Abstract
Motivation
The identification of signal peptides in protein sequences is an important step toward protein localization and function characterization.
Results
Here, we present DeepSig, an improved approach for signal peptide detection and cleavage-site prediction based on deep learning methods. Comparative benchmarks performed on an updated independent dataset of proteins show that DeepSig is the current best performing method, scoring better than other available state-of-the-art approaches on both signal peptide detection and precise cleavage-site identification.
Availability and implementation
DeepSig is available as both standalone program and web server at https://deepsig.biocomp.unibo.it. All datasets used in this study can be obtained from the same website.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Protein sorting and compartmentalization are complex biological mechanisms, often guided by specific sequence signals present in the nascent protein. Signal peptides are short sequence segments located at the N-termini of newly synthesized proteins that are sorted toward the secretory pathway (von Heijne, 1990). Proteins endowed with a signal peptide include proteins resident in endoplasmic reticulum and Golgi apparatus, secreted proteins and proteins inserted in the plasma membrane. Identifying signal peptides in the protein sequence is a prerequisite to unveil protein destination and function.
Several computational methods have been trained on available experimental data to detect the signal sequence in the N-terminus of a query protein. The most successful methods are based on machine learning models. Artificial Neural Networks and Support Vector Machines learn directly from the available experimental data the signal sequence features (Nugent and Jones, 2009; Petersen et al., 2011). Other methods (Bagos et al., 2010; Käll et al., 2005; Reynolds et al., 2008; Tsirigos et al., 2015; Viklund et al., 2008) adopt Hidden Markov Models to define regular grammars. They explicitly model the modular architecture of the signal sequence, consisting of three regions: the positively charged N-region, the central hydrophobic H-region and the polar uncharged C-region containing the cleavage site (Martoglio and Dobberstein, 1998).
A major challenge in signal peptide prediction is discriminating between true signal sequences and other hydrophobic regions, and, in particular, N-terminal transmembrane helices. The accurate prediction of the cleavage site is also challenging, mainly due to the high variability of the signal sequence length and the absence of sequence motifs that unambiguously mark the position of the cutting site.
In this paper, we present DeepSig, a new method that takes advantage of Deep Learning advancement and improves the state-of-the-art performance. DeepSig is designed for both detecting signal peptides and finding their cleavage sites in protein sequences. The predictor consists of two consecutive building blocks: a deep neural network architecture and a probabilistic method that incorporates the current biological knowledge of the signal peptide structure.
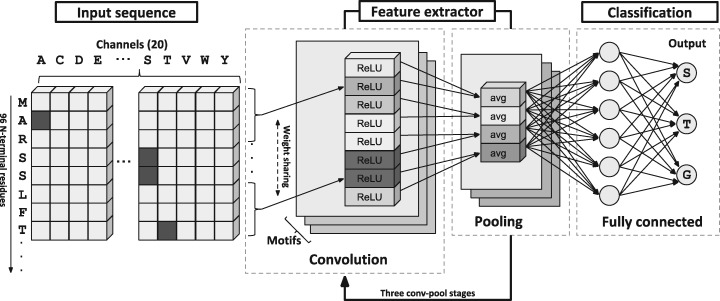
In the first stage, the N-terminus of a query protein sequence is analysed to assess the presence of a signal peptide. For this purpose, we designed a Deep Convolutional Neural Network (DCNN) (LeCun et al., 2015) architecture (Fig. 1), specifically tuned to recognize signal peptide sequences. DCNNs are very powerful deep learning architectures that achieve very high performance in several applications (Alipanahai et al., 2015; Krizhevsky et al., 2012; Zhou and Troyanskaya, 2015). Here, we devise a DCNN comprising three cascading convolution-pooling stages that process the N-terminus of the query protein, sorting out three classes: signal peptides, transmembrane regions and ‘anything else.’
Fig. 1.
