Abstract
Motivation
Correct and rapid determination of Mycobacterium tuberculosis (MTB) resistance against available tuberculosis (TB) drugs is essential for the control and management of TB. Conventional molecular diagnostic test assumes that the presence of any well-studied single nucleotide polymorphisms is sufficient to cause resistance, which yields low sensitivity for resistance classification.
Summary
Given the availability of DNA sequencing data from MTB, we developed machine learning models for a cohort of 1839 UK bacterial isolates to classify MTB resistance against eight anti-TB drugs (isoniazid, rifampicin, ethambutol, pyrazinamide, ciprofloxacin, moxifloxacin, ofloxacin, streptomycin) and to classify multi-drug resistance.
Results
Compared to previous rules-based approach, the sensitivities from the best-performing models increased by 2-4% for isoniazid, rifampicin and ethambutol to 97% (P < 0.01), respectively; for ciprofloxacin and multi-drug resistant TB, they increased to 96%. For moxifloxacin and ofloxacin, sensitivities increased by 12 and 15% from 83 and 81% based on existing known resistance alleles to 95% and 96% (P < 0.01), respectively. Particularly, our models improved sensitivities compared to the previous rules-based approach by 15 and 24% to 84 and 87% for pyrazinamide and streptomycin (P < 0.01), respectively. The best-performing models increase the area-under-the-ROC curve by 10% for pyrazinamide and streptomycin (P < 0.01), and 4–8% for other drugs (P < 0.01).
Availability and implementation
The details of source code are provided at http://www.robots.ox.ac.uk/~davidc/code.php.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Tuberculosis (TB) was one of the leading 10 causes of death worldwide, ranking above HIV/AIDS as the prominent cause of death from infectious disease. Drug-resistant TB has emerged as a substantial concern for public health and threatens global TB control. The World Health Organisation (WHO) reported in 2017 (WHO, 2017) that an estimated 4.1% (95% confidence interval [CI]: 2.8–5.3%) of new cases and 19% (95% CI: 9.8–27%) of previously treated cases had multi-drug resistant TB (MDR-TB), defined as being resistant to isoniazid INH and rifampicin RIF) or rifampicin-resistant TB (RR-TB). WHO now recommends that all RR-TB cases be treated with an MDR-TB treatment regime (WHO, 2016). The emergence and spread of drug-resistant TB have been reported to be related to vulnerabilities of current TB-control efforts (Cohen et al., 2015), biological factors (Casali et al., 2014), treatment-related risk factors (Ford et al., 2013), compensatory evolution of bacteria (Comas et al., 2011), population displacement and political instability (Eldholm et al., 2016).
One critical challenge in tackling the global TB epidemic is timely diagnosis and correct treatment. Rapid molecular diagnostic tests help to ensure early detection and prompt treatment; these tests assume the presence of a single nucleotide polymorphism (SNP), which is one of the previously identified SNPs, is sufficient to cause resistance. Such tests are effective with the most common mutations causing resistance to drugs, but their underpinning technology restricts them to a relatively small number of targets per drug. Research to date has focused on the identification of the described multivariate associations (Coll et al., 2015; Farhat et al., 2013; Georghiou et al., 2012; Walker et al., 2015; Zhang et al., 2013). However, methods for identifying such multivariate association can be of limited utility in light of the fact that association is still poorly understood for some anti-TB drugs. Compounding this effect, the genetic basis of drug resistance is more complex than anticipated: resistance-related genes are likely to contain nonsynonymous SNPs associated with drug resistance as a result of drug pressure (Zhang et al., 2013); and mutations that interact in a complex manner could produce high-level resistance to a specific drug (Safi et al., 2013). In countries with the highest incidence of MDR-TB, it was found that more than 30% of MDR clinical isolates had compensatory mutations (Comas et al., 2011).
Multivariate association between genetic variants can be explored with predictive models based on machine learning. Previous studies have adopted a number of such algorithms to predict Mycobacterium tuberculosis (MTB) resistance; e.g. logistic regression (Zhang et al., 2013) and random forests (?). However, a thorough evaluation of potentially applicable methods for the classification of MTB drug-resistance has not been reported. In this paper, using a large collection of MTB isolates, we evaluated the ability of different models to classify drug resistance for the four first-line drugs, several second-line drugs and MDR-TB. Our models achieved comparable or better classification of drug resistance in comparison to the direct association that depends solely on any resistance-determinants previously identified in the literature. Our results validate the use of machine learning algorithms for the identification of MTB resistance, and support the hypothesis that previously unknown multivariate associations and interactions contribute to resistance to several anti-TB drugs.
