Abstract
Down syndrome (DS) is caused by the presence of part or an entire extra copy of chromosome 21, a phenomenon that can cause a wide spectrum of clinically defined phenotypes of the disease. Most of the clinical signs of DS are typical of the aging process including dysregulation of immune system. Beyond the causative genetic defect, DS persons display epigenetic alterations, particularly aberrant DNA methylation patterns that can contribute to the heterogeneity of the disease. In the present work, we investigated the levels of 5-hydroxymethylcytosine and of the Ten-eleven translocation dioxygenase enzymes, which are involved in DNA demethylation processes and are often deregulated in pathological conditions as well as in aging. Analyses were carried out on peripheral blood mononuclear cells of DS volunteers enrolled in the context of the MARK-AGE study, a large-scale cross-sectional population study with subjects representing the general population in eight European countries. We observed a decrease in 5-hydroxymethylcytosine, TET1, and other components of the DNA methylation/demethylation machinery in DS subjects, indicating that aberrant DNA methylation patterns in DS, which may have consequences on the transcriptional status of immune cells, may be due to a global disturbance of methylation control in DS.
Keywords: 5-hydroxymethylcytosine, TET enzymes, DNA methylation, aging
Down Syndrome (DS) is the most common genetic autosomal disorder compatible with life in humans. In the majority of cases, DS is a whole-chromosome trisomy due to meiotic nondisjunction of chromosome 21. The phenotypical features of the disease are multiple and heterogeneous, and they mainly comprise intellectual disability, growth defects, and endocrine disorders. Moreover, DS individuals are characterized by an acceleration of the aging process, affecting particularly the immune and central nervous system. In fact, individuals with DS show alterations of innate and adaptive immunity cell subsets and are more prone to develop dementia and Alzheimer’s disease (1–3). Accordingly, we recently demonstrated that neuropsychological functions and adaptive skills show an age-associated decline in persons affected by DS, including those recruited in the MARK-AGE study and further studied in this work (4,5). Moreover, an accelerated aging was also revealed in DS when N-glycans were analyzed (6).
Recently, much effort has been invested into the investigation of epigenetic signatures in DS persons (7,8). Attention has mainly been focused on DNA methylation for its prominent role in the control of gene expression both in physiological and pathological conditions. In mammals, DNA methylation originates from the addition of a methyl group to cytosine bases, generally located at the CpG dinucleotides, by the DNA methyltransferase (DNMT) family enzymes (DNMT1, DNMT3A, and DNMT3B) to form 5-methylcytosine (5mC). DNA methylation patterns are established and maintained to define transcriptional networks associated with cell-type identity. Several proteins are involved in the regulation DNA methylation dynamics including chromatin readers of 5mC, methylation-sensitive transcription factors, and, particularly, the Ten-eleven translocation (TET) family of DNA hydroxylases. In fact, TET enzymes (TET1, TET2 and TET3) catalyze the oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC), which can act as distinct epigenetic marks or represent intermediates of DNA demethylation. In particular, TET-mediated active DNA demethylation entails the thymine DNA glycosylase (TDG)-mediated DNA repair pathway for the removal of unnecessary oxidized 5mC (9).
A number of epigenome-wide analyses have shown differences in DNA methylation patterns between DS and control persons in brain, blood, buccal epithelial cells, and extra-embryonic tissues (8,10,11). These studies unambiguously demonstrated that in DS aberrantly methylated regions are distributed across all chromosomes and not limited to chromosome 21. Both hypermethylation and hypomethylation events were found in DS samples, although the incidence of hypermethylation was generally higher in brain regions and placenta (8,10,11). The accelerated aging phenotype of DS is mirrored by epigenetic alterations, as the disease increases DNA methylation age in whole blood (on average, 4.6 years), in leukocytes (on average, 3.9 years), and in brain (on average, 11.5 years) (12). Moreover, DNA methylation changes were already detectable in fetal and early postnatal development (8).
The contribution of 5hmC in DS pathology, however, is less characterized. Down-regulation of TET genes was suggested to be associated with DNA hypermethylation in placenta from DS samples (11), whereas hypomethylation of a TET1 gene regulative region (ie TET1 CpG island 3′-shore) was demonstrated to occur in whole blood from DS persons compared with their siblings (10). Only recently, a report has highlighted that 5mC and 5hmC additively account for most of the alterations of DNA methylation in DS cerebellum. A part of these alterations, however, were at the expense of 5hmC only; in that case, the levels of 5hmC were mainly decreased in DS samples (8). Notably, global changes in 5hmC levels have been identified in several neurodegenerative conditions (eg Alzheimer’s disease and multiple sclerosis), and loss of 5hmC is considered a hallmark of cancer. Moreover, we and others have recently demonstrated that blood cells undergo a gradual loss of 5hmC in aging (13,14). In particular, we demonstrated that the reduction of 5hmC was accompanied by decreased expression of TET1 and TET3 (14). Our results were obtained on peripheral blood mononuclear cells (PBMCs) deriving from volunteers enrolled in the Europe-wide cross-sectional population study “MARK-AGE.” In this context, also a cohort of DS persons was recruited because DS shares similarities with progeroid diseases, due to the precocious occurrence of age-associated features (1,3). In order to assess the role of DNA hydroxymethylation in DS immune cells we analyzed 5hmC levels and the expression of TET genes in PBMCs from DS persons compared with control individuals.
Materials and Methods
Study Population, Recruitment, Data, and Blood Collection
PBMC samples used in the present work derived from an Italian cohort of DS blood donors (n = 50; age mean = 40 ± 12; 48% female) and control volunteers from Italy (n = 45; age mean = 49 ± 12; 43% female) that have been recruited in the context of the MARK-AGE project (5,15,16). A group of young (n = 52; age mean = 38 ± 4; 58% female) individuals and another one of elders (n = 49; age mean = 78 ± 2; 45% female), enrolled in the same project from different European countries and previously tested (14), were also used for comparison with DS samples. The recruitment procedure and PBMC isolation details have been published (17,18). Briefly, PBMCs were isolated from EDTA-whole blood, obtained by phlebotomy after an overnight fasting, by discontinuous density gradient centrifugation in Percoll. Isolated cells were subsequently cryopreserved, stored in liquid nitrogen, and shipped to the MARK-AGE Biobank located at the University of Hohenheim, Stuttgart, Germany. From the Biobank, coded samples were subsequently sent to the Sapienza University of Rome on dry ice, where they were stored in liquid nitrogen.
