Abstract
Several reviews and case reports have described how information derived from the analysis of genomes are currently included in electronic health records (EHRs) for the purposes of supporting clinical decisions. Since the introduction of this new type of information in EHRs is relatively new (for instance, the widespread adoption of EHRs in the United States is just about a decade old), it is not surprising that a myriad of approaches has been attempted, with various degrees of success. EHR systems undergo much customization to fit the needs of health systems; these approaches have been varied and not always generalizable. The intent of this article is to present a high-level view of these approaches, emphasizing the functionality that they are trying to achieve, and not to advocate for specific solutions, which may become obsolete soon after this review is published. We start by broadly defining the end goal of including genomics in EHRs for healthcare and then explaining the various sources of information that need to be linked to arrive at a clinically actionable genomics analysis using a pharmacogenomics example. In addition, we include discussions on open issues and a vision for the next generation systems that integrate whole genome sequencing and EHRs in a seamless fashion.
Introduction
Genome analysis is relatively new to the practicing clinician, and thus its inclusion in electronic health records (EHRs) is not yet standardized. Most genome analyses performed in Clinical Laboratory Improvement Amendments (CLIA) certified laboratories in the United States today consist of targeted panels for pharmacogenomics or cancer-related genes. However, there are an increasing number of analyses of exome and/or whole genome sequencing (WGS). This article discusses genome analyses as represented in the EHRs today and how we envision it will be included when WGS becomes mainstream to clinical medicine.
WGS-derived Information: Where Is It in the EHR?
The goal of a clinical interpretation of WGS analysis is to identify individual variations in DNA (e.g. substitutions, insertions, deletions, duplications etc.) that have clinical significance. By clinical significance we mean a phenotype that is strongly associated with the variation. Clinicians often refer to these as ‘actionable’ variations, whose usefulness is not limited to assisting in prescribing (e.g. initiation or adjustment of dosing for a certain medication), but also help in diagnosing and managing a disease or other health conditions.
The EHR is a legal document designed to assist in documentation and billing, in addition to facilitating clinical care. It is difficult to determine what type of genome information to include in the EHR because of the evolving nature of genetic test interpretations. It is also challenging to determine how to best use this information at the point of care. Similarly, as in other types of clinical decision support, the development and maintenance of a knowledge base reflecting rules that are implemented in EHRs to support clinician decisions is costly and, if not carefully managed, prone to obsolescence and errors that may inadvertently harm, rather than help, clinical decision making. As an illustration of how heterogeneous the solutions have been, consider for example how clinicians enter and receive information from WGS analyses: as clinicians move around different health systems, they may find WGS-derived information in different sections of the EHR. Furthermore, the recommended actions for an identical genome analysis may vary greatly depending on how the health system adjudicates clinical evidence [e.g. by internal committees or by adoption of recommendations from a particular specialty society, typically in the United States the American College of Medical Genetics or Association of Clinical Pathology (ACP), or guidelines from experts such as the Clinical Pharmacogenetics Implementation Consortium] (1,2).
A review of the recent literature (3) indicates that the final results or interpretations of WGS analyses, such as the presence of pathogenic, clinically significant, variants in germline genomes, are commonly entered in the EHRs as ‘allergies’ (so that clinicians are alerted accordingly for the purposes of prescribing certain medications) or ‘problems’ embedded in problem lists that typically contain several entries. Human leukocyte antigen class I-B (HLA-B) and abacavir is one such gene–drug pair example enlisted in an EHR allergies section. Individuals carrying an HLA-B*57: 01 allele are reported to be at high risk for developing hypersensitivity to abacavir, a nuclease reverse transcriptase inhibitor used to treat HIV disease. In the Mayo EHR system, this allergy information is shown to a physician at the time of prescribing, with suggestions to change orders toward improving patient safety (4). In contrast, Boston Children’s Hospital stores the relevant variant information, including the HLA-B*57:01 above, in the EHR problem list (5). Another example is thiopurine methyltransferase. The SNP rs1800460 (chr6: 18138997 C/T) in thiopurine S-methyltransferase (TPMT) gene is one of example of a relevant gene–drug pair (6). The risk allele T is known to be associated with deficiency in thiopurine efficacy and leukopenia as an adverse drug effect. If a patient has CT or TT alleles instead of CC, a wild-type, the phrase ‘TPMP enzyme deficiency’ is inserted to the EHR problem list (6). Clearly, while the type of information is very different, the reason why these variants are placed in the ‘problem’ or ‘allergy’ list relates to the easy implementation of EHR alerts and reminders. By placing this information in these lists, EHR users are simply trying to make sure the information is quickly available for the next provider, instead of buried in narrative clinical notes. Although practical from a care point of view, when these allergy and problem lists are used for research and for the learning healthcare system, they need to be carefully curated.