2 Materials and methods
2.1 Specimen and laboratory phenotyping
We included 1839 samples from Walker et al. (2015). On all study isolates, DST was performed for each drug through an initial screen for resistance in liquid culture, which was then confirmed using Lowenstein Jensen methods. Up to 11 drugs were assayed, including isoniazid (INH), rifampicin (RIF), ethambutol (EMB), pyrazinamide (PZA), amikacin (AK), capreomycin, (CAP), ciprofloxacin (CIP), kanamycin (KAN), moxifloxacin (MOX), ofloxacin (OFX) and streptomycin (SM).
2.2 DNA sequencing
The details of DNA sequencing refer to Walker et al. (2015). Nucleotide bases were called using standard filters on sequencing and alignment quality, as well as the number of reads for each base. After filtering, the nucleotide bases at certain positions that could not be called with confidence were denoted as null calls and not used in our analysis.
2.3 Genomic data pre-processing
23 candidate genes and their 100 base-pair upstream regions were targeted in this study (As described in Supplementary Material A). We limited our investigation to these genes because each has at least one previously described drug-resistance mutation [the source papers related to these genes were summarized in Walker et al. (2015)], allowing us to focus our investigation upon those areas of the genome in which we have a high prior belief of involvement. M.tuberculosis lineage was assigned based on polymorphisms described in the literature (Feuerriegel et al., 2014; Stucki et al., 2012) and corroborated by a previously published phylogeny (Walker et al., 2015). We reported every nucleotide site that differed from the reference genome, identified the corresponding amino acid substitution where there was one, and differentiated between different amino acids at those variant sites. We therefore considered the variant sites with amino acid substitution as SNP. The presence of a SNP in the isolate was represented by a binary variable, with 1 indicating the presence of the SNP and 0 indicating absence. The average number of SNPs per isolate was 6, ranging between 1 and 47. Null calls were considered to be SNPs if the base with the highest percentage of reads did not agree with the reference. In total across the 1839 isolates, 2629 SNPs were found in the 23 candidate genes.
3 Results
3.1 Phenotype
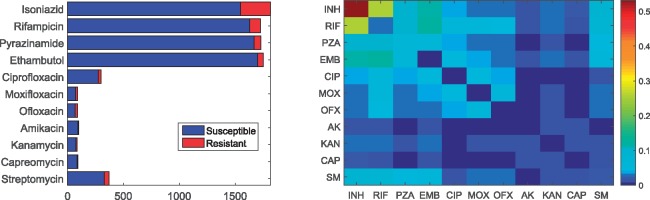
Our study included 1839 M.tuberculosis isolates, representing all the major TB clades [the phylogenetic tree is given in Supplementary Fig. S1 by Walker et al. (2015)]. Each isolate underwent culture-based drug-susceptibility testing to a maximum of 11 anti-TB drugs. Not every isolate was tested against all drugs. The four first-line anti-TB drugs were tested on the majority of isolates (Fig. 1, left panel). Of the 1811 isolates that were tested against INH, 266 (15%) were resistant and 1545 (85%) were susceptible; the ratio of the number of the resistant and susceptible isolates was approximately 1:5.7 for INH. In the case of EMB, RIF and PZA, the numbers of resistant isolates were only 47, 97 and 59, representing 3%, 6% and 3% of the total number, respectively. Correspondingly, the ratio of the two classes declined to 1:36, 1:16.8 and 1:28.2, respectively. Regarding the second-line drugs, no more than 400 isolates were tested against individual drugs, meanwhile the number of the resistant versus susceptible isolates was approximately 1:10. Since there were few AK, KAN and CAP-resistant isolates, these three drugs were not be investigated in the following analysis.
Fig. 1.
