Abstract
The combination of electronic health records (EHRs) with genetic data has ushered in the next wave of complex disease genetics. Population-based biobanks and other large cohorts provide sufficient sample sizes to identify novel genetic associations across the hundreds to thousands of phenotypes gleaned from EHRs. In this review, we summarize the current state of these EHR-linked biobanks, explore ongoing methods development in the field and highlight recent discoveries of genetic associations. We enumerate the many existing biobanks with EHRs linked to genetic data, many of which are available to researchers via application and contain sample sizes >50 000. We also discuss the computational and statistical considerations for analysis of such large datasets including mixed models, phenotype curation and cloud computing. Finally, we demonstrate how genome-wide association studies and phenome-wide association studies have identified novel genetic findings for complex diseases, specifically cardiometabolic traits. As more researchers employ innovative hypotheses and analysis approaches to study EHR-linked biobanks, we anticipate a richer understanding of the genetic etiology of complex diseases.
Introduction
The increased adoption of electronic health records (EHRs) in clinical settings has created a rich resource for the genetics research community (1). Variation in the human phenome, the set of physical characteristics and diseases (phenotypes) expressed in humans, is measurable using billing codes, narrative notes, death certificates and laboratory values from EHRs. As the cost of high throughput genotyping and sequencing continues to fall, EHRs coupled with genetic data from biobank samples are now available for hundreds of thousands of people.
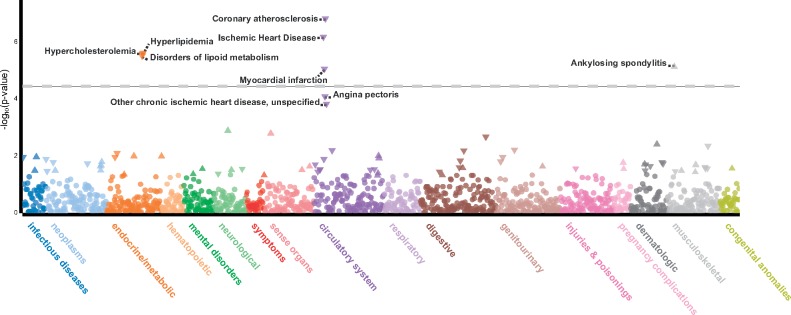
Historically, large cohorts of cases and controls were amassed to study only one specific phenotype of interest, or a few closely related phenotypes [e.g. coronary artery disease (CAD) and blood lipid levels] in a genome-wide association study (GWAS; few phenotypes analyzed at many variants). Variants significantly associated with one phenotype were then tested for association with additional phenotypes in a phenome-wide association study (PheWAS) to more fully understand cross-phenotype associations. The first PheWAS (2), analysis of many phenotypes for a few variants, was published in 2010 and researchers are continuing to increase the number of phenotypes examined. Today, EHR-linked DNA biobanks with large sample sizes enable GWAS on millions of variants to be performed for thousands of phenotypes resulting in a phenome-wide GWAS which we refer to as PheGWAS (many phenotypes analyzed at many variants). An example PheGWAS is available at University of Michigan’s PheWeb which hosts genetic association results for 28 million variants across 1403 ICD-based traits (http://pheweb.sph.umich.edu:5003) identified in 400 000 individuals (3) (Fig. 1).
Figure 1.
Locus zoom plot of the lead variant (rs116843064) in ANGPTL4 from the PheGWAS available at University of Michigan’s PheWeb. The variant is associated with coronary atherosclerosis (P-value <1.6e−7) in 20 023 cases and 377 103 controls in UKBB. The variant is also associated with other phenotypes at phenome-wide significance (P-value < 5e−5) including hypercholesterolemia and ischemic heart disease as expected. Notably, this variant is also associated with ankylosing spondylitis—a form of arthritis affecting the spine and large joints. While ankylosing spondylitis is seemingly pathologically different than CAD, a link between the two has been reported previously (51). The constellation of associations across circulatory, metabolic and musculoskeletal systems provides evidence for pleiotropy or shared pathways for disease pathogenesis.
In this review, we focus primarily on recent research involving GWAS, PheWAS and PheGWAS in cohorts combining EHRs and genetic data. Many types of additional omics data may exist in cohorts employing EHRs (e.g. transcriptomics, metabolomics, epigenomics) although the results from such studies are outside the scope of this review. Here, we summarize the current state of EHR-linked biobanks, explore ongoing methods development and considerations in the field and highlight recent discoveries of genetic associations in EHR-linked biobanks.