Other groups have considered WGS a ‘procedure’ (5), due to the multiple ways in which a WGS analysis workflow can be conducted, and the importance of understanding the trade-offs due to different types of analyses. The Substitutable Medical Applications & Reusable Technologies (SMART) on Fast Health Interoperability Resource (FHIR) Genomics Resource team proposed to add the <SequencingLab> extension to the <Procedure> resource to store the information about sequencing protocols and variant calling algorithms, similar to other <Procedure> examples such as surgical procedure, endoscopic procedure and biopsy procedure (5). In contrast, the observed sequence (A/C/T/G) data generated by above ‘procedure’ goes to the <Sequence> resource and genotype-phenotype information is saved in the <Observation> resource, which also includes other observed elements like body weight and temperature, glomerular filtration rate, bone density, EKG data and tobacco use.
Even the notion of a clinically significant variant is fluid. Whether a variant is pathogenic or benign is often determined by an organized body of experts using their clinical experience and available literature (7,8). However, an important tenet of precision medicine (and thus WGS interpretation) is that not all variants have the same effects in all patients). Therefore, describing which tools were used to identify variants (i.e. the basis for considering a DNA base uncommon) and what evidence was used to arrive at a clinical significance interpretation for that variant or structural variation (i.e. to name a particular variant as ‘clinically significant’) is important, hence the advocacy for classifying WGS as a ‘procedure’. The rationale is that, just as important to understand whether a mass was found by MRI or x-ray, it is important to know how a pathogenic variant was determined. Workflows for WGS analyses that are CLIA-compliant have been recently proposed by the ACP (9,10). However, a side effect of considering WGS results ‘procedures’ is that designing alerts and reminders from procedures in major EHR systems is not as easy as defining them from ‘allergy’ or ‘problem’ lists.
Conceptually, WGS interpretation could be considered a laboratory test in which the interpretation could change given the findings in the literature and the accumulation of evidence in the learning healthcare system (11). The similarities with a liver enzyme test, in which daily reference ranges are produced at each laboratory, are several: the results cannot be interpreted without the reference, there are thresholds to flag results as abnormal or borderline, and they are used in the context of other findings for the patient. In essence, the analytical procedures applied to the WGS to arrive at variant detection are an inextricable part of the WGS test, much like reference ranges are part of a specific laboratory result. There are important differences too: unlike liver enzymes, sequencing of germline genomes does not need to be repeated as the phenotype changes (unless technical improvements warrant re-sequencing of the genome), and the ‘reference’ changes not just by range, but by variant or variation as new evidence accumulates. That is, unlike a liver enzyme test, which can be documented as ‘elevated 3x from the reference normal range on 2/27/18,’ a WGS interpretation constitutes millions of ‘tests’ and can only be documented as ‘clinically significant variant ABC found on 2/27/18, considering evidence from the Nth version of the XYZ resource’ [e.g. ClinVar (12)]. Additionally, the next time a WGS interpretation is ‘ordered’, portions of the analytical pipeline may need to be re-run, but not the ‘test’.
The Mayo Clinic puts its variant findings in the EHR Lab ‘Results’ section (4). Unlike Boston Children’s Hospital, Mayo puts the same rs1800460 in TPMT and thiopurine pair in pharmacogenomics lab results. The Mayo system reminds clinicians of the necessity of reactive genotype testing. For example, TPMT for azathioprine has linked genotype–phenotype documentation either in the problem list or allergy section, which links to suggestions for possible prescription modifications such as alternative drug or reduced dosage (3).
Nevertheless, regardless of where WGS interpretations are placed in the EHR, they remain critical data for patient-centered targeted therapy. The goal is to assist clinicians in using genetic information to diagnose, counsel, treat and prognosticate. WGS results should be placed in an EHR such that they can be integrated into complex decision making that maximizes the use of actionable information while preserving efficiency in busy clinical practices. The components of this ecosystem that could interact with an integrated EHR system are described next.
Components of a WGS-Based Clinical Decision Support System
We will henceforth consider WGS analysis outputs as test results and describe the components of a WGS-based clinical decision support system using a common pharmacogenomics example. We divide decision support components into (i) ordering of the WGS test, (ii) generating the interpretation at the ‘laboratory’, (iii) importing results into the EHR, (iv) triggering of alerts and reminders and (v) evaluating outcomes.
Ordering of WGS test
Clinician provider order entry (CPOE) systems have been in place for decades, and CDS associated with the orders has been shown to be effective in reducing unnecessary or inappropriate testing and costs in many studies, particularly when the CPOE system is integrated with the EHR system. Decision support in CPOE ranges from drug–drug interaction checks to guidelines for requesting imaging studies. Although many studies show benefits of CPOE, some show no difference and others show that sub-optimal implementation of CPOE can have opposite effects (13,14). No studies have evaluated large implementations of WGS CPOE to date, but the expectation is that they will show a similar trend as studies involving other types of CPOE. For example, if a clinician is interested in prescribing abacavir to a patient, the system should suggest first ordering a ‘test’ for screening HLA-B*5701, known to be associated with hypersensitivity to this medication. Conversely, if WGS is already available, the system should be able to query this particular genotype. Although this is an example in which the association is well known and the test is recommended by the US Department of Health and Human Services, examples involving variants of unknown significance also exist.