Phenotype of 1839 isolates. left: bar plot of phenotype availability for the different drugs. right: heatmap quantifying the number of instances of co-occurrence of resistance between drugs normalized by total number of isolates resistant to at least one drug. Off-diagonal elements show co-occurrence of resistance between different drugs; on-diagonal elements show cases which are resistant to a single drug
Co-occurrence of resistance was frequently observed for the tested drugs in our cohort (Fig. 1, rightmost). Within the 11 drugs, single-drug-resistant isolates only existed for INH, RIF, PZA and streptomycin (SM) (only cells that correspond to INH, RIF, PZA and SM on the diagonal are non-zero entries). Of 320 isolates that were resistant to at least one of the eleven drugs, 65% was mono-resistant to single drug, where 170 (53%), 8 (2.5%), 20 (6%) and 2 (0.6%) were mono-resistant to INH, RIF, EMB and PZA, respectively. Of these 320 isolates, 81 (25%) was resistant to both NIH and RIF, 19 (6%) was both EMB and PZA resistance. In other cases of resistance to any two first-line drugs, the number of the isolates was similar (10–12% of the 320 isolates); they were the same for the resistance to both SM and any first-line drug. For all other two-drug resistance co-occurrence, the number of the isolates was no more than 16 (5%).
3.2 Clustering
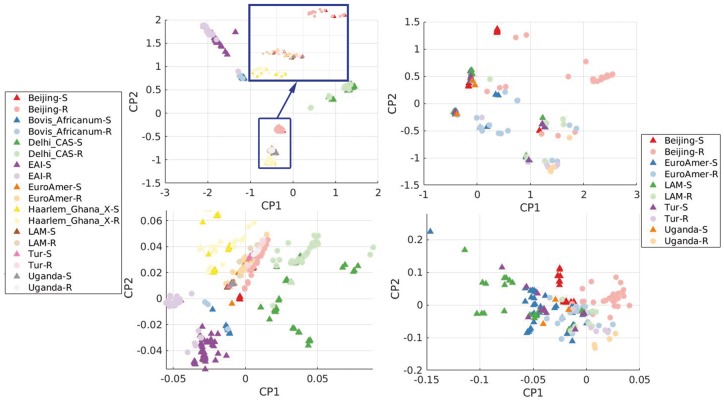
We performed both principal component analysis (PCA) and a sparse logistic version (SL-PCA) to explore the underlying structure of genetic variation within our cohort of bacterial isolates. For these 1839 isolates, 2629 SNPs were found in 23 genes in which previous studies had identified one or more known resistance-conferring mutation. Both the PCA and SL-PCA reduced the dimensionality of 2629 to 2. Figure 2 shows the resulting structure, with resistance shown in terms of INH-resistance and sub-lineage, a separate annotation obtained by phylogeny analysis introduced in Section 3.3, independent of phenotype. The use of conventional PCA results in compact structure, with little variation for each cluster and a ‘Y’ shape in the subspace spanned by the first two principal components (PCs), which separates the various cluster. Comparatively, SL-PCA produces structure that reveals variation for all clusters and the resistant/susceptible phenotype. Using the first two logistic PCs, four clusters (EAI, Haarlem_Ghana_X, Delhi_CAS and Bovis_Africanum) were well-separated and positioned around a central cluster. We term this cluster C1, which is composed of isolates from the Beijing, EuroAmer, LAM, Tur and Uganda clades. Within this C1 cluster, conventional PCA shows poor separability between INH-resistant and susceptible classes. Overall, SL-PCA is more informative than PCA: SL-PCA can be used to identify clusters and provide better separation for resistant and susceptible classes within the cluster C1. We performed classification analysis both on entire dataset and on several selected clusters; the latter is provided in Supplementary Material I.
Fig. 2.
PCA (upper row) and SL-PCA (lower row) for all clades [Clades are defined based on the whole genome sequences (not just resistance genes). Interested readers are referred to Benavente et al. (2015).] (left plots) and cluster C1 (right plots) in terms of INH resistance (C1: Beijing, Euro, LAM, Tur and Uganda) (Color version of this figure is available at Bioinformatics online.)
3.3 Direct association
Existing methods classify drug resistance based on the presence of any determinant from a library of such determinants that has been assembled from the literature; we term this method ‘direct association’ (DA). We examined the use of two such libraries: (i) the ‘Dream TB’ database and (ii) those described in an existing study (Walker et al., 2015). We term the methods using these two libraries DA-D and DA-L, respectively (as listed in Supplementary Material B and C, respectively). To classify resistance against a given drug, we applied an ‘OR’ rule: if any of the mutations associated with a given drug in the library were present for a given isolate, the isolate was labelled as being resistant to that drug. Full results for resistance classification by the DA methods are provided in Supplementary Material F.