Established EHR-Linked Biobanks
The earliest population-wide biobank is Iceland’s deCODE genetics which started in 1996 as a private company with government support (4) and is currently owned by Amgen. One of the first institution-wide biobanks is Vanderbilt University’s bioVU which utilized de-identified leftover blood samples from clinical blood draws (5). Biobanks typically feature opt-in consent and protections for personal health information (PHI) allowing prospective phenotype updates. Since 2007 the National Institutes of Health (NIH) has funded the Electronic Medical Records and Genomics (eMERGE) Network which links biobanks to EHRs at multiple sites to perform genomic research and establish best practices (6). Additional academic centers host large studies combining EHR-linked biobanks such the University of Michigan’s Michigan Genomics Initiative, the Mount Sinai BioMe biobank, and the Estonian Genome Center at the University of Tartu’s biobank, among others. In recent years, private companies in the United States’ health care and insurance industries [e.g. Kaiser Permanente (7)] have begun their own studies building on EHRs of customers that consent to research. Because drug mechanisms with genetic evidence in humans are twice as likely to successfully move from phase 1 trials to approval (8), the pharmaceutical industry is also increasingly investing in EHR-linked biobanks. This is evidenced by the DiscovEHR cohort, a collaboration between Regeneron Genetics Center and Geisinger Health System and the largest existing collection of EHRs linked to sequencing data. In November 2017, Geisinger announced its National Precision Health Initiative which is an expansion of the MyCode Community Health Initiative which has consented the 50 726 patients in DiscovEHR. In the summer of 2017, the United Kingdom Biobank (UKBB) released genotype and phenotype data for 488 377 individuals which is an unprecedented amount of genetic data freely available to researchers via an application process (9). In January 2018, it was announced that exome sequencing for all UKBB participants will be completed by 2019 funded by Regeneron Pharmaceuticals and several life science companies (10).
FinnGen was announced in December 2017 with the goal of linking GWAS data to clinical data for 500 000 participants consented for recall appointments to perform more detailed clinical examination of individuals with genetic variants of uncertain significance (11). Notably, countries like Finland with nationalized health systems are uniquely poised to study genetics at a population scale using nationally connected EHRs linked to biobanks. These studies are additionally benefitted by nationalized pharmaceutical and cause of death registries that provide useful information for phenotype curation. Moving toward ever larger sample sizes, the Million Veteran Program (MVP) aims to partner with one million U.S. armed services veterans receiving care through the Veterans Affairs Healthcare system (12,13), and NIH’s All of Us cohort (part of the Precision Medicine Initiative) will open to nationwide enrollment of one million participants in early 2018. The large sample sizes (Table 1) that are available in these studies aid in the discovery of genetic associations for both rare mutations causing Mendelian disease and common complex diseases with causal variants of smaller effect.
Table 1.
Selected biobanks with linked EHRs and genetic data in ≥50 000 participants listed in descending order of sample size with available genetic data
| Cohort | Country | Institution or companya | Cohort sizeb,c | Samples with matched EHR and genetic data availableb,d | Access | References |
|---|---|---|---|---|---|---|
| UK BioBank (UKBB) http://www.ukbiobank.ac.uk | United Kingdom | UK Biobank charity | 500 000 | 488 377 genotyped | Application for bona fide researcher | (9,52) |
| DeCODE Genetics https://www.decode.com | Iceland | Amgen | >350 000 | >350 000 | Contact to collaborate | (4,53,54) |
| Million Veteran Program (MVP) https://www.research.va.gov/mvp/ | USA | Department of Veterans Affairs | >500 000 | >350 000 | Contact to collaborate | (12,13) |
| BioBank Japan Project http://www.pgrn.org/biobank-japan.html | Japan | Pharmacogenomics Research Network | 200 000 | 162 255 genotyped | Contact to collaborate | (34,55) |
| China Kadoorie Biobank http://www.ckbiobank.org/site/ | China | University of Oxford, Chinese Academy of Medical Sciences | 510 000 | >130 000 | Application for bona fide researcher | (56,57) |
| Kaiser Permanente Research Bank https://researchbank.kaiserpermanente.org/ our-research/for-researchers/ | USA | Kaiser Permanente | 270 570 | 102 998 genotyped | Application for bona fide researcher | (7,58) |
| eMerge Network https://emerge.mc.vanderbilt.edu | USA | NHGRI | 105 325 | 83 717 | Application for eMERGE affiliate membership | (6,18) |
| Danish Biobank Register http://www.biobankdenmark.dk | Denmark | Danish National Biobank | 5.7 million | >70 000 | Application for bona fide researcher | (59) |
| Nord Trondelag Health Study (HUNT) https://www.ntnu.edu/hunt | Norway | Norwegian University of Science and Technology | 120 000 | 69 037 genotyped | Application and collaboration with PI affiliated with a Norwegian research institute | (15,60) |
| DiscovEHR http://www.discovehrshare.com | USA | Geisenger Health System, Regeneron Genetics Center | 50 000 | >50 000 exome sequences | Contact to collaborate | (43,61) |
Main institution responsible for the resource, many other institutions may provide funding or support.