The architecture of the DCNN processing an input protein sequence to detect signal peptides. Feature extraction involves the application of three convolution-pooling (conv-pool) stages. The final classification is performed by a standard fully-connected neural network
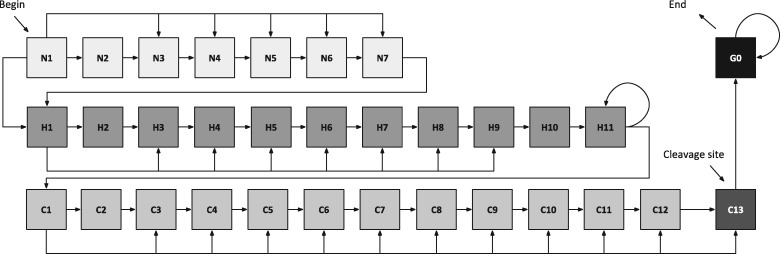
If a signal peptide is detected, the protein is passed to the next prediction stage where the precise position of the cleavage site is identified (Fig. 2). This task is tackled in DeepSig as a sequence labelling problem, where each residue is labelled as signal-peptide (S) or not (N). In particular, we adopted a probabilistic sequence labelling model (Fariselli et al., 2009) similar to the regular grammars adopted by other HMM-based approaches (Käll et al., 2004).
Fig. 2.
The signal-peptide GRHCRF model capturing the modular structure of the signal peptide. States labeled with N, H, and C represents the positively charged N-region, the hydrophobic H-region and the cleavage C-region, respectively (see Section 2.4 for further details)
For improving cleavage site detection, we also applied the Deep Taylor Decomposition (Montavon et al., 2017) to compute how relevant each residue at the N-terminus is for the recognition of the signal sequence. This score is used as additional feature for the sequence labelling model to improve cleavage-site prediction.
We trained the DeepSig predictor on the dataset of proteins adopted by SignalP, one of the best performing methods developed so far (Petersen et al., 2011). It comprises 10 303 non-redundant proteins extracted from UniprotKB and belonging to three different organism classes: Eukaryotes, Gram-positive and Gram-negative bacteria.
Comparative benchmarks were performed on a new independent validation dataset comprising 1707 sequences with experimental annotations in UniprotKB and not included in the training set. In all experiments, DeepSig outperforms other state-of-the-art approaches in both signal peptide detection and cleavage site prediction. Interestingly, when restricting the negative dataset to the most challenging cases (N-terminal transmembrane regions), DeepSig outperforms state-of-the-art predictors, specifically in the case of Eukaryotic proteins.
2 Materials and methods
2.1 Datasets
2.1.1 The SignalP4.0 dataset
The first dataset used in this work was generated to train and test the well-known SignalP method (Petersen et al., 2011). Data were extracted from UniProtKB/SwissProt release 2010_05 including proteins from Eukaryotes, Gram-positive and Gram-negative bacteria. Only proteins with experimentally annotated signal peptide cleavage sites were retained. Negative sets (i.e. proteins lacking a signal peptide) were chosen from two different subsets: (i) proteins experimentally annotated as cytosolic and/or nuclear (ii) proteins experimentally annotated as single- or multi-pass membrane proteins, with a transmembrane segment annotated in the first 70 positions. All data were homology-reduced in order to obtain non-redundant datasets for each of the three organism classes. Two eukaryotic proteins were considered as similar if a local alignment between them included more than 17 identical residues out of 70 N-terminal residues. A threshold of 21 residues was instead used for bacterial proteins. See Table 1 for a summary of the SignalP4.0 dataset.
Table 1.
Statistics of the three datasets adopted in this study
| Dataset | Organism | SP | T | N/C | Total |
|---|---|---|---|---|---|
| SignalP4.0 | Eukaryotes | 1640 | 987 | 5133 | 7760 |
| Gram-positive | 208 | 117 | 360 | 685 | |
| Gram-negative | 423 | 523 | 912 | 1858 | |
| SPDS17 | Eukaryotes | 46 | 323 | 689 | 1058 |
| Gram-positive | 9 | 189 | 240 | 438 | |
| Gram-negative | 23 | 89 | 99 | 211 | |
| E.coli | – | 573 | 1024 | 4375 | 5972 |
Note: SP, signal-peptide proteins; T, transmembrane proteins (with a single alpha helix in the N-terminal region); N/C, Nuclear and/or Cytosolic proteins (proteins without signal peptide); Total, total sum.
2.1.2 The SPDS17 blind dataset
We generated a new benchmark dataset to compare different approaches on signal peptide detection and cleavage-site prediction. We selected proteins from UniprotKB (rel. 04_2017) released after June 2015. This allowed to exclude any protein already included in the SignalP dataset used for training.
Positive data were separately collected for Eukaryotes, Gram-negative and Gram-positive (in constructing this set we considered only proteins from Actinobacteria and Firmicutes phyla) by extracting proteins endowed with an experimentally annotated cleavage site for the signal peptide.