3.4 Classification using machine learning methods
We investigated seven machine learning classifiers, including include logistic regression with L1 and L2 regularisation (LR-L1 and LR-L2), support vector machine with L2 regularisation and a radial basis function kernel (SVM-L2 and SVM-RBF), random forest (RF), a product-of-marginals model (PM) and a class-conditional Bernoulli mixture model (CBMM) (the methodology details and learning scheme are provided in Supplementary Material E and D, respectively). While evaluating all methods, we repeatedly and randomly selected the susceptible isolates from susceptible class in each experiment, to give the same number as in the resistant class in terms of individual drugs. The detail regarding dataset generation is illustrated in Supplementary Material D.
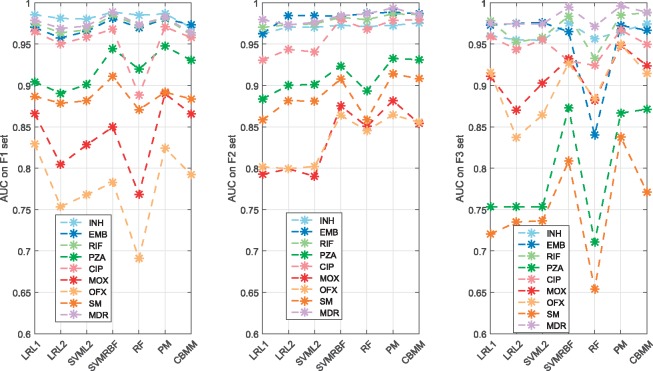
Three sets of features, F1–F3, were considered, to evaluate their performances. Feature set F1 is the baseline feature set (all SNPs found within 23 candidate genes, Supplementary Material A). Feature set F2 contains only those SNPs that were previously suspected of being resistance-determinants [108 SNPs reported in (Walker et al., 2015)]. Feature set F3 is a subset of F1 given a particular drug (where genes with only resistance-determinants to that drug are included). F3 therefore reflects what could be considered as direct determinants, whereas F2 incorporates the possibility of using resistance co-occurrence (Fig. 1) within all direct determinants for 11 drugs to inform prediction, and F1 interprets resistance co-occurrence on a larger scale.
Figure 3 shows comparisons in AUC performance for the seven machine learning classifiers, for the three feature sets, using eight drugs [amikacin (AK), clarithromycin (CAP) and Kanamycin (KAN) were excluded due to under 10 resistant isolates]. The drugs can be classified into three groups: i) the drugs where almost all classifiers were robust on the three feature sets (INH, RIF, EMB and MDR); ii) the drugs where the classifiers were better with feature sets F1 and F2 (PZA and SM); iii) the drugs where the classifiers were better with feature set F3 (MOX and OFX). Detection of resistance to INH, RIF, EMB and MDR may be seen to be robust to the choice of classifier, with all classifiers achieving at least 93% AUC for all feature sets (except for the RF with F1 and F3 in the case of CIP). The AUC performances of all classifiers for PZA and SM were improved at least 10% with F1 and F2 compared with F3, respectively (the SNPs that were statistically important for predicting PZA and SM resistance based on PM model, which outputs probability for presence or absence of a SNP given a Beta prior, are listed in Supplementary Material J). MOX and OFX resistance identification was most challenging, with all classifiers achieving AUC values below 90% for feature sets F1 and F2. In particular, the average performance with F3 was noticeably better than those with either F1 or F2 with at least 85% mean AUC. Based on variable importance measures in RF, the SNPs within the suspect genes given the investigated drugs are listed in Supplementary Material K.
Fig. 3.
Classification performance in AUC for seven classifiers across eight anti-TB drugs and MDR-TB with the F1, F2 and F3 feature sets. While the horizontal axis is discrete, dashed lines are shown between data for ease of viewing (Color version of this figure is available at Bioinformatics online.)