Sample size as of January 2018. In situations where up to date sample sizes were difficult to find, sample sizes from recent publications were used.
Unique number of participants with some type of data available (52–61).
Actual samples available for analysis may be less due to quality control. Number includes both sequencing and genotyping with the type of data described when possible.
Methods Developments
Avoiding data-driven bias
Large, longitudinal, population-based studies with EHR-linked biobanks present many challenges in areas of data curation and analysis, most of which are areas of current methods development. In longitudinal studies, epidemiological survey questionnaires are often revised and updated between biobank enrollments which introduces missing data and highlights the importance of thoughtful and consistent study design when possible. Because of differing enrollment strategies some biobanks contain more complete EHRs than others. For example, Geisinger Health System provides comprehensive care in a rural area resulting in ‘cradle to grave’ records while academic biobanks may see patients only for specialized care resulting in fragmented EHRs but higher rates of more serious cases. Longitudinal studies with multiple enrollment periods can be prone to batch effects as technology or protocol changes introduce confounders. Ascertainment bias remains a concern even in population-based cohorts, with studies like UKBB being, on average, younger and healthier and with more female participants than the British population at large. These possible confounding factors should be accounted for in analyses, for example with birth year and sex as covariates in a linear regression model.
As study sample sizes continue to increase so does the number of family members contained in a given population-based cohort, and this phenomenon has inspired current method development efforts. One approach is to analyze only an unrelated subset of samples from a population-based cohort (14). However, removing related individuals from the analysis may decrease sample size, and therefore statistical power, particularly in highly related populations such as the Nord Trøndelag Health Study (HUNT) (15) in which 81% of the cohort has at least a third degree relative that is also in the study. Even in the metropolitan UKBB, 81 000 samples are removed when analyzing the unrelated subset (9). Both relatedness and population substructure may be addressed using single variant association testing with linear mixed models (16). While it is important to perform GWAS in populations of diverse ancestries, population-based biobanks with a mix of ancestries are vulnerable to false positive findings from population stratification between cases and controls. Currently, most GWAS of binary traits in UKBB are performed using only the subset of samples with self-reported and principal component confirmed white British ancestry.
When very few cases for a given phenotype exist in a cohort, an unbalanced case–control ratio may inflate type I error in GWAS results (17). A novel method, SAIGE (3), allows for analysis of binary traits with unbalanced case–control ratio in large sample sizes while accounting for sample relatedness. It is important to note that removing related individuals from a cohort while preferentially retaining cases may ameliorate extreme case–control imbalance for some phenotypes.
Phenotype curation
Phenotype curation from EHRs is an ongoing area of research with the eMERGE Network largely spearheading the effort (18). International Classification of Diseases (ICD) codes are a main feature of EHRs and are typically used in national hospital registries and as health insurance billing codes in medical practice. ICD codes may not always indicate a true diagnosis of a disease (e.g. an ICD code may be listed as a hypothetical reason for a laboratory test) (2). Broad or ambiguous ICD codes may lead to a heterogeneous definition of cases. Therefore, false positives or false negatives may arise when only ICD codes are used in phenotype definitions. Recent work compared groupings of EHR ICD-based billing codes to demonstrate the superiority of manually curated phecodes (19,20) for defining phenotypes from EHRs. Researchers should also consider which subset of a cohort to use as healthy controls. For example, patients with Type 1 diabetes would be inappropriate controls for a study of Type 2 diabetes. The phenotype definitions of cases and healthy controls are critical for accurate genetic studies, and the optimal approach may depend on the specific cohort and data at hand.
Challenges with big data
The sample size of many cohorts poses computational challenges including (i) data transfer, (ii) time and memory resources required for analysis and (iii) storage space necessary for the terabytes of raw phenotype and genotype data and the resulting association results. Therefore, many of the large biobanks and groups analyzing biobank-based data have started to use remote or cloud environments for data storage and analysis (21). NHLBI’s Trans-Omics for Precision medicine (TOPMed) hosts a TOPMed Cloud Analysis Pilot called Encore (https://encore.sph.umich.edu) which provides a simple web-based interface to allow investigators to run large-scale association analysis without requiring specific technical computing skills. Encore handles splitting up jobs and distributing requests to available computing resources, and provides interactive plots and summaries for exploration of association results.
Another challenge regarding the analysis of large number of samples from a biobank is the sample relatedness which can increase type I error of the analysis. As described above, this can be overcome using linear mixed models, which are usually computationally intensive. Even when using a cloud environment for the computation, BOLT-LMM (16) and SAIGE (3) are the only existing mixed model association methods computationally feasible for analysis of large sample sizes (N > 20 000).