Next, analogously to the SignalP4.0 dataset, for each organism class, two negative sets were generated: (i) proteins with a membrane-spanning segment in the first 70 residues and (ii) proteins localized into the nucleus and/or the cytoplasm. To generate these sets, we retained only proteins with experimental or manually curated annotation (corresponding to the UniProtKB evidence codes ECO: 0000269 and ECO: 0000305, respectively).
The set redundancy was reduced to 25% sequence identity by running the blastclust algorithm and retaining a representative sequence from each cluster. Furthermore, we excluded all proteins sharing more than 25% sequence identity with any protein in the SignalP dataset. The blastp program with e-value threshold set to 1e-3 was adopted to search for similar proteins. Table 1 contains a summary of the SPDS17 dataset.
2.1.3 The Escherichia coli proteome
We assessed DeepSig proteome-wide performance using the entire proteome of Escherichia coli (strain K12). From release 11_2017 UniprotKB we downloaded all the 5972 reviewed entries. The sequences endowed with signal peptide are 573; 1024 have a transmembrane segment annotated in the first 70 residues.
2.2 Deep convolutional neural networks for signal peptide prediction
Deep Convolutional Neural Networks (DCNNs) (LeCun et al., 2015) are powerful deep learning models devised to process multi-channel input data. Several data types fall in this category. The main application domain of DCNN is image processing (e.g. image object recognition or segmentation), where each pixel of a 2-Dimensional image is encoded by a vector of three intensity channels.
Here, we apply DCNNs to protein sequence analysis. In this case, the input domain is a 1-dimensional signal, where each position in a sequence is represented by a multi-channel (i.e. multi-dimensional) vector encoding the residue type at each position of a protein, one channel for each residue type.
Signal-peptide prediction is a special task of protein classification where the goal is to detect the presence/absence of the signal sequence in the N-terminus of the protein. Figure 1 summarizes the architecture of the DCNN defined in this paper for signal peptide prediction, comprising two basic modules: the feature extraction and the classification.
2.2.1 Feature extraction module
The feature extraction module consists of several hierarchical convolution (conv) and pooling (pool) layers which collectively compute a feature representation of the input protein sequence. Convolutional layers can be seen as sequence motif detectors used to scan the input sequence. A convolutional layer is mainly characterized by the number of motifs (or filters) it applies and by the motif length. Each motif detector slides along the input sequence, and computes the positional score for the motif at any sequence position. The scores are stored in the convolution neurons. Motif parameters are learnt during training and, routinely, parameter sharing is enforced (i.e. the same motif weights are applied to all positions during sequence scanning). After convolution, pooling layers are applied to aggregate neighbour convolution neurons into a single output neuron, with a consequent reduction of dimensionality. Typical pooling operations include max or average functions, computed over short non-overlapping slices of convolution neurons. The main parameter of a pooling layer is the width of the slice adopted. Iterative applications of convolution-pooling (conv-pool) operations are performed to extract a complex feature representation of the input sequence. In fact, a hierarchical feature extraction protocol is adopted where low-level motifs are progressively aggregated to model higher level inter-motif interactions. Adding conv-pool layers to the network allows extracting complex patterns of interaction through motifs, though increasing the complexity of the network.
More formally, an input protein sequence is defined as a matrix where is the sequence length and is the number of different residue types. Here, protein sequences are shortened to the 96 N-terminal residues, hence .
A motif detector of odd-sized width in the first convolution layer is defined as a weight matrix of dimension . If different motif detectors are applied, the output of the convolution layer is a matrix , where the element is computed as:
| (1) |
where is -th motif weight matrix and is the rectified linear unit (ReLU) activation function. Using ReLUs instead of other activation functions (such as tanh or sigmoid) speeds up the training process, particularly in networks with many layers (LeCun et al., 2015).
The role of the pooling layer with pool size equal to , applied to each motif of matrix , is to reduce its dimensionality by merging together neighbour convolutional neurons into one. Although other schemes are possible, here average pooling is applied to adjacent pairs of convolution neurons, leading to dimensionality reduction from to . The pooling layer computes a matrix defined as follows:
| (2) |
where i ranges between 1 and m=l/2. Overall, a single conv-pool application transforms an input sequence of dimension to a non-linear feature representation of dimension . Hence, a series of conv-pool stages, stacked together, extracts a non-linear feature space representation of dimension , with being the number of motif detectors in the last conv layer.
In our final network, we apply three cascading conv-pool stages. Different architectures were tried and selected through cross-validation, varying the number of motif detectors and motif width on each conv layer.
