Abstract
Structural neuroimaging measures based on magnetic resonance imaging have been at the forefront of imaging genetics. Global efforts to ensure homogeneity of measurements across study sites have enabled large-scale imaging genetic projects, accumulating nearly 50K samples for genome-wide association studies (GWAS). However, not many novel genetic variants have been identified by these GWAS, despite the high heritability of structural neuroimaging measures. Here, we discuss the limitations of using heritability as a guidance for assessing statistical power of GWAS, and highlight the importance of discoverability—which is the power to detect genetic variants for a given phenotype depending on its unique genomic architecture and GWAS sample size. Further, we present newly developed methods that boost genetic discovery in imaging genetics. By redefining imaging measures independent of traditional anatomical conventions, it is possible to improve discoverability, enabling identification of more genetic effects. Moreover, by leveraging enrichment priors from genomic annotations and independent GWAS of pleiotropic traits, we can better characterize effect size distributions, and identify reliable and replicable loci associated with structural neuroimaging measures. Statistical tools leveraging novel insights into the genetic discoverability of human traits, promises to accelerate the identification of genetic underpinnings underlying brain structural variation.
Introduction
Identifying the genetic underpinning of structural variation in the human brain is the first step in elucidating the complex dynamic processes of both normal and abnormal brain development. Enabled by a global effort to standardize magnetic resonance image (MRI) acquisition, automatic image processing and quantification of MRI measures, the research community has now amassed sufficient neuroimaging data to search genome-wide for the genetic variants associated with neuroimaging measures. Large-scale projects, such as PING (1), IMAGEN (2), ENIGMA (3–5), UK Biobank Imaging Study (6) and the recently launched ABCD (7), have collectively accumulated nearly 50K structural MRI samples, in the hope of finding common genetic loci that account for variation in human brain phenotypes. The confluence of imaging and genetics is thus poised to provide important insights into genetic influences on in vivo human brain variability.
Despite this global effort, however, few genetic loci have been found to be consistently associated with neuroimaging measures (3–5). This was unanticipated by the imaging community. Genes appear to explain a large proportion of the variability in structural neuroimaging measures, i.e. they exhibit high heritability (8–10), while the number of causal loci is presumably fewer than complex diagnostic traits, such as schizophrenia (11). Given that the power of detecting novel loci using genome-wide association studies (GWAS) is the product of sample size, effect size distribution and number of causal loci (12), GWAS of structural neuroimaging measures were expected to identify more novel genetic loci than studies of complex diagnostic traits (13). The apparent lack of power for identifying significant loci in imaging genetics has stimulated debate about whether neuroimaging measures as phenotypes are closer to the biological processes than complex diagnostic traits (3), prompting reconsideration of the genetic architectures of structural neuroimaging measures to guide the development of better analytical tools.
It is now well appreciated in quantitative genetics that high heritability does not guarantee power to detect novel loci in GWAS, as the effect size distribution of causal loci can vary greatly across traits with similar heritability (12). To emphasize the need for a statistical framework to improve discovery of novel genetic loci in imaging genetics, we used the term discoverability, to represent the number of genetic variants that can be consistently and reproducibly detected with GWAS for a given phenotype. In this review, we discuss this concept for imaging genetics and demonstrate methods that have been developed to improve the discoverability in GWAS of structural imaging measures.
This review is structured as following. We briefly revisit the heritability of structural imaging measures and discuss why the concept of discoverability is important. We then focus our review on two new approaches. First, we demonstrate that by leveraging enrichment priors from either genic annotations or other GWAS we can identify single nucleotide polymorphisms (SNPs) robustly associated with structural imaging measures that were below the traditional significance threshold in GWAS. Then we show that by re-defining imaging measures, we can take biological processes into account and, thus, improve the discoverability of neuroimaging GWAS.