Generating the interpretation at the ‘laboratory’
Application programming interfaces (APIs) that allow communication between EHR systems and knowledge bases such as ClinVar (12) and PharmGKB (15) are increasingly being made available by laboratories and major EHR vendors. These APIs hold promise in making the EHR systems less insular, allowing integration of external software to enhance EHR functions (5,16). The goal is to have an automated screening system in a patient’s EHR that helps clinicians make decisions. The current bottleneck is not necessarily the API technology, but the acceptance of these knowledge bases at the clinical sites. Reports from CLIA labs only contain information considered important at the time they were created and may not have explicit information about variants that were later considered clinically significant. It is therefore inconsistent that the remainder of a WGS would be considered not ‘CLIA-certified.’ This may be because the variant calling algorithm would need to be adjusted.
Importing results into the EHR
Unfortunately, just like many pathology and radiology reports, many CLIA-certified WGS results still consist of raw, unstandardized, narrative interpretations that are inserted in the EHRs as pdf documents, making it more difficult to compute. Parsing XML results from major laboratories is relatively simple, but XML schemas vary widely across multiple vendors and querying for specific variants is not within scope. Re-analysis of WGS, which is critically important given the fluid nature of evidence (17), is not currently available. Furthermore, effectively interfacing genomics and other data in the EHR (e.g. diagnoses, medications, laboratory test results) to generate accurate algorithms and clinically relevant recommendations (e.g. drug–gene interactions and monitoring) will greatly enhance personalized medicine practice at the point of care.
Triggering of alerts and reminders
The design, implementation and evaluation of rules for an EHR-based CDS system have been studied in many contexts outside WGS analysis. A few case studies report on frameworks for implementation at a single healthcare system (18). The complexity of these tasks for WGS data is higher when these data are added to other clinical data: not only is the pace of evidence generation higher, but new discoveries due to ancestry and environment (in the case of somatic variants) are being reported daily. Because evidence from the literature is not yet easily computable, the need for curating the literature is high and non-scalable solutions are still the norm. Whether outsourcing to a scientifically-abled ‘crowd’ (19) or the emergence of computable literature (20) will be feasible solutions remains to be seen.
Evaluating outcomes
Many questions remain on how to evaluate the outcomes of WGS-based CDS systems. For example, if the intended outcome is not achieved for a treatment, would re-interpretation of WGS be warranted? Or should a reinterpretation be updated ‘continuously’ and clinicians and/or patients alerted when new findings become relevant to them? (17) If all existing evidence comes from populations that are quite distinct than that of the patient, would WGS results be a mismatch? Should recommendations based on these results be avoided? What will constitute WGS interpretation malpractice? How effective (or disruptive) are genomics-based alerts and reminders? Unfortunately, the field is too immature for these questions to be answered. Additionally, the current applications of WGS-based CDS have been mostly limited to academic medical centers; these issues have yet to be prioritized by administrators and policy makers.
Standards for Representing Genomic Variants
Once the determination is made to acquire and store WGS in an EHR record, the next important decision is what to store. There are a number of formats that have been used to represent genomic information. For variants, the optimal representation would include the common ‘name’ as it is more readily recognized, easier to read and elicits knowledge of its clinical impact. For example, the PML–RARA fusion gene, present in 98% of promyelocytic leukemia patients, is a target of all-trans retinoic acid and arsenic trioxide and thus predictive of a favorable outcome due to sensitivity of these molecular therapies (21,22). Recording its common name, ‘PML–RARA’, is simpler to interpret but provides limited computability. Instead, it may be preferable to record multiple forms of the information, such as the chromosomal translocation t(9; 22)(q34; q11.2), the resulting RNA changes, as well as the resulting oncoprotein. This allows multiple uses of these patient data. For example, having the variant represented in a common structured nomenclature enables the querying of external knowledge bases for updated information on its clinical relevance. Using the short name in clinical notes allows for better readability and transfer of this information among clinicians caring for the patient. The Molecular Pathology Resource Committee of the College of American Pathologists has highlighted some of the challenges in this area and issued recommendations on how to optimally provide information to maximize utility for the clinician (23).
As mentioned, if a DNA variant is recorded using a standard nomenclature, it allows one to build links to knowledge bases to inform clinicians about the impact of a particular variant. This would require standardization to a common nomenclature and structured format by the EHR and the knowledge base. Unfortunately, the most common way variants from WGS are reported today is via narrative text reports, typically in PDF format, and without a common standard vocabulary or structured format. Standards to enable interoperability and standardized exchange of clinical genomic data are emerging, but are not yet widely adopted.