As most of the currently available biobank data is genotyped using existing genotyping chips or custom chips to capture whole genome variation, imputation of the genotype data is suggested to increase the number of markers available for association testing. Not only is imputation one of the most computationally intensive components of a GWAS analysis pipeline, but the choice of imputation panel greatly affects the quality and the number of variants that are well-imputed (22–24). In the usual case where there is no population-specific imputation panel available for the dataset, imputation of variants available from emerging resources such as TOPMed (25) or the Haplotype Reference Consortium (26) may be worthwhile. The Michigan Imputation Server (27) (https://imputationserver.sph.umich.edu) and Sanger Imputation Service (26) (https://imputation.sanger.ac.uk) provide remote computational resources for free genotype imputation with up to date reference panels.
Historically GWAS studies have considered a P-value of 5e−8 as the genome-wide significance threshold for European-descent GWAS which adjusts for 1–2 million independent tests (28–30). As the number of variants assayed increases due to imputation with larger reference panels, it is an active area of discussion whether a more stringent threshold should now be considered. Recent work in UKBB a data demonstrated the validity of CAD GWAS signals meeting a less stringent threshold for genome-wide significance at a false discovery rate of 5% (31). As datasets continue to increase in size, more research is needed to establish best practices of cloud-based computing and appropriate statistical rigor in analyses.
Novel approaches for data analysis
Population-based EHR-linked biobanks usually allow for definition of hundreds to thousands of different phenotypes and outcomes which facilitates the usage of new analysis methods, such as large-scale heritability analyses (32). Another type of analysis that is highly efficient in datasets with EHRs is the analysis of genetic correlations (33) which can be used to find variants with possible pleiotropic effects. Recent work in the Biobank Japan Project identified 313 pleiotropic loci across 53 quantitative traits (34). Both of these methods can be used to prioritize phenotypes for more concentrated genetic studies.
EHR-linked biobanks can also be used to identify and prioritize possible drug targets. Because of the large number of samples in population-based datasets, the chance to find individuals with homozygous loss-of-function (LOF) mutations for specific genes is much higher which makes the search for human knock-outs feasible. This, combined with the availability of wide variety of phenotypes, allows for studies of possible side-effects of gene inhibition. As an example, homozygous carriers of PCSK9 LOF mutations were analyzed against a wide variety of outcomes to find possible negative lifetime effects of low PCKS9 levels, similar to that of PCKS9 gene inhibition effect. The study showed that homozygous carriers of PCSK9 LOF mutations had lower levels of low-density lipoprotein cholesterol levels and increased risk for Type 2 diabetes (35), spina bifida, osteoporosis and fractures, suggesting that the long term usage of PCSK9 inhibitors may have negative implications (36).
EHRs in combination with other registry-based data (e.g. pharmaceutical, death registry or cancer registry data) and epidemiological surveys allow for creation of novel phenotypes that can be used in GWAS and PheWAS. Finnish researchers demonstrated a YODA Score, representing Years of Drugs Applied, can be calculated from national registries of prescription drug purchase history. The presented YODA score combines purchase information for selected drugs studied in FINRISK and was found to associate with polygenic risk score for CAD (37). The association is mainly driven by the CAD related drugs and demonstrates proof of concept. Both YODA and another registry-based measure, cumulative months of hospitalization periods, could potentially be used to predict mortality.
For certain traits of interest which are rare or late-onset there may be few cases available for study even in large cohorts. To analyze these traits, epidemiological survey data can be utilized to identify unaffected first degree relatives of affected individuals (e.g. proxy-cases) to perform genome-wide association by proxy (38,39). A GWAS on family history of Alzheimer’s disease (AD) in 300 000 individuals from the UKBB allowed the study of 32 222 cases of maternal AD and 16 613 cases of paternal AD that when meta-analyzed with an existing cohort identified 6 novel loci (40). EHRs also provide information such as age of onset which allows for a more granular study of cases. For example, a recent GWAS stratified by age of onset showed genetic susceptibility to major depressive disorder (MDD) is different between early and adult onset MDD (41). In summary, data-mining of EHR-linked biobanks provides the opportunity for novel analysis approaches that build upon discoveries from GWAS and PheWAS analyses.
Selected Findings
GWAS and PheWAS in large biobanks have yielded novel genetic findings for a wide variety of cardio-metabolic traits and increased our understanding of the clinical and translational value of these genetic discoveries. Recently, about 50 000 individuals with whole exome sequence data available from DiscovEHR cohort were screened for variants that cause familial hypercholesterolemia (FH). The study group found that 1 in 256 people carry an FH variant and only 24% of the carriers had an FH diagnosis and 42% of carriers were not currently on statins (42). This study demonstrated by large-scale sequencing that many FH individuals are not identified through standard clinical practice, and a large number of individuals would benefit from additional screening and treatment with statins to reduce the risk of heart disease. The same exome sequence dataset from DiscovEHR, together with other cohorts, has also been used for study of ANGPTL4 (43) (Fig. 1) and LPL (44) inactivating and protein altering mutations and their connection to lipid metabolisms and risk of CAD. In these studies, an association between ANGPTL4 inactivating mutations and decreased risk of CAD was observed, whereas the association of LPL disruptive mutations with CAD was in the opposite direction. These results highlight ANGPTL4, which also blocks the inhibition of LPL, as a possible drug target for future clinical trials.
