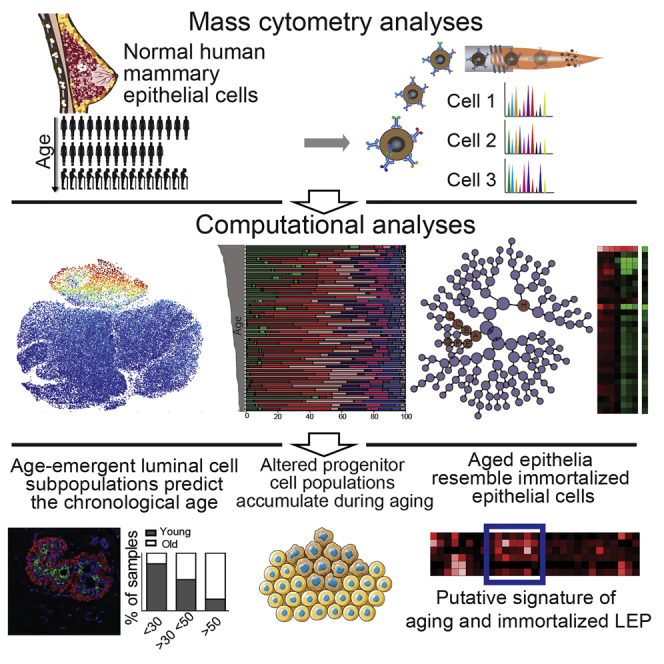
Highlights
-
•
CyTOF analysis reveals human mammary epithelial heterogeneity with age
-
•
Age-emergent luminal cells share phenotypes with candidate breast cancer cells of origin
-
•
Classification models correctly assign tissue samples to their age group
-
•
Age-related changes are conserved between mammary epithelial tissue and primary cells
Vatter et al. find that single-cell mass cytometry of human mammary epithelial cells from 57 women, from 16 to 91 years old, depicts an in-depth phenotyping of aging mammary epithelia. Subpopulations of altered luminal and progenitor cells that accumulate with age may be at increased risk for oncogenic transformation.
Introduction
Adult tissue stem and progenitor epithelial cells generate differentiated daughter cells for tissue remodeling and homeostasis (Biteau et al., 2008, Mansilla et al., 2011). Evidence suggests skewed stem cell function contributes to diseases of aging (Sharpless and DePinho, 2007). Human breast epithelium, comprised of apical luminal epithelium (LEP) and basal myoepithelium (MEP) cell layers surrounded by a basement membrane, undergoes remarkable growth and remodeling between puberty and menopause and during lactation, supported by stem and progenitor cells. The greatest risk factor for breast cancer is age and exclusively genetic explanations are inadequate (LaBarge et al., 2016, Stephens et al., 2012). Differentiation-defective progenitor cells accumulate and tumor-suppressive MEPs decline with age, while older LEPs display basal properties, such as nuclear-localized YAP and MEP gene expression (Chen et al., 2014, Garbe et al., 2012, Pelissier et al., 2014, Skibinski et al., 2014). We hypothesize that these age-associated changes elevate cancer risk. Congruently, LEPs from women with high cancer risk (e.g., BRCA1/2 carriers) show basal characteristics, and luminal progenitors with a basal phenotype are suggested cells of origin for murine mammary adenocarcinoma (Lim et al., 2009, Molyneux et al., 2010, Proia et al., 2011). We therefore sought to gain insight into molecular changes in the mammary epithelium during aging and comprehensively catalog age-emergent phenotypic diversity using mass cytometry (Bandura et al., 2009) in samples from women aged 16–91 years old.
Results
High-Dimensional Analysis of Cellular Heterogeneity within Human Mammary Epithelia
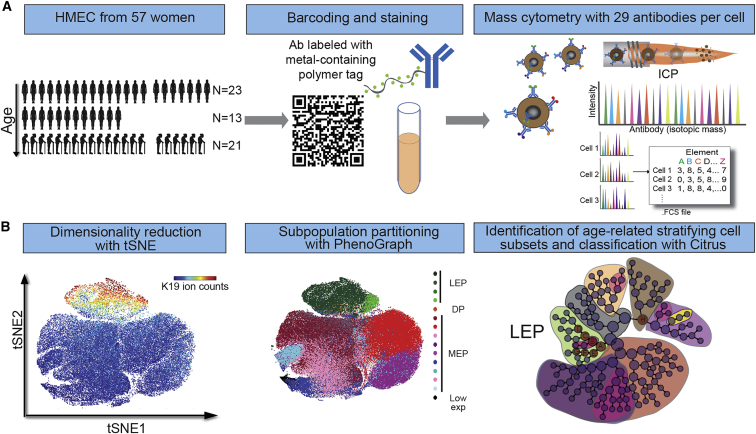
To measure age-emergent phenotypic diversity in the human breast, we used mass cytometry to obtain single-cell proteomic profiles of cryopreserved normal primary human mammary epithelial cell (HMEC) strains at passage four, from 44 women of ages 16 to 91 years old (Figure 1; Table S1). A 29-antibody panel recognizing human mammary epithelial lineage markers and intracellular signaling proteins was used to establish high-dimensional phenotypes of single HMECs (Figure S1; Table S2) (dos Santos et al., 2013, LaBarge et al., 2007, Lim et al., 2009, Regan et al., 2012, Taylor-Papadimitriou et al., 1989, Villadsen et al., 2007). Simultaneous analysis of ≥20,000 HMECs from each of the women from three age groups (<30 years, n = 16; >30 years < 50 years, n = 13; and >50 years, n = 15) measured 29 protein epitope dimensions (Ds) (Figure 1A). Non-linear dimensionality reduction, t-distributed stochastic neighbor embedding (tSNE) (Amir et al., 2013), created a 2D map of the entire dataset at single-cell resolution with similar phenotypes proximal to each other (Figure 1B). Distinct LEP (K19+/K7+/K8/18+/CD133+) and MEP (K14+/K5/6+) cell populations were distinguishable on the tSNE map (Figure 2A). Several signaling markers showed lineage dependence, especially in MEPs; CD44, YAP, phospho-epidermal growth factor receptor (pEGFR), pStat1, pS6, and p-Phospholipase C Gamma 2 (pPLCγ2), previously implicated in myoepithelial function and contractility (Pasic et al., 2011), were prevalent in the MEP cell population (Figures 2A, S1B, and S1C). The partial superposition of K14+ and K5/6+ cell population revealed the high degree of heterogeneity. A small subpopulation of cells with low marker expression also was noted, which may have corresponded to the K14−/K19− epithelia previously described (Villadsen et al., 2007), and it was not further examined.
Figure 1.
Mass Cytometry Analysis of Human Mammary Epithelial Cells
(A) Summary of experimental design. 57 samples of HMECs from women aged 16 to 91 years old (comprising 13 uncultured breast epithelia samples and 44 primary cultured HMEC strains at passage 4 [p4]) were barcoded and stained using a panel of 29 antibodies labeled with isotope tags and analyzed using mass cytometry.
(B) Strategy to analyze high-dimensional single-cell data and identify lineage and age-related phenotypic divergence.
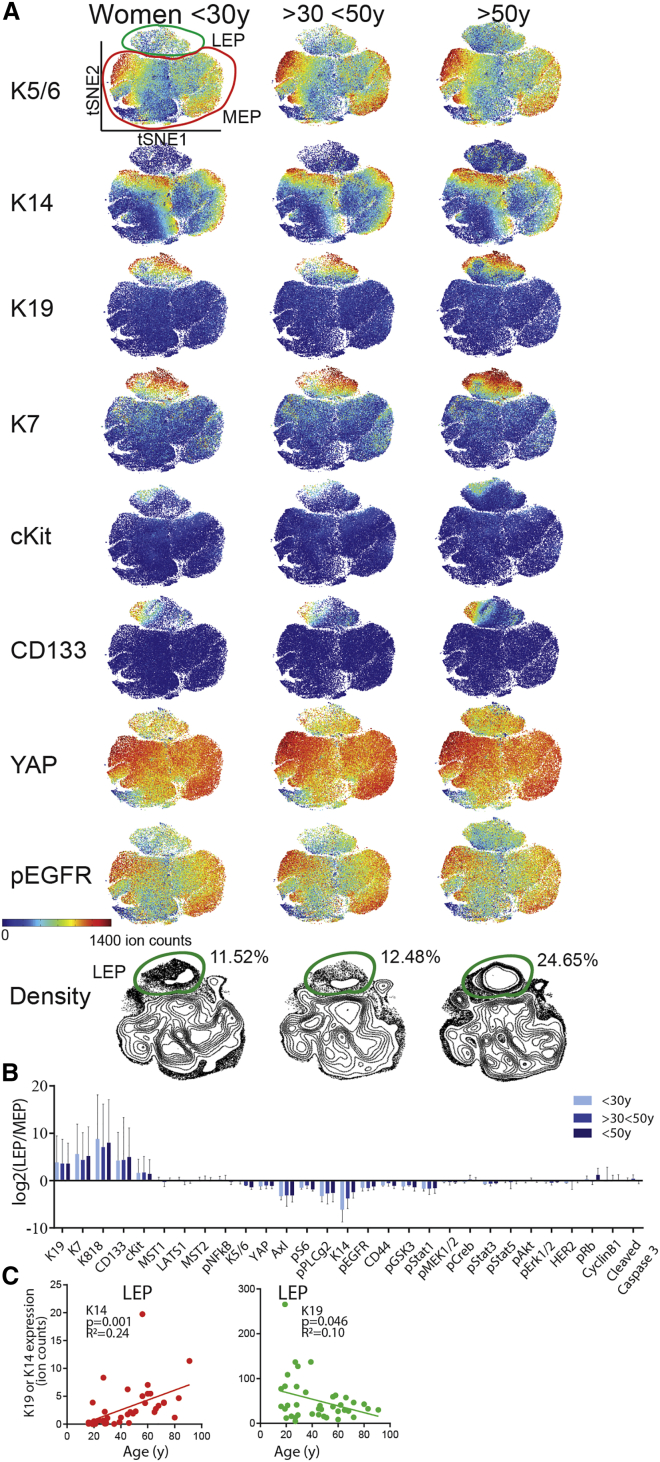
Figure 2.