Recommendations on nomenclature and a tiered-approach to clinical significance have been recently issued by professional societies relevant to clinical genomics and are a step towards providing EHR and clinical genomic testing centers common standards for interoperability (7). The proposed consensus is that variants should be described using standards published by the Human Genome Variation Society (HGVS) which joined forces with the Human Genome Organization Gene Nomenclature Committee and the Human Variome Project (24) to produce standards for documenting reference sequences for genomic DNA, coding DNA, non-coding DNA, mitochondrial DNA, RNA and the resulting protein (25). Logical Observation Identifiers Names and Codes implements the HGVS syntax to represent variants of interest (26) and provides and expanding array codes for reporting clinical genomic tests and results (27). Health Level 7 has also issued an implementation guide for structuring genetic test results into the EHR (28). As these specifications continue to improve through the standards process, they will hopefully be adopted by both clinical genomics testing laboratories and EHR vendors. So far, the science has far outpaced genomics healthcare data standards. This has resulted in a significant lost opportunity in being able to bring the full complement of modern computing to support clinicians in what is an increasingly complex decision-making environment.
Linking to Knowledge and Tools for Interpretation of WGS
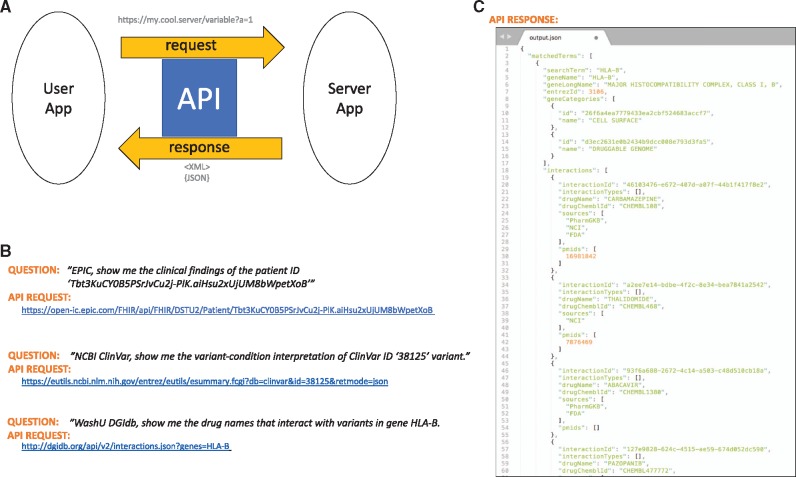
Knowledge about WGS-derived variants is increasing rapidly and will quickly overwhelm a clinician’s ability to have relevant information brought to bear at the point of care. There are a number of resources and databases available for secondary analysis and for obtaining contemporary information about a particular variant. Access from the EHR to these decision support systems will be critical as this knowledge continues to expand. Linkages from the EHR to external knowledge bases through APIs will be a key factor in maximizing the benefit of WGS for patients. An API is a set of definitions about program instructions, protocols, and tools for application software. It enables communication between different software applications (5,16). An application makes a request to a server, which returns a response back to the requesting system. Typically, a user of the requesting system triggers the process. Today, the most common way to implement APIs is to use the hypertext transfer protocol (HTTP) using a Representational State Transfer (REST) architectural style, frequently referred to as Representational State Transfer-ful Web Services. The Global Alliance for Genomics and Health initiated the Beacon Project (29) with the request query https://beacon-network.org/#/search?pos=32936732&chrom=13&allele=C&ref=G&rs=GRCh37 for a genetics question, ‘UC Santa Cruz, do you have a genome that has allele ‘C’ at position 32936732 on chromosome 13 in the human genome build GRCh37?’ Then UCSC transfers an XML file with ‘true’ as a response. The same question was asked to Broad Institute, and the response was ‘false’. Figure 1 shows more working API examples from the Epic system, ClinVar archive and the drug–gene interaction database called DBIdb (30). The questions in words are translated into machine-readable APIs. Figure 1B demonstrates that clinical findings, such as a patient’s vital sign (e.g. systolic blood pressure), can also be retrieved in XML/JSON format by requesting the data from the EHR system (e.g. Epic) via a FHIR API. The second example in Figure 1B is making a request to a resource database, NCBI ClinVar (12,31), for a variant-condition interpretation. The third request in Figure 1B asks for a specific gene–drug interaction. A JSON text file, as shown in Figure 1C, is returned as a result, with pathogenicity and breast/ovarian cancers as ‘related conditions’. APIs thus enable data sharing and integration of genomic variants and clinical phenotypes. Through APIs, different apps can communicate efficiently in a fully automated way. APIs are flexible because they communicate at the application layer. APIs can connect two apps seamlessly and are easily customizable, as illustrated by the multiple FHIR Apps (5) released at Healthcare Information and Management Systems Society (HIMSS) annual conference 2014 and by Duke University (16). Currently, genomics APIs are still under development and recently Google Genomics, SMART Genomics and 23andMe released their early versions (32).
Figure 1.
Schematic illustration of API for genomics and EHR. (A) Generic user-server communication using an API. (B) Translation of words into API request URLs against real servers in EPIC, NIH and WashU. (C) An example response, or output, from DGIdb in JSON format. Given a gene, HLA-B, drug ABACAVIR is returned based on PharmGKB and FDA data, as described in this article.