Heritability Does Not Predict Discoverability
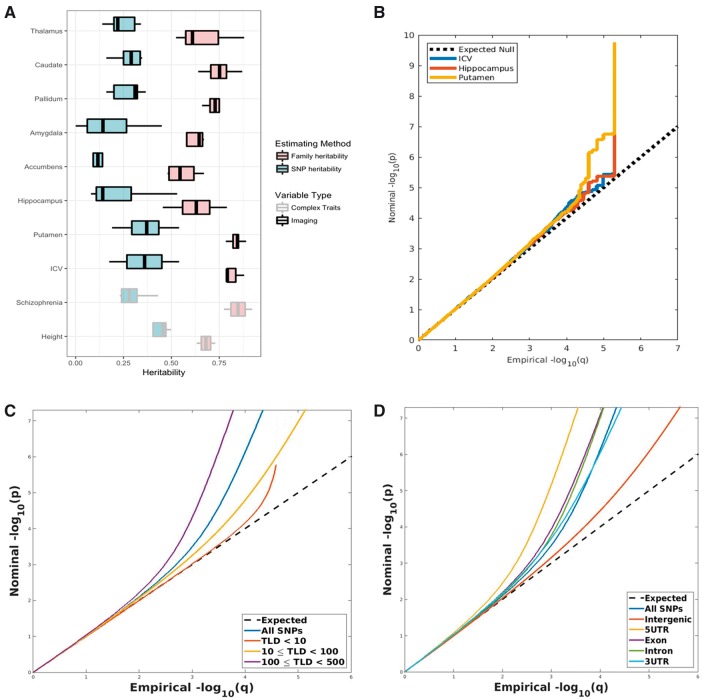
From twin/family studies, the variance explained by genetic effects in structural neuroimaging measures, such as volume of hippocampus, volume of putamen, area of cortical surface and thickness of cortical ribbon, is estimated to be from 40 to 80% (8,9,14–17). The SNP-based heritability of these structural neuroimaging measures, estimated based on genotyped genetic relationships among unrelated individuals, is also significant, up to 54% for subcortical volumes (18) and 45% for cortical surface area (19). Though heritability estimates from SNP-based methods are generally less than from the twin/family designs, the patterns are consistent across these analytic strategies. For example, intra-cranial volume has the highest heritability in twin studies and its SNP-based heritability is also comparable to some highly heritable complex traits, such as height and schizophrenia (20,21). The heritability estimates mentioned above are summarized in Figure 1A.
Figure 1.
Heritability and effect size distribution of structural neuroimaging measures. (A) Heritability estimates based on family (9,14,16) or SNP data (18,51,52). For reference purpose, the heritability of two complex traits, height and schizophrenia, were also provided here. (B) Quantile–quantile plot of effect size distribution in imaging GWAS, including intracranial volumes (ICV), volumes of putamen, and volumes of hippocampus. Flex upward in the tail region represent higher than expected effect sizes in the GWAS findings. (C). Stratified quantile–quantile plot of GWAS for putamen. The effect sizes of putamen GWAS are further stratified according to how well the SNP were tagged, i.e. LD. (D) Stratified quantile–quantile plot of GWAS for putamen, given genic annotation. The effect sizes of putamen GWAS are stratified according to the regulatory regions where SNP located.
Mirroring earlier results with large scale GWAS of other human complex traits (22), high heritability has not consistently been associated with detection of a large number of novel loci for imaging phenotypes. In an analysis of 37 000 individuals, ENIGMA uncovered only one genetic locus contributing to variation in intracranial volume based on T1 images (15,23). In contrast, three to five predictive genetic loci were identified and replicated (15,24) for neuroimaging measures with compatible or lower heritability estimates, such as putamen (mean SNP heritability: 0.36) and hippocampus (mean SNP heritability: 0.27). Figure 1B demonstrates the effect size distributions from the ENIGMA GWAS, using quantile–quantile plots. A flex upward of the tail distribution of effect sizes indicates that large effect size SNPs occurred more often than expected by chance, which is apparent for the hippocampus and putamen GWAS (Fig. 1B). Although many factors can contribute to variability in the success of GWAS to detect specific causal loci, such as measurement error, this unexpected pattern across measures highlights the need to develop alternatives to heritability estimates for assessing the power to detect novel genetic loci. This is particularly important for imaging genetics, as the inherent high dimensionality of neuroimaging measures imposes a large toll on power associated with multiple comparisons. Blindly searching for causal loci for all possible neuroimaging measures without proper selection criteria could reduce the chance of identifying novel loci contributing to brain variation and increase false positive rates. Thus we have emphasized the advantage of assessing the power to detect novel loci, or discoverability of genetic factors, over heritability per se, for selecting imaging phenotypes.