2.2.2 Classification module
The output classification is performed by means of a module implementing a conventional fully connected feed-forward neural network, comprising a single hidden-layer with neurons. The number of neurons in the hidden layer was varied and optimized through cross-validation, separately for each organism class. Firstly, the computed feature representation is flattened into a column vector that encodes the input of the feed-forward network.
Each neuron in the hidden layer computes a non-linear transformation, defined as follows:
| (3) |
where and are, respectively, the weight vector and bias of the hidden neuron (again, the ReLU activation is used).
Finally, the hidden layer output vector is mapped to the -th output neuron as follows:
| (4) |
where and are, respectively, the weight vector and the bias of the output neuron , and the function is the function, allowing a probabilistic interpretation of the network output.
The final output of our DCNN comprises three output neurons accounting for three different output classes: signal peptide (S), transmembrane segment (T) or other (N). This three-class schema allows to reduce the misclassification between transmembrane regions and signal peptides (Section Results). An input protein sequence is classified into the class with the highest predicted probability, namely:
| (5) |
Given a training set of protein sequences with true output targets, network parameters are optimized by minimizing the average cross-entropy loss function on the training set, defined as:
| (6) |
where is the -th network output when the -th sequence is provided in input.
2.3 Evaluating residue positional relevance with deep Taylor decomposition
The DCNN described in the previous section is designed to provide a prediction of the presence/absence of the signal peptide sequence in the N-terminus of an input protein. In general, when such predictions are performed with DCNN, some of the elements of an input sequence (i.e. individual residues) may be more determinant than others in driving the model classification toward one specific class or another. An important question is then how this piece of information can be extracted from the analysis of the internal neuronal activity of DCNN.
Many methods are available to analyse the complex behaviour of non-linear classifiers in the attempt of quantifying the importance of basic elements in the input data with respect to the task at hand (Bach et al., 2015; Montavon et al., 2017; Simonyan et al., 2013). For instance, in image classification, one wants to identify a subset of relevant pixels that are responsible for the recognition of an object in the image (Bach et al., 2015; Montavon et al., 2017; Simonyan et al., 2013; Szegedy et al., 2013). In the context of signal peptide detection, given an input protein sequence in which a signal peptide has been recognized, we want to identify residue positions along the sequence that are more relevant for the global recognition of the signal.
Available methods can be roughly classified into two different categories: functional approaches look at networks as function approximators and highlight the most relevant input features by analysing the prediction function (Simonyan et al., 2013); message passing approaches exploit the network as a computational graph and propagate prediction values throughout the different layers back to input variables (Bach et al., 2015).
Here, we adopt the deep Taylor decomposition (Montavon et al., 2017), a hybrid functional/message passing approach that has been recently introduced for the analysis of deep neural networks. The method focuses on image classification, but it can be easily extended to other types of prediction scenarios, such as protein sequence classification. We briefly describe here its main aspects and refer to the original paper for a comprehensive mathematical description of the method (Montavon et al., 2017) and to our Supplementary Material for a description of how this method can be applied to our signal-peptide DCNN.
Let be an input protein sequence of length where each is a -channel vector representing a residue in the sequence. is the scalar function implemented with a DCNN and evaluated on the input . The function quantifies the evidence (or score) that a signal peptide is present in the N-terminus of the sequence . We want to assign to each residue a relevance score that quantifies the individual contribution of that residue to the total predicted evidence function .
Operatively, deep Taylor decomposition proceeds by assigning to each neuron in a deep network a relevance score which is a measure of the contribution of the neuron to the total predicted score . Neuron relevance scores are computed by establishing local, connectivity-dependent functional mappings between neuron activation values and propagated relevance values from upper-layers. Taylor expansions of these local mappings at neuron-specific root points are then computed. Depending on the functional form of the mappings and on the nature of the input domain, different relevance propagation rules are defined (for details, see Supplementary Material).
We apply this procedure to our signal peptide DCNN to evaluate the contribution of each residue position to the detection of the signal sequence. The result for a sequence in input of length is a vector:
| (7) |
where the component is the relevance of the residue in position .
2.4 Prediction of the signal peptide cleavage site
When a signal peptide is detected with the DCNN, the protein sequence passes to the second prediction stage which identifies the location of the cleavage site. In particular, each residue of a positively-predicted sequence is assigned to one of two classes: signal peptide (S) or non-signal region (N).
