Abstract
The field of pharmacogenomics is an area of great potential for near-term human health impacts from the big genomic data revolution. Pharmacogenomics research momentum is building with numerous hypotheses currently being investigated through the integration of molecular profiles of different cell lines and large genomic data sets containing information on cellular and human responses to therapies. Additionally, the results of previous pharmacogenetic research efforts have been formulated into clinical guidelines that are beginning to impact how healthcare is conducted on the level of the individual patient. This trend will only continue with the recent release of new datasets containing linked genotype and electronic medical record data. This review discusses key resources available for pharmacogenomics and pharmacogenetics research and highlights recent work within the field.
Introduction
The field of pharmacogenomics (PGx1) seeks to elucidate the interactions between genetic variation and the metabolism/mechanism of pharmaceutical compounds. In the United States, greater than 70% of visits to clinics and hospitals involving drug therapy (1), leading to approximately 59% of adults having at least one drug prescription and 39% of adults aged 65 or older using five or more prescription drugs (2). The prevalence of prescription drugs highlights the need for a better understanding of drug response, especially with regards to drug–gene, drug–drug, and drug–drug–gene interactions.
The goal of PGx research is to further the understanding of these interactions. Within the PGx field, the pathways of drug response are divided into two categories: pharmacokinetics (PK) and pharmacodynamics (PD). PK focuses primarily on the absorption, distribution, metabolism and excretion (ADME) of drugs, the movement and lifecycle of a drug within the body. The actions and interactions (i.e. binding, activation, deactivation, upregulation, downregulation) of a drug with biological molecules in order to alter the physiology of disease or symptoms toward normalcy is the focus of PD. At the center of the interaction between drugs and genes are pharmacogenes. Pharmacogenes are genes that have been found to play a role in the PD, PK or both aspects of drug response. PGx research is primarily conducted through in vitro and in vivo studies that aim to quantify the effects of a drug at the molecular, cellular and whole organism level. Currently within the PGx field, most big genomic datasets have been derived from cell line data or have been the reutilization of data sets created without regard to PGx specifically (Fig. 1, left). Thus far, these data sets have facilitated discoveries regarding drug PK and PD, as well as drug positioning and repurposing efforts.