RNA Extraction and cDNA Synthesis
Samples were thawed by incubation at 37°C, followed by dropwise addition of RPMI containing 10% FCS to a final dilution of 1:20. After centrifugation, cells were processed for RNA extraction by using RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions and subjected to DNase I digestion using RNase-free DNase (Qiagen). RNA concentration, purity, and integrity were evaluated as previously described (19). Reverse transcription was carried out using the SuperScript VILO cDNA Synthesis Kit (Life technologies) on equal amounts of total RNA.
Real-Time Quantitative RT-PCR
The expression of candidate genes was determined by quantitative PCR using Taqman Gene Expression Assays (Applied Biosystems) following the manufacturer’s protocol on the iCycler IQ detection system (Bio-Rad). Each set of primers and probe showed an efficiency of 90%–100%. Assays were performed in duplicate with 30 ng of reverse transcribed RNA. Gene expression analysis was performed by the relative calibrator normalized quantification method using the expression level of the β-glucuronidase gene (GUSB) as reference (19). An inter-run calibration sample was used in each plate to correct for technical variance between runs and to compare results from different plates. The calibrator consisted of cDNA prepared from HEK293T cells. Taqman Gene Expression Assays IDs for each set of primers and probe were as follows: Hs00286756_m1 (TET1); Hs00758658_m1 (TET2); Hs00379125_m1 (TET3); Hs00702322_s1 (TDG); Hs00154749_m1 (DNMT1); Hs01027166_m1 (DNMT3A); Hs00171876_m1 (DNMT3B); and Hs99999908_m1 (GUSB).
Dot Blot Assay
DNA was extracted from PBMCs with DNeasy Blood & Tissue Kit (QIAGEN), denatured in 0.4 M NaOH, 10 mM EDTA at 95°C for 10 minutes, and then neutralized by adding an equal volume of ice-cold 4 M ammonium acetate (pH 7.0). Two-fold dilutions of DNA samples (250 ng) were spotted on the positively charged nylon membranes Hybond-N+ (Amersham Biosciences) in an assembled Bio-Dot apparatus (Bio-Rad). After vacuum-mediated blotting, the membrane was washed with 2× SSC buffer, air-dried, and blocked with 5% nonfat dry milk followed by incubation with primary polyclonal anti-5hmC antibody (Active Motif). After incubation with HRP-conjugated secondary antibody, signals were visualized by chemiluminescence (Amersham ECL Western Blotting detection reagents). To correct for technical variance between replicates and to compare results from different experiments, control DNA obtained from HEK293T cells was used in all assays. Filters were stained with 0.02% methylene blue (MB) in 0.3 M sodium acetate (pH 5.2) to monitor DNA loading. Densitometric analysis was performed by Quantity One Software (Bio-Rad Laboratories) according to manufacturer’s instructions.
EpiTYPER Assay for Quantitative DNA Methylation Analysis
The EpiTYPER assay (Sequenom) was used to quantitatively assess the DNA methylation state of TET1 CpG island. DNA (1 μg) was bisulfite-converted using the EZ-96 DNA Methylation Kit (Zymo Research) with the following modifications: incubation in CT buffer for 21 cycles of 15 minutes at 55°C and 30 seconds at 95°C, elution of bisulfite-treated DNA in 100 μl of water. PCR was performed on 10 ng of converted DNA using TET1 CGI_Fw: aggaagagagGGTTTTTAGTTTTAA GTTTGTATTAGTTTT and TET1 CGI_Rev: cagtaatacgactcactataGGGAGAAGGC TATCATACAACCCTACCTACCTCTCC primers.
Statistical Analysis
For continuous variables, normal distribution of data was verified by Kolmogorov–Smirnov and Shapiro–Wilk normality tests (data not shown). Log transformation of data was used, as some variables were not normally distributed. When log-transformed data displayed normal distribution parametric tests were applied, otherwise, we used nonparametric tests. In particular, the identification of potential critical variables that can affect the variable under examination was performed by Mann–Whitney U-test and Student’s t-test for two-group comparisons. One-way ANOVA followed by Tukey’s post hoc test was used for more than two independent groups. When a significant p-value was found, a multiple comparison test by generalized linear models (GLMs) was performed and pairwise comparisons (adjusted for multiple comparisons by Dunn’s and Bonferroni’s methods for the Kruskal–Wallis and the GLM tests, respectively) were used to identify significant differences between percentile groups of each categorized variable. The Bonferroni adjustment for multiple comparisons was used to confirm differences between subgroups. GLMs were also used to investigate the influence of confounding variables (tested as categorized and continuous variables) on group-related changes of analyzed parameters. Pearson correlations were used to assess associations between defined variables. All statistical analyses were carried out using SPSS software (SPSS Inc., Chicago, IL; Version 22.0).