3.5 Comparing existing methods with machine-learning methods
We compared DA resistance classification with the seven machine learning classifiers. We additionally performed an experiment in which we removed a set of four commonly occurring SNPs (gyrA_E21, gyrA_S95, gyrA_G668, katG_R463) that are known not to be causally involved with resistance from feature set F1. As shown in Table 1, the best-performing machine learning classifier yielded higher AUC values in comparison to the baseline DA method for all examined drugs (P < 0.01). Our models yielded results with improved mean sensitivity in resistance classification for all drugs in comparison to DA-L (P < 0.01), while maintaining mean specificity above 90%.
Table 1.
Comparing performance between best classifier and DA-L for resistance prediction with 8 drugs and MDR-TB
| Drug | DA-L |
Best classifier |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Sens | Spec | AUC | Classifier (Feature set) | Sens | Spec | AUC | Classifier with F1* | AUC | |
| INH | 93 ± 0.3 | 99 ± 0.1 | 96 ± 0.0 | RF(F1) | 97† ± 0.3 | 94† ± 0.4 | 99† ± 0.0 | RF(F1) | 98† ± 0.0 |
| EMB | 95 ± 0.7 | 97 ± 0.6 | 96 ± 0.1 | CBMM(F2) | 97† ± 1.0 | 96† ± 0.6 | 99† ± 0.1 | PM(F2) | 99† ± 0.1 |
| RIF | 94 ± 0.5 | 98 ± 0.3 | 96 ± 0.1 | CBMM(F3) | 97† ± 0.4 | 97 ± 0.4 | 99† ± 0.1 | CBMM(F3) | 99† ± 0.1 |
| PZA | 69 ± 1.4 | 100 ± 0.0 | 85 ± 0.0 | PM(F1) | 84† ± 1.2 | 90† ± 1.1 | 95† ± 0.2 | SVM-RBF(F1) | 95† ± 0.2 |
| CIP | 87 ± 1.0 | 99 ± 0.4 | 94 ± 0.1 | PM(F2) | 96† ± 0.9 | 98 ± 0.4 | 98† ± 0.3 | PM(F2) | 98† ± 0.3 |
| MOX | 83 ± 1.4 | 93 ± 0.8 | 87 ± 0.1 | PM(F3) | 95† ± 1.4 | 93 ± 1.0 | 95† ± 0.4 | PM(F3) | 94† ± 0.5 |
| OFX | 81 ± 1.5 | 95 ± 0.9 | 87 ± 0.3 | PM(F3) | 96† ± 1.4 | 92 ± 1.3 | 95† ± 0.5 | PM(F3) | 95† ± 0.6 |
| SM | 63 ± 1.8 | 98 ± 0.6 | 81 ± 0.1 | SVM-RBF(F2) | 87† ± 1.5 | 90† ± 1.0 | 91† ± 0.3 | PM(F2) | 92† ± 0.2 |
| MDR | 90 ± 0.7 | 100 ± 0.2 | 95 ± 0.0 | PM(F3) | 96† ± 0.6 | 98† ± 0.5 | 100† ± 0.1 | PM(F3) | 100† ± 0.0 |
Note: ‘D.SNPs’ refers to those SNPs known not to be causally involved with resistance mechanisms, and which are removed from F1 feature set in one experiment. Sensitivity (sens) and specificity (spec) are shown with AUC, where results are reported as mean and standard error.
P-value is lower than 0.01 (P < 0.01). The P-value of performance measurement of the examined classifier compared to the DA-L was obtained by Wilcoxon signed-rank test. Feature set F1* denotes the feature set F1 without D.SNPs.
Compared to DA-L method, the sensitivities from the best-performing models increased by 2–4% for isoniazid, rifampicin and ethambutol to 97% (P < 0.01), respectively; for ciprofloxacin and multi-drug resistant TB, they increased to 96%. For moxifloxacin and ofloxacin, sensitivities increased by 12 and 15% from 83 and 81% based on existing known resistance alleles to 95% and 96% (P < 0.01), respectively. Particularly, our models improved sensitivities compared to the previous rules-based approach by 15 and 24% to 84 and 87% for pyrazinamide and streptomycin (P < 0.01), respectively. The best-performing models increase the area-under-the-ROC curve by 10% for pyrazinamide and streptomycin (P < 0.01), and 4–8% for other drugs (P < 0.01). Meanwhile, the specificities for all drugs dropped by 4–10%. Removing the known disassociated mutations (D.SNPs) altered the mean AUC by no more than 0.8% for all drugs. In addition, for several interesting subclades (e.g. EAI, CAS, Beijing and C1), the best classifiers also improved the mean sensitivity and AUC for INh, EMB, RIF, PZA and MDR (Supplementary Material I).