Boosting SNP Discovery in Imaging Genetics
When sample size is fixed, the power to detect novel loci using GWAS is determined by the effect size distribution of true causal loci (12). The larger the effect size per true causal locus, the more discoverable the locus is. Given similar heritability, when there are more true causal loci (i.e. higher polygenicity) more signals diffuse into the background, making SNP effects harder to detect (25). Furthermore, the correlation among SNPs, i.e. linkage-disequilibrium (LD), mingles the diffused background with true signals from causal SNPs, making the signals even harder to differentiate (25). To empirically characterize this phenomenon, we used a mixture of normal models to estimate the effect size distribution from GWAS of neuroimaging measures (26). By modeling mixture between null/non-null SNP effects due to LD (26), the mixture model approximates well the true effect size distribution from GWAS (Fig. 1B) and therefore the power to detect novel loci can be better characterized for each trait. Importantly, the discoverability framework reveals which neuroimaging measures are more likely to yield novel genetic loci with a given GWAS sample size, guiding future study design of the imaging genetics.
The flexibility of mixture modeling provides opportunities to increase SNP discovery in imaging genetics. Because effect sizes from GWAS are approximated by mixtures of distributions, enrichment priors can be incorporated into the mixture components to improve the accuracy of effect size estimation (27). By better estimating the true effect size of each SNP, we can improve discoverability without relying on significance thresholds defined by assuming all SNPs are equal (28–30). Enrichment methodologies have been extensively explored in the quantitative genetics field and a full discussion of these approaches is beyond the scope of this review. Interested readers are referred to our previous review focusing on polygenic enrichment methodologies for complex diseases (31). Here, we summarize the enrichment methods in Table 1 and focus on recent innovations and their application to imaging genetics.
Table 1.
Summary of different enrichment methods for improving loci discovery
| Method | Year | Citation | Applied to structural neuroimaging measures |
|---|---|---|---|
| Conditional FDR | 2013 | (53) | Yes |
| Conjunction FDR | 2013 | (53) | Yes |
| Association Summary Bayes Factor | 2014 | (54) | No |
| Bayesian colocalization (coloc) | 2014 | (39) | No |
| Covariate Modulated local FDR (cmlocFDR) | 2014 | (30) | No |
| fGWAS | 2014 | (41) | No |
| Genetic analysis incorporating Pleiotropy and Annotation (GPA) | 2014 | (55) | No |
| SNP Effect Concordance Analysis (SECA) | 2014 | (56) | Yes |
| Bayesian Conditional FDR | 2015 | (57) | No |
| informed GWAS (iGWAS) | 2015 | (58) | No |
| Covariate-Modulated Mixture Modeling (CM3) | 2016 | (29) | No |
| Genome Wide Association Prioritizer (GenoWAP) | 2016 | (59) | No |
| Pairwise analysis of GWAS (GWAS-PW) | 2016 | (60) | No |
| Weighted Bonferroni Correction | 2016 | (61) | No |
| Genetic analysis incorporating Pleiotropy and Annotation with graphical models (graph-GPA) | 2017 | (62) | No |
| Scalable Functional Bayesian Association (SFBA) | 2017 | (63) | No |
| Multi-trait analysis of GWAS summary statistics (MTAG) | 2018 | (64) | No |
One of the key insights motivating the use of enrichment methods is that genetic variants have attributes that predict their effect sizes across many complex traits (29,32,33). To name a few, GWAS associations are systematically overrepresented in coding and regulatory regions and depleted in intergenic regions (21,32,34–36). SNPs with a higher level of LD with nearby regions tend to have larger effect sizes in GWAS, regardless of the phenotype of interest (32,37). Further, SNPs found to be associated with one phenotype tend to have larger effect sizes in another GWAS of a closely related phenotype (32,38). Leveraging those attributes, and borrowing strength from other sources of prior information, has lead to better estimation of effect sizes for GWAS, resulting in increased probability to detect and replicate causal loci (29,30,39–41). However, until recently, it was unclear if imaging genetics could benefit from these enrichment approaches.