Results
Analysis of 5hmC Levels in PBMCs From DS Samples
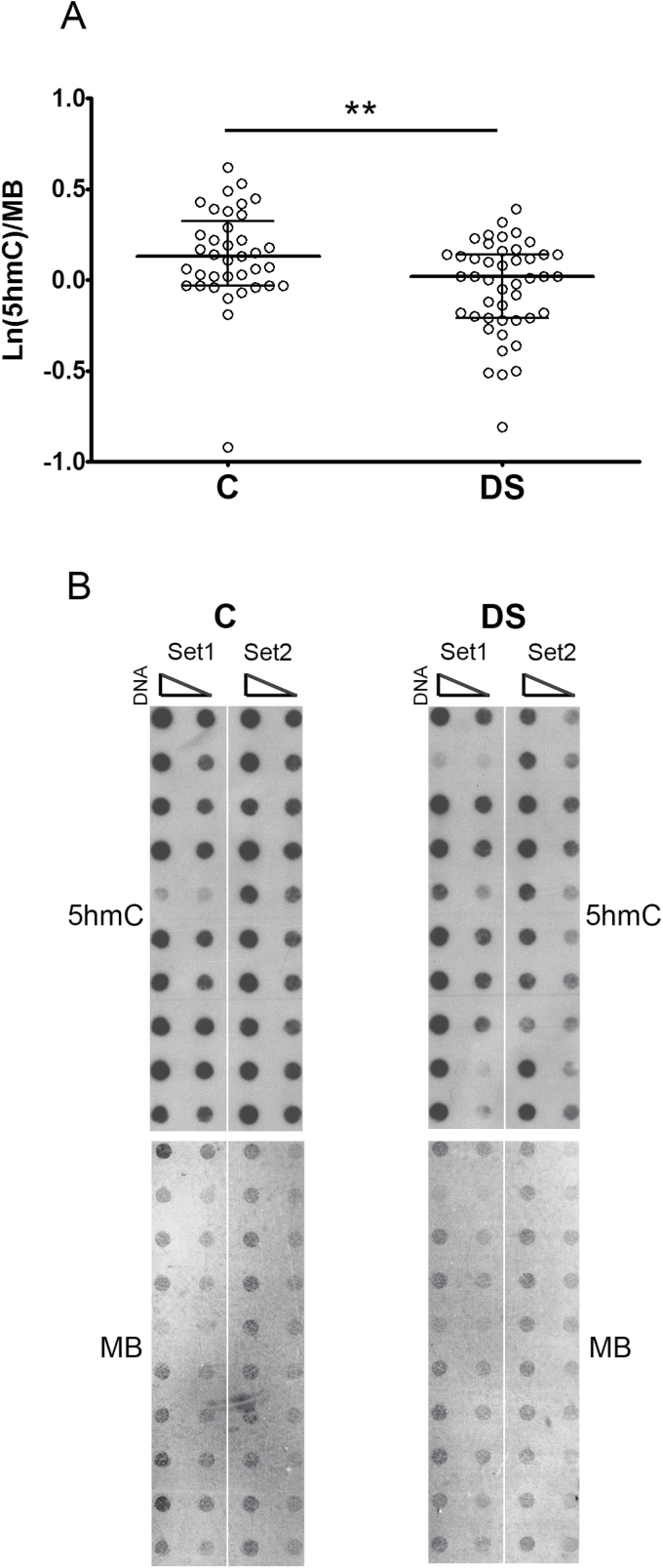
The content of 5hmC was measured by dot-bot assay on DNA from PBMCs obtained from DS persons and healthy controls. As shown in Figure 1, the levels of 5hmC are lower in DS samples compared with controls, as determined by Mann–Whitney U-test (p = .0037). To determine the influence of the covariates gender, age, and leukocyte composition (lymphocyte/monocyte ratio) on the 5hmC levels, the multivariate tests and the GLM procedures were computed (Supplementary Tables 1 and 2). These analyses demonstrated that the difference in 5hmC content between DS and controls was independent of any of those variables.
Figure 1.
5-hydroxymethylcytosine levels are decreased in DS PBMCs. (A) Scatter plot showing median and interquartile ranges of 5hmC levels in DS and control (C) subjects. Statistical analysis was performed by Mann–Whitney U-test. (B) Representative dot blot of DNA from DS and control subjects performed by using anti-5hmC antibody (left panel) and methylene blue (MB) staining (right panel) as a DNA loading control, **p < .01.
Expression of TET Genes in PBMCs From DS Samples
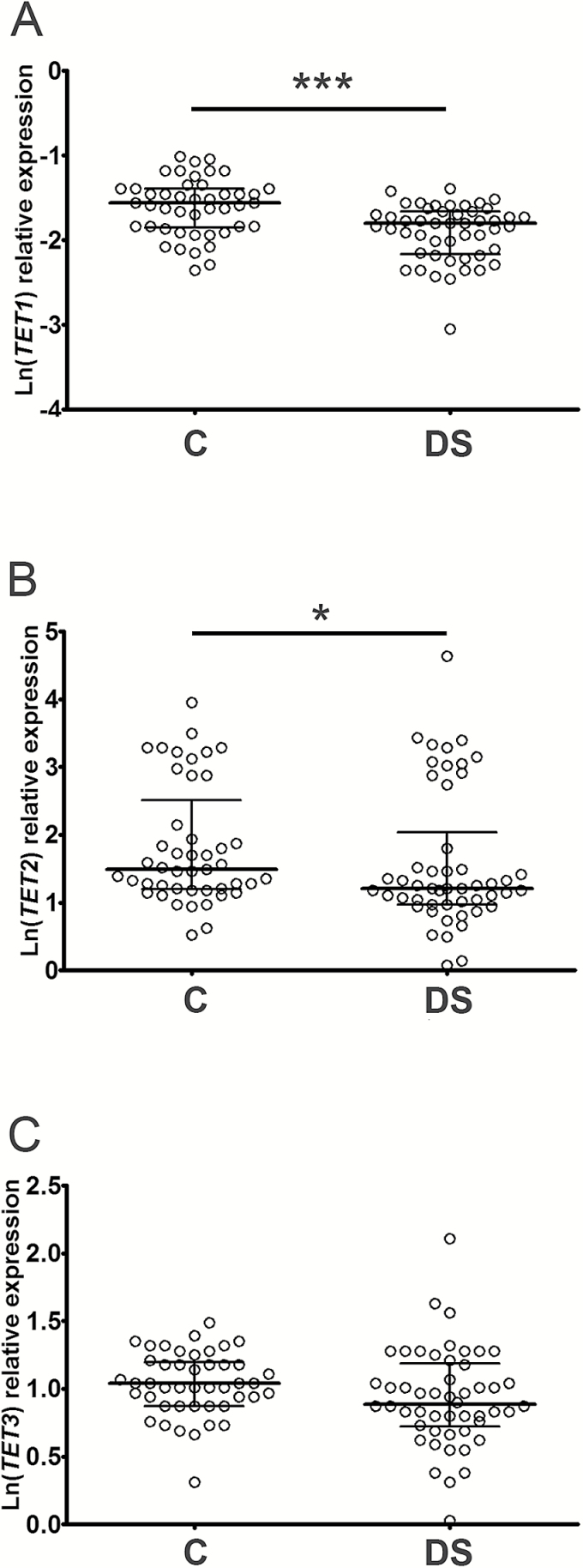
Subsequently, analyses were performed to see if the reduction of 5hmC levels in DS samples was associated with a deregulation of the enzymes involved in 5hmC formation. Expression of the 5mC-hydroxylase enzymes TET1, TET2, and TET3 was assessed by the real-time PCR analyses demonstrating that the levels of TET1 and TET2 were reduced in DS samples with respect to controls (TET1 t-test: p < .001; TET2 Mann–Whitney U-test: p = .0295; Figure 2). However, multivariate test analysis demonstrated that the expression of TET2 was affected by confounding factors while TET1 expression was independent of age, gender and lymphocyte/monocyte ratio, as also highlighted by GLM analysis (Supplementary Tables 1 and 3). The significant decrease in TET1 expression in DS sample was also confirmed by a generalized mixed linear model introducing batch effects as random factor (Supplementary Table 4). As concerns TET3, its expression was slightly reduced in DS samples, but this result was not statistically significant (TET3 t-test: p = .0926; Figure 2) and affected by covariates (Supplementary Table 5).