Collective tSNE Analysis Distinguishes Major Luminal and Myoepithelial Lineages
(A) The raw data have been transformed with arcsinh with the cofactor of 5. tSNE maps from HMECs at p4 from women <30 years old (merged and subsampled at 50,000 cells, n = 16), >30 < 50 years old (n = 13), and >50 years old (n = 15).
(B) Log2 fold change of marker expression of LEP over MEP manually gated from tSNE projection map in HMECs from women <30 years old, >30 < 50 years old, and >50 years old. Data are log2 of ratio of median ± SD.
(C) K19 and K14 expression in LEP as a function of age. 250MK, 90P, 245AT, 173T, and an outlier 42P were excluded from the analysis.
See also Figures S1–S3.
To address how aging affects the human mammary epithelium, we first demarcated LEP and MEP populations using a manual K19 gate (Figure 2A). K19, K7, K8/18, CD133, and cKit expression was high in LEPs, while K14 and K5/6 expression was higher in MEPs (Figure 2B). LEPs exhibited higher phospho-nuclear factor κB (pNF-κB), which is implicated in mouse mammary epithelial proliferation and branching (Brantley et al., 2001). MEPs had increased expression of basal markers (Axl, pS6, pPLGγ2, pEGFR, CD44, pGsk3, and pStat1) involved in myoepithelial homeostasis (Pasic et al., 2011). The phenotypic space projections on the tSNE maps were similar among the three age groups (Figure 2A), however, the expression levels of a number of markers changed significantly with age (Figures S2 and S3). The most prominent age-related difference was observed in the LEP population, where K14 and YAP expression increased and K19 and K7 decreased with age (Figures 2C and S3B). Overall these results revealed remarkable phenotypic heterogeneity within the mammary epithelia.
Intra-lineage and Age-Related Phenotypic Divergence in HMECs
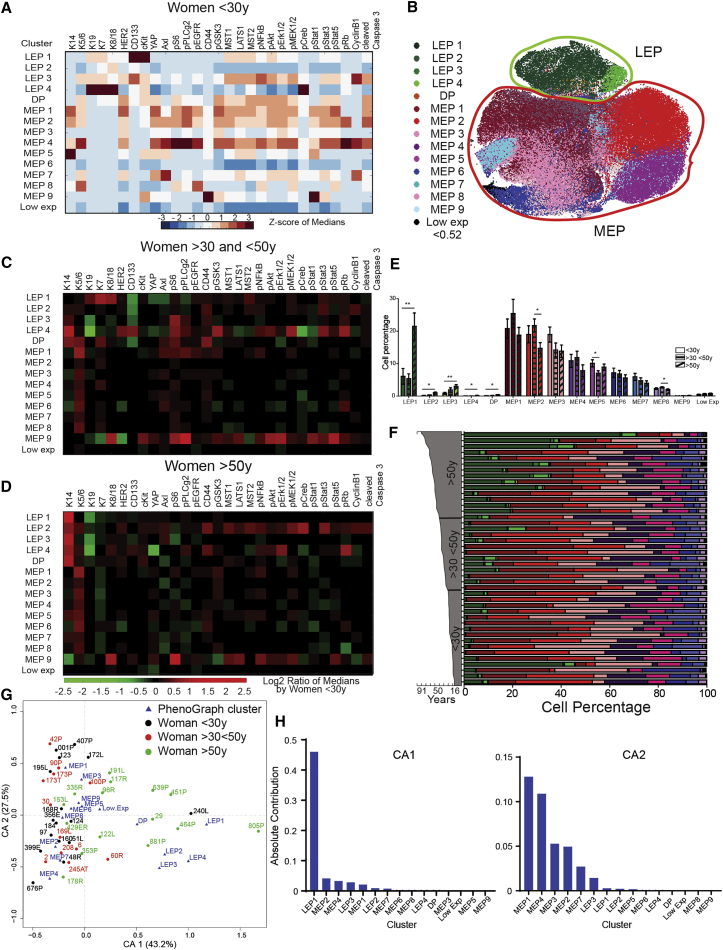
The tSNE map displayed regions of cell density (Figure 2A) and K14, K5/6, pRb, and CyclinB1 expression within the MEP lineage, indicative of distinct cellular subpopulations (Figures 2A and S1C). Intra-lineage subpopulations were identified as distinct clusters of cells with shared phenotypes using PhenoGraph (Levine et al., 2015) (Figures 3A and S4A). There were four LEP (LEP1–LEP4) and nine MEP (MEP1–MEP9) clusters (Figure 3B). One subpopulation, denoted double positive (DP, between 120 and 503 non-doublet cells), co-expressed K14 and K19 in a separate phenotypic space between the LEP and MEP populations. This DP population likely comprised epithelial progenitors (Villadsen et al., 2007). A small subpopulation of cells with low marker expression was not further examined (<0.52 ion counts per cell). The LEP3 cluster showed high levels of pRb and CyclinB1, indicative of higher proliferation compared to the other LEP subpopulations (reviewed in Giacinti and Giordano, 2006). Clusters MEP4 and MEP7 expressed higher levels of CyclinB1 that correlated with higher DNA content (iridium intercalator counts; Figure S4B) compared to the other MEP subpopulations.
Figure 3.
Age-Related Phenotypic Divergence in the Landscape of HMECs
(A) Heatmaps of marker expression in PhenoGraph clusters of HMECs from women <30 years old (Z score scale, merged, n = 16) (excluding 250MK, 90P and 245AT, 173T).
(B) tSNE projection of the PhenoGraph clusters identified with PhenoGraph identified in (A), colored by cluster.
(C and D) Heatmaps of marker expression in each PhenoGraph cluster in HMECs from (C) women >30 and <50 years old and (D) women >50 years old, normalized to values from <30-year-old women.
(E) Plots of cell percentage in each PhenoGraph cluster (excluding 250MK, 90P and 245AT, 173T). Data are mean ± SEM.
(F) Intra-sample heterogeneity for each woman is represented graphically by a horizontal bar in which segment lengths represent the proportion of the sample assigned to each cluster, colored accordingly (excluding 250MK).
(G) The first two components of correspondence analysis (CA), accounting for 70% of the co-association structure between PhenoGraph subpopulations and different strains. Proximity among women and among clusters indicates similarity, however, only a small angle connecting a woman and a cluster to the origin indicates an association. The angle between women <50 years old and LEP was statistically smaller than the angle between women <30 years old and women >30 and <50 years old and LEP (t test, p < 0.001). PhenoGraph subsets are displayed as triangles and HMEC samples as circles.
(H) Contributions of the PhenoGraph subpopulations to CA-1 and CA-2.
See also Figure S4.
Age-related changes in marker expression were observed mainly within the LEP subpopulations. Heatmaps of marker expression in each PhenoGraph cluster, in HMECs from women >30 and <50 years old (Figure 3C) and women >50 years old (Figure 3D), were normalized to values from <30-year-old women to highlight age-related changes. Increased K14 and decreased K19 expression was observed with age in LEP2, LEP3, and LEP4 clusters from women >30 and <50 years old and in all LEP subpopulations from women >50 years old. In addition to phenotypic changes with age, the abundance of the LEP clusters significantly increased, whereas abundance of MEP2, MEP5, and MEP8 clusters significantly decreased with age (Figure 3E). This trend was observed at the individual level, with high inter-sample heterogeneity (Figure 3F). We previously reported age-related changes in LEP and MEP cells in vivo based on K14/K19 staining, and 4 lineage markers (Garbe et al., 2012) did not discern the degree of heterogeneity apparent in this new analysis. Prominent changes in marker expression and abundance occurred in three of four LEP types as early as middle age, and all four types change beyond 50 years. Indeed, the abundance of LEP1 increased more than 3-fold. Decreased abundance of MEP also was type specific.