4 Discussion
A machine learning approach towards MTB resistance classification is both viable and, particularly when the underlying biologic mechanisms are less well-studied, offers improvement upon the current clinical state-of-the-art. It is typically thought that sensitivity is most important in our application, because failure to identify resistance can harm patients: in such cases, treatment would proceed using drugs that do not affect the (resistant) bacteria. Comparatively, the DA methods are generally very specific, but not as sensitive as machine learning methods. In our study, specificity often decreased with the machine learning methods, which could be due to some susceptible isolates containing determinants classified as causing resistance, but were labeled as drug susceptible due to limitations in the phenotypic methods used to assess drug resistance.
The best classifiers (with feature set F1), offer improvement upon the baseline DA method for INH and PZA, potentially because there are additional mutations to reported resistance-determinants, or because there may be multivariate associations (i.e. co-occurrence of resistance) or interactions between mutations within the 23 genes considered in this study. The fact that machine learning methods did disproportionately better with PZA could also because that there is a large number of contributory variants for PZA resistance, compared to most of the other drugs investigated. In the case of the best classifiers with feature set F2, the improvement regarding to EMB, CIP and SM could result from resistance co-occurrence, upon which the machine learning models capitalize to improve its results. For RIF, MOX and OFX, the improvement from using the best classifiers with feature set F3 suggests there may be unknown associations within genes suspected to be related to resistance. The likely interpretation associated with F3 being the best classifier for some drugs, is that additional mutations to reported resistance-determinants or combinations of mutations (i.e. interactions) in the suspected genes are more sensitive for identifying resistance to the considered drug. Relatively higher performance obtained by the best classifier with feature set F3 for MDR-TB detection may illustrates there is potential pattern related to drug resistance co-occurrence. The additional SNP candidates to resistance-determinants within the 23 genes are listed in Supplementary Material H.
In general, classifier performance with the F1 feature set removing known D.SNPs gave results that still outperformed DA in improving AUC for all drugs. Our methods achieved at least 95% AUC, except for MOX, OFX and SM, where poorer performance might also be due to a result of there being small numbers of resistant isolates for these drugs. This indicates that the best classifiers are robust to the removal of D.SNPs, which supports the potential application in whole genome sequencing especially when the resistance mechanisms for some drugs remains incompletely understood. It would be interesting to explore in future work whether performance further improves, or degrades, if either synonymous SNPs [which can rarely be associated with resistance Ando et al. (2014)] or SNPs across the entire genome (not just in genes previously associated with drug resistance) are considered. Both would vastly expand the size of the feature sets.
The machine learning approaches that we have investigated provide the greatest improvements in classification performance for those drugs in which the mechanisms of resistance are less well-understood. For PZA and SM, the baseline DA prediction achieved relative high specificity, but very low sensitivity (58 and 63%, respectively). For the same drugs, the machine learning methods improved overall AUC by 10% (attaining 86 and 89% sensitivity, respectively, while retaining >87% specificity). This improvement is likely to be caused by patterns of resistance to multiple drugs that the machine learning models can exploit. For instance, if an isolate is resistant to PZA, it is also likely to be resistant to INH or RIF. This can also be used to explain why all machine learning methods with F1 and F2 were better than F3 for both PZA and SM in Figure 3. While these co-occurrences may obscure the true mechanistic basis of resistance, they are still practically useful when designing a treatment plan for a patient.
Among the machine learning classifiers, PM and SVM-RBF rank as the top two best-performing classifiers overall. The former integrates prior knowledge of resistance determinant explicitly (though prior knowledge about susceptibility determinants, lineage defining SNPs and compensatory mutations was not included), and the latter suggests that the method’s nonlinear model is suitable for unveiling nonlinear relationships among mutations in terms of resistance association for INH, CIP and SM. We acknowledge our dataset was subset of the dataset in Walker et al. (2015), from which the library of DA-L was derived. It is expected that the DA-L would over-perform DA-D, however, this is not true for EMB and second-line drugs (Supplementary Material F).