Figure 2.
TET1 expression levels are decreased in DS PBMCs. Scatter dot plot showing median and interquartile ranges of TET1 (A), TET2 (B), and TET3 (C) mRNA levels in DS and control (C) subjects. Statistical analysis was performed by unpaired t-test for TET1 and Mann–Whitney U-test for TET2, *p < .05; ***p < .001.
We then analyzed the correlation between 5hmC levels and expression of TET genes demonstrating a positive association with TET1 (r = 0.2729, p = .0131) and also a positive trend, albeit not significant, with TET2 (r = 0.1959, p = .0778; Supplementary Figure 1). A possible coregulation between TET enzymes was also tested by correlation analysis and a relevant association of TET2 with TET3 was observed (r = 0.3595, p = .0003), but not of TET1 with both of them (r(TET2) = 0.131, p = .2081; r(TET3)= −0.0413, p = .6711).
DNA Methylation Analysis of TET1 CpG Island
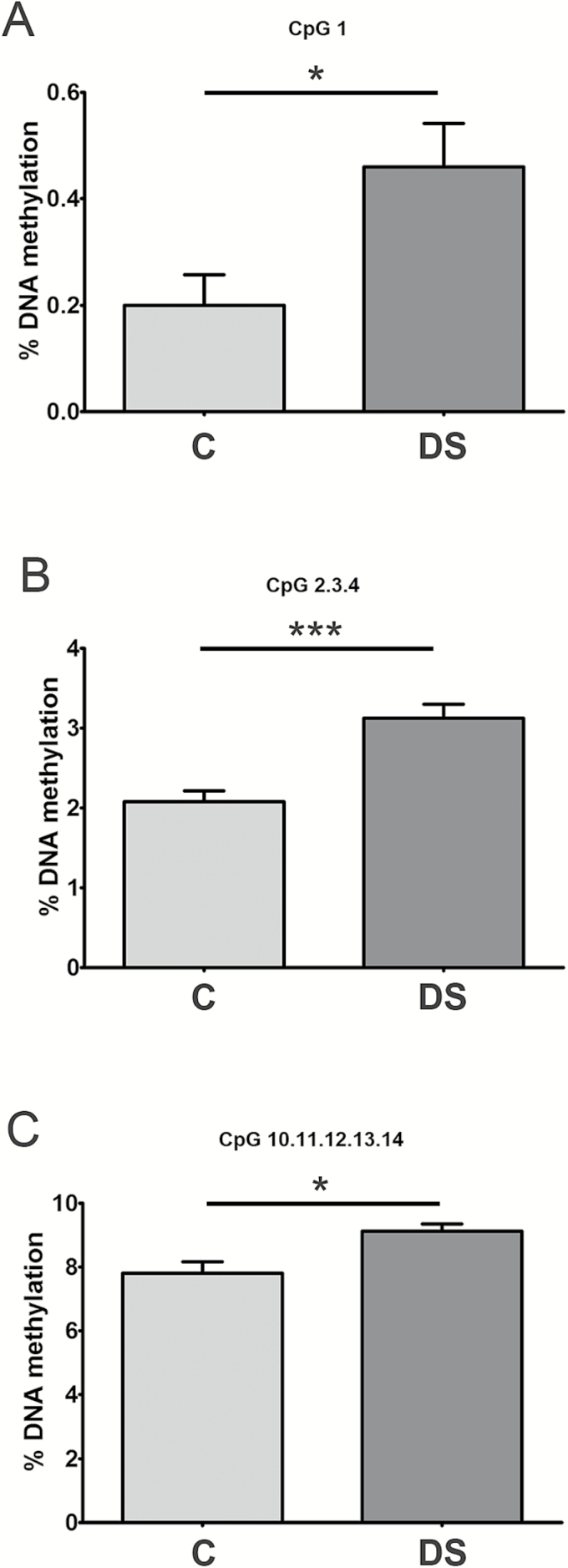
To investigate whether DNA methylation is responsible of TET1 down-regulation in PBMCs from DS persons, quantitative analysis of DNA methylation and identification of differentially methylated CpG sites at the CpG island of TET1 gene were assessed by using MassARRAY EpiTYPER. The analysis was performed with primers encompassing a 215-bp DNA region mapped to chr10:70,320,251-70,320,466. The comparison of the epigenetic profile between DS and control healthy individuals demonstrated that few CpGs present at the TET1 CpG island were slightly but significantly hypermethylated in DS PBMC samples (Figure 3 and Supplementary Figure 2A). As some of those CpGs were also shown to undergo hypermethylation with aging, we compared the DNA methylation levels at TET1 CpG island in DS with a group of elderly population (age range: 69–74 years), and in this case, no difference was observed at any CpG (Supplementary Figure 2B).
Figure 3.
DNA methylation levels at the TET1 CpG island in DS and control PBMCs. Graphs represent the CpGs at TET1 CpG island showing difference in DNA methylation between DS and control (C) subjects. DNA methylation analysis was performed by epiTYPER assay. Statistical significance was determined by unpaired t-test, *p < .05; ***p < .001.