Correspondence analysis (CA) provided a global understanding of the relationships between all PhenoGraph clusters and the age factor (Härdle and Simar, 2007). CA reduces high-dimensional observations to a smaller set of explanatory components, allowing visualization of data on each woman and PhenoGraph subsets in the same space (Figure 3G). Women >50 years old were associated with LEP1–4 subsets and women <30 years old were associated with MEP1–9 subsets, probably reflecting the relative abundance of those lineages with age. The DP subset, which represents progenitor cells, was associated mainly with older women. The first component, contributing 43.2% and comprising mainly LEP1, captured the tendency of older women to have more LEP (Figures 3G and 3H). The second component (27.5%) provided a different ordering. Altogether, there was a significant association between an age-dependent luminal subset and the chronological age of the primary epithelial cells.
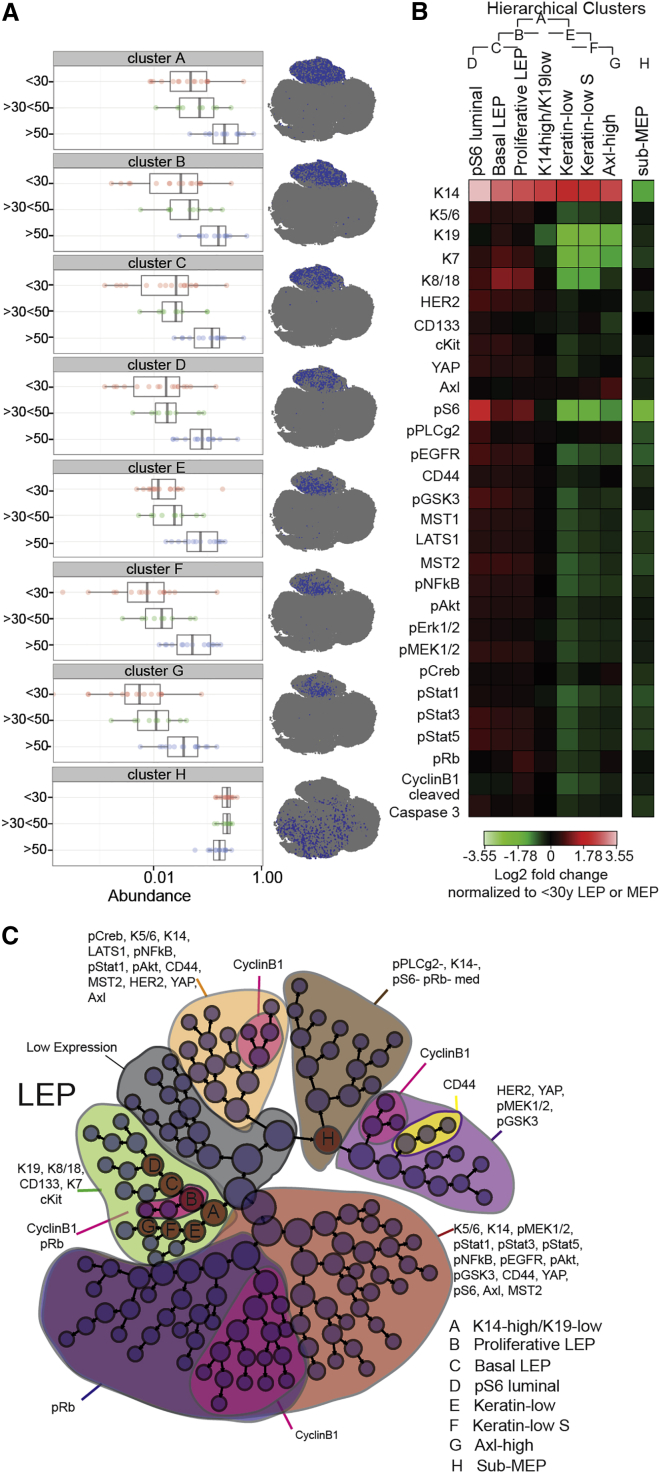
Unsupervised agglomerative hierarchical clustering (Citrus) was used to examine age-dependent changes in an orthogonal manner. Multidimensional single-cell data were distilled to a hierarchy of marker expression-related clusters, and cluster-specific cell frequency changes were determined (Bruggner et al., 2014). Seven clusters were identified (Figures 4A–4C) that were significantly more abundant with age (prediction error of 26% as estimated by cross-validation and a p value < 0.05 using a Student’s t test) (Figure 4A; Figure S4D), all of which represented the LEP compartment. Figure S4C illustrates the agglomerative clustering. The LEP subpopulations that showed age-dependent changes had specific marker expression signatures consistent with acquired MEP/basal-like characteristics (Figures 4A and 4B; Figure S4G). The age-emergent LEP clusters were all higher in K14 compared with the <30-year LEP. Cluster A, residing at the apex of the hierarchy, was K19low and K14high (Figures S4E and S4G). Clusters B, C, and D showed higher YAP, HER2, cKit, Axl, pS6, pPLCγ2, pEGFR, CD44, pGSK3, pNF-κB, pAkt, pERK1/2, pMEK1/2, pStat1, pStat3, and pStat5 expression than <30-year LEP. Most of these markers are associated with proliferation and migration and are mainly expressed in young MEP. Each cluster had defining characteristics, e.g., cluster B had the highest pRb and CyclinB1 expression that correlated with higher DNA content (Figure 4B; Figure S4F). Only cluster H decreased in abundance with age (Figures 4A, 4C, and S4D), and it mapped to the MEP compartment of the tSNE landscape. Those cells expressed low levels of K14, pS6, CyclinB1, and pRb (Figure S4F), possibly indicative of a quiescent, terminally differentiated MEP.
Figure 4.
Evidence of Age-Dependent Phenotypic Divergence in the Luminal Population
The Citrus algorithm was applied to identify cell populations by hierarchical clustering of phenotypically similar cells from an aggregate dataset from all samples (excluding 250MK, 90P and 245AT, 173T). A defining characteristic of each cluster is denoted as follows: cluster A, K14high/K19low; cluster B, proliferative LEP; cluster C, basal LEP; cluster D, pS6 luminal; cluster E, Keratinlow; cluster F, Keratin-low S; cluster G, Axlhigh; and cluster H, sub-MEP.
(A) Boxplots of cell abundance in each age-related cluster and its representative tSNE phenotypic projection. Each data point on these graphs represents the proportion of the cluster cell number compared to the total cell number in a single sample. The log10 scale represents an abundance of cells from 0 to 1.
(B) Heatmaps of marker expression of each cluster normalized to LEP from <30-year-old women for clusters A to G and MEP from <30-year-old women for cluster H.
(C) Hierarchical tree of agglomerative clusters obtained with the Citrus analysis. Node sizes are scaled on the basis of frequency of cells in each cluster.
See also Figure S4.
Collectively, these results indicated that a subset of LEP acquires a basal phenotype and accumulates while a subset of MEP decreases in abundance with age.
Age-Emergent Epithelial Cells in Primary Breast Epithelia
To confirm our findings, we conducted mass cytometry profiling using the 29-antibody panel on epithelial cells (≥10,000 cells per sample) derived from uncultured breast epithelia samples obtained from 13 women of different ages (<30 years, n = 7 and >50 years, n = 6; Table S1). As predicted by the HMEC analysis, the tSNE map revealed extensive phenotypic heterogeneity in the breast epithelia (Figure 5A; Figure S5A). Unsupervised clustering identified four distinct phenotypes of LEP (LEP1–4), seven types of MEP (MEP1–7), a DP subpopulation (between 13 and 719 non-doublet cells), and a low-expressing cell phenotype (<18.32 ion counts per cell) (Figure 5B). Protein expression patterns were consistent with these phenotypic designations and with the HMEC analysis (Figure 5C). K19, K7, K8/18, CD133, and cKit expression was high in LEP, while MEP showed higher expression of basal markers (K14, K5/6, Axl, pS6, CD44, pEGFR, and pStat1). The abundance of the LEP1 subpopulation significantly increased, whereas the abundance of MEP2 significantly decreased with age (Figure 5D), a trend also observed at the individual level (Figure 5E). Citrus analysis identified three clusters that were significantly more abundant with age (Figure 5F; Figure S5B), all residing within the LEP compartment of the tSNE phenotypic landscape. All three LEP clusters (A, B, and C) showed age-dependent changes in specific marker expression signatures, consistent with acquired MEP/basal-like characteristics (Figure 5G; Figure S5C). To quantify the extent of acquired MEP-like/basal phenotype, we calculated the geometric distance between the breast epithelia or HMEC LEP Citrus clusters and their respective MEP populations. This demonstrated that the LEP-MEP phenotypic distance was reduced by 26.7% and 32% in the breast epithelia and HMECs with age, respectively (Figure 5H). Collectively, this finding supports the notion that age-emergent epithelial cells derived from uncultured breast epithelia samples showed phenotypes that matched those identified in the primary cultured HMECs.
Figure 5.
Age-Related Phenotypic Divergence in Uncultured Breast Epithelia
(A) tSNE maps from dissociated uncultured breast epithelia from women <30 years old (merged and subsampled at 50,000 cells, n = 7) and >50 years old (merged and subsampled at 50,000 cells, n = 6). The pGsk3 channel was removed from the analysis due to a technical issue.