The recent release of publicly available UKBB data led to a wave of genetic association studies, and two such studies for cardiometabolic traits have already been performed. The first is an association study of CAD (45) that identified 64 new CAD associated loci by combining the new UKBB dataset with an existing public dataset from CARDIoGRAMplusC4D Consortium. Another example is a recent study of atrial fibrillation (AF) (46), where data from the UKBB was combined with other EHR and GWAS datasets in a meta-analysis that comprised more than one million samples including 60 000 cases. Using this enormous dataset, the authors were able to identify a total of 111 loci associated with AF. With time, some of the AF-associated genes may become new drug targets for arrhythmia disorders.
While analysis of the large biobanks is more concentrated on disease endpoints, quantitative traits are still mainly studied in worldwide consortia combining data from smaller datasets with a meta-analysis approach. In the field of cardiometabolic genetics there are multiple consortia each with a focus on different trait(s). Examples of such are the Genetic Investigation of ANthropometric Traits (GIANT), Global Lipids Genetics Consortium (GLGC), Consortia for echocardiographic trait genetics (EchoGen) and International Consortium for Blood Pressure (ICBP). The latest publication from the ICBP (47) was a meta-analysis combining data from a total of 380 000 samples which found 6 novel loci associated with blood pressure traits. From EchoGen, the latest meta-analysis (48) combined echocardiographic data from up to 30 000 individuals and found 10 new loci associated to left ventricular structure, and systolic and diastolic function. GLGC and GIANT consortia are currently concentrating on rare, low-frequency and coding variation. GIANT identified 14 coding variants associated with body mass index (BMI) (49) which had on average 10 times higher effect sizes compared to common variants associated with BMI, and GLGC identified 75 new loci associated with blood lipids using an Exome Chip genotyped dataset which also allowed for fine-mapping of 131 previously known loci (50).
Conclusion
EHRs allow a shift from purpose-built cohorts centered around a particular phenotype to large cohorts where the entire phenome can be studied through PheGWAS. Methods development to handle the computational and statistical complexities of such large datasets is ongoing, but new data handling and analysis methods including mixed models and robust EHR-derived phenotype definitions are already being employed. The next wave of genetic analysis in hundreds or thousands of phenotypes, enabled by population-based EHR-linked biobanks, has only just begun. We have already seen the importance of vast phenotypic information in large datasets through recent studies of putative drug targets such as PCSK9 and ANGPTL4. These studies are, however, just the tip of the iceberg. The high information content of EHR datasets allows for innovative new hypotheses and analyses which are poised to become the driving force of complex disease genetics.
Acknowledgements
The authors wish to apologize in advance to authors of outstanding work that is not acknowledged in this review due to space constraints. The authors wish to thank Wei Zhou, Matthew Flickinger, Matthew Zawistowski, and Paavo Häppölä for insights provided.
Conflict of Interest statement. None declared.
Funding
National Institutes of Health (R01-HL127564 and R35-HL135824 to C.J.W. and I.S.); National Science Foundation Graduate Research Fellowship Program (DGE 1256260 to B.N.W.).
References
- 1. Kohane I.S. (2011) Using electronic health records to drive discovery in disease genomics. Nat. Rev. Genet., 12, 417. [DOI] [PubMed] [Google Scholar]
- 2. Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. (2010) PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics, 26, 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Elvestad M.B., Wolford B.N., LeFaive J., VandeHaar P., Gifford A., Bastarache L.A.. et al. (2017) Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. bioRxiv, 212357 https://doi.org/10.1101/212357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gulcher J., Stefansson K. (1999) An Icelandic saga on a centralized healthcare database and democratic decision making. Nat. Biotechnol., 17, 620.. [DOI] [PubMed] [Google Scholar]
- 5. Pulley J., Clayton E., Bernard G.R., Roden D.M., Masys D.R. (2010) Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin. Transl. Sci., 3, 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCarty C.A., Chisholm R.L., Chute C.G., Kullo I.J., Jarvik G.P., Larson E.B., Li R., Masys D.R., Ritchie M.D., Roden D.M.. et al. (2011) The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genom., 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kvale M.N., Hesselson S., Hoffmann T.J., Cao Y., Chan D., Connell S., Croen L.A., Dispensa B.P., Eshragh J., Finn A.. et al. (2015) Genotyping informatics and quality control for 100, 000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics, 200, 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Floratos A., Tipney H., Painter J.L., Whittaker J.C., Shen J., Wang J., Cardon L.R., Nelson M.R., Li M.J., Sham P.C.. et al. (2015) The support of human genetic evidence for approved drug indications. Nat. Genet., 47, 856. [DOI] [PubMed] [Google Scholar]
- 9. Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O'Connell J.. et al. (2017) Genome-wide genetic data on ∼500 000 UK Biobank participants. bioRxiv, 166298 https://doi.org/10.1101/166298. [Google Scholar]
- 10. UK Biobank. (2018). Regeneron announces major collaboration to exome sequence UK Biobank genetic data more quickly. Retrieved from http://www.ukbiobank.ac.uk/2018/01/regeneron-announces-major-collaboration-to-exome-sequence-uk-biobank-genetic-data-more-quickly/; date last accessed March 14, 2018.