Expression of DNMTs and TDG in PBMCs From DS Samples
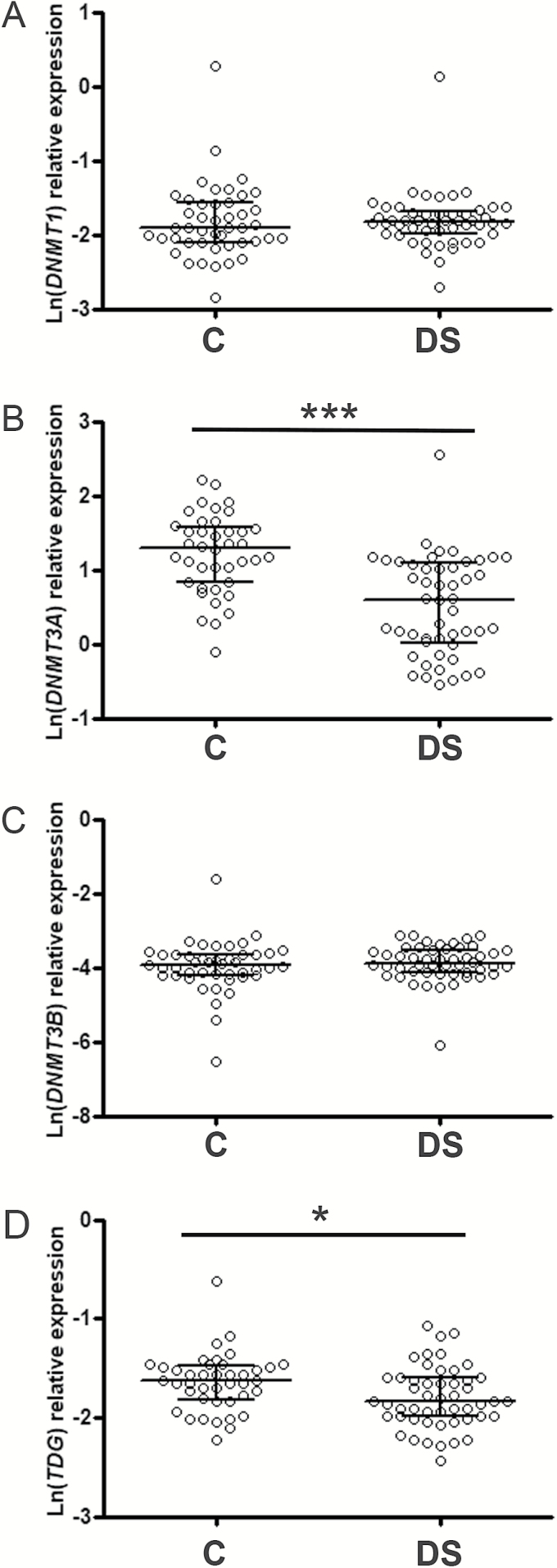
To better characterize changes in relevant epigenetic enzymes connected with 5hmC in DS samples, the expression of the active DNA methyltransferase enzymes (DNMT1, DNMT3A, and DNMT3B) and of TDG, the main glycosylase involved in TET-mediated DNA demethylation, was also assessed. Results demonstrated that the levels of DNMT1 and DNMT3B mRNA were similar between DS and control samples (DNMT1 Mann–Whitney U-test: p = .4808; DNMT3B Mann–Whitney U-test: p = .2829), whereas DNMT3A and TDG genes were down-regulated in PBMCs from DS persons (DNMT3A Mann–Whitney U-test: p < .0001; TDG t-test: p = .0136; Figure 4). These differences were confirmed by multivariate tests and GLM analysis (Supplementary Tables 1, 6, and 7), which also highlighted that the expression of DNMT3A was partially influenced by the leukocyte composition of PBMCs (Supplementary Table 6). In particular, the impact of lymphocyte/monocyte ratio on DNMT3A expression was mainly ascribable to control individuals, as a relevant positive correlation was observed in them (r = 0.4710, p = .0033) but not in DS persons (r = −0.01432, p = .9222; Supplementary Figure 3). In addition, due to the clear-cut reduction of DNMT3A in DS, we tested a possible association with the levels of 5hmC and TET genes demonstrating a positive correlation with TET2 (r = 0.2302, p = .03) and TET3 (r = 0.3895, p = .0002) but not with TET1 (r = 0.1259, p = .2396) and 5hmC (r = 0.07261, p = .5275; Supplementary Figure 4).
Figure 4.
The expression levels of DNMT3A and TDG are decreased in DS PBMCs. Scatter plot showing median and interquartile ranges of DNMT1 (A), DNMT3A (B), DNMT3B (C), and TDG (D) levels in DS and control (C) subjects. Statistical analysis was performed by Mann–Whitney U-test for DNMT3A and unpaired t-test for TDG, *p < .05; ***p < .001.