(B) tSNE projection of the PhenoGraph clusters. The tSNE projection (right panel) of women <30 years old (blue) and women >50 years old (green) is shown.
(C) Heatmaps of Z score of marker expression in PhenoGraph clusters of uncultured breast epithelia from women <30 years old (merged, n = 7).
(D) Plots of cell percentage in each PhenoGraph cluster. Data are mean ± SEM.
(E) Intra-sample heterogeneity for each woman is represented graphically by a horizontal bar in which segment lengths represent the proportion of the sample assigned to each cluster, colored accordingly.
(F) Boxplots of cell abundance in each age-related Citrus cluster and its representative tSNE phenotypic projection.
(G) Heatmaps of marker expression of each cluster normalized to LEP from <30-year-old women for clusters A to C and MEP from <30-year-old women for clusters D to G.
(H) The geometric distance was calculated using the square root of the sum of the squared differences between the median of each marker for each subpopulation.
(I) Representative human breast sections immunostained for K14 (red), K19 (green), and DAPI (blue) from a 17-year-old, 36-year-old, and 58-year-old woman (left to right, respectively). Scale bar represents 100 μm.
(J) Plots show classification performance of 171 breast sections from 50 women (<30 years n = 52, >30 < 50 years n = 86, and >50 years n = 33), analyzed using morphometric context with increasing training set size.
(K) Plot shows Citrus classification performance using a training set of 10 women (<30 years n = 5, >50 years n = 5). The black and white circles indicate whether an incorrectly assigned sample was from a peripheral non-tumor mastectomy (P), milk (MK), a tumor (T), or a tissue with no history, respectively. Data are mean ± SEM.
See also Figures S5 and S6.
Age-Emergent Phenotypes Predict Breast Tissue Age In Vivo
As the K14highK19low clusters from both breast tissue and HMECs formed the apex of the age-dependent cluster hierarchy, we hypothesized that the expression pattern of these cytokeratins could be used to predict the approximate age of normal breast tissue. Human breast sections were stained with anti-K14 and anti-K19 (<30 years, n = 52 [10 women]; >30 < 50 years, n = 86 [25 women]; and >50 years, n = 33 [15 women]) (Figure 5I), and a classification model was built using morphometric context (Chang et al., 2013). At least 1,000 cells per section were analyzed. This computational approach relied on automated cell segmentation, with manual curation, to define different epithelial cells prior to quantification of single-cell K14 and K19 levels and morphometric features. The machine learning-based classification model correctly assigned more than 50% of the samples into their correct age group, as compared with a random guess of 33.3% (Figure 5J), based on the higher level of K14 and lower level of K19 in LEP with age, as observed on the tissue sections (Figure 5I). These data validated predictions from the mass cytometry data, and the in situ analysis demonstrated quantifiable age-related changes in LEP in breast tissue.
Next, we used age-dependent phenotypic divergence to build a second classification model to test the hypothesis that age-related changes in marker expression from our statistical analysis would generalize to an independent dataset. This second model was based on the totality of the mass cytometry data, and it was not restricted to K14 and K19. In general, classification models use cross-validation to avoid testing hypotheses suggested by the data (type III errors). Using a training set of 5 HMECs each from women in the <30-year and >50-year age groups, we successfully assigned 13/16 women <30 years old and 12/15 women >50 years old (Figure 5K). The classification performance was increased with the number of training samples (Figure S6A). Strikingly, the HMEC strains that were incorrectly classified as “old” were derived from tissue peripheral to a breast tumor, harbored a known high cancer risk mutation, or had LEP proportions above the mean, while a >50-year HMEC strain with decreased LEP proportion was incorrectly assigned as “young.” Hence, the classification model validated the hypothesis that subsets of luminal cells change with age and can be used as age predictors, and it suggested that this information could be a relevant indicator for cancer risk.
Age-Emergent Luminal Cells Acquire Increased Basal Function
Next we investigated the functional consequences associated with the age-related basal phenotypic changes in the mammary luminal compartment. MEPs form cell-cell contacts with both LEPs and other MEPs and adhere to the basement membrane (Bergstraesser et al., 1995, Pitelka et al., 1973). These adhesive properties are characteristic of MEP, whereas LEP-extracellular matrix interactions are relatively minimal (Cerchiari et al., 2015). We hypothesized that the increased basalness of older LEPs would affect their cell adhesion and migratory capacity. Cell migration kinetics was measured by real-time impedance in LEP and MEP cells, sorted via fluorescence-activated cell sorting (FACS), from 6 different primary HMEC strains. LEPs isolated from women <30 years old migrated faster than isogenic MEPs (Figures S6B and S6C). In contrast, LEPs from women >50 years old migrated much slower and at a rate comparable to the cognate MEPs (Figures S6B and S6C), consistent with increased basal adhesion properties.
In addition, EGFR-mitogen-activated protein kinase (MAPK) pathway activation (pEGFR, pMEK, pErk, and pAkt) was higher in MEPs than in LEPs (Figure 2A; Figure S1), and it was higher in the LEP clusters that changed in abundance with age (Figure 4B). Therefore, we evaluated age-dependent differences in HMEC responses to EGF. HMECs from 3 women <30 years old and 3 women >50 years old were treated with EGF combined with vanadate, and signal transduction was measured by mass cytometry (Figures S6D–S6F). Both LEPs and MEPs exhibited EGFR pathway activation within the time course (Figure S1B); however, pStat, pEGFR, pErk, pMEK, and pPLCγ2 levels were increased in LEPs from women >50 years old compared to younger women. tSNE analysis revealed a subpopulation of HMECs with activated EGFR (Figure S6E). LEPs derived from older women were more prevalent in this population, consistent with increased EGF-signaling capacity (Figure S6F). Thus, older LEPs acquired myoepithelial-like adhesion and migration characteristics, as well as an increased EGF signaling.
Age-Dependent Phenotypic Divergence in the Luminal Progenitor Population
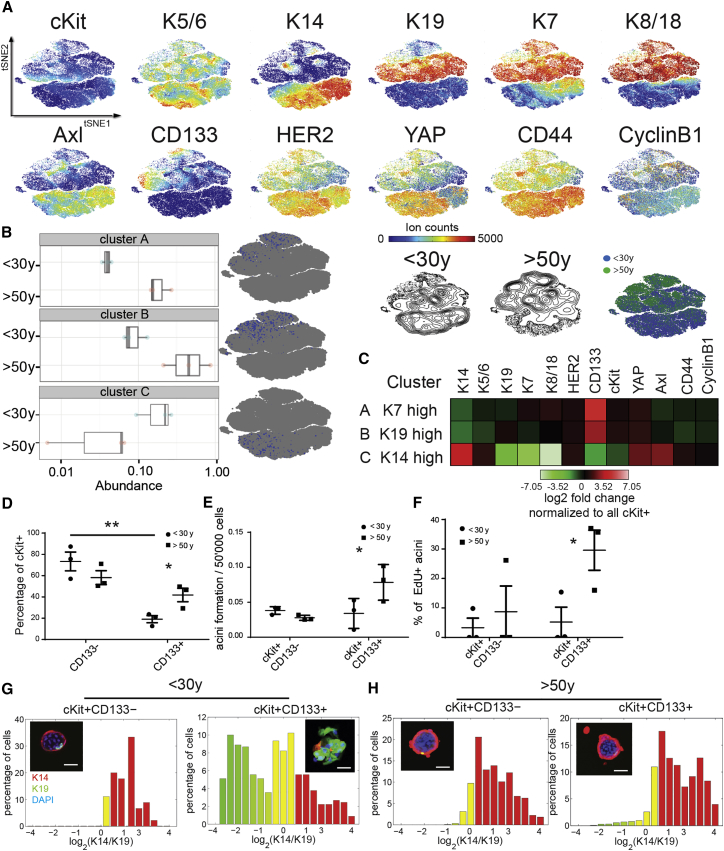
We next asked whether the prominent age-related changes in LEPs were the result of changes in luminal progenitors (LPs). Luminal-biased progenitors expressing cKit (Lim et al., 2009) were FACS enriched from HMEC strains derived from 3 women <30 years old and 3 women >50 years old at passage 4, and they were analyzed by mass cytometry using 12 mammary epithelial progenitor and lineage markers (Figure 6A). Unsupervised agglomerative clustering identified two LP clusters that were more abundant (A and B) and one cluster that was less abundant (C) with age (Figure 6B; Figure S7A). These clusters localized within the luminal compartment of the tSNE phenotypic space, and they displayed specific marker signatures (Figure 6C; Figure S7D): cluster A was K7high while cluster B was K19high. Both of these clusters displayed high expression of CD133, cKit, HER2, and YAP, establishing an age-emergent LP marker signature; thus, cKit+/CD133− and cKit+/CD133+ LP cells were FACS enriched from HMECs at passage 4. A higher proportion of cKit+/CD133+ LPs were present in older women (Figure 6D). The older cKit+/CD133+ LPs generated more acini in a Matrigel/collagen 3D embedding assay (Figure 6E), which incorporated more 5-ethynyl-2′-deoxyuridine (EdU), a proxy for cell proliferation (Figure 6F), compared to the corresponding younger LPs. In addition, cKit+CD133+ LPs were more luminally biased than cKit+CD133− LPs in younger women (Figure 6G; Figure S7B), as demonstrated by a higher proportion of K19+/K14− cells in the organoids. In contrast, older cKit+CD133+ LPs showed a higher proportion of basal K14+/K19− cells (Figure 6H; Figure S7C). Overall, the older cKit+/CD133+ LPs had higher clonogenic activity in vitro and gave rise to cells with more MEP/basal-like characteristics, which is consistent with the interpretation that altered LPs give rise to the LEPs that bear the phenotypic hallmarks of aging mammary epithelial cells.