- 11. University of Hesinki. (2017). FinnGen, a global research project focusing on genome data of 500 000 Finns, launched. Retrieved from https://www.eurekalert.org/pub_releases/2017-12/uoh-fag121917.php; date last accessed March 14, 2018.
- 12. Klarin S.D., Cho S., Duvall G., Peloso K.M., Chang J., Huang J., Lynch Y.L., Ho D., Liu D., Saleheen S., et al. (2017) Genetic analysis of lipids in >300 000 participants in the Million Veteran Program; #1869. Presented at the 67th Annual meeting of American Society of Human Genetics. October 20, 2017, Orlando, FL.
- 13. Gaziano J.M., Concato J., Brophy M., Fiore L., Pyarajan S., Breeling J., Whitbourne S., Deen J., Shannon C., Humphries D.. et al. (2016) Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol., 70, 214–223. [DOI] [PubMed] [Google Scholar]
- 14. Abraham K.J., Diaz C. (2014) Identifying large sets of unrelated individuals and unrelated markers. Source Code Biol. Med., 9, 6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krokstad S., Langhammer A., Hveem K., Holmen T.L., Midthjell K., Stene T.R., Bratberg G., Heggland J., Holmen J. (2013) Cohort profile: the HUNT Study, Norway. Int. J. Epidemiol., 42, 968–977. [DOI] [PubMed] [Google Scholar]
- 16. Loh P.-R., Tucker G., Bulik-Sullivan B.K., Vilhjálmsson B.J., Finucane H.K., Salem R.M., Chasman D.I., Ridker P.M., Neale B.M., Berger B.. et al. (2015) Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet., 47, 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dey R., Schmidt E.M., Abecasis G.R., Lee S. (2017) A fast and accurate algorithm to test for binary phenotypes and its application to PheWAS. Am. J. Hum. Genet., 101, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gottesman O., Kuivaniemi H., Tromp G., Faucett W.A., Li R., Manolio T.A., Sanderson S.C., Kannry J., Zinberg R., Basford M.A.. et al. (2013) The electronic medical records and genomics (eMERGE) network: past, present, and future. Genet. Med., 15, 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei W.-Q., Bastarache L.A., Carroll R.J., Marlo J.E., Osterman T.J., Gamazon E.R., Cox N.J., Roden D.M., Denny J.C. (2017) Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS ONE, 12, e0175508.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Denny J.C., Bastarache L., Ritchie M.D., Carroll R.J., Zink R., Mosley J.D., Field J.R., Pulley J.M., Ramirez A.H., Bowton E.. et al. (2013) Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol., 31, 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dinov I.D. (2016) Methodological challenges and analytic opportunities for modeling and interpreting Big Healthcare Data. Gigascience, 5, 12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou W., Fritsche L.G., Das S., Zhang H., Nielsen J.B., Holmen O.L., Chen J., Lin M., Elvestad M.B., Hveem K.. et al. (2017) Improving power of association tests using multiple sets of imputed genotypes from distributed reference panels. Genet. Epidemiol., 41, 744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang J., Howie B., McCarthy S., Memari Y., Walter K., Min J.L., Danecek P., Malerba G., Trabetti E., Zheng H.-F.. et al. (2015) Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat. Commun., 6, 8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gudbjartsson D.F., Helgason H., Gudjonsson S.A., Zink F., Oddson A., Gylfason A., Besenbacher S., Magnusson G., Halldorsson B.V., Hjartarson E.. et al. (2015) Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet., 47, 435–444. [DOI] [PubMed] [Google Scholar]
- 25. NHLBI (2018). Whole Genome Sequencing in the NHLBI Trans-Omics for Precision Medicine. Retrieved from https://www.nhlbiwgs.org; date last accessed March 14, 2018.