Here, we adopt a Grammar-Restrained Hidden Conditional Random Field (GRHCRF) (Fariselli et al., 2009; Indio et al., 2013; Savojardo et al., 2013; Savojardo et al., 2017). Like HMMs, a GRHCRF can be represented as a finite state automaton whose state structure and transitions reflect a regular grammar describing the problem at hand (Fariselli et al., 2009). Each state of the model is associated to a label that can be assigned to each element of a sequence. Model parameters are weights that score the compatibility between input sequences included in the training set and their true labelling. Once the model has been trained, sequence labelling is performed by assigning labels corresponding to the most probable state path in the model. The optimal state path is computed by means of Posterior-Viterbi decoding (Fariselli et al., 2009).
The GHRCRF model is defined on top of the grammar depicted in Figure 2 as a finite-state automaton. The model defines different states organized to capture the modular structure of a typical signal peptide: 7 states to model the initial positively charged N-region (states N1–N7), 11 states for the hydrophobic H-region (states H1–H11) and 13 states for the cleavage C-region (states C1–C13). State transitions are defined such that minimal and maximal lengths for each sub-region are enforced. In particular, N-regions can be from two up to seven residues long. In contrast, the H-region has a minimal length of four residues with no upper bound. Finally, C-regions comprise between 3 and 13 residues. The remaining mature protein portion is modelled through a single recursive state (G0). The cleavage site corresponds to the position of the residue assigned to state C13.
Training of the GRHCRF is performed on a training set of protein sequences endowed with signal peptides. Also in this case, sequences are reduced to the first 96 N-terminal residues. Each residue is encoded using a 21-dimensional feature vector consisting of:
20 positions of the vector correspond to the usual residue encoding described above;
the relevance score of the residue computed from the DCNN and deep Taylor decomposition as described in Section 2.3.
2.5 Model optimization and implementation
All the models are trained on the SignalP4.0 dataset using a nested 5-fold cross-validation procedure as done in Petersen et al. (2011). Three different optimization runs are performed on Eukaryotic, Gram-positive and Gram-negative, respectively.
Firstly, the entire dataset is randomly split into five subsets containing broadly the same number of proteins. Random splits are computed so that the balancing between signal-peptide, transmembrane and other proteins is maintained on each subset and it is similar to the one observed in the whole dataset.
Secondly, a nested cross-validation procedure is performed as follows: one subset is kept out and used for testing while a full inner 4-fold cross-validation is performed on the remaining four subsets. In each run of this inner procedure, three subsets are used for training and one for validation. The inner cross-validation is used to optimize the network parameters and architecture. In fact, we retain the top-performing network as evaluated on the inner validation sets. The procedure is repeated leaving out each time a different subset for testing. In summary, 20 different networks are obtained (four optimal networks that are identical in parameters and architecture but have been trained on different inner training sets for each one of the five main subsets). When performance is evaluated on the testing set, outputs of the four inner networks are averaged to give the final score. In the final version of our DeepSig predictor, we average the output of all the 20 optimal networks.
The same procedure and data split was applied to train/test the cleavage site predictor based on the GRHCRF model.
The DCNN is implemented using the Keras Python package (https://keras.io) (Chollet et al., 2015) with the Tensorflow (https://www.tensorflow.org) (Abadi et al., 2015) backend. The categorical cross-entropy loss minimization is carried-out with the standard error back-propagation procedure and the stochastic gradient descent algorithm. Default hyper-parameters were used for training the networks (Default hyper-parameters for network training were set to 0.01 for learning rate and to 0 for momentum and weight decay).
2.6 Scoring measures
Signal-peptide detection is scored using the following measures:
- Matthews Correlation Coefficient (MCC), defined as:
where TP and TN are the correct predictions in the positive and negative classes, respectively, and FN and FP are the number of under- and over-predictions in the signal peptide class(8) - False positive rate computed on transmembrane proteins, defined as:
where FPT is the number of transmembrane proteins misclassified as having a signal peptide and NT is the total number of transmembrane proteins.(9) - Cleavage-site prediction is scored by the cleavage-site F1 measure defined as:
namely, the harmonic mean between Cleavage Site Sensitivity, and Cleavage Site Precision, , where Ncorr is the number of correctly identified cleavage sites and N and NP are, respectively, the true number of signal peptides and the number of predicted signal peptides.(10)
3 Results
3.1 Performance on the SignalP4.0 dataset
We firstly evaluate the performance of our DeepSig predictor on the dataset adopted to train and test SignalP4.0 (Petersen et al., 2011) with the same nested cross-validation procedure described in Section 2.5.