Figure 6.
Evidence of Age-Dependent Phenotypic Divergence in HMEC Progenitors
(A) tSNE maps from FACS-enriched HMEC cKit+ progenitors from 3 women <30 years old and 3 women >50 years old (merged and subsampled at 50,000 cells). The lower right tSNE map shows the spatial projection of women <30 years old (blue) and women >50 years old (green).
(B) Boxplots of cell abundance of age-dependent clusters identified with Citrus and their representative tSNE spatial projection.
(C) Heatmaps of marker expression of each cluster compared to the background.
(D) Proportions of cKit+CD133− and cKit+CD133+ as a function of age (n = 3; t test, young cKit+CD133− versus cKit+CD133+ p = 0.0046, and young cKit+CD133+ versus old cKit+CD133+ p = 0.037).
(E) Acini formation potential of cKit+CD133− and cKit+CD133+ in Matrigel/collagen I gels as a function of age (n = 3; t test, p = 0.0123).
(F) Proportions of acini that were incorporating EdU as a function of age. Data are means ± SEM. An acinus was quantified as EdU positive if at least one cell was incorporating EdU (n = 3; t test, p = 0.0452). Data are means ± SEM.
(G and H) Histograms represent log2-transformed ratios of K14 to K19 protein expression in single cells of acini (G) from a representative woman <30 years old (240L, 19 years) and (H) from a representative woman >50 years old (029, 68 years). Histograms are heat mapped to indicate the phenotypes of K14−/K19+ LEP (green), K14+/K19+ progenitors (yellow), and K14+/K19− MEP (red). Insets show representative HMEC organoids immunostained for K14 (red), K19 (green), and DAPI (blue). Scale bar represents 50 μm.
See also Figure S7.
Aged Epithelial Cells Resemble Immortalized Epithelial Cells
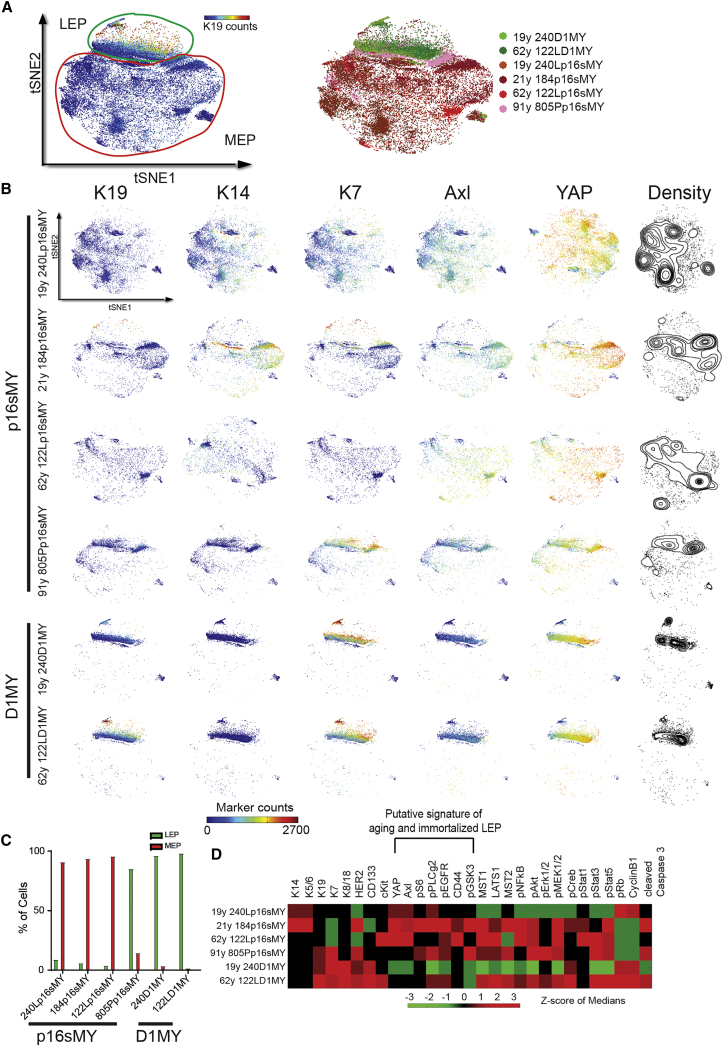
Normal epithelial cells must bypass tumor-suppressive barriers to give rise to malignancies. Pre-stasis HMECs can be efficiently immortalized in a two-step process that bypasses Rb function (by CCND1 expression or CDKN2A knockdown) and reactivates telomerase activity (indirectly by MYC expression), while incurring no gross genomic changes (Garbe et al., 2014). Mass cytometry analysis was conducted on 6 immortalized HMEC cell lines and visualized by tSNE. Immortalization via CCND1 overexpression to bypass stasis was associated with a luminal subtype, whereas knockdown of CDKN2A was associated with a basal subtype, and age >60 years, independent of CDKN2A, favored more luminal subtypes to emerge, consistent with our previous report (Lee et al., 2015) (Figures 7A–7C). Strikingly, five of the six immortal cell lines exhibited high expression of the basal markers YAP, Axl, pS6, pPLCγ2, pEGFR, CD44, and pGSK3 (Figures 7D and S7E), which is the same protein cluster observed in the subset of LEPs that accumulated with age (Figures 4B and 5G). Thus, the specific marker expression signatures found in age-emergent LEP subpopulations resemble the immortalized derivatives of older HMECs. This is consistent with the hypothesis that accumulation of altered LPs and LEPs with basal traits during mammary gland aging reflects a breast cancer susceptibility phenotype.
Figure 7.
Aged Epithelia Resemble Immortalized Epithelial Cells
(A) tSNE map of immortalized HMECs (left, merged, 6,000 cells per sample, n = 6). Right: each color represents a strain.
(B) Five selected markers are shown (K19, K4, K7, Axl, and YAP), with knockdown of CDKN2A: p16sMY and overexpression of CCND1: D1MY.
(C) Plots show percentage of LEP and MEP in each strain according to the gating strategy.
(D) Heatmap of Z score of median of marker expression of each strain.
See also Figure S7.
Discussion
At single-cell resolution, these data show the dynamic phenotypic heterogeneity of human mammary epithelia spanning eight decades of life. Phenotypic diversity is present within all cell populations, and en bloc-averaged behavior may not represent that of individual cells (Altschuler and Wu, 2010). Mapping the normal diversity of cellular phenotypes within an adult tissue is key to understanding organ-level function and cell-level functionality. Using unbiased computational analyses of 29-parameter mass cytometry, we interrogated epithelial cell lineage diversity in HMEC and uncultured human breast epithelia samples from 57 individuals. We show that the mammary epithelium comprises a complex population of cells residing in phenotypically and functionally diverse states that change with age, and it is more dynamic and heterogeneous than previously perceived (Santagata and Ince, 2014, Taylor-Papadimitriou et al., 1989, Villadsen et al., 2007). This unique data resource provides a repository of single-cell proteomic data combined with cell functional and in situ tissue validation to better understand the aging process in mammary epithelia. An important outcome of the high-dimensional comparison, between breast epithelia and primary cultures of HMEC strains, was the excellent correspondence between lineage representation and phenotypes of aging. In general, a challenge of aging research in human tissues is that age-dependent changes can be cataloged (e.g., transcriptional, proteomic, and biochemical), but it is difficult to study the functional consequences of those changes. As shown here, age-related functional changes in migration, EGF signaling, and 3D colony formation, which were predicted by measurements of the 29 epitopes in breast epithelia, could be explored using primary cultures. Further development of these types of approaches, which combine careful primary HMEC culture with advanced analytical instrumentation and computational analysis, represents a means of accessing human tissue biology in a new and meaningful way.
Older women accumulate mammary multipotent progenitors, the putative cancer cell of origin, with altered differentiation potential, which we and others hypothesize is one of the mechanisms that underlies increased susceptibility to breast cancer with age (Choudhury et al., 2013, Garbe et al., 2012, LaBarge et al., 2016, Proia et al., 2011). Their LEP daughters are not fully lineage committed and acquire a basal phenotype. By using a test set of appropriate size and machine-learning tools, we identified a unique subset of LEPs that accumulate with age, which have skewed differentiation and a basal-like phenotype. These age-emergent LEPs are more adherent to the extracellular matrix (ECM), and they exhibit increased EGF signaling compared to younger LEPs. An unsupervised classification model based on these age-related markers validates the hypothesis that specific age-related changes in LEP subpopulations are good age group predictors. Concomitant age-related changes in LP cells support the concept that the altered stem/progenitor cell populations that accumulate during aging give rise to the age-dependent LEP populations.