It turns out that enrichment patterns observed for many complex human phenotypes hold for structural neuroimaging measures. Figure 1C and D demonstrates the stratified effect size distribution for ENIGMA putamen GWAS. SNPs with higher LD have increased effect sizes compared to those with lower LD (Fig. 1C). SNPs located in regulatory regions have increased effect sizes compared to those located in intergenic regions (Fig. 1D). Exploiting these properties, we can model the effect size distribution given SNP attributes, and characterize the mixture of null/non-null SNPs according to their expected effect size distribution. With this framework, a recent study found six more novel loci from the ENIGMA GWAS associated with volumes of putamen (42).
Leveraging the genetic overlap between similar traits also improves discoverability in imaging genetics. Using SNP effect sizes obtained from large-scale GWAS of schizophrenia as an SNP attribute (43), a recent analysis discovered more genetic variants consistently associated with volumes of hippocampus, putamen and intra-cranial volumes (44). The analysis yielded six additional loci jointly associated with schizophrenia and structural neuroimaging measures that were below the significance threshold in the original GWAS of the imaging measures. Several of those identified loci reached genome-wide significance in subsequent larger GWASs of these neuroimaging measures, demonstrating the increased power and validity of this mixture with conditional priors approach (23,24).
Defining Complex Imaging Phenotypes Based on Genetics
In addition to better characterizing effect sizes using SNP attributes, another way to boost discoverability is by redefining neuroimaging measures. The anatomical delineations of structural neuroimaging measures used in large-scale GWAS are, to a certain degree, arbitrary, based on traditional anatomical conventions rather than genomic evidence. Anatomically based segmentation involves expert-defined regions of interest, such as hippocampus, that contain many subfields and subpopulations of neuronal and non-neuronal cells. Thus, the summed tissue volume across that region may well be as heterogeneous and polygenic as other complex traits, such as human height (20,42). Analyses of thousands of conventional neuroimaging measures that may produce noisy and highly correlated distributions of genetic effects might not be optimal for discovery, as the multiple comparisons further restrict the power for detecting novel loci.
The curse of dimensionality and the need to define better measures have actually plagued the imaging research community for a long time. This challenge has prompted the community to improve methods for meaningful dimension reduction. Imaging researchers have embraced statistical learning approaches, such as independent component analysis or learning sparseness (45), to define quantitative measures beyond expert-defined anatomical criteria. With the increasing availability of neuroimaging genetic data, some researchers have begun to explore the joint space between genes and images, trying to find a better way to quantify relevant brain variability (46–48).
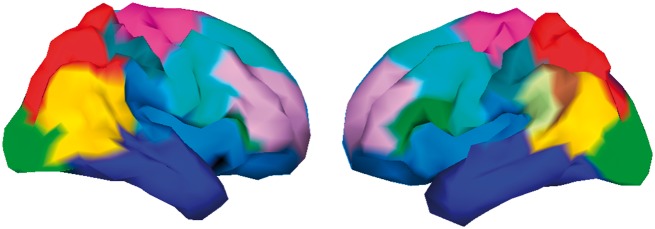
One strategy is to scan through fine-resolution imaging measures and group these based on estimated genetic effects. Genetic correlations estimated from twin studies have been used to cluster T1 imaging measures (49) and show that the clustering patterns are highly consistent with gene expression and SNP correlations (19,50). Figure 2 demonstrates an extension of this approach, using summary statistics of cortical surface GWAS to identify clusters that shared consistent GWAS signals.
Figure 2.
Clustering results based on effect size distribution of vertexwise GWAS. Clustering based on summary statistics from GWAS of each vertex of cortical surface can form nearly identical patterns with clusters derived from twin/family analysis.
Methods that form clusters based on heritability estimates from genotyping data have also been explored. For example, using a kernel estimator to quickly scan through T1 thickness measures across the cortical surface, brain regions with high levels of heritability were identified that did not reflect conventional anatomical landmarks (48). However, as high heritability is not equated to discoverability, it would be interesting to see if similar framework can be applied to the estimation on discoverability.
