An 18-gene RNA signature detects severe cases among young Cambodian secondary-infected dengue patients and yields insights into the underlying pathogenesis. We present evidence that the detection is robust for peripheral blood mononuclear cells and whole blood and different experimental techniques.
Keywords: dengue, severity, blood, RNA, transcriptome, biomarker, prognosis, risk, signature
Abstract
Background
Early detection of severe dengue can improve patient care and survival. To date, no reliable single-gene biomarker exists. We hypothesized that robust multigene signatures exist.
Methods
We performed a prospective study on Cambodian dengue patients aged 4 to 22 years. Peripheral blood mononuclear cells (PBMCs) were obtained at hospital admission. We analyzed 42 transcriptomic profiles of patients with secondary dengue infected with dengue serotype 1. Our novel signature discovery approach controls the number of included genes and captures nonlinear relationships between transcript concentrations and severity. We evaluated the signature on secondary cases infected with different serotypes using 2 datasets: 22 PBMC samples from additional patients in our cohort and 32 whole blood samples from an independent cohort.
Results
We identified an 18-gene signature for detecting severe dengue in patients with secondary infection upon hospital admission with a sensitivity of 0.93 (95% confidence interval [CI], .82–.98), specificity of 0.67 (95% CI, .53–.80), and area under the receiver operating characteristic curve (AUC) of 0.86 (95% CI, .75–.97). At validation, the signature had empirical AUCs of 0.85 (95% CI, .69–1.00) and 0.83 (95% CI, .68–.98) for the PBMCs and whole blood datasets, respectively.
Conclusions
The signature could detect severe dengue in secondary-infected patients upon hospital admission. Its genes offer new insights into the pathogenesis of severe dengue.
Dengue is the most widespread mosquito-borne viral infection worldwide. Currently, 40%–50% of the world population lives in areas at risk for dengue virus (DENV) transmission [1]. If the majority of dengue cases are uncomplicated, it is estimated that each year 500000 cases, mostly children, progress to severe dengue and require hospitalization. According to the World Health Organization (WHO), approximately 2.5% of those affected by severe dengue requiring hospitalization are still dying from complications [1]. The recent explosive spread of the related Zika virus might further increase this burden. Indeed, the complications associated with severe dengue are more common after secondary infection than after primary infection [2], and recent studies both in vitro and in vivo have highlighted the potential of anti-Zika immunity to trigger dengue enhancement [3]. As recently highlighted by the WHO, robust and early detection of severe dengue, along with access to proper medical care, would not only decrease the fatality rate to 1% but also reduce healthcare costs and economic burden of the disease [1].
Although diagnosis methods for dengue infection are well established, there are no prognostic tests to help the clinician evaluate the risk of infection progressing to severe dengue. A number of signatures that use clinical variables for detecting severe cases of dengue infection have been proposed for adults and/or children [4–9]. Nevertheless, none of the signatures we found in the literature have been replicated on independent datasets. In addition to these studies, others have aimed to identify molecular biomarkers, based on either messenger RNA expression or on protein or cytokine levels. A number of genome-wide expression profiling studies have also been performed in Nicaragua, Cambodia, Thailand, and Vietnam [10–14]. Every study uncovered differentially expressed genes associated with severe dengue. Many of these genes have functions associated with innate immunity, vascular permeability, coagulation, neutrophil-derived antimicrobial resistance, inflammation, and lipid metabolism. However, their capacity to detect severe cases among dengue patients was not evaluated [6–8, 10–13], or the studies excluded children [9].
We hypothesized that a simple combination of a small number of gene expression markers may be robust enough to establish reproducible detection of severe cases among newly admitted dengue patients. With this in mind, we attempted to develop a signature discovery algorithm—one that allowed for not only linear but also more general monotonic relationships among features, meaning more complex, but still easily interpretable, relationships among genes.
MATERIALS AND METHODS
Cohorts Studied
We conducted a prospective study in the Kampong Cham referral hospital in Cambodia during a 3-year period (2011–2013). Patients with suspected dengue infection were identified on the date of admission to participate in the study and were enrolled after obtaining agreement and written informed consent from the patient or from parents or guardians.