In addition to technical considerations, the inclusion of genomics in the EHRs must also consider ethical, social and legal implications (33). Complicated issues include interpretability of results and availability of trained personnel to answer questions, particularly in an era where patient portals (34) and open notes (35) are increasingly being implemented to promote transparency in the EHRs, and patient preferences towards data sharing and privacy need to be addressed in parallel with technical advances (36,37). Furthermore, full integration into the clinical workflow is needed. An illustrative use case relates to pharmacogenomics and peri-operative workflows.
A major challenge in executing pharmacogenomics tailored programs includes more clinician buy-in. Obtaining buy-in from a medical community is a complex process. As an example, the use of pharmacogenomics in perioperative medicine has much potential. During a patient’s perioperative experience, healthcare providers are tasked with the management of multimodal pharmacotherapy. Anesthetic and pain management during the intra- and post-operative period may be challenging, as several classes of medications have varying degrees of efficacy and toxicity. The effects are even more unpredictable due to inherent genetic variability. Effective opioid-sparing strategies are important for successful enhanced recovery after surgery (ERAS) protocols (38). These protocols are aimed to outline the appropriate steps of perioperative care for major surgery; however, all of the potential factors affecting optimal care are not fully known because of genetic variability across patients. Having access to a patient’s pharmacogenomics information would allow healthcare providers to practice personalized medicine, thereby driving more effective outcomes. Several studies have linked an association of genotype with opioid metabolism (39–44). Although the field of pharmacogenomics has been longstanding, its widespread implementation into perioperative care is still at its infancy. The operating room is a fast-paced clinical environment, therefore, inclusion of pharmacogenomics into clinical decision workflows need to be well integrated without disrupting the time required to maintain operating room efficiency.
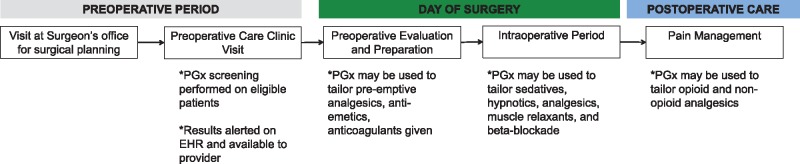
Figure 2 illustrates the basic workflow of the perioperative experience for a patient: (i) preoperative period, in which patients are screened by surgeons and anesthesiologists to determine their surgical candidacy and whether they are medically optimized; (ii) intraoperative period, in which patients undergo a surgical procedure requiring a variety of pharmacological agents to provide anesthesia and analgesia; and (iii) postoperative period, in which patients recover from surgery and need to meet various discharge milestones. Patient consenting (45) and education regarding pharmacogenomics should be performed during the preoperative period (i.e. at the preoperative care clinic where the anesthesiologist prepares patients for surgery or at the surgery clinic). There should be enough time from requesting genotype data to day of surgery so that the results are available in a timely manner. Once pharmacogenomics results are available, perioperative providers (i.e. anesthesiologists, surgeons, nurses and pharmacists) should be notified regarding which surgical patients have available results in the EHR. Based on the genes tested, providers should be given information regarding potential drug responses to different pharmacological categories including: anticoagulants, beta-blockers, sedatives, anti-emetics, hypnotics, muscle relaxants, analgesics (opioid and non-opioids) and volatile anesthetics. On the day of surgery, providers should have the information to integrate the pharmacogenomics results into their practice in order to optimize personalized care during the pre-, intra- and post-operative periods (including acute pain management during the first week following surgery). Pharmacogenomics integrated into intra- and post-operative management has potential to reduce total opioid consumption. Such tools may be implemented in ERAS programs and Acute Pain Services. Patients undergoing surgeries associated with higher opioid use—such as joint arthroplasty, complex spine surgery and major abdominal surgery—may benefit. Much more studies are needed to design protocols that optimize dosing and medication changes based on genetic risk and associated outcomes. As these studies are produced, these protocols should be easily integrated into a perioperative pharmacogenomics service.
Figure 2.
Workflow of the perioperative process and integration of pharmacogenomics. PGx, pharmacogenomics.
The use of pharmacogenomics holds great promise to the personalization of perioperative care; however, there are several challenges that need to be addressed: (i) the plethora of results from pharmacogenomics screening (especially when hundreds of genes are tested) will make user-friendly presentation of the results to clinicians challenging; (ii) the barrier to physician buy-in needs to be overcome with evidence of patient outcomes improvement; (iii) disruption to usual clinical flow must be minimized (i.e. will adding another test and report to a physician’s daily workflow be deemed cumbersome?); and (iv) clinical decision support must be provided (e.g. What are the associated meaningful phenotypes to the reported genotypes? What should healthcare providers actually do with the data?). For pharmacogenomics to become widely adopted in the perioperative space, all of these challenges need to be met adequately.