The unique constellation of protein levels and modification states that enable classification of mammary epithelia according to age constitutes a signature of aging in the mammary gland. A primary component that distinguishes LEP clusters by age is increased expression of the basal cytokeratin K14 with decreased luminal K19 expression. In situ validation of the mass cytometry-derived classification model, by K14 and K19 quantification in tissue sections of normal breast, robustly assigned most breast biopsy samples into their correct age group, further supporting the observation that increased basalness of the LEP compartment is a hallmark characteristic of aging breast tissue. The reduction of LEP migration rate with age, approaching that of MEP, was consistent with increased ECM engagement. Change in the LEP-ECM binding energy is predicted to impair the ability to maintain normal epithelial bilayers (Cerchiari et al., 2015). Indeed, LEP-ECM interactions in older LEP could inhibit the Hippo pathway (Cordenonsi et al., 2011) and activate YAP/TAZ (Naylor et al., 2005), which may help explain our previous observation of increased nuclear YAP in post-menopausal LEP in vivo (Pelissier et al., 2014).
Accumulation of stem/progenitor cells with skewed differentiation and function is a hallmark of aging in a number of tissues (Encinas et al., 2011, Garbe et al., 2012, Lugert et al., 2010), and it may confer increased susceptibility to oncogenic events. BRCA1 basal-like breast cancers may originate from cKit-expressing progenitors (Lim et al., 2009, Molyneux et al., 2010). The key changes in the subpopulations of LPs that accumulated with age involved a specific marker signature where cKit, CD133, YAP, and HER2 expression was increased and CD44 expression was decreased. These findings are congruent with the observation that cKit overexpression prevents normal differentiation in murine mammary epithelial progenitors (Regan et al., 2012), and they may explain the accumulation of cKit+ LPs with age. CD133 (Prominin 1), another putative LP marker (Hilton et al., 2014, Raouf et al., 2008), changed the most with age (increased 18.25- and 6.36-fold and decreased 5-fold in protein abundance in each respective cluster compared to <30-year cKit+). In some breast cancers CD133 positivity correlates with a restricted subgroup of tumor stem cells in BRCA1-deficient mammary tumors (Wright et al., 2008). YAP expression in LPs might result in incompletely differentiated LEPs with basal traits (Pelissier et al., 2014). Amplification of HER2 has been shown to play an important role in the development and progression of certain aggressive types of breast cancer (Ménard et al., 2000). Hence, these age-related phenotypes correspond with the cancer-relevant hypothetical effects of cKit, CD133, HER2, and YAP marker expression signature. Older cKit+CD133+ LPs formed basal proliferative colonies in 3D Matrigel/collagen gels, resembling the reported activity of LPs from BRCA1 carriers (Lim et al., 2009). Moreover, hormone treatment in luminal breast cancer reduced estrogen receptor α levels and promoted a cancer stem cell phenotype (CD133high and CD44low) (Sansone et al., 2016). This observation correlates well with the fact that augmented CD133high and CD44low cell proportion is associated with an increase in malignancy (Pece et al., 2010) and probably breast cancer risk (Garbe et al., 2012). The cKit+CD133+ LPs are considered potential cells of origin for breast cancer, and we propose that their accumulation with age represents an important facet of age-related susceptibility to breast cancer. Moreover, based on the involvement of these signature proteins (i.e., cKit, CD133, YAP, HER2, and CD44) in a number of breast cancer-relevant contexts, it is tempting to speculate that their dysregulation in older LPs also is related to increased susceptibility to breast cancer with age.
We identified shared expression signatures between age-emergent luminal cell subpopulations and immortalized derivatives of older HMECs, which have overcome at least two major barriers to tumor progression. Five of the six immortal cell lines exhibited high expression of the core markers (YAP, Axl, pS6, pPLCγ2, pEGFR, CD44, and pGSK3 [inactivated when phosphorylated]), which defines a putative signature of transformed HMECs that was also part of the core changes in normally aging LEPs. All of those core markers were reported to have a role in breast cancer progression (Gjerdrum et al., 2010, Kassis et al., 1999, Kim et al., 2015, Ko et al., 2016, Louderbough and Schroeder, 2011, Masuda et al., 2012, Yanai et al., 2015). Machine-learning algorithms that we used to validate the aging signature incorrectly classified nine young and middle-aged primary strains as old. Six of those strains were from women with mutations in BRCA1 or ATM, or they were derived from normal-appearing tissue that was peripheral to a tumor, or they were the normal cells cultured out directly from a tumor. It is tempting to speculate that epithelia in women who are innately at higher risk will exhibit an effective age that is older than the chronological age. A similar idea was proposed by epidemiologists that took into account breast tissue age, using only measures of hormones and childbirths (Pike et al., 1983). Here we provide evidence of a cell- and molecular-level manifestation of this concept. A biological explanation of advanced effective age in epithelia could be that molecular states that are associated with risk exert field effects (Deng et al., 1996), which alter the epithelia through a dynamic and reciprocal communication between stroma and epithelia. Ultimately, further investigation may lead to the development of novel approaches for prevention, patient stratification, and therapeutic interventions to combat age-associated breast cancers.
Experimental Procedures
Cell Culture and Uncultured Breast Epithelia
All cell culture was in M87A medium with 0.1 nM oxytocin (X) and cholera toxin (CT) at 0.5 ng/mL (Garbe et al., 2009). Primary HMEC strains were generated and maintained as described (Labarge et al., 2013). All tissues were obtained with proper oversight from the Lawrence Berkeley National Laboratory institutional review board. Breast tissue from reduction mammoplasty was manually dissected to enrich for gland-containing material. Stromal tissue was separated from epithelial fragments using a brief treatment with collagenase. The uncultured breast epithelia samples were dissociated as single cells with trypsin. All the pre-stasis HMEC strains were used at fourth passage (Table S1). Fibroblasts were removed and collected separately by differential trypsinization during the first passage. During the functional assay, HMECs were treated with EGF (Sigma E-9644, 0.1 μg/mL) and sodium orthovanadate (Sigma 13721-39-6, 12.5 mM) for 1 hr. Samples were harvested with TrypLE, fixed with 1.6% paraformaldehyde (PFA) for 10 min at room temperature (RT), and frozen as a pellet at −80°C for further analysis.
Antibodies Used for Mass Cytometry Analysis
Antibodies were obtained in carrier protein-free PBS and then prepared using the MaxPAR antibody conjugation kit (Fluidigm), according to the manufacturer’s protocol. After determining the percentage yield by measurement of absorbance at 280 nm, the metal-labeled antibodies were diluted in Candor PBS Antibody Stabilization solution (Candor Bioscience) for long-term storage at 4°C (Table S2). Antibodies were titrated and validated beforehand using both positive and negative cell controls (Table S2; Figure S1). Extensive antibody validation has been performed and published previously (Chevrier et al., 2017, Giesen et al., 2014).
Cell Barcoding and Antibody Staining
HMEC strains were incubated with cisplatin (WR International, Cat# 89150-634, 25 μM) for 1 min to assess cell viability (Fienberg et al., 2012), fixed in 1.6% PFA for 10 min at RT, and washed once with Cell Staining Media (CSM, PBS with 0.5% BSA and 0.02% NaN3 with 0.03% saponin). The cells were then resuspended in PBS, and DMSO stocks of the barcoding reagent were added as described (Bodenmiller et al., 2012, Zivanovic et al., 2014). The cells were incubated at RT for 30 min, washed three times with CSM, and then pooled into a single FACS tube for staining with metal-labeled antibodies for 1 hr at RT. A staining volume of 800 μL was used (∼30 × 106 cells/mL). After antibody staining, the cells were washed twice with CSM and once with PBS, and then incubated for 20 min at RT or overnight at 4°C with an iridium-containing intercalator (DVS Sciences) in PBS with 1.6% PFA. The cells were then washed three times with CSM and once with PBS, diluted with water to ∼106 cells/mL, and filtered through a 40-μm membrane just before analysis by mass cytometry.
Data Analysis
The scale used before analysis is the arcsinh with the cofactor of 5 (x_transf = asinh(x/5)). After gating out viable and iridium-labeled events, the data were analyzed by applying tSNE. This non-linear dimensionally reduction technique is implemented via Barnes-Hut approximations in the MATLAB toolbox cyt (Amir et al., 2013). We used the default parameters (initial dimensions, 110; perplexity, 30; and theta, 0.5). Each sample contained 20,000 cells, when merged, 320,000 cells from HMEC <30 years, 260,000 cells from HMEC >30 < 50 years, and 300,000 cells from HMEC >50 years. In tSNE, each cell is represented as a point in high-dimensional space. Each dimension is one parameter (the expression level of each protein in our case).