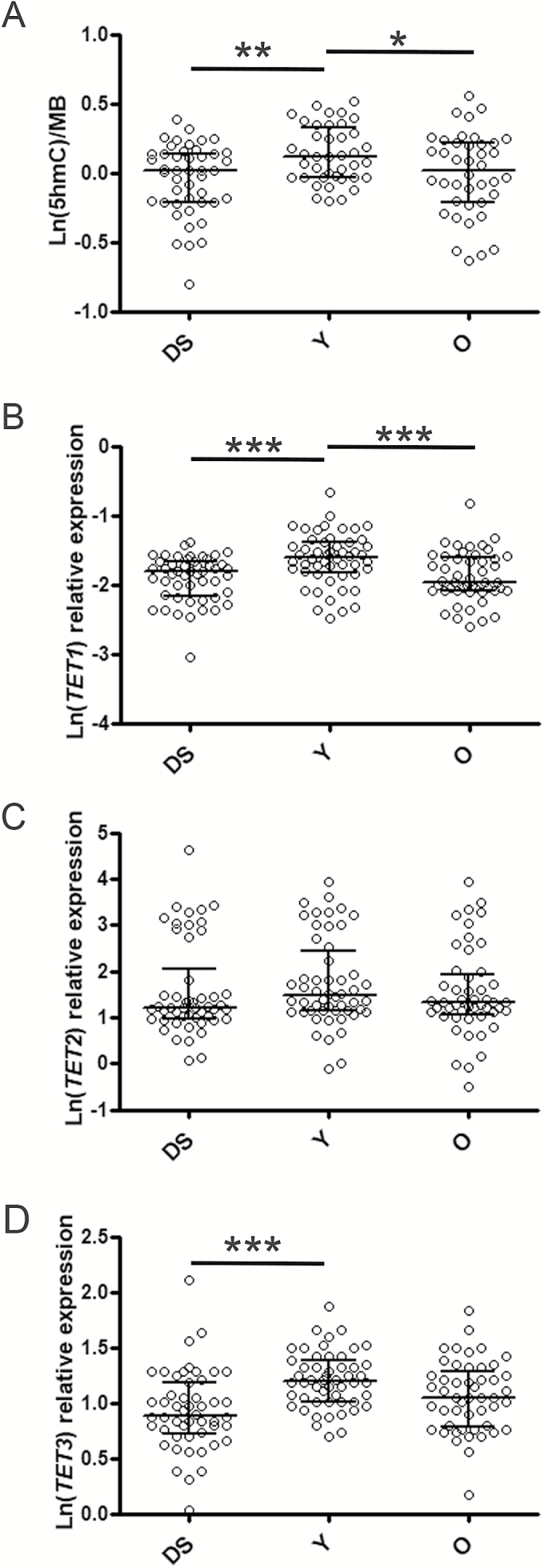
Analysis of 5hmC and DNA Methylation/Demethylation Machinery in DS, Young Persons, and Elders
As DS is considered a disease with premature aging features, we decided to compare the variables so far analyzed in DS with groups of young (age range: 31–45 years) and elderly people (age range: 69–74 years) previously assessed (14). Notably, we observed that levels of 5hmC, TET1, and TET3 in DS were significantly lower than young persons but not different from the elderly group (Figure 5), also when considering gender, country of origin and leukocyte composition as covariates in GLM analysis (Supplementary Tables 8–10). On the contrary, DNMT1 expression was lower in the elderly population than both DS and young persons (Supplementary Figure 5), but GLM analysis did not confirm the result due to a relevant influence of recruitment center (Supplementary Table 11). A difference between groups was obtained for TDG by GLM analysis independently of covariates but the pairwise comparison was statistically significant only for young and old people groups (Supplementary Figure 5 and Supplementary Table 12). No change was observed for TET2 and DNMT3B expression (Figure 5, Supplementary Figure 5, and Supplementary Tables 13 and 14), whereas DNMT3A was not tested as no data were available for the young and old populations. Moreover, it is noteworthy that most of variables (ie 5hmC, TET1, TET3, TDG, and DNMT1) showed a negative correlation with age when considering young and old people together, but this association was lost when DS persons were included. Only the negative correlation of DNMT1 with aging was retained when all samples were grouped (Supplementary Table 15).
Figure 5.
5hmC levels as well as TET1 and TET3 expression were similar between DS samples and an elderly population. Scatter plot showing median and interquartile ranges of 5hmC (A), TET1 (B), TET2 (C), and TET3 (D) levels in DS persons, young (Y) persons (age: 31–45 years) and old (O) persons (age: 69–74 years). Statistical analysis was performed by one-way ANOVA with post hoc Tukey’s test, *p < .05; **p < .01; ***p < .001.
Discussion
The presence of an extra-copy of genes located in chromosome 21 has long been considered the only driving cause for the DS pathogenesis. However, DS does not only originate from aneuploidy of the entire or fragmented chromosome 21 itself, but different DNA regions have been identified to be affected by trisomy and associated with different phenotypes. Along with the gene dosage theory, studies on both murine and human models have suggested that several other mechanisms determine the DS phenotype. In fact, the complexity of DS also depends on the trans effects that each karyotype can induce on distal genomic loci by physical interactions due to higher-order structure of chromatin. Moreover, much importance has been attributed to epigenetics in the pathogenesis of DS as numerous papers have highlighted trans effects of trisomy 21 also on DNA methylation patterns (7,8). Regions that are differentially methylated in healthy and DS individuals are largely distributed along the entire genome in several tissues. Most of the DNA methylation differences are tissue specific, but a certain degree of overlap has been shown in brain and T-cells and largely corresponding to specific transcription factor binding sites (8).
The contribution of 5hmC in DS was investigated in brain (8), the tissue with the highest levels of DNA hydroxymethylation, in which differentially hydroxymethylated regions were observed in genes largely related to brain development and function. In addition, hyperhydroxymethylation has been shown to characterize the internal promoter region of the PR domain containing 8 (PRDM8) gene in PBMCs of DS samples, in which an up-regulation of the associated transcript variant 2 was observed (20).
Our present work is the first one describing alterations of 5hmC in blood cells from DS persons. In particular, we provide evidence of decreased 5hmC levels in DS, an event frequently occurring in many multifactorial pathological conditions, such as cancer and neurodegeneration (21,22). Notably, 5hmC reduction has also been observed in another chromosomal disorder known as constitutional trisomy 8 mosaicism (23). The drop of 5hmC in DS samples is mainly associated with the down-regulation of TET1. Moreover, we show hypermethylation of few CpGs at TET1 promoter in DS samples when compared with age-matched controls, but this slight increase in 5mC is unlikely to explain by itself the silencing of TET1. Notably, we observed no difference with the methylation profile of a group of elderly population enrolled in the context of the same project (14). This result together with the evidence that 5hmC, TET1, and TET3 in DS show similar level of the elderly population and lower than the young one supports the view of DS as a disorder characterized by premature aging features.
Alteration of DNA methylation is by now a well-established phenomenon occurring in DS (8,24), but, at least in blood, no information exists about the levels of DNMT enzymes. Our analysis of the transcripts of the active methyltransferases demonstrated a down-regulation of DNMT3A in DS PBMCs. A reduction of DNMT3A expression along with DNMT3B has been recently reported in DS brain (25), whereas no difference for DNMT3A was observed in induced pluripotent stem cells generated from the twins discordant for DS (26). In addition, deregulation of DNMT3A as well as of other DNMTs has been documented in several tissues during aging (27–29).