Are We There Yet? Missing Links to Realize the Vision of Genomic Medicine
Cholesterol measurements took over 20 years to be standardized for clinical use; their widespread ordering was primarily tied to research findings from Framingham cohort studies (46). WGS has been around for over 10 years; only recently has the price per genome dropped sufficiently to make it a potentially viable ‘test’ for a small percentage of the world’s population. For example, coverage from health insurance in the United States is still low and limited to specific gene panels. It is thus not surprising that standardization of WGS results and standardization of how they are included in the EHR have not been prioritized. Additionally, cholesterol measurements are limited to a few tests, as opposed to WGS-based results, which may involve thousands of ‘tests’. In both cases, a large cohort of individuals needs to provide data and samples so that associations can be determined and inferences can be made. It is thus premature to suggest that WGS-based analyses will transform the way medicine is practiced overnight, especially since large scale studies that compare the incremental benefit of WGS-based ‘tests’ over traditional family history data have not been conducted in the context of clinical care settings. Nevertheless, it would also be premature to declare that WGS-based testing will not be widely adopted because of the difficulties in storing the sequences in a protected environment, analyzing the computationally complex data and the constantly moving basis for interpretation.
Initiatives such as the All of Us (47) Research program, which will collect clinical data, participant-provided information, and samples from over a million individuals from diverse backgrounds over a decade, will boost discoveries and provide a good setting for investigating the feasibility of incorporating WGS-based results into healthcare, and consequently, into EHRs. Over time EHRs are expected to become more modernized, provide more open APIs for a variety of uses, and have lower frequencies of missing data for important variables such as social determinants of health, ancestry and environment. Aside from achieving personalized medical goals based on genomics data, futuristic goals of generating new knowledge and clinically relevant discoveries using population-based genomics data can someday be achieved by using EHRs. The confluence of integrating genomics data with new analytical methods to answer questions specific to an individual and policies that promote data sharing while preserving the privacy of individuals and institutions has already started. Just as monitoring child growth, we should keep in mind how frequently it makes sense to perform measurements of genomic medicine success. Progress is happening daily. If the scientific community addresses issues of standards and evidence generation now, genomic medicine will be a reality that will make future generations wonder how medicine was practiced before the human genome was sequenced.
Acknowledgments
Conflict of Interest statement. None declared.
Funding
J.K. and L.O.-M. are partially funded by National Institutes of Health (grants R01HG008802, R01HL136835) and VA IIR (12–068). L.O.-M. and M.H. are partially funded by PCORI (contract CDRN-1306–04819) and National Institutes of Health (award OTOD025462). L.O.-M. is partially funded by National Institutes of Health (grant R01GM118609).
References
- 1. Caudle K.E., Rettie A.E., Whirl-Carrillo M., Smith L.H., Mintzer S., Lee M.T., Klein T.E., Callaghan J.T. (2014) Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA-B genotypes and phenytoin dosing. Clin. Pharmacol. Ther., 96, 542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leckband S.G., Kelsoe J.R., Dunnenberger H.M., George A.L. Jr, Tran E., Berger R., Muller D.J., Whirl-Carrillo M., Caudle K.E., Pirmohamed M. (2013) Clinical Pharmacogenetics Implementation Consortium guidelines for HLA-B genotype and carbamazepine dosing. Clin. Pharmacol. Ther., 94, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caraballo P.J., Bielinski S.J., St Sauver J.L., Weinshilboum R.M. (2017) Electronic medical record-integrated pharmacogenomics and related clinical decision support concepts. Clin. Pharmacol. Ther., 102, 254–264. [DOI] [PubMed] [Google Scholar]