The unsupervised PhenoGraph algorithm in cyt has been used to group cells that are phenotypically similar and cluster these subpopulations using modularity optimization (Levine et al., 2015). tSNE and PhenoGraph were performed only on surface markers. A number of neighbors of 800 was selected. This parameter was chosen based on prior knowledge of the underlying cell types. Lower values for nearest neighbors result in an overclustering and higher values an underclustering.
The Citrus toolbox in R was used to identify clusters that changed in abundance with age (Bruggner et al., 2014) in an unsupervised manner. Therefore, clusters were identified using a hierarchical clustering and linked to clinical data for characterization. The minimal selected cluster size was 0.1% of the total analyzed data. Stratifying clusters were learned by using regularized unsupervised learning methods. Heatmaps were obtained with MATLAB and Cytobank. The results were reproduced with strains obtained from reduction mammoplasties (RMs) only.
Classification
Citrus was implemented in the PAM package for R and used nearest shrunken centroids as a predictive model to identify properties that are predictive of sample class. The prediction model was based on the initial training data model. Therefore, new samples were mapped and later assigned to the initial clusters for prediction. Using a training set of 5 HMECs from women <30 years old and 5 women >50 years old (n = 10), Citrus efficiently assigned most of the test set to young or old. The training set was changed to n = 8 and to n = 12 with similar efficiency. After randomization of the training set, the classification failed. The R code is found in the Supplemental Experimental Procedures.
Classification Using Morphometric Context
Each image was represented as its Cellular Morphometric Context (Chang et al., 2013), which was constructed as the histogram of cellular morphometric subtypes derived from the cellular morphometric features (K14/K19 signals) through K-Means (dictionary size = 1,024). Homogeneous kernel map (Vedaldi and Zisserman, 2012) was then applied on the Cellular Morphometric Context representation, so that linear support vector machine (SVM) (Yang et al., 2009) could be adopted for efficient and effective differentiation among age groups.
Statistical Analysis
GraphPad Prism, R, and MATLAB were used for all statistical analyses. Standard linear regression and t tests were used. Grouped analyses were performed with Bonferroni-Holm correction for multiple comparisons. Significance was established when ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Acknowledgments
We thank Dr. Thibault Vatter, Dr. Robert Bruggner, and Dr. Adeeb Rahman for expert advice. The imaging and FACS were performed at the Molecular Imaging Center (MIC) at the University of Bergen. F.A.P.V. was supported by a University of Bergen pre-doctoral fellowship and travel grants. D.S. was supported by the Forschungskredit of the University of Zurich (grant FK-74419-01-01) and the BioEntrepreneur-Fellowship of the University of Zurich (BIOEF-17-001). J.B.L. is supported by grants from the Research Council of Norway (240130), Centres of Excellence funding scheme (223250), and Helse Vest Health Authority (911794). M.A.L. is supported by NIH R00AG033176 and R01AG040081 and a Congressionally Directed Medical Research Programs Breast Cancer Research Program Era of Hope Scholar Award BC141351. B.B. is supported by the Swiss National Science Foundation (SNSF) R’Equip grant 316030-139220, a SNSF Assistant Professorship grant PP00P3-144874, the PhosphonetPPM SystemsX grant (UC4 DK108132), and funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement 336921. D.S. was supported by a Forschungskredit Fellowship of the University of Zurich.
Author Contributions
F.A.P.V., J.B.L., B.B., and M.A.L. designed the research. F.A.P.V. performed experiments. F.A.P.V., D.S., H.C., A.D.B., J.K.L., and B.P. analyzed the data and provided conceptual input. M.R.S., A.D.B., and M.A.L. provided cell strains, tissue sections, and other key reagents. B.B. provided the CyTOF platform. F.A.P.V., D.S., B.B., J.B.L., and M.A.L. wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: April 24, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.114.
Contributor Information
Mark A. LaBarge, Email: mlabarge@coh.org.
Bernd Bodenmiller, Email: bernd.bodenmiller@imls.uzh.ch.
James B. Lorens, Email: jim.lorens@uib.no.
Data and Software Availability
The accession number for the CyTOF data reported in this paper is Mendeley Data: https://doi.org/10.17632/j7mrbgt3hh.1.
Supplemental Information
References
- Altschuler S.J., Wu L.F. Cellular heterogeneity: do differences make a difference? Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir E.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., Pe’er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandura D.R., Baranov V.I., Ornatsky O.I., Antonov A., Kinach R., Lou X., Pavlov S., Vorobiev S., Dick J.E., Tanner S.D. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- Bergstraesser L.M., Srinivasan G., Jones J.C., Stahl S., Weitzman S.A. Expression of hemidesmosomes and component proteins is lost by invasive breast cancer cells. Am. J. Pathol. 1995;147:1823–1839. [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Hochmuth C.E., Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B., Zunder E.R., Finck R., Chen T.J., Savig E.S., Bruggner R.V., Simonds E.F., Bendall S.C., Sachs K., Krutzik P.O., Nolan G.P. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantley D.M., Chen C.L., Muraoka R.S., Bushdid P.B., Bradberry J.L., Kittrell F., Medina D., Matrisian L.M., Kerr L.D., Yull F.E. Nuclear factor-kappaB (NF-kappaB) regulates proliferation and branching in mouse mammary epithelium. Mol. Biol. Cell. 2001;12:1445–1455. doi: 10.1091/mbc.12.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggner R.V., Bodenmiller B., Dill D.L., Tibshirani R.J., Nolan G.P. Automated identification of stratifying signatures in cellular subpopulations. Proc. Natl. Acad. Sci. USA. 2014;111:E2770–E2777. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchiari A.E., Garbe J.C., Jee N.Y., Todhunter M.E., Broaders K.E., Peehl D.M., Desai T.A., LaBarge M.A., Thomson M., Gartner Z.J. A strategy for tissue self-organization that is robust to cellular heterogeneity and plasticity. Proc. Natl. Acad. Sci. USA. 2015;112:2287–2292. doi: 10.1073/pnas.1410776112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H., Borowsky A., Spellman P., Parvin B. Proceedings of the Conference on Computer Vision and Pattern Recognition. IEEE; 2013. Classification of Tumor Histology via Morphometric Context; pp. 2203–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhang N., Gray R.S., Li H., Ewald A.J., Zahnow C.A., Pan D. A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev. 2014;28:432–437. doi: 10.1101/gad.233676.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier S., Levine J.H., Zanotelli V.R.T., Silina K., Schulz D., Bacac M., Ries C.H., Ailles L., Jewett M.A.S., Moch H. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell. 2017;169:736–749.e18. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S., Almendro V., Merino V.F., Wu Z., Maruyama R., Su Y., Martins F.C., Fackler M.J., Bessarabova M., Kowalczyk A. Molecular profiling of human mammary gland links breast cancer risk to a p27(+) cell population with progenitor characteristics. Cell Stem Cell. 2013;13:117–130. doi: 10.1016/j.stem.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M., Zanconato F., Azzolin L., Forcato M., Rosato A., Frasson C., Inui M., Montagner M., Parenti A.R., Poletti A. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- Deng G., Lu Y., Zlotnikov G., Thor A.D., Smith H.S. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274:2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- dos Santos C.O., Rebbeck C., Rozhkova E., Valentine A., Samuels A., Kadiri L.R., Osten P., Harris E.Y., Uren P.J., Smith A.D., Hannon G.J. Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proc. Natl. Acad. Sci. USA. 2013;110:7123–7130. doi: 10.1073/pnas.1303919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas J.M., Michurina T.V., Peunova N., Park J.H., Tordo J., Peterson D.A., Fishell G., Koulakov A., Enikolopov G. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566–579. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fienberg H.G., Simonds E.F., Fantl W.J., Nolan G.P., Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81:467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe J.C., Bhattacharya S., Merchant B., Bassett E., Swisshelm K., Feiler H.S., Wyrobek A.J., Stampfer M.R. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 2009;69:7557–7568. doi: 10.1158/0008-5472.CAN-09-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe J.C., Pepin F., Pelissier F.A., Sputova K., Fridriksdottir A.J., Guo D.E., Villadsen R., Park M., Petersen O.W., Borowsky A.D. Accumulation of multipotent progenitors with a basal differentiation bias during aging of human mammary epithelia. Cancer Res. 2012;72:3687–3701. doi: 10.1158/0008-5472.CAN-12-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe J.C., Vrba L., Sputova K., Fuchs L., Novak P., Brothman A.R., Jackson M., Chin K., LaBarge M.A., Watts G. Immortalization of normal human mammary epithelial cells in two steps by direct targeting of senescence barriers does not require gross genomic alterations. Cell Cycle. 2014;13:3423–3435. doi: 10.4161/15384101.2014.954456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacinti C., Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- Giesen C., Wang H.A., Schapiro D., Zivanovic N., Jacobs A., Hattendorf B., Schüffler P.J., Grolimund D., Buhmann J.M., Brandt S. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- Gjerdrum C., Tiron C., Høiby T., Stefansson I., Haugen H., Sandal T., Collett K., Li S., McCormack E., Gjertsen B.T. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc. Natl. Acad. Sci. USA. 2010;107:1124–1129. doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härdle W., Simar L. Volume 22007. Springer; 2007. (Applied multivariate statistical analysis). [Google Scholar]