On the same day, the patient or his/her parents were interviewed and stated the day of fever onset. Patients were processed as follows: plasma was used for dengue confirmatory diagnostics, including serology and molecular diagnostics, as described elsewhere [15], and blood clots and peripheral blood mononuclear cells (PBMCs) were kept for later analyses (see description below). Briefly, plasma was tested for dengue infection by NS1 rapid diagnostic test at the hospital and sent, on the same day, to Institut Pasteur Cambodia for DENV reverse transcription polymerase chain reaction (RT-PCR) confirmation and DENV serotyping. The RT-PCR result was sent back to the hospital. A second blood sample was taken at the day of discharge from the hospital and sent to Institut Pasteur Cambodia. Both samples were tested for immunoglobulin M by M antibody-captured enzyme-linked immunosorbent assay and total antibodies by hemagglutination inhibition assay. Primary or secondary immune status of DENV infections was determined by hemagglutination inhibition assay according to WHO criteria [16] and was confirmed by comparing the hemagglutination inhibition assay titer between the 2 samples. We subsequently used only the transcriptome of the first blood sample collected at hospital admission for multigene signature discovery.
Although 438 patients were initially enrolled, only a fraction donated an amount of blood compatible with the biochemistry and serological tests, as well as the isolation of PBMCs for transcriptomic analysis. Out of these, only 1 case was a primary infection, and few were non–DENV-1-infected. To maximize the likelihood for obtaining a meaningful RNA signature, we further limited our training set to samples from secondary, DENV-1–infected patients. This training dataset consisted of 42 patients (15 with severe dengue, 27 with nonsevere dengue). From the remaining patients, 22 samples (7 with severe dengue, 15 with nonsevere dengue, from secondary patients with different serotypes) contained sufficient amounts of RNA for quantitative real-time polymerase chain reaction (qRT-PCR). These 22 samples were used as a validation dataset. The mean ages of these 2 datasets were 9.0 ± 3.8 years (7.7 ± 2.4 y for severe patients; 9.8 ± 4.2 y for nonsevere patients) and 8.8 ± 3.1 years (7.0 ± 1.5 y for severe patients; 9.7 ± 4.3 for nonsevere patients) (Table 1 and Supplementary Material 1).
Table 1.
Two Cohorts and Training and Validation Subcohorts
| Case characteristics | Clinical cohort | Devignot et al[11] cohort | ||
|---|---|---|---|---|
| Entire clinical cohort | PBMC microarray subcohort | PBMC qRT-PCR subcohort | Whole blood microarray subcohort | |
| Training set | Validation set | Validation set | ||
| Suspected dengue cases | 438 | 42 | 22 | 32 |
| Age, y | 8.8 ± 3.3 | 9.0 ± 3.8 | 8.8 ± 3.1 | 7.8 ± 2.5 |
| Sex, % female | 51 | 52 | 60 | 66 |
| Mean day of blood sampling after onset of fever | 3.3 ± 1.5 | 3.1 ± 1.9 | 3.6 ± 1.3 | 5.1 ± 1.0 |
| Confirmed dengue | 316 | 42 | 22 | 32 |
| DENV-1 | 265 | 42 | 10 | 4 |
| DENV-2 | 15 | 0 | 6 | 3 |
| DENV-3 | 0 | 0 | 0 | 16 |
| DENV-4 | 28 | 0 | 6 | 1 |
| Unknown | 8 | 0 | 0 | 8 |
| Secondary infection | 183 | 42 | 22 | 32 |
| Classification according to WHO 2009 criteria | ||||
| Nonsevere dengue, with or without warning signs | 236 | 27 | 15 | 14 |
| Severe dengue | 80 | 15 | 7 | 18 |
Abbreviations: DENV, dengue virus; PBMC, peripheral blood mononuclear cells; qRT-PCR, quantitative real-time polymerase chain reaction; WHO, World Health Organization.
For this PBMC cohort, disease severity was classified according to the 2009 WHO criteria [16] using clinical and biological data recorded at admission and throughout the entire hospitalization period. Additionally, an independent publicly available dataset was used for a second validation [11]. In that dataset, gene expression had been quantified globally using microarrays from Cambodian blood samples. Disease severity was classified according to the description in the section “Signature Discovery” below. Table 1 provides additional summary information across all datasets we used.
Ethics Statement
The study was approved by the Cambodian National Ethics Committee for Health Research (approval no. 087NECHR /2011 and no. 063NECHR/2012). Before enrollment, written consent signed by the participant or by a legal representative for participants aged <16 years was obtained.
RNA Preparation, Microarray Hybridization, and Quantitative Real-Time Polymerase Chain Reaction Validation
RNA was extracted from PBMCs stored in RNA protect cell reagent (Qiagen, Hilden, Germany) with a miRNeasy kit (Qiagen), and RNA quality was checked on a BioAnalyzer 2100 (Agilent, Santa Clara, CA).