- 4. Caraballo P.J., Hodge L.S., Bielinski S.J., Stewart A.K., Farrugia G., Schultz C.G., Rohrer-Vitek C.R., Olson J.E., St Sauver J.L., Roger V.L.. et al. (2017) Multidisciplinary model to implement pharmacogenomics at the point of care. Genet. Med., 19, 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alterovitz G., Warner J., Zhang P., Chen Y., Ullman-Cullere M., Kreda D., Kohane I.S. (2015) SMART on FHIR Genomics: facilitating standardized clinico-genomic apps. J. Am. Med. Inform. Assoc., 22, 1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Manzi S.F., Fusaro V.A., Chadwick L., Brownstein C., Clinton C., Mandl K.D., Wolf W.A., Hawkins J.B. (2017) Creating a scalable clinical pharmacogenomics service with automated interpretation and medical record result integration - experience from a pediatric tertiary care facility. J. Am. Med. Inform. Assoc., 24, 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li M.M., Datto M., Duncavage E.J., Kulkarni S., Lindeman N.I., Roy S., Tsimberidou A.M., Vnencak-Jones C.L., Wolff D.J., Younes A.. et al. (2017) Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: a Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists . J. Mol. Diagn., 19, 4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E.. et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med., 17, 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai E.A., Shakbatyan R., Evans J., Rossetti P., Graham C., Sharma H., Lin C.F., Lebo M.S. (2016) Bioinformatics workflow for clinical whole genome sequencing at partners healthcare personalized medicine. J. Pers. Med., 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghosh R., Oak N., Plon S.E. (2017) Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol., 18, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kho A.N., Rasmussen L.V., Connolly J.J., Peissig P.L., Starren J., Hakonarson H., Hayes M.G. (2013) Practical challenges in integrating genomic data into the electronic health record. Genet. Med., 15, 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harrison S.M., Dolinsky J.S., Knight Johnson A.E., Pesaran T., Azzariti D.R., Bale S., Chao E.C., Das S., Vincent L., Rehm H.L. (2017) Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet. Med., 19, 1096–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Amato M.G., Salazar A., Hickman T.T., Quist A.J., Volk L.A., Wright A., McEvoy D., Galanter W.L., Koppel R., Loudin B.. et al. (2017) Computerized prescriber order entry-related patient safety reports: analysis of 2522 medication errors. J. Am. Med. Inform. Assoc., 24, 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kannampallil T.G., Abraham J., Solotskaya A., Philip S.G., Lambert B.L., Schiff G.D., Wright A., Galanter W.L. (2017) Learning from errors: analysis of medication order voiding in CPOE systems. J. Am. Med. Inform. Assoc., 24, 762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barbarino J.M., Whirl-Carrillo M., Altman R.B., Klein T.E. (2018) PharmGKB: a worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med., doi:10.1002/wsbm.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bloomfield R.A. Jr, Polo-Wood F., Mandel J.C., Mandl K.D. (2017) Opening the Duke electronic health record to apps: implementing SMART on FHIR. Int. J. Med. Inform., 99, 1–10. [DOI] [PubMed] [Google Scholar]
- 17. Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L.. et al. (2015) ClinGen–the clinical genome resource. N. Engl. J. Med., 372, 2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peterson J.F., Bowton E., Field J.R., Beller M., Mitchell J., Schildcrout J., Gregg W., Johnson K., Jirjis J.N., Roden D.M.. et al. (2013) Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet. Med., 15, 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Griffith M., Spies N.C., Krysiak K., McMichael J.F., Coffman A.C., Danos A.M., Ainscough B.J., Ramirez C.A., Rieke D.T., Kujan L.. et al. (2017) CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet., 49, 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wei C.H., Harris B.R., Kao H.Y., Lu Z. (2013) tmVar: a text mining approach for extracting sequence variants in biomedical literature. Bioinformatics, 29, 1433–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nasr R., Lallemand-Breitenbach V., Zhu J., Guillemin M.C., de The H. (2009) Therapy-induced PML/RARA proteolysis and acute promyelocytic leukemia cure. Clin. Cancer Res., 15, 6321–6326. [DOI] [PubMed] [Google Scholar]
- 22. Nagai S., Takahashi T., Kurokawa M. (2010) The impact of molecularly targeted therapies upon the understanding of leukemogenesis and the role of hematopoietic stem cell transplantation in acute promyelocytic leukemia. Curr. Stem Cell Res. Ther., 5, 372–378. [DOI] [PubMed] [Google Scholar]
- 23. Gulley M.L., Braziel R.M., Halling K.C., Hsi E.D., Kant J.A., Nikiforova M.N., Nowak J.A., Ogino S., Oliveira A., Polesky H.F.. et al. (2007) Clinical laboratory reports in molecular pathology. Arch. Pathol. Lab. Med., 131, 852–863. [DOI] [PubMed] [Google Scholar]
- 24. Burn J., Watson M. (2016) The Human Variome Project. Hum. Mutat., 37, 505–507. [DOI] [PubMed] [Google Scholar]
- 25. den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.F., Smith T., Antonarakis S.E., Taschner P.E. (2016) HGVS recommendations for the description of sequence variants: 2016 update. Hum. Mutat., 37, 564–569. [DOI] [PubMed] [Google Scholar]
- 26. Campbell J.R., Talmon G., Cushman-Vokoun A., Karlsson D., Scott Campbell W. (2016) An extended SNOMED CT concept model for observations in molecular genetics. AMIA Annual Symposium Proceedings, vol. 2016, pp. 352–360. [PMC free article] [PubMed] [Google Scholar]