This allows a direct and accurate comparison of DeepSig with SignalP4.0. In particular, three different versions of SignaP are scored: SignalP-TM, the version of the method optimized to distinguish signal peptides from transmembrane regions; SignalP-noTM which is not optimized and SignalP4.0 which is a combination of the two methods above.
Our DeepSig predictor is also evaluated in two versions, either using the relevance profile as feature for cleavage-site prediction (Section 2.4) or not (‘no relevance’ in Table 2).
Table 2.
Performance of different versions of SignalP and DeepSig on signal peptide detection and cleavage site prediction in 5-fold cross-validation on the SignalP4.0 dataset (Petersen et al., 2011)
| Method | Eukaryotes |
Gram-positive |
Gram-negative |
||||||
|---|---|---|---|---|---|---|---|---|---|
| MCC | FPRT | F1cs | MCC | FPRT | F1cs | MCC | FPRT | F1cs | |
| SignalP 4.0a | 0.874 | 6.1 | 67.1 | 0.851 | 2.6 | 77.8 | 0.848 | 1.5 | 68.0 |
| SignalP-TMa | 0.871 | 3.3 | 67.2 | 0.851 | 2.6 | 77.8 | 0.815 | 1.1 | 67.7 |
| SignalP-noTMa | 0.674 | 38.1 | 54.6 | 0.556 | 47.9 | 49.4 | 0.497 | 35.8 | 67.7 |
| DeepSig (no relevance) | 0.910 | 2.6 | 71.1 | 0.878 | 5.9 | 69.7 | 0.900 | 1.5 | 83.5 |
| DeepSig | 0.910 | 2.6 | 73.3 | 0.878 | 5.9 | 72.3 | 0.900 | 1.5 | 86.2 |
Note: MCC, Matthews Correlation Coefficient; FPRT, False Positive Rate on transmembrane proteins; F1cs, The harmonic mean between precision and recall on cleavage-site detection. No relevance = without relevance profile as feature for cleavage-site prediction (Section 2.4).
Data taken from Petersen et al. (2011).
Comparative results of both signal peptide detection and cleavage site prediction are reported in Table 2. Methods are trained and scored separately on each organism class: Eukaryotes, Gram-positive and Gram-negative. Results for SignalP are derived from the original paper (Petersen et al., 2011).
The first aspect evaluated is the detection of the signal peptide (with the MCC and FPRT scoring indexes). DeepSig outperforms SignalP (considering the MCC values on Table 2) on all the three datasets (Eukaryotes, Gram-positive and Gram-negative proteins).
Specifically, on the Eukaryote dataset our method shows a lower false positive rate on proteins with a transmembrane segment annotated in the first 70 residues (Table 2, FPRT). It is well known that the ability to distinguish true signal peptides from N-terminal transmembrane regions is one of the main challenges for signal-peptide detection methods, due the similar physical-chemical profiles (Petersen et al., 2011). In this respect, DeepSig scores with a false positive rate of 2.6%, lower than that of SignalP-TM (3.3%) and a higher MCC value. In absolute terms, DeepSig and SignalP-TM produce 20 and 27 false positive predictions out of 787 transmembrane proteins, respectively.
On the two other datasets (Gram-positive and Gram-negative bacteria) DeepSig scores on transmembrane proteins with a false positive rate of 5.9% and 1.5%, respectively. On Gram-positive bacteria, the DeepSig false positive rate is higher compared to that reported SignalP-TM (Table 2). It is possible that low number of transmembrane proteins of Gram-positives hampers the ability of the DCNN to discriminate true signal sequence from transmembrane regions.
The second aspect evaluated is the ability to identify the correct location of the cleavage site. As described in Section 2.4, our method is based on a probabilistic sequence labelling approach which makes use of the relevance profile computed by means of deep Taylor decomposition. For this reason, we are interested in quantifying the impact of this additional feature on the cleavage-site prediction performance. As highlighted in Table 2, considering the F1cs values of all the three protein sets, the inclusion of the relevance profile leads to a better F1 score in cleavage-site prediction of DeepSig. This demonstrates that the relevance profile, when incorporated into the probabilistic sequence labelling method, provides additional information that, in conjunction with primary sequence, helps in identifying the correct extent of the signal sequence.
Comparing results in Table 2, we can conclude that the cleavage site position is better predicted by DeepSig than SignalP, with the exception of Gram positive bacteria. The improvement ranges from 2% to 4%.