For microarray analysis of the training cohort, gene expression in PBMCs was analyzed using Affymetrix Human Transcriptome Array 2 (HTA2) GeneChips. The HTA2 chips were prepared, hybridized, and scanned according to the manufacturer’s instructions. For qRT-PCR of the PBMC validation cohort, 200 ng of RNA were reverse-transcribed with SuperScript VILO cDNA synthesis kit (Invitrogen, Life Technologies, Carlsbad, CA) using a combination of random hexamer and Oligo(dT)12–18 primers. TaqMan Gene Expression Assays (Life Technologies) were used for each candidate gene according to the manufacturer’s instructions. Relative expression was calculated with the 2-ΔΔCt method, using beta-glucuronidase as an endogenous control for normalization and a calibrator sample as a comparator for every sample.
Machine Learning Methodology
Our signature was created using a machine learning approach based on monotonic regression on a training cohort (Supplementary Material 3–6). Briefly, new predictions made by the signature are based on 0/1 (nonsevere/severe) predictions (“votes”) derived from pairs of transcripts in the signature. Measured concentrations for any given transcript pair are turned into a binary vote using a 2-dimensional monotonic function [17], a generalization of a linear function that monotonically increases or decreases with the concentration of each transcript. The final consensus prediction is “severe” if the mean of all votes is above a suitably chosen threshold t, and “nonsevere” otherwise. The performance of individual transcript pairs on future patients is estimated using cross-validation.
Initially, a set of transcript pairs with optimal performance estimate is determined. Using a permutation test, those genes that do not confer a statistical performance advantage over the performance of their partner alone are eliminated. The resulting model represents a unique combination of lower- and higher-complexity features tailored toward the discovery of complex disease signatures. The monotonic model represents a generalization of linear models. Nevertheless, the resulting predictors can still be visually and intuitively understood. Controlling the number of transcripts in the signature allows different trade-offs between performance, robustness, and assay cost (Supplementary Material 9).
The signature is rescaled to different datasets by mapping transcripts to genes and quantile-normalizing the expression values (Supplementary Material 6). In cases where only 1 of 2 transcript measurements is available, the missing variable is replaced by a duplicate of the variable that is available.
Signature Discovery
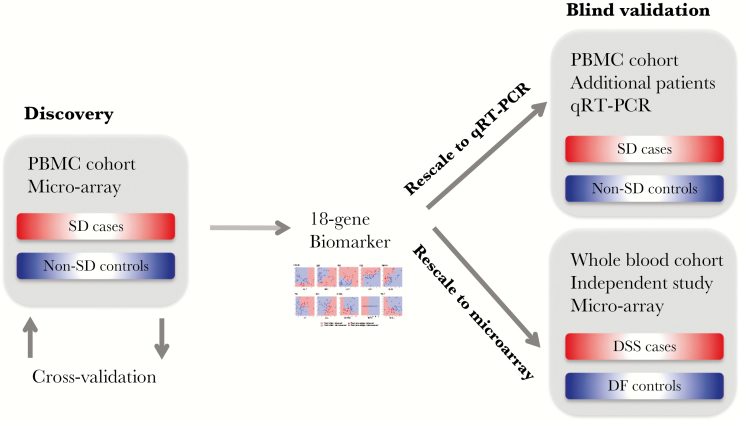
We applied the above machine learning approach to microarray transcriptomes of the PBMC training cohort, leading to an initial assessment of its performance via rigorous cross-validation. After applying quantile normalization (Supplementary Material 6), we evaluated this signature on 2 other datasets (Figure 1).
Figure 1.
Discovery and validation of the severe dengue signature.
The first validation dataset consisted of 22 patients (7 with severe dengue, 15 with nonsevere dengue) from the PBMC validation cohort whose gene expression was measured using qRT-PCR.
The second validation dataset was an independent, publicly available, Cambodian whole blood dataset selected for its large size and high quality [11]. It consisted of whole blood transcriptome data from 48 dengue-infected patients. At the time of that study, phenotype was still established according to the 1997 WHO classification: DSS (dengue shock syndrome), DHF (dengue hemorrhagic fever), and DF (dengue fever) [18]. To make phenotype data comparable, we reclassified the disease severity as well as possible in terms of the 2009 WHO classification. We considered all 18 DSS patients as severe dengue and all 14 DF patients as nonsevere, considering that DF patients that are reclassified as severe dengue in the 2009 WHO classification are rare. The DHF patients could not be classified without additional clinical information that was unavailable to us and were thus excluded.
Performance Evaluation
We summarized the empirical performance of our signature using receiver operating characteristic curves, which consist of the different combinations of empirical true- and false-positive rates that are obtained by varying the above threshold t between 0 and 1. For evaluations of the performance of state-of-the-art machine learning methods, we used the implementations from the Python sklearn package [19].