- 27. Deckard J., McDonald C.J., Vreeman D.J. (2015) Supporting interoperability of genetic data with LOINC. J. Am. Med. Inform. Assoc., 22, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bosca D., Marco L., Burriel V., Jaijo T., Millan J.M., Levin A., Pastor O., Robles M., Maldonado J.A. (2013) Genetic testing information standardization in HL7 CDA and ISO13606. Stud. Health Technol. Inform., 192, 338–342. [PubMed] [Google Scholar]
- 29. Health T.G.A. f.G.a. (2016) A federated ecosystem for sharing genomic, clinical data. Science, 352, 1278–1280. [DOI] [PubMed] [Google Scholar]
- 30. Cotto K.C., Wagner A.H., Feng Y.Y., Kiwala S., Coffman A.C., Spies G., Wollam A., Spies N.C., Griffith O.L., Griffith M. (2017) DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res., doi:10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gelb B.D., Cave H., Dillon M.W., Gripp K.W., Lee J.A., Mason-Suares H., Rauen K.A., Williams B., Zenker M., Vincent L.M. (2018) ClinGen’s RASopathy Expert Panel consensus methods for variant interpretation. Genet. Med., doi:10.1038/gim.2018.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Swaminathan R., Huang Y., Moosavinasab S., Buckley R., Bartlett C.W., Lin S.M. (2016) A Review on Genomics APIs. Comput. Struct. Biotechnol. J., 14, 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hazin R., Brothers K.B., Malin B.A., Koenig B.A., Sanderson S.C., Rothstein M.A., Williams M.S., Clayton E.W., Kullo I.J. (2013) Ethical, legal, and social implications of incorporating genomic information into electronic health records. Genet. Med., 15, 810–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wolff J.L., Kim V.S., Mintz S., Stametz R., Griffin J.M. (2017) An environmental scan of shared access to patient portals. J. Am. Med. Inform. Assoc., 25, 408–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wolff J.L., Darer J.D., Berger A., Clarke D., Green J.A., Stametz R.A., Delbanco T., Walker J. (2017) Inviting patients and care partners to read doctors’ notes: openNotes and shared access to electronic medical records. J. Am. Med. Inform. Assoc., 24, e166–e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mafi J.N., Mejilla R., Feldman H., Ngo L., Delbanco T., Darer J., Wee C., Walker J. (2016) Patients learning to read their doctors’ notes: the importance of reminders. J. Am. Med. Inform. Assoc., 23, 951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perzynski A.T., Roach M.J., Shick S., Callahan B., Gunzler D., Cebul R., Kaelber D.C., Huml A., Thornton J.D., Einstadter D. (2017) Patient portals and broadband internet inequality. J. Am. Med. Inform. Assoc. 24, 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ljungqvist O., Scott M., Fearon K.C. (2017) Enhanced recovery after surgery: a review. JAMA Surg., 152, 292–298. [DOI] [PubMed] [Google Scholar]
- 39. Fujita K., Ando Y., Yamamoto W., Miya T., Endo H., Sunakawa Y., Araki K., Kodama K., Nagashima F., Ichikawa W.. et al. (2010) Association of UGT2B7 and ABCB1 genotypes with morphine-induced adverse drug reactions in Japanese patients with cancer. Cancer Chemother. Pharmacol., 65, 251–258. [DOI] [PubMed] [Google Scholar]
- 40. Linares O.A., Daly D., Linares A.D., Stefanovski D., Boston R.C. (2014) Personalized oxycodone dosing: using pharmacogenetic testing and clinical pharmacokinetics to reduce toxicity risk and increase effectiveness. Pain Med., 15, 791–806. [DOI] [PubMed] [Google Scholar]
- 41. Linares O.A., Fudin J., Schiesser W.E., Daly Linares A.L., Boston R.C. (2015) CYP2D6 phenotype-specific codeine population pharmacokinetics. J. Pain Palliat. Care Pharmacother., 29, 4–15. [DOI] [PubMed] [Google Scholar]
- 42. Seripa D., Latina P., Fontana A., Gravina C., Lattanzi M., Savino M., Gallo A.P., Melchionda G., Santini S.A., Margaglione M.. et al. (2015) Role of CYP2D6 polymorphisms in the outcome of postoperative pain treatment. Pain Med., 16, 2012–2023. [DOI] [PubMed] [Google Scholar]
- 43. Takashina Y., Naito T., Mino Y., Yagi T., Ohnishi K., Kawakami J. (2012) Impact of CYP3A5 and ABCB1 gene polymorphisms on fentanyl pharmacokinetics and clinical responses in cancer patients undergoing conversion to a transdermal system. Drug Metab. Pharmacokinet., 27, 414–421. [DOI] [PubMed] [Google Scholar]
- 44. Wu X., Yuan L., Zuo J., Lv J., Guo T. (2014) The impact of CYP2D6 polymorphisms on the pharmacokinetics of codeine and its metabolites in Mongolian Chinese subjects. Eur. J. Clin. Pharmacol., 70, 57–63. [DOI] [PubMed] [Google Scholar]
- 45. Howard H.C., Joly Y., Avard D., Laplante N., Phillips M., Tardif J.C. (2011) Informed consent in the context of pharmacogenomic research: ethical considerations. Pharmacogenomics J., 11, 155–161. [DOI] [PubMed] [Google Scholar]
- 46. Tsao C.W., Vasan R.S. (2015) Cohort Profile: the Framingham Heart Study (FHS): overview of milestones in cardiovascular epidemiology. Int. J. Epidemiol., 44, 1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sankar P.L., Parker L.S. (2017) The Precision Medicine Initiative’s All of Us Research Program: an agenda for research on its ethical, legal, and social issues. Genet. Med., 19, 743–750. [DOI] [PubMed] [Google Scholar]