3.2 Performance on the SPDS17 independent dataset
Five state-of-the-art predictors are benchmarked toghether with DeepSig on an independent and blind SPDS17 validation set. The predictors are: SignalP4.1 (Petersen et al., 2011), TOPCONS2.0 (Tsirigos et al., 2015), SPOCTOPUS (Viklund et al., 2008), PolyPhobius (Käll et al., 2005), Philius (Reynolds et al., 2008) and PRED-TAT (Bagos et al., 2010), all based on different and well established methods. Again, predictions were generated separately on Eukaryote, Gram-positive and Gram-negative data, either launching the sequences on the respective web-servers or running in-house the standalone versions. Three complementary aspects are compared: the efficiency of the signal peptide detection evaluated with the Matthews correlation coefficient (MCC), the precision of the discrimination between signal peptides and N-terminal transmembrane regions, and the performance on the prediction of the cleavage-site, measured with the F1 score.
For all the organism classes and for all the considered aspects, DeepSig reports the best performances (Table 3). The MCCs of the signal peptide detection are 2 to 4 percentage points higher than the state-of-the art SignalP4.1. When restricting the negative dataset to the most challenging cases (N-terminal transmembrane regions), DeepSig reports the best false positive rate, outperforming SignalP4.1 by 1.5% in the case of eukaryotic proteins. Moreover, DeepSig gives a more exact prediction of the cleavage site in all the three organisms, as highlighted by the cleavage-site F1 values.
Table 3.
Comparative benchmark of different methods in signal peptide detection and cleavage site prediction on the SPDS17 independent dataset
| Method | Eukaryotes |
Gram-positive |
Gram-negative |
||||||
|---|---|---|---|---|---|---|---|---|---|
| MCC | FPRT | F1cs | MCC | FPRT | F1cs | MCC | FPRT | F1cs | |
| SPOCTOPUS | 0.54 | 16.7 | 0.20 | 0.28 | 20.2 | 0.37 | 0.63 | 14.3 | 0.12 |
| PRED-TAT | 0.55 | 9.3 | 0.33 | 0.26 | 2.2 | 0.72 | 0.82 | 9.9 | 0.14 |
| Philius | 0.62 | 6.5 | 0.46 | 0.31 | 3.4 | 0.72 | 0.87 | 7.4 | 0.22 |
| PolyPhobius | 0.73 | 7.4 | 0.42 | 0.44 | 11.2 | 0.53 | 0.80 | 7.9 | 0.06 |
| TOPCONS2.0 | 0.74 | 5.3 | 0.27 | 0.49 | 4.5 | 0.60 | 0.91 | 2.6 | 0.08 |
| SignalP4.1 | 0.82 | 4.0 | 0.69 | 0.50 | 0.0 | 0.79 | 0.93 | 4.2 | 0.33 |
| DeepSig | 0.86 | 2.5 | 0.72 | 0.54 | 0.0 | 0.82 | 0.95 | 2.6 | 0.36 |
Note: MCC, Matthews Correlation Coefficient; FPRT, False Positive Rate on transmembrane proteins; F1cs, The harmonic mean between precision and recall on cleavage-site detection.
3.3 Proteome-wide scanning and detection of TAT-type signal peptides
As a final benchmark, we assessed the performance of DeepSig on the entire proteome of E.coli (strain K12).
DeepSig scores with a MCC value of 0.81, which is in line with the results obtained on other benchmarks. A very low false positive rate on transmembrane proteins was also registered: only 4 out of 1024 transmembrane proteins were incorrectly classified as signal peptides, corresponding to a FPRT of 0.39%. Furthermore, the method was also able to recover the correct cleavage site for 340 signal peptides, corresponding to a F1cs value of 69%. Specifically, the set contains 138 sequences with an experimentally detected signal peptide: DeepSig correctly identifies 126 sequences and correctly places cleavage sites of 116.
Interestingly, even if DeepSig has not been trained to explicitly recognize Twin-Arginine Translocation (TAT-type) signal sequences (Berks, 2015), the method correctly detects 18 out of 32 Tat-type signals that were annotated on E.coli sequences (sensitivity is 56%).
To further investigate the performance of DeepSig on detecting TAT-type signal sequences, we downloaded from UniprotKB/SwissProt all reviewed sequences carrying this kind of signal. We ended up with 553 bacterial proteins, 466 of which were from Gram-negative and 71 from Gram-positive bacteria. Running DeepSig on these sequences, we were able to recover 330 out of 553 TAT signals, corresponding to a sensitivity of about 60%.