RESULTS
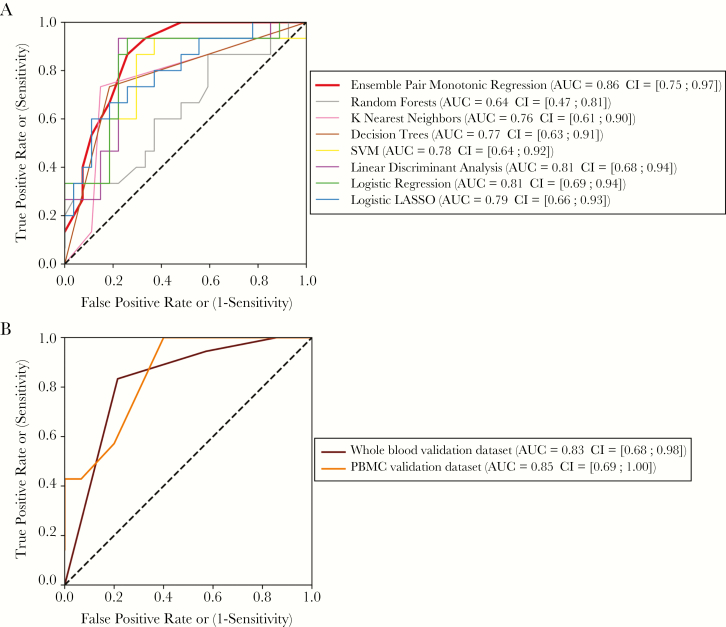
Our automated machine learning approach for signature discovery resulted in an 18-gene signature that allows the detection of severe dengue from a blood sample taken from dengue patients upon hospital arrival. We obtained area under the receiver operating characteristic curve (AUC) values of 0.86, 0.83, and 0.85 for the training set and 2 validation datasets with widely different characteristics, respectively (Figure 2).
Figure 2.
Evaluation of classification performance. A, Cross-validated training data, various methods. B, Validation data, our signature.
Examples of empirical sensitivity-specificity combinations are shown in Table 2. The difference in the threshold required for PBMCs and whole blood may be attributable to the difference in composition between these 2 sample types. Twelve of the 18 genes in the signature are immune-related (Table 3). Certain genes have already been associated with severe dengue.
Table 2.
Empirically Observed Combinations of Sensitivity and Specificity Across Datasets
| Sensitivity (95% CI) | Specificity (95% CI) | Thresholda | |
|---|---|---|---|
| PBMC microarray training dataset | 0.93 (.82–.98) | 0.67 (.53–.80) | 0.40 |
| PBMC qRT-PCR validation dataset |
0.86 (.67–.96) | 0.67 (.48–.84) | 0.40 |
| Whole blood microarray validation dataset |
0.83 (.67–.93) | 0.79 (.63–.90) | 0.91 |
Abbreviations: CI, confidence interval; PBMC, peripheral blood mononuclear cells; qRT-PCR, quantitative real-time polymerase chain reaction.
aThresholds were selected to balance specificity and sensitivity.
Table 3.
Constitutive Gene Pairs of the RNA Signature
| Gene | Gene Name | Description | Over/ under (+/−) expressed in severe dengue |
Known link with severe dengue in literature |
|---|---|---|---|---|
| E2F7 | E2F transcription factor 7 | Transcription factor implicated in angiogenesis, polyploidization of specialized cells and DNA damage response. Acts as a negative regulator of keratinocyte differentiation | + | … |
| ENKUR | Enduring, TRPC channel interacting protein | Calcum-mediated signaling | − | … |
| ARG1 | Arginase 1 | Controls arginine metabolism in neutrophils, hence controlling NO production (iNOS pathway) moderator of T cell function. | + | [13] |
| JUNB | JunB proto-oncogene, AP-1 TF subunit | Transcription factor involved in regulating gene activity following the primary growth factor response. Expressed in neutrophils. Part of the iNOS pathway | − | … |
| E2F7 | E2F transcription factor 7 | Transcription factor implicated in angiogenesis, polyploidization of specialized cells, and DNA damage response. Acts as a negative regulator of keratinocyte differentiation | + | … |
| MPO | Myeloperoxydase | Produced mainly by neutrophils. This enzyme produces hypohalous acids central to the microbicidal activity of neutrophils. | + | [11, 13] |
| LRP1 | Low-density lipoprotein receptor-related protein 1 | Endocytic receptor involved in endocytosis and in phagocytosis of apoptotic cells. Involved in the plasma clearance of chylomicron remnants and activated LRPAP1 (alpha 2-macroglobulin) | − | … |
| PGD | Phosphogluconate dehydrogenase | Enzyme involved in the pentose phosphate pathway, hence producing more NADPH. NADPH is a cofactor used in anabolic reactions, such as lipid and nucleic acid synthesis, which require NADPH as a reducing agent. | + | … |
| EGR3 | Early growth response 3 | This gene encodes a transcriptional regulator that belongs to the EGR family of C2H2-type zinc-finger proteins. It is an immediate-early growth response gene that is induced by mitogenic stimulation. The protein encoded by this gene participates in the transcriptional regulation of genes in controlling biological rhythm. It may also play a role in a wide variety of processes including endothelial cell growth. | − | … |