Another approach is to learn a lower dimensional representation from heritable disorders themselves. Given a disorder with a strong genetic component, especially when associated with a relatively homogeneous pathological process, learning a disease specific representation may be closer to the biological function than an expert-defined region of interest. Using this approach, sparse canonical correlation was used to learn a lower dimensional representation of imaging phenotypes based on an Alzheimer’s disease cohort (46). Meanwhile, we recently used penalized regressions to extract Williams Syndrome specific anatomical scores, finding that lower dimensional representation of imaging measures can boost the statistical power for both phenotypic and genetic associations (47).
Redefining the imaging phenotypes through various dimension reduction approaches is not without limitations. First, the solutions can be highly unstable given that the sample sizes can be considerably smaller than the number of imaging measures included. Even with carefully crafted penalty functions to prevent over-fitting, small genetic effect sizes and limited MRI samples can reduce the utility of this approach. Second, the computational cost for deriving lower dimensional representations grows exponentially with the number of measures included. Nevertheless, improving discoverability in large-scale imaging genetics by redefining the imaging phenotypes is still an active research field with great opportunities for innovations.
Conclusion
The goal of imaging genetics, after all, is to gain insight into genetic regulation of the human brain. Although heritability is a useful metric for the aggregate of genetic signals, it does not reflect the power to detect novel genetic loci contributing to the phenotype. With the dawn of large-scale imaging genetic studies, we are now positioned for breakthrough discoveries of specific gene effects on the human brain. Estimates of discoverability can provide good guidance for designing analyses, accelerating the pace of these discoveries.
Conflict of Interest statement. None declared.
Funding
NIH grant R01MH100351.
References
- 1. Jernigan T.L., Brown T.T., Hagler D.J., Akshoomoff N., Bartsch H., Newman E., Thompson W.K., Bloss C.S., Murray S.S., Schork N.. et al. (2016) The Pediatric Imaging, Neurocognition, and Genetics (PING) data repository. NeuroImage, 124, 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schumann G., Loth E., Banaschewski T., Barbot A., Barker G., Buchel C., Conrod P.J., Dalley J.W., Flor H., Gallinat J.. et al. (2010) The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol. Psychiatry, 15, 1128–1139. [DOI] [PubMed] [Google Scholar]
- 3. Franke B., Stein J.L., Ripke S., Anttila V., Hibar D.P., van Hulzen K.J., Arias-Vasquez A., Smoller J.W., Nichols T.E., Neale M.C.. et al. (2016) Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept. Nat. Neurosci., 19, 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Erp T.G., Hibar D.P., Rasmussen J.M., Glahn D.C., Pearlson G.D., Andreassen O.A., Agartz I., Westlye L.T., Haukvik U.K., Dale A.M.. et al. (2016) Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol. Psychiatry, 21, 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brouwer R.M., Panizzon M.S., Glahn D.C., Hibar D.P., Hua X., Jahanshad N., Abramovic L., de Zubicaray G.I., Franz C.E., Hansell N.K.. et al. (2017) Genetic influences on individual differences in longitudinal changes in global and subcortical brain volumes: results of the ENIGMA plasticity working group. Hum. Brain Mapp., 38, 4444–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller K.L., Alfaro-Almagro F., Bangerter N.K., Thomas D.L., Yacoub E., Xu J., Bartsch A.J., Jbabdi S., Sotiropoulos S.N., Andersson J.L.R.. et al. (2016) Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci., 19, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adolescent Brain Cognitive Development study (ABCD), https://abcdstudy.org
- 8. Eyler L.T., Chen C.H., Panizzon M.S., Fennema-Notestine C., Neale M.C., Jak A., Jernigan T.L., Fischl B., Franz C.E., Lyons M.J.. et al. (2012) A comparison of heritability maps of cortical surface area and thickness and the influence of adjustment for whole brain measures: a magnetic resonance imaging twin study. Twin Res. Hum. Genet., 15, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. den Braber A., Bohlken M.M., Brouwer R.M., van 't Ent D., Kanai R., Kahn R.S., de Geus E.J., Hulshoff Pol H.E., Boomsma D.I. (2013) Heritability of subcortical brain measures: a perspective for future genome-wide association studies. Neuroimage, 83, 98–102. [DOI] [PubMed] [Google Scholar]