4 Conclusion
In this paper, we present DeepSig, a novel approach to predict signal peptides in proteins based on deep learning and sequence labelling methods. The proposed approach was evaluated and compared with other available predictors, including the top-performing SignalP (Petersen et al., 2011). In all the benchmarks, DeepSig reported performances that were comparable and even superior to other state-of-the-art methods.
The method is available as web server and as a standalone program (https://deepsig.biocomp.unibo.it). The standalone version of the program is very fast and easy to install. It takes only 40 min to process the entire human proteome containing some 70 000 protein sequences (test executed by running DeepSig in parallel using four CPU cores). All this suggests that DeepSig is a premier candidate for proteome-scale assessment of protein sub-cellular localization (where high precision is crucial) as well as for single-protein analyses where one is interested in the accurate identification of the signal sequence and cleavage site.
Conflict of Interest: none declared.
Supplementary Material
References
- Abadi M. et al. (2015) Tensorflow: Large-Scale Machine Learning on Heterogeneous Systems. Software available online: https://www.tensorflow.org.
- Alipanahi B. et al. (2015) Predicting the sequence specificities of DNA- and RN-binding proteins by deep learning. Nat. Biotechnol., 33, 831–838. [DOI] [PubMed] [Google Scholar]
- Bach S. et al. (2015) On pixel-wise explanations for non-linear classifier decision by layer-wise relevance propagation. PLoS One, 10, e0130140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagos P.G. et al. (2010) Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics, 26, 2811–2817. [DOI] [PubMed] [Google Scholar]
- Berks B.C. (2015) The twin-arginine protein translocation pathway. Annu. Rev. Biochem., 84, 843–864. [DOI] [PubMed] [Google Scholar]
- Chollet F. et al. (2015) Keras. Software available online: https://keras.io.
- Fariselli P. et al. (2009) Grammatical-restrained hidden conditional random fields for bioinformatics applications. Algorithms Mol. Biol., 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indio V. et al. (2013) The prediction of organelle targeting peptides in eukaryotic proteins with Grammatical Restrained Hidden Conditional Random Fields. Bioinformatics, 29, 981–988. [DOI] [PubMed] [Google Scholar]
- Käll L. et al. (2004) A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol., 338, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Käll L. et al. (2005) An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics, 21, i251–i257. [DOI] [PubMed] [Google Scholar]
- Krizhevsky A. et al. (2012) Imagenet classification with deep convolutional neural networks. In: Pereira F. (ed.), Advances in Neural Information Processing Systems, pp. 1097–1105. [Google Scholar]
- LeCun Y. et al. (2015) Deep learning. Nature, 521, 436–444. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Dobberstein B. (1998) Signal sequences: more than just greasy peptides. Trends Cell Biol., 8, 410–415. [DOI] [PubMed] [Google Scholar]
- Montavon G. et al. (2017) Explaining nonlinear classification decisions with deep Taylor decomposition. Pattern Recogn., 65, 211–222. [Google Scholar]
- Nugent T., Jones D.T. (2009) Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics, 10, 159.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T.N. et al. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Reynolds S.M. et al. (2008) Transmembrane topology and signal peptide prediction using dynamic Bayesian networks. PLoS Comput. Biol., 4, e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savojardo C. et al. (2013) BETAWARE: a machine-learning tool to detect and predict transmembrane beta-barrel proteins in Prokaryotes. Bioinformatics, 29, 504–505. [DOI] [PubMed] [Google Scholar]
- Savojardo C. et al. (2017) ISPRED4: interaction site PREDiction in protein structures with a refining grammar model. Bioinformatics, 33, 1656–1663. [DOI] [PubMed] [Google Scholar]
- Simonyan K. et al. (2013) Deep inside convolutional networks: visualizing image classification models and saliency maps. Comput. Res. Repository, 1312.6034. [Google Scholar]
- Szegedy C. et al. (2013) Intriguing properties of neural networks. Comput. Res. Repository, 1312.6199. [Google Scholar]
- Tsirigos K.D. et al. (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res., 43, W401–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund H. et al. (2008) SPOCTOPUS: a combined predictor of signal peptides and membrane protein topology. Bioinformatics, 24, 2928–2929. [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1990) The signal peptide. J. Membr. Biol., 115, 195–201. [DOI] [PubMed] [Google Scholar]
- Zhou J., Troyanskaya O.G. (2015) Predicting effects of noncoding variants with deep learning-based sequence model. Nature Methods, 12, 931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.