| MGAM | Maltase-glucoamylase | This gene encodes maltase-glucoamylase, which plays a role in the final steps of digestion of starch. | + | … |
| HP | Haptoglobin | Binds free plasma hemoglobin, antimicrobial activity | + | [11, 13, 14] |
| MYB | Myeloblastosis proto-oncogene, transcription factor | Transcriptional activator, implicated in B-cell lymphoma | + | … |
| IGKC | Immunoglobulin kappa constant | Codes for the constant region of antibody light chains. Antibodies are produced and secreted by plasma cells and contribute to the elimination of the pathogen. |
+ | … |
| PPBP | Pro-platelet basic protein | Platelet-derived growth factor of the CXC family. It is a potent chemoattractant and activator of neutrophils and has antimicrobial properties. | − | … |
| CD40LG | CD40 ligand | This gene is expressed on the surface of T cells. It regulates B-cell function by engaging CD40 on the B-cell surface. A defect in this gene results in an inability to undergo immunoglobulin class switch and is associated with hyper immunoglobulin M syndrome. | − | … |
| OX40L | OX40 ligand | The protein functions in T-cell antigen-presenting cell interaction and mediates adhesion of activated T cells to endothelial cells. | − | … |
| SDPR | Serum deprivation response | Participates in the formation of caveolae. | − | [20] |
| TCF7 | Transcription factor 7 (T-cell specific, HMG-box) | This gene is expressed predominantly in T cells and plays a critical role in the development of natural killer cells and innate lymphoid cells. The encoded protein forms a complex with beta-catenin and activates transcription through a Wnt/ beta-catenin signaling pathway. | − | … |
| ASAP2 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 | The protein is localized in the Golgi apparatus and at the plasma membrane. The protein forms a stable complex with PYK2 in vivo. | − | … |
Genes are grouped into pairs and singletons.
Abbreviations: iNOS, inducible nitric oxide synthase; NADPH, reduced form of nicotinamide adenine dinucleotide phosphate; NO, nitric oxide.
To determine whether the inclusion of a larger number of genes or the restriction to a linear state-of-the-art variable selection model would have increased classification accuracy, we estimated the performance of several well-known classification methods (Figure 2A). Even though all pairs of confidence intervals overlap, the AUC estimate of our method was among the highest. This was also the case for the public whole blood dataset (Supplementary Material 10). For the PBMC qRT-PCR dataset, we only measured transcripts from our signature and therefore could not compare results with the performance of other methods on this dataset, except logistic regression with a lasso penalty (“logistic lasso”). Contrary to our method, logistic lasso generated a classifier whose performance was not better than random on the PBMC qRT-PCR dataset (Supplementary Material 11).
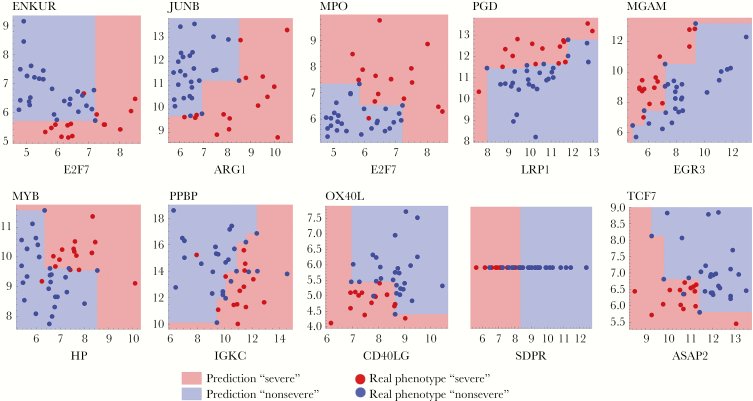
Figure 3 provides a visualization of the models associated with the transcript pairs of the signature. The panels show the points corresponding to transcripts from the PBMC training cohort. Different monotonic functions capture different types of gene–gene interactions. For example, for the second pair of transcripts (JUNB and ARG1), patients have a severe phenotype when JUNB expression is high or ARG1 expression is low. For the OX40L/CD40LG pair, OX40L and CD40LG both are underexpressed in the severe patients. For the EGR3/MGAM pair, the lower the EGR3 expression and the higher the MGAM expression, the more likely the patient is to be predicted severe.