Among the clinical manifestations typical of both aging and DS, 5hmC and TET deregulation in PBMCs may contribute to the decline of immune function, which is associated with increased susceptibility to infection and autoimmune diseases. New-borns with DS already show an abnormal structure of thymus, the central organ of T lymphocytes differentiation, with a concomitantly decreased production of mature T cells in the periphery (30). Notably, dynamic changes of 5hmC have been characterized during development and differentiation of T-cell lineage. Thymus-specific enhancers and gene bodies of highly transcribed genes showed enrichment of 5hmC. Moreover, higher levels of 5hmC were detected in active genes of precursor cells, suggesting the functional importance of 5hmC in T-cell specification (31). 5hmC-driven active demethylation has also been implicated in the thymic development of regulatory T cells (Tregs), a small subpopulation of T cells with fundamental roles in immune homeostasis control and thus in the suppression of autoimmunity. In fact, TET1 and TET2 activities were demonstrated to mediate the demethylation-dependent activation of FOXP3 gene, the master regulator of Tregs development and function (32,33). In addition, DS persons also have a significantly higher risk of developing leukemia, particularly acute lymphoblastic leukemia and acute megakaryoblastic leukemia (34), the onset of which has been shown to be accompanied by sequential epigenetic changes in DS (35). Notably, loss of 5hmC is a typical feature of blood cell malignancies and of solid tumors as well (36). TET1 expression is frequently deregulated in solid tumors, but a prominent role in the regulation of B-cell malignancies has been demonstrated, and evidence of its mutations exists in T-acute lymphoblastic leukemia (37–39). TET2 is a master regulator of normal hematopoiesis, and mutations of the TET2 gene have been frequently identified in multiple spectra of myeloid malignancies (40,41). Similarly, among DNMT enzymes, DNMT3A represents one of the most frequently mutated genes in diverse types of hematological cancers (41). Finally, it is worth noting that DNMT3A and TET2 mutations are also frequently observed in age-related clonal hematopoiesis (42), suggesting a central role of these two enzymes in age-related phenotypes and tumor development.
In conclusion, our analysis of DNA hydroxymethylation in PBMCs from DS patients demonstrated a decrease in 5hmC levels and components of the relative enzymatic machinery. The positive correlation between TET1 expression and 5hmC levels may suggest a causative role of TET1 down-regulation in DNA hypermethylation in DS patients. Viewed together, our results further strengthen the evidence that multiple pathological states are associated with deregulation of 5hmC that can contribute to the establishment of disease-associated DNA methylation patterns. Apart from being an intermediate in DNA demethylation processes, 5hmC directly controls gene expression programs acting as an epigenetic mark in its own right, recognized by specific chromatin readers. Therefore, the epigenetic consequences downstream of 5hmC alteration in blood cells can induce age-related clinical impairment of DS immune system (eg immunosenescence), with higher occurrence of infections, hematological malignancies, and autoimmune diseases in DS persons.
Supplementary Material
Supplementary data is available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Funding
This work was supported by the European Union’s Seventh Framework Program [grant HEALTH-F4-2008–200880 MARK-AGE].
Conflicts of Interest Statement
None declared.
Supplementary Material
Acknowledgments
The authors thank the MARK-AGE subjects for participating in this study, Gudrun von Scheven for excellent technical assistance, and all members of the MARK-AGE Consortium for fruitful discussion and collaboration.
References
- 1. Patterson D, Cabelof DC. Down syndrome as a model of DNA polymerase beta haploinsufficiency and accelerated aging. Mech Ageing Dev. 2012;133:133–137. doi: 10.1016/j.mad.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 2. Devenny DA, Wegiel J, Schupf N et al. . Dementia of the Alzheimer’s type and accelerated aging in Down syndrome. Sci Aging Knowledge Environ. 2005;2005:dn1. doi: 10.1126/sageke.2005.14.dn1. [DOI] [PubMed] [Google Scholar]
- 3. Zigman WB. Atypical aging in Down syndrome. Dev Disabil Res Rev. 2013;18:51–67. doi: 10.1002/ddrr.1128. [DOI] [PubMed] [Google Scholar]
- 4. Ghezzo A, Salvioli S, Solimando MC et al. . Age-related changes of adaptive and neuropsychological features in persons with Down syndrome. PLoS One. 2014;9:e113111. doi: 10.1371/journal.pone.0113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capri M, Moreno-Villanueva M, Cevenini E et al. . MARK-AGE population: from the human model to new insights. Mech Ageing Dev. 2015;151:13–17. doi: 10.1016/j.mad.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 6. Borelli V, Vanhooren V, Lonardi E et al. . Plasma N-glycome signature of Down syndrome. J Proteome Res. 2015;14:4232–4245. doi: 10.1021/acs.jproteome.5b00356. [DOI] [PubMed] [Google Scholar]
- 7. Do C, Xing Z, Yu YE, Tycko B. Trans-acting epigenetic effects of chromosomal aneuploidies: lessons from Down syndrome and mouse models. Epigenomics. 2017;9:189–207. doi: 10.2217/epi-2016-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mendioroz M, Do C, Jiang X et al. . Trans effects of chromosome aneuploidies on DNA methylation patterns in human Down syndrome and mouse models. Genome Biol. 2015;16:263. doi: 10.1186/s13059-015-0827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 10. Bacalini MG, Gentilini D, Boattini A et al. . Identification of a DNA methylation signature in blood cells from persons with Down syndrome. Aging (Albany, NY). 2015;7:82–96. doi: 10.18632/aging.100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jin S, Lee YK, Lim YC et al. . Global DNA hypermethylation in down syndrome placenta. PLoS Genet. 2013;9:e1003515. doi: 10.1371/journal.pgen.1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horvath S, Garagnani P, Bacalini MG et al. . Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015;14:491–495. doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiong J, Jiang HP, Peng CY et al. . DNA hydroxymethylation age of human blood determined by capillary hydrophilic-interaction liquid chromatography/mass spectrometry. Clin Epigenetics. 