We note that our analysis was limited by low resistance, even for INH and RIF there were not so many resistance cases. It is acknowledged that there is trade-off between bias and variance in the machine learning methods. We attempted to use cross-validation to manage the trade-off for small dataset and only reported average performance on testing set instead of training set. In the analysis of selected clusters (Supplementary Material I), all methods resulted in higher variance than that in the entire dataset. DA method was more biased in the clusters than in the entire data (classification sensitivity of DA was higher in clusters for INH and PZA up to 98%, but lower for RIF, EMB and MDR-TB down to 81%); while the machine learning classifiers gave similar performance with that obtained when using the data as a whole.
5 Conclusion
We investigated several classifiers for resistance classification that demonstrated the potential to model genetic data (e.g. SVM_RBF) and take into account the prior knowledge and latent subgroup structure (e.g. PM and CBMM). We applied the classifiers to three different feature sets and the best-performing model outperformed the baseline method (DA) in terms of sensitivity to resistance classification. This work showed great potentials of machine learning in improving resistance classification given high-dimensional genetic data, especially when the underlying biological resistance mechanism is poorly understood for many drugs. Use of the best model examined in this paper to predict MTB resistance is promising to improve patient outcomes and reduce risk of acquiring multi-drug resistance. The validation on global samples will be the future work. Limitation of our work is that the examined methods considered ALL polymorphisms (and resistance-determining) to be the same even though some polymorphisms might have different effects.
Supplementary Material
Acknowledgements
Y.Y. gratefully acknowledges the support of a K.C.Wong Education Foundation. K.E.N. acknowledges funding from the Rhodes Trust and the RCUK Digital Economy Programme grant number EP/G036861/1 (Center for Doctoral Training in Healthcare Innovation). D.A.C. was supported by the Royal Academy of Engineering, the EPSRC via a ‘Grand Challenge’ award. This project was supported by the Bill & Melinda Gates Foundation, Wellcome Trust and the NIHR Senior Investigators. D.J.W. is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (Grant 101237/Z/13/Z).
Funding
This project was funded by a K.C. Wong Postdoctoral Fellowship, the Rhodes Trust, the RCUK Digital Economy Programme (Grant EP/G036861/1), an EPSRC Grand Challenge award (EP/N020774/1), the Bill & Melinda Gates Foundation, the Wellcome Trust, NIHR Senior Investigatorships, and the Royal Society (Grant 101237/Z/13/Z).
Conflict of Interest: none declared.
References
- Ando H. et al. (2014) A silent mutation in maba confers isoniazid resistance on Mycobacterium tuberculosis. Mol. Microbiol., 91, 538–547. [DOI] [PubMed] [Google Scholar]
- Benavente E.D. et al. (2015) Phytb: Phylogenetic tree visualisation and sample positioning for M. tuberculosis. BMC Bioinformatics, 16, 155.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali N. et al. (2014) Evolution and transmission of drug resistant tuberculosis in a Russian population. Nat. Genet., 46, 279.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen K.A. et al. (2015) Evolution of extensively drug-resistant tuberculosis over four decades: whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med., 12, e1001880.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll F. et al. (2015) Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med., 7, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I. et al. (2011) Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in rna polymerase genes. Nat. Genet., 44, 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldholm V. et al. (2016) Armed conflict and population displacement as drivers of the evolution and dispersal of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA, 113, 13881–13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat M.R. et al. (2013) Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet., 45, 1183–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerriegel S. et al. (2014) Phylogenetic polymorphisms in antibiotic resistance genes of the Mycobacterium tuberculosis complex. J. Antimicrob. Chemother., 69, 1205–1210. [DOI] [PubMed] [Google Scholar]
- Ford C.B. et al. (2013) Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet., 45, 784–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georghiou S.B. et al. (2012) Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PloS One, 7, e33275.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global tuberculosis report (2017). Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO.
- Safi H. et al. (2013) Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-[beta]-d-arabinose biosynthetic and utilization pathway genes. Nat. Genet., 45, 1190–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki D. et al. (2012) Two new rapid snp-typing methods for classifying Mycobacterium tuberculosis complex into the main phylogenetic lineages. PLoS One, 7, e41253.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T.M. et al. (2015) Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect. Dis., 15, 1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO treatment guidelines for drug-resistant tuberculosis (2016 update). (2016). Geneva: World Health Organization. [PubMed]
- Zhang H. et al. (2013) Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet., 45, 1255–1260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.