Figure 3.
Visual representation of the RNA signature.
DISCUSSION
We have identified and independently validated an RNA signature for the detection of severe cases among young, secondary-dengue patients from blood samples taken upon arrival at the hospital. Previous attempts have addressed the complexity of dengue infection by measuring multiple molecules. Nhi et al identified 19 plasma proteins exhibiting significantly different relative concentrations (P < .05) on 16 patients (6 with severe dengue, 10 with nonsevere dengue) [20]. Among them, a combination of antithrombin III and angiotensin had strong power to detect the 6 severe dengue patients (AUC = 0.87). Pang et al developed a signature combining transcript, protein, and clinical markers, mostly linked to innate immunity and coagulation, that was able to detect patients with warning signs and needing to be hospitalized with sensitivity of 96% and specificity of 54.6% on a validation cohort that was chosen to match the learning cohort [9]. However, none of the signatures have been replicated on an independent, publicly available cohort.
Our objective was to identify an RNA signature to detect severe dengue cases from blood samples taken upon patient admission to hospital. We used data from a prospective study in Cambodia, in which young patients with suspected dengue gave blood samples upon hospital admission. Severe dengue cases were identified according to the WHO 2009 criteria using data at admission and during hospital stay. Because in our cohort only 1 severe patient appeared to have primary dengue and most had DENV-1 serotype, we limited our analysis to the 42 high-quality samples from secondary DENV-1–infected cases. We then globally quantified gene expression profiles of PBMCs using transcriptome microarrays.
Our novel signature discovery approach models linear and nonlinear monotonic interactions between transcript levels with controlled complexity and preserves interpretability and applicability for small input datasets. To our knowledge, the resulting RNA signature comprising 18 genes represents the first signature to detect severe cases in dengue patients with a demonstrated high performance across several validation datasets.
The consistency of the empirical performance estimates of our RNA signature may indicate a high degree of robustness across a number of sample characteristics. First, blood sample type: training and first validation datasets were generated from PBMCs, but the second validation dataset originates from whole blood. Second, DENV serotype: the signature was derived from DENV-1 samples, but the validation datasets were a mixture of different serotypes. Third, measurement technologies: both array-based and qRT-PCR technologies were used to measure mRNA concentrations. Further heterogeneity arose from the fact that, even though dengue is considered a pediatric disease in Cambodia, the age of patients varied. In addition, our data are from a single time sample from the day of hospital admission (ie, at variable times after fever onset). The above sources of heterogeneity point to possibilities to obtain refined, and potentially higher-performing, signatures with tighter performance estimates in studies with larger and more homogeneous cohorts, in particular for different age groups. Finally, one important source of heterogeneity that we did not address is immune status. All patients in our training and validation sets had secondary infection, and the question about the performance of the signature in the presence of primary severe dengue samples, more typical of regions in which dengue transmission is low, remains to be addressed.
Genes used in robust, high-performing signatures may also represent robust pointers into the biology of severe dengue. The genes OX40L and CD40L that comprise the first gene pair of our signature are both underexpressed in severe cases (Figure 3). OX40L and CD40L are membrane proteins expressed by dendritic cells and by activated T cells, respectively, that are essential to mount an efficient adaptive immune response. OX40L binds to its co-receptor OX40 and allows T cells to survive after clonal expansion. Stimulation of B cells by T cells through CD40L is necessary for class switching and somatic hypermutation, and hence both genes are required to produce potent neutralizing antibodies [21]. In the context of dengue infection, OX40L has been shown to be downregulated in human monocyte-derived dendritic cells after in vitro infection, supporting a role of the costimulatory molecule in dengue infection [22]. In addition, we have observed a differential regulation of the expression of the OX40 signaling pathway in asymptomatic dengue cases compared with clinical cases [23].
The role of CD40L expression during dengue infection is less clear; on the one hand, CD40L has been described as an enhancer of viral particle production by infected dendritic cells by providing survival signals [24], but, on the other hand, CD40L is upregulated in dengue-specific CD4+ T cells and is important for protection against the virus through an antibody-independent pathway [25].