2015;7:72. doi: 10.1186/s13148-015-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Valentini E, Zampieri M, Malavolta M et al. . Analysis of the machinery and intermediates of the 5hmC-mediated DNA demethylation pathway in aging on samples from the MARK-AGE Study. Aging (Albany, NY). 2016;8:1896–1922. doi: 10.18632/aging.101022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bürkle A, Moreno-Villanueva M, Bernhard J et al. . MARK-AGE biomarkers of ageing. Mech Ageing Dev. 2015;151:2–12. doi: 10.1016/j.mad.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 16. Baur J, Kötter T, Moreno-Villanueva M et al. . The MARK-AGE extended database: data integration and pre-processing. Mech Ageing Dev. 2015;151:31–37. doi: 10.1016/j.mad.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 17. Moreno-Villanueva M, Capri M, Breusing N et al. . MARK-AGE standard operating procedures (SOPs): A successful effort. Mech Ageing Dev. 2015;151:18–25. doi: 10.1016/j.mad.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 18. Moreno-Villanueva M, Kötter T, Sindlinger T et al. . The MARK-AGE phenotypic database: Structure and strategy. Mech Ageing Dev. 2015;151:26–30. doi: 10.1016/j.mad.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 19. Zampieri M, Ciccarone F, Guastafierro T et al. . Validation of suitable internal control genes for expression studies in aging. Mech Ageing Dev. 2010;131:89–95. doi: 10.1016/j.mad.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 20. Lu Z, Liu Y, Ren Z, Yan J. PRDM8 internal promoter hyperhydroxymethylation correlates with increased expression of the corresponding transcript in Down syndrome. Mol Med Rep. 2016;14:1227–1234. doi: 10.3892/mmr.2016.5375. [DOI] [PubMed] [Google Scholar]
- 21. Sherwani SI, Khan HA. Role of 5-hydroxymethylcytosine in neurodegeneration. Gene. 2015;570:17–24. doi: 10.1016/j.gene.2015.06.052. [DOI] [PubMed] [Google Scholar]
- 22. Jeschke J, Collignon E, Fuks F. Portraits of TET-mediated DNA hydroxymethylation in cancer. Curr Opin Genet Dev. 2016;36:16–26. doi: 10.1016/j.gde.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 23. Davidsson J, Veerla S, Johansson B. Constitutional trisomy 8 mosaicism as a model for epigenetic studies of aneuploidy. Epigenetics Chromatin. 2013;6:18. doi: 10.1186/1756-8935-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerkel K, Schupf N, Hatta K et al. . Altered DNA methylation in leukocytes with trisomy 21. PLoS Genet. 2010;6:e1001212. doi: 10.1371/journal.pgen.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El Hajj N, Dittrich M, Böck J et al. . Epigenetic dysregulation in the developing Down syndrome cortex. Epigenetics. 2016;11:563–578. doi: 10.1080/15592294.2016.1192736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sailani MR, Santoni FA, Letourneau A et al. . DNA-Methylation Patterns in Trisomy 21 Using Cells from Monozygotic Twins. PLoS One. 2015;10:e0135555. doi: 10.1371/journal.pone.0135555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Balada E, Ordi-Ros J, Serrano-Acedo S, Martinez-Lostao L, Rosa-Leyva M, Vilardell-Tarrés M. Transcript levels of DNA methyltransferases DNMT1, DNMT3A and DNMT3B in CD4+ T cells from patients with systemic lupus erythematosus. Immunology. 2008;124:339–347. doi: 10.1111/j.1365-2567.2007.02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Casillas MA Jr, Lopatina N, Andrews LG, Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. [DOI] [PubMed] [Google Scholar]
- 29. Ciccarone F, Malavolta M, Calabrese R et al. . Age-dependent expression of DNMT1 and DNMT3B in PBMCs from a large European population enrolled in the MARK-AGE study. Aging Cell. 2016;15:755–765. doi: 10.1111/acel.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kusters MA, Verstegen RH, Gemen EF, de Vries E. Intrinsic defect of the immune system in children with Down syndrome: a review. Clin Exp Immunol. 2009;156:189–193. doi: 10.1111/j.1365-2249.2009.03890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsagaratou A, Aijo T, Lio CW et al. . Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. In: Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E3306–E3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toker A, Engelbert D, Garg G et al. . Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J Immunol. 2013;190:3180–3188. doi: 10.4049/jimmunol.1203473. [DOI] [PubMed] [Google Scholar]
- 33. Yang R, Qu C, Zhou Y et al. . Hydrogen sulfide promotes Tet1- and Tet2-mediated Foxp3 demethylation to drive regulatory T cell differentiation and maintain immune homeostasis. Immunity. 2015;43:251–263. doi: 10.1016/j.immuni.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mateos MK, Barbaric D, Byatt SA, Sutton R, Marshall GM. Down syndrome and leukemia: insights into leukemogenesis and translational targets. Transl Pediatr. 2015;4:76–92. doi: 10.3978/j.issn.2224-4336.2015.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malinge S, Chlon T, Doré LC et al. . Development of acute megakaryoblastic leukemia in Down syndrome is associated with sequential epigenetic changes. Blood. 2013;122:e33–e43. doi: 10.1182/blood-2013-05-503011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 2014;30:464–474. doi: 10.1016/j.tig.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li L, Li C, Mao H et al. . Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers. Sci Rep. 2016;6:26591. doi: 10.1038/srep26591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao Z, Chen L, Dawlaty MM et al. . Combined Loss of Tet1 and Tet2 Promotes B Cell, but Not Myeloid Malignancies, in Mice. Cell Rep. 2015;13:1692–1704. doi: 10.1016/j.celrep.2015.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cimmino L, Dawlaty MM, Ndiaye-Lobry D et al. . TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16:653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bowman RL, Levine RL. TET2 in normal and malignant hematopoiesis. Cold Spring Harb Perspect Med. 2017;7:a026518 doi: 10.1101/cshperspect.a026518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sato H, Wheat JC, Steidl U, Ito K. DNMT3A and TET2 in the pre-leukemic phase of hematopoietic disorders. Front Oncol. 2016;6:187. doi: 10.3389/fonc.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jaiswal S, Fontanillas P, Flannick J et al. . Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.