- 26., McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P.,et al. (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet., 48, 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M.. et al. (2016) Next-generation genotype imputation service and methods. Nat. Genet., 48, 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. International HapMap Consortium (2005) A haplotype map of the human genome. Nature, 437, 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Risch N., Merikangas K. (1996) The future of genetic studies of complex human diseases. Science, 273, 1516–1517. [DOI] [PubMed] [Google Scholar]
- 30. Hoggart C.J., Clark T.G., De Iorio M., Whittaker J.C., Balding D.J. (2008) Genome-wide significance for dense SNP and resequencing data. Genet. Epidemiol., 32, 179–185. [DOI] [PubMed] [Google Scholar]
- 31. Nelson C.P., Goel A., Butterworth A.S., Kanoni S., Webb T.R., Marouli E., Zeng L., Ntalla I., Lai F.Y., Hopewell J.C.. et al. (2017) Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet., 49, 1385–1391. [DOI] [PubMed] [Google Scholar]
- 32. Ge T., Chen C.-Y., Neale B.M., Sabuncu M.R., Smoller J.W. (2017) Phenome-wide heritability analysis of the UK Biobank. PLoS Genet., 13, e1006711.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.-R., Duncan L., Perry J.R.B., Patterson N., Robinson E.B.. et al. (2015) An atlas of genetic correlations across human diseases and traits. Nat. Genet., 47, 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanai M., Akiyama M., Takahashi A., Matoba N., Momozawa Y., Ikeda M., Iwata N., Ikegawa S., Hirata M., Matsuda K.. et al. (2018) Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. doi:10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 35. Schmidt A.F., Swerdlow D.I., Holmes M.V., Patel R.S., Fairhurst-Hunter Z., Lyall D.M., Hartwig F.P., Horta B.L., Hyppönen E., Power C.. et al. (2017) PCSK9 genetic variants and risk of type 2 diabetes: a Mendelian randomisation study. Lancet Diabetes Endocrinol., 5, 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jerome R.N., Pulley J.M., Roden D.M., Shirey-Rice J.K., Bastarache L.A., Bernard G.R., Ekstrom L.B., Lancaster W.J., Denny J.C. (2017) Using human ‘experiments of nature’ to predict drug safety issues: an example with PCSK9 inhibitors. Drug Saf., 41, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ripatti S., Havulinna A., Kiiskinen T., Helkkula P., Hautakangas H., Häppölä P., Ruotsalainen S., Koskela J., Kurki M., Surakka I.. et al. (2017) Phenomewide association study of life course health events: Analyzing 50 yaers of hospitalization, prescription drug use and death data; #372. Presented at the 67th Annual meeting of American Society of Human Genetics. October 21, 2017, Orlando, FL.
- 38. Joshi P.K., Fischer K., Schraut K.E., Campbell H., Esko T., Wilson J.F. (2016) Variants near CHRNA3/5 and APOE have age- and sex-related effects on human lifespan. Nat. Commun., 7, 11174.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J.Z., Erlich Y., Pickrell J.K. (2017) Case-control association mapping by proxy using family history of disease. Nat. Genet., 49, 325–331. [DOI] [PubMed] [Google Scholar]
- 40. Marioni R., Harris S.E., McRae A.F., Zhang Q., Hagenaars S.P., Hill W.D., Davies G., Ritchie C.W., Gale C., Starr J.M.. et al. (2018) GWAS on family history of Alzheimer's disease. bioRxiv, 246223 https://doi.org/10.1101/246223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Power R.A., Tansey K.E., Buttenschøn H.N., Cohen-Woods S., Bigdeli T., Hall L.S., Kutalik Z., Lee S.H., Ripke S., Steinberg S.. et al. (2017) Genome-wide association for major depression through age at onset stratification: major depressive disorder working group of the Psychiatric Genomics Consortium. Biol. Psychiatry, 81, 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abul-Husn N.S., Manickam K., Jones L.K., Wright E.A., Hartzel D.N., Gonzaga-Jauregui C., O'Dushlaine C., Leader J.B., Lester Kirchner H., Lindbuchler D.A.M.. et al. (2016) Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science, 354, aaf7000. [DOI] [PubMed] [Google Scholar]
- 43. Dewey F.E., Gusarova V., O'Dushlaine C., Gottesman O., Trejos J., Hunt C., Van Hout C.V., Habegger L., Buckler D., Lai K.-M.V.. et al. (2016) Inactivating variants in ANGPTL4 and risk of coronary artery disease. N. Engl. J. Med., 374, 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Khera A.V., Won H.-H., Peloso G.M., O’Dushlaine C., Liu D., Stitziel N.O., Natarajan P., Nomura A., Emdin C.A., Gupta N.. et al. (2017) Association of rare and common variation in the lipoprotein lipase gene with coronary artery disease. JAMA, 317, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van der Harst P., Verweij N. (2018) The identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ. Res., 122, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nielsen J.B., Thorolfsdottir R.B., Fritsche L.G., Zhou W., Skov M.W., Graham S.E., Herron T.J., McCarthy S., Schmidt E.M., Sveinbjornsson G.. et al. (2018) Genome-wide association study of 1 million people identifies 111 loci for atrial fibrillation. bioRxiv, 242149 https://doi.org/10.1101/242149. [Google Scholar]