The second gene pair of our signature, ARG1 and JUNB, controls inflammation. Both genes are expressed in neutrophils and are known to regulate the production of reactive nitrogen species. ARG1 degrades the substrate of inducible nitric oxide synthase (iNOS) [26]. JUNB transcriptionally regulates the expression of iNOS [27]. Hence, these genes together control the inflammatory status of the main blood component. Moreover, it has been found that JUNB is a key transcriptional modulator of macrophage expression. It activates the expression of ARG1 in the presence of interleukin 4 [28]. The role of ARG1 in flavivirus infection has been extensively described; in the case of dengue, the production of reactive nitrogen species (RNS) is required to inhibit viral replication during the early phases of infection. However, an overproduction of RNS in the late phases of the disease leads to the inhibition of coagulation, leading to dengue-typical bleeding. ARG1 is therefore required to reduce the amount of RNS and bleeding during dengue infection [29].
Although the RNA signature presented here may help diagnose severity across different cohorts, different technological platforms, and blood sample types, the practical application assumes a correct normalization between severe and nonsevere subgroups and a possibly adjusted decision threshold, both of which can be established using a new set of patients with known severe/nonsevere status. Once this has been established, the application of our RNA signature to new patients requires only a quantitative measurement of the expression level of 18 specific genes from a blood sample. Using multiplex qRT-PCR technology, this can be done in a few hours for the 18 transcripts, and future studies may focus on technologies with even faster processing times [30]. Such a protocol could be used in the clinic, sequentially or in parallel with a diagnostic test, allowing efficient monitoring of individuals with high risk for severe disease. Furthermore, because the concentrations of most proteins may be linearly related to RNA concentrations [31], a protein-level implementation is conceivable. The signature could be especially useful in nonendemic regions where physicians often do not have extensive experience in dengue diagnosis and management. In future studies, its potential as a prognostic signature for an even earlier detection of the risk of evolution toward severe dengue may also be evaluated on samples at an earlier stage of the disease.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgement. We thank all the patients who participated in the study. We acknowledge all of the Virology and Epidemiology Units’ staff at Institut Pasteur in Cambodia for their contribution. We thank the doctors and nurses of the 3 hospitals in Kampong Cham province for patient enrollment and sample collection and H. Rekol and the team from the National Dengue Control Program. We gratefully acknowledge 3 anonymous reviewers.
V. D., P. Bu., and A. S. designed the studies. V. D., S. L., P. Bu., and P. D. collected the samples and clinical data. E. S. L., I. C., M. P., and F. K. did the transcriptome quantification experiments. I. N., B. S., and K. B. designed the methods. I, N., U. C., and B. S. analyzed the data. P. Bo. and T. C. interpreted the data. I. N., P. B., I. C., V. D., P. D., E. S. L., A. S., and B. S. wrote the manuscript. All authors read the manuscript and approved its submission.
Disclosure. P. Bu. is currently an employee of GlaxoSmithKline Vaccines.
Financial support. Data generation was funded by the European Union Seventh Framework Programme (FP7/2007/2011) under grant agreement 282 378 (DENFREE) and Biomérieux. I. N. was supported by Labex IBEID, the Frontières Du Vivant doctoral school, and Institut OpenHealth.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Dengue and severe dengue Available at: http://www.who.int/mediacentre/factsheets/fs117/en/
- 2. Halstead SB. Dengue antibody-dependent enhancement: knowns and unknowns. Microbiol Spectr 2014; 2: 1–18. doi: 10.1128/microbiolspec.AID-0022-2014. [DOI] [PubMed] [Google Scholar]
- 3. Stettler K, Beltramello M, Espinosa DA et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 2016; 353:823–6. [DOI] [PubMed] [Google Scholar]
- 4. Tuan NM, Nhan HT, Chau NVV, Hung NT, Manh H. An evidence-based algorithm for early prognosis of severe dengue in the outpatient setting. Clin Infect Dis 2017; 64:656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee IK, Liu JW, Chen YH et al. Development of a simple clinical risk score for early prediction of severe dengue in adult patients. PLoS One 2016; 11:e0154772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. John DV, Lin YS, Perng GC. Biomarkers of severe dengue disease—a review. J Biomed Sci 2015; 22:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soundravally R, Agieshkumar B, Daisy M, Sherin J, Cleetus CC. Ferritin levels predict severe dengue. Infection 2015; 43:13–9. [DOI] [PubMed] [Google Scholar]