- Hilton H.N., Santucci N., Silvestri A., Kantimm S., Huschtscha L.I., Graham J.D., Clarke C.L. Progesterone stimulates progenitor cells in normal human breast and breast cancer cells. Breast Cancer Res. Treat. 2014;143:423–433. doi: 10.1007/s10549-013-2817-2. [DOI] [PubMed] [Google Scholar]
- Kassis J., Moellinger J., Lo H., Greenberg N.M., Kim H.G., Wells A. A role for phospholipase C-gamma-mediated signaling in tumor cell invasion. Clin. Cancer Res. 1999;5:2251–2260. [PubMed] [Google Scholar]
- Kim H.M., Jung W.H., Koo J.S. Expression of Yes-associated protein (YAP) in metastatic breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:11248–11257. [PMC free article] [PubMed] [Google Scholar]
- Ko H.W., Lee H.H., Huo L., Xia W., Yang C.C., Hsu J.L., Li L.Y., Lai C.C., Chan L.C., Cheng C.C. GSK3β inactivation promotes the oncogenic functions of EZH2 and enhances methylation of H3K27 in human breast cancers. Oncotarget. 2016;7:57131–57144. doi: 10.18632/oncotarget.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBarge M.A., Petersen O.W., Bissell M.J. Of microenvironments and mammary stem cells. Stem Cell Rev. 2007;3:137–146. doi: 10.1007/s12015-007-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarge M.A., Garbe J.C., Stampfer M.R. Processing of human reduction mammoplasty and mastectomy tissues for cell culture. J. Vis. Exp. 2013;(71):50011. doi: 10.3791/50011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBarge M.A., Mora-Blanco E.L., Samson S., Miyano M. Breast Cancer beyond the Age of Mutation. Gerontology. 2016;62:434–442. doi: 10.1159/000441030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.K., Garbe J.C., Vrba L., Miyano M., Futscher B.W., Stampfer M.R., LaBarge M.A. Age and the means of bypassing stasis influence the intrinsic subtype of immortalized human mammary epithelial cells. Front. Cell Dev. Biol. 2015;3:13. doi: 10.3389/fcell.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir E.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E., Vaillant F., Wu D., Forrest N.C., Pal B., Hart A.H., Asselin-Labat M.L., Gyorki D.E., Ward T., Partanen A., kConFab Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Louderbough J.M., Schroeder J.A. Understanding the dual nature of CD44 in breast cancer progression. Mol. Cancer Res. 2011;9:1573–1586. doi: 10.1158/1541-7786.MCR-11-0156. [DOI] [PubMed] [Google Scholar]
- Lugert S., Basak O., Knuckles P., Haussler U., Fabel K., Götz M., Haas C.A., Kempermann G., Taylor V., Giachino C. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Mansilla E., Díaz Aquino V., Zambón D., Marin G.H., Mártire K., Roque G., Ichim T., Riordan N.H., Patel A., Sturla F. Could metabolic syndrome, lipodystrophy, and aging be mesenchymal stem cell exhaustion syndromes? Stem Cells Int. 2011;2011:943216. doi: 10.4061/2011/943216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H., Zhang D., Bartholomeusz C., Doihara H., Hortobagyi G.N., Ueno N.T. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res. Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard S., Tagliabue E., Campiglio M., Pupa S.M. Role of HER2 gene overexpression in breast carcinoma. J. Cell. Physiol. 2000;182:150–162. doi: 10.1002/(SICI)1097-4652(200002)182:2<150::AID-JCP3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Molyneux G., Geyer F.C., Magnay F.A., McCarthy A., Kendrick H., Natrajan R., Mackay A., Grigoriadis A., Tutt A., Ashworth A. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Naylor M.J., Li N., Cheung J., Lowe E.T., Lambert E., Marlow R., Wang P., Schatzmann F., Wintermantel T., Schüetz G. Ablation of beta1 integrin in mammary epithelium reveals a key role for integrin in glandular morphogenesis and differentiation. J. Cell Biol. 2005;171:717–728. doi: 10.1083/jcb.200503144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasic L., Eisinger-Mathason T.S., Velayudhan B.T., Moskaluk C.A., Brenin D.R., Macara I.G., Lannigan D.A. Sustained activation of the HER1-ERK1/2-RSK signaling pathway controls myoepithelial cell fate in human mammary tissue. Genes Dev. 2011;25:1641–1653. doi: 10.1101/gad.2025611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S., Tosoni D., Confalonieri S., Mazzarol G., Vecchi M., Ronzoni S., Bernard L., Viale G., Pelicci P.G., Di Fiore P.P. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Pelissier F.A., Garbe J.C., Ananthanarayanan B., Miyano M., Lin C., Jokela T., Kumar S., Stampfer M.R., Lorens J.B., LaBarge M.A. Age-related dysfunction in mechanotransduction impairs differentiation of human mammary epithelial progenitors. Cell Rep. 2014;7:1926–1939. doi: 10.1016/j.celrep.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike M.C., Krailo M.D., Henderson B.E., Casagrande J.T., Hoel D.G. ‘Hormonal’ risk factors, ‘breast tissue age’ and the age-incidence of breast cancer. Nature. 1983;303:767–770. doi: 10.1038/303767a0. [DOI] [PubMed] [Google Scholar]
- Pitelka D.R., Hamamoto S.T., Duafala J.G., Nemanic M.K. Cell contacts in the mouse mammary gland. I. Normal gland in postnatal development and the secretory cycle. J. Cell Biol. 1973;56:797–818. doi: 10.1083/jcb.56.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proia T.A., Keller P.J., Gupta P.B., Klebba I., Jones A.D., Sedic M., Gilmore H., Tung N., Naber S.P., Schnitt S. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf A., Zhao Y., To K., Stingl J., Delaney A., Barbara M., Iscove N., Jones S., McKinney S., Emerman J. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Regan J.L., Kendrick H., Magnay F.A., Vafaizadeh V., Groner B., Smalley M.J. c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene. 2012;31:869–883. doi: 10.1038/onc.2011.289. [DOI] [PubMed] [Google Scholar]
- Sansone P., Ceccarelli C., Berishaj M., Chang Q., Rajasekhar V.K., Perna F., Bowman R.L., Vidone M., Daly L., Nnoli J. Self-renewal of CD133(hi) cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer. Nat. Commun. 2016;7:10442. doi: 10.1038/ncomms10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S., Ince T.A. Normal cell phenotypes of breast epithelial cells provide the foundation of a breast cancer taxonomy. Expert Rev. Anticancer Ther. 2014;14:1385–1389. doi: 10.1586/14737140.2014.956096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless N.E., DePinho R.A. How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Skibinski A., Breindel J.L., Prat A., Galván P., Smith E., Rolfs A., Gupta P.B., LaBaer J., Kuperwasser C. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep. 2014;6:1059–1072. doi: 10.1016/j.celrep.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P.J., Tarpey P.S., Davies H., Van Loo P., Greenman C., Wedge D.C., Nik-Zainal S., Martin S., Varela I., Bignell G.R., Oslo Breast Cancer Consortium (OSBREAC) The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Papadimitriou J., Stampfer M., Bartek J., Lewis A., Boshell M., Lane E.B., Leigh I.M. Keratin expression in human mammary epithelial cells cultured from normal and malignant tissue: relation to in vivo phenotypes and influence of medium. J. Cell Sci. 1989;94:403–413. doi: 10.1242/jcs.94.3.403. [DOI] [PubMed] [Google Scholar]
- Vedaldi A., Zisserman A. Efficient additive kernels via explicit feature maps. IEEE Trans. Pattern Anal. Mach. Intell. 2012;34:480–492. doi: 10.1109/TPAMI.2011.153. [DOI] [PubMed] [Google Scholar]
- Villadsen R., Fridriksdottir A.J., Rønnov-Jessen L., Gudjonsson T., Rank F., LaBarge M.A., Bissell M.J., Petersen O.W. Evidence for a stem cell hierarchy in the adult human breast. J. Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M.H., Calcagno A.M., Salcido C.D., Carlson M.D., Ambudkar S.V., Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai A., Inoue N., Yagi T., Nishimukai A., Miyagawa Y., Murase K., Imamura M., Enomoto Y., Takatsuka Y., Watanabe T. Activation of mTOR/S6K But Not MAPK Pathways Might Be Associated With High Ki-67, ER(+), and HER2(-) Breast Cancer. Clin. Breast Cancer. 2015;15:197–203. doi: 10.1016/j.clbc.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Yang J., Yu K., Gong Y., Huang T. Proceedings of the Conference on Computer Vision and Pattern Recognition. IEEE; 2009. Linear Spatial Pyramid Matching Using Sparse Coding for Image Classification; pp. 1794–1801. [Google Scholar]
- Zivanovic N., Jacobs A., Bodenmiller B. A practical guide to multiplexed mass cytometry. Curr. Top. Microbiol. Immunol. 2014;377:95–109. doi: 10.1007/82_2013_335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.