- 47. Wain L.V., Vaez A., Jansen R., Joehanes R., van der Most P.J., Erzurumluoglu A.M., O’Reilly P.F., Cabrera C.P., Warren H.R., Rose L.M.. et al. (2017) Novel blood pressure locus and gene discovery using genome-wide association study and expression data sets from blood and the kidney. Hypertension, 70, e4–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wild P.S., Felix J.F., Schillert A., Teumer A., Chen M.-H., Leening M.J.G., Völker U., Großmann V., Brody J.A., Irvin M.R.. et al. (2017) Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J. Clin. Invest., 127, 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Turcot V., Lu Y., Highland H.M., Schurmann C., Justice A.E., Fine R.S., Bradfield J.P., Esko T., Giri A., Graff M.. et al. (2018) Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet., 50, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu D.J., Peloso G.M., Yu H., Butterworth A.S., Wang X., Mahajan A., Saleheen D., Emdin C., Alam D., Alves A.C.. et al. (2017) Exome-wide association study of plasma lipids in >300, 000 individuals. Nat. Genet., 49, 1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ungprasert P., Srivali N., Kittanamongkolchai W. (2015) Risk of coronary artery disease in patients with ankylosing spondylitis: a systematic review and meta-analysis. Ann. Transl. Med., 3, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Littlejohns T.J., Sudlow C., Allen N.E., Collins R. (2017) UK Biobank: opportunities for cardiovascular research. Eur. Heart J. https://doi.org/10.1093/eurheartj/ehx254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gulcher J., Kong A., Stefansson K. (2001) The genealogic approach to human genetics of disease. Cancer J., 7, 61–68. [PubMed] [Google Scholar]
- 54. Kong A., Thorleifsson G., Frigge M.L., Vilhjalmsson B.J., Young A.I., Thorgeirsson T.E., Benonisdottir S., Oddsson A., Halldorsson B.V., Masson G.. et al. (2018) The nature of nurture: effects of parental genotypes. Science, 359, 424–428. [DOI] [PubMed] [Google Scholar]
- 55. Nagai A., Hirata M., Kamatani Y., Muto K., Matsuda K., Kiyohara Y., Ninomiya T., Tamakoshi A., Yamagata Z., Mushiroda T.. et al. (2017) Overview of the BioBank Japan Project: study design and profile. J. Epidemiol., 27, S9–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen Z., Chen J., Collins R., Guo Y., Peto R., Wu F., Li L. (2011) China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int. J. Epidemiol., 40, 1652–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Millwood I.Y., Bennett D.A., Walters R.G., Clarke R., Waterworth D., Johnson T., Chen Y., Yang L., Guo Y., Bian Z.. et al. (2016) A phenome-wide association study of a lipoprotein-associated phospholipase A2 loss-of-function variant in 90 000 Chinese adults. Int. J. Epidemiol., 45, 1588–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jorgenson E., Thai K.K., Hoffmann T.J., Sakoda L.C., Kvale M.N., Banda Y., Schaefer C., Risch N., Mertens J., Weisner C.. et al. (2017) Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol. Psychiatry, 22, 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Agerbo E., Sullivan P.F., Vilhjálmsson B.J., Pedersen C.B., Mors O., Børglum A.D., Hougaard D.M., Hollegaard M.V., Meier S., Mattheisen M.. et al. (2015) Polygenic risk score, parental socioeconomic status, family history of psychiatric disorders, and the risk for schizophrenia: a Danish population-based study and meta-analysis. JAMA Psychiatry, 72, 635–641. [DOI] [PubMed] [Google Scholar]
- 60. Nielsen J.B., Fritsche L.G., Zhou W., Teslovich T.M., Holmen O.L., Gustafsson S., Gabrielsen M.E., Schmidt E.M., Beaumont R., Wolford B.N.. et al. (2018) Genome-wide study of atrial fibrillation identifies seven risk loci and highlights biological pathways and regulatory elements involved in cardiac development. Am. J. Hum. Genet., 102, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dewey F.E., Murray M.F., Overton J.D., Habegger L., Leader J.B., Fetterolf S.N., O’Dushlaine C., Hout C.V.V., Staples J., Gonzaga-Jauregui C.. et al. (2016) Distribution and clinical impact of functional variants in 50, 726 whole-exome sequences from the DiscovEHR study. Science, 354, aaf6814. [DOI] [PubMed] [Google Scholar]