- 10. Eyler L.T., Prom-Wormley E., Panizzon M.S., Kaup A.R., Fennema-Notestine C., Neale M.C., Jernigan T.L., Fischl B., Franz C.E., Lyons M.J.. et al. (2011) Genetic and environmental contributions to regional cortical surface area in humans: a magnetic resonance imaging twin study. Cereb. Cortex, 21, 2313–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meyer-Lindenberg A., Weinberger D.R. (2006) Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat. Rev. Neurosci., 7, 818–827. [DOI] [PubMed] [Google Scholar]
- 12. Park J.-H., Wacholder S., Gail M.H., Peters U., Jacobs K.B., Chanock S.J., Chatterjee N. (2010) Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet., 42, 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ripke S., O'Dushlaine C., Chambert K., Moran J.L., Kähler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M.. et al. (2013) Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet., 45, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kremen W.S., Prom-Wormley E., Panizzon M.S., Eyler L.T., Fischl B., Neale M.C., Franz C.E., Lyons M.J., Pacheco J., Perry M.E.. et al. (2010) Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. Neuroimage, 49, 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hibar D.P., Stein J.L., Renteria M.E., Arias-Vasquez A., Desrivieres S., Jahanshad N., Toro R., Wittfeld K., Abramovic L., Andersson M.. et al. (2015) Common genetic variants influence human subcortical brain structures. Nature, 520, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blokland G.A., de Zubicaray G.I., McMahon K.L., Wright M.J. (2012) Genetic and environmental influences on neuroimaging phenotypes: a meta-analytical perspective on twin imaging studies. Twin Res. Hum. Genet., 15, 351–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gillespie N.A., Neale M.C., Hagler D.J. Jr, Eyler L.T., Fennema-Notestine C., Franz C.E., Lyons M.J., McEvoy L.K., Dale A.M., Panizzon M.S.. et al. (2017) Genetic and environmental influences on mean diffusivity and volume in subcortical brain regions. Hum. Brain Mapp., 38, 2589–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Toro R., Poline J.B., Huguet G., Loth E., Frouin V., Banaschewski T., Barker G.J., Bokde A., Buchel C., Carvalho F.M.. et al. (2015) Genomic architecture of human neuroanatomical diversity. Mol. Psychiatry, 20, 1011–1016. [DOI] [PubMed] [Google Scholar]
- 19. Chen C.-H., Peng Q., Schork A.J., Lo M.-T., Fan C.-C., Wang Y., Desikan R.S., Bettella F., Hagler D.J., Westlye L.T.. et al. (2015) Large-scale genomics unveil polygenic architecture of human cortical surface area. Nat. Commun., 6, 7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W.. et al. (2010) Common SNPs explain a large proportion of the heritability for human height. Nat. Genet., 42, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang J., Manolio T.A., Pasquale L.R., Boerwinkle E., Caporaso N., Cunningham J.M., de Andrade M., Feenstra B., Feingold E., Hayes M.G.. et al. (2011) Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet., 43, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J. (2017) 10 Years of GWAS Discovery: biology, Function, and Translation. Am. J. Hum. Genet., 101, 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adams H.H., Hibar D.P., Chouraki V., Stein J.L., Nyquist P.A., Renteria M.E., Trompet S., Arias-Vasquez A., Seshadri S., Desrivieres S.. et al. (2016) Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci., 19, 1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hibar D.P., Adams H.H., Jahanshad N., Chauhan G., Stein J.L., Hofer E., Renteria M.E., Bis J.C., Arias-Vasquez A., Ikram M.K.. et al. (2017) Novel genetic loci associated with hippocampal volume. Nat. Commun., 8, 13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J., Weedon M.N., Purcell S., Lettre G., Estrada K., Willer C.J., Smith A.V., Ingelsson E., O'Connell J.R., Mangino M.. et al. (2011) Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet., 19, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holland D., Fan C.-C., Frei O., Shadrin A.A., Smeland O.B., Sundar V.S., Andreassen O.A., Dale A.M. (2017) Estimating phenotypic polygenicity and causal effect size variance from GWAS summary statistics while accounting for inflation due to cryptic relatedness. bioRxiv, 133132.