- 8. Thanachartwet V, Oer-Areemitr N, Chamnanchanunt S et al. Identification of clinical factors associated with severe dengue among Thai adults: a prospective study. BMC Infect Dis 2015; 15:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pang J, Lindblom A, Tolfvenstam T et al. Discovery and validation of prognostic biomarker models to guide triage among adult dengue patients at early infection. PLoS One 2016; 11:e0155993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwissa M, Nakaya HI, Onlamoon N et al. Dengue virus infection induces expansion that stimulates plasmablast differentiation. Cell Host Microbe 2014; 16:115–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Devignot S, Sapet C, Duong V et al. Genome-wide expression profiling deciphers host responses altered during dengue shock syndrome and reveals the role of innate immunity in severe dengue. PLoS One 2010; 5:e11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Popper SJ, Gordon A, Liu M, Balmaseda A, Harris E, Relman DA. Temporal dynamics of the transcriptional response to dengue virus infection in Nicaraguan children. PLoS Negl Trop Dis 2012; 6:e1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoang LT, Lynn DJ, Henn M et al. The early whole-blood transcriptional signature of dengue virus and features associated with progression to dengue shock syndrome in Vietnamese children and young adults. J Virol 2010; 84:12982–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simmons CP, Popper S, Dolocek C et al. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis 2007; 195:1097–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duong V, Lambrechts L, Paul RE et al. Asymptomatic humans transmit dengue virus to mosquitoes. Proc Natl Acad Sci U S A 2015; 112:14688–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. World Health Organization. Dengue: guidelines for diagnosis, treatment, prevention and control. 2009. World Health Organization, ISBN-13: 978-92-4-154787-1. Available at: https://www.ncbi.nlm.nih.gov/books/NBK143157/ [PubMed] [Google Scholar]
- 17. Stout QF. Isotonic regression via partitioning. Algorithmica 2013; 66:93–112. [Google Scholar]
- 18. World Health Organization. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control. 2nd ed Vol 40 Geneva, Switzerland: World Health Organization;1997:103–17. [Google Scholar]
- 19. Pedregosa F, Varoquaux G, Gramfort A et al. Scikit-learn: machine learning in python. J Mach Learn Res 2012; 12:2825–30. [Google Scholar]
- 20. Nhi DM, Huy NT, Ohyama K et al. A proteomic approach identifies candidate early biomarkers to predict severe dengue in children. PLoS Negl Trop Dis. 2016; 10:e0004435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev 2009; 229:152–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gandini M, Reis SR, Torrentes-Carvalho A et al. Dengue-2 and yellow fever 17DD viruses infect human dendritic cells, resulting in an induction of activation markers, cytokines and chemokines and secretion of different TNF-α and IFN-α profiles. Mem Inst Oswaldo Cruz 2011; 106:594–605. [DOI] [PubMed] [Google Scholar]
- 23. Simon-Lorière E, Duong V, Tawfik A et al. Increased adaptive immune responses and proper feedback regulation protect against clinical dengue. Sci Transl Med 2017; 9:eaal5088. [DOI] [PubMed] [Google Scholar]
- 24. Sun P, Celluzzi CM, Marovich M et al. CD40 ligand enhances dengue viral infection of dendritic cells: a possible mechanism for T cell-mediated. J Immunol 2016; 177:6497–503. [DOI] [PubMed] [Google Scholar]
- 25. Yauch LE, Prestwood TR, May MM et al. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J Immunol 2010; 185:5405–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Munder M, Mollinedo F, Calafat J et al. Arginase I is constitutively expressed in human granulocytes and participates in fungicidal activity. Immunobiology 2005; 105:2549–56. [DOI] [PubMed] [Google Scholar]
- 27. Ratajczak-Wrona W, Jablonska E, Garley M, Jablonski J, Radziwon P, Iwaniuk A. Role of AP-1 family proteins in regulation of inducible nitric oxide synthase (iNOS) in human neutrophils. J Immunotoxicol 2013; 10:32–9. [DOI] [PubMed] [Google Scholar]
- 28. Fontana MF, Baccarella A, Pancholi N, Pufall MA, Herbert DR, Kim CC. JUNB is a key transcriptional modulator of macrophage activation. J Immunol 2015; 194:177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burrack KS, Morrison TE. The role of myeloid cell activation and arginine metabolism in the pathogenesis of virus-induced diseases. Front Immunol 2014; 5:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Islam N, Masud MK, Haque H et al. RNA biomarkers: diagnostic and prognostic potentials and recent developments of electrochemical biosensors. Small Methods 2017; 1:1700131. [Google Scholar]
- 31. Edfors F, Danielsson F, Hallström BM et al. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol Syst Biol 2016; 12:883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.