- 27. Gianola D. (2013) Priors in whole-genome regression: the Bayesian alphabet returns. Genetics, 194, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thompson W.K., Wang Y., Schork A.J., Witoelar A., Zuber V., Xu S., Werge T., Holland D. Schizophrenia Working Group of the Psychiatric Genomics, C. Andreassen O.A.. et al. (2016) An empirical Bayes mixture model for effect size distributions in genome-wide association studies. PLoS Genet., 11, e1005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y., Thompson W.K., Schork A.J., Holland D., Chen C.H., Bettella F., Desikan R.S., Li W., Witoelar A., Zuber V.. et al. (2016) Leveraging genomic annotations and pleiotropic enrichment for improved replication rates in Schizophrenia GWAS. PLoS Genet., 12, e1005803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zablocki R.W., Schork A.J., Levine R.A., Andreassen O.A., Dale A.M., Thompson W.K. (2014) Covariate-modulated local false discovery rate for genome-wide association studies. Bioinformatics, 30, 2098–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schork A.J., Wang Y., Thompson W.K., Dale A.M., Andreassen O.A. (2016) New statistical approaches exploit the polygenic architecture of schizophrenia–implications for the underlying neurobiology. Curr. Opin. Neurobiol., 36, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schork A.J., Thompson W.K., Pham P., Torkamani A., Roddey J.C., Sullivan P.F., Kelsoe J.R., O'Donovan M.C., Furberg H., Schork N.J.. et al. (2013) All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet., 9, e1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K.. et al. (2015) Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet., 47, 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J.. et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science, 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. U. S. A., 106, 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gusev A., Lee S.H., Trynka G., Finucane H., Vilhjalmsson B.J., Xu H., Zang C., Ripke S., Bulik-Sullivan B., Stahl E.. et al. (2014) Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet., 95, 535–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J. Schizophrenia Working Group of the Psychiatric Genomics, C. Patterson N., Daly M.J., Price A.L., Neale B.M. (2015) LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet., 47, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu J.Z., Hov J.R., Folseraas T., Ellinghaus E., Rushbrook S.M., Doncheva N.T., Andreassen O.A., Weersma R.K., Weismuller T.J., Eksteen B.. et al. (2013) Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat. Genet., 45, 670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet., 10, e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kichaev G., Yang W.-Y., Lindstrom S., Hormozdiari F., Eskin E., Price A.L., Kraft P., Pasaniuc B. (2014) Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet., 10, e1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pickrell J.K. (2014) Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet., 94, 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen C.H., Wang Y., Lo M.T., Schork A., Fan C.C., Holland D., Kauppi K., Smeland O.B., Djurovic S., Sanyal N.. et al. (2017) Leveraging genome characteristics to improve gene discovery for putamen subcortical brain structure. Sci. Rep., 7, 15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ripke S., Neale B., Corvin A., Walters J., Farh K., Holmans P., Lee P. (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smeland O.B., Wang Y., Frei O., Li W., Hibar D.P., Franke B., Bettella F., Witoelar A., Djurovic S., Chen C.H.. et al. (2017) Genetic overlap between schizophrenia and volumes of hippocampus, putamen, and intracranial volume indicates shared molecular genetic mechanisms. Schizophrenia Bull., sbx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hastie T., Tibshirani R., Friedman J. (2009) The Elements of Statistical Learning. Springer, New York. [Google Scholar]
- 46. Du L., Huang H., Yan J., Kim S., Risacher S.L., Inlow M., Moore J.H., Saykin A.J., Shen L. and Initiative, f.t.A.s.D.N. (2016) Structured sparse canonical correlation analysis for brain imaging genetics: an improved GraphNet method. Bioinformatics, 32, 1544–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fan C.C., Brown T.T., Bartsch H., Kuperman J.M., Hagler D.J., Schork A., Searcy Y., Bellugi U., Halgren E., Dale A.M. (2017) Williams syndrome-specific neuroanatomical profile and its associations with behavioral features. NeuroImage: Clin., 15, 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ge T., Nichols T.E., Lee P.H., Holmes A.J., Roffman J.L., Buckner R.L., Sabuncu M.R., Smoller J.W. (2015) Massively expedited genome-wide heritability analysis (MEGHA). Proc. Natl Acad. Sci., 112, 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen C.H., Gutierrez E.D., Thompson W., Panizzon M.S., Jernigan T.L., Eyler L.T., Fennema-Notestine C., Jak A.J., Neale M.C., Franz C.E.. et al. (2012) Hierarchical genetic organization of human cortical surface area. Science, 335, 1634–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peng Q., Schork A., Bartsch H., Lo M.-T., Panizzon M.S., Westlye L.T., Kremen W.S., Jernigan T.L., Le Hellard S., Steen V.M.. et al. (2016) Conservation of distinct genetically-mediated human cortical pattern. PLoS Genet., 12, e1006143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zheng J., Erzurumluoglu A.M., Elsworth B.L., Kemp J.P., Howe L., Haycock P.C., Hemani G., Tansey K., Laurin C., Pourcain B.S.. et al. (2017) LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics, 33, 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roshchupkin G.V., Gutman B.A., Vernooij M.W., Jahanshad N., Martin N.G., Hofman A., McMahon K.L., van der Lee S.J., van Duijn C.M., de Zubicaray G.I.. et al. (2016) Heritability of the shape of subcortical brain structures in the general population. Nat. Commun., 7, 13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Andreassen O.A., Thompson W.K., Schork A.J., Ripke S., Mattingsdal M., Kelsoe J.R., Kendler K.S., O'Donovan M.C., Rujescu D., Werge T.. et al. (2013) Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet., 9, e1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Iversen E.S., Lipton G., Clyde M.A., Monteiro A.N. (2014) Functional annotation signatures of disease susceptibility loci improve SNP association analysis. BMC Genomics, 15, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chung D., Yang C., Li C., Gelernter J., Zhao H. (2014) GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet., 10, e1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nyholt D.R. (2014) SECA: sNP effect concordance analysis using genome-wide association summary results. Bioinformatics, 30, 2086–2088. [DOI] [PubMed] [Google Scholar]
- 57. Liley J., Wallace C. (2015) A pleiotropy-informed Bayesian false discovery rate adapted to a shared control design finds new disease associations from GWAS summary statistics. PLoS Genet., 11, e1004926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fortney K., Dobriban E., Garagnani P., Pirazzini C., Monti D., Mari D., Atzmon G., Barzilai N., Franceschi C., Owen A.B.. et al. (2015) Genome-wide scan informed by age-related disease identifies loci for exceptional human longevity. PLoS Genet., 11, e1005728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lu Q., Yao X., Hu Y., Zhao H. (2016) GenoWAP: gWAS signal prioritization through integrated analysis of genomic functional annotation. Bioinformatics, 32, 542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pickrell J.K., Berisa T., Liu J.Z., Segurel L., Tung J.Y., Hinds D.A. (2016) Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet., 48, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sveinbjornsson G., Albrechtsen A., Zink F., Gudjonsson S.A., Oddson A., Masson G., Holm H., Kong A., Thorsteinsdottir U., Sulem P.. et al. (2016) Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat. Genet., 48, 314–317. [DOI] [PubMed] [Google Scholar]
- 62. Chung D., Kim H.J., Zhao H. (2017) graph-GPA: a graphical model for prioritizing GWAS results and investigating pleiotropic architecture. PLoS Comput. Biol., 13, e1005388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang J., Fritsche L.G., Zhou X., Abecasis G. and International Age-Related Macular Degeneration Genomics, C. (2017) A scalable Bayesian method for integrating functional information in genome-wide association studies. Am. J. Hum. Genet., 101, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Turley P., Walters R.K., Maghzian O., Okbay A., Lee J.J., Fontana M.A., Nguyen-Viet T.A., Wedow R., Zacher M., Furlotte N.A.. et al. (2018) Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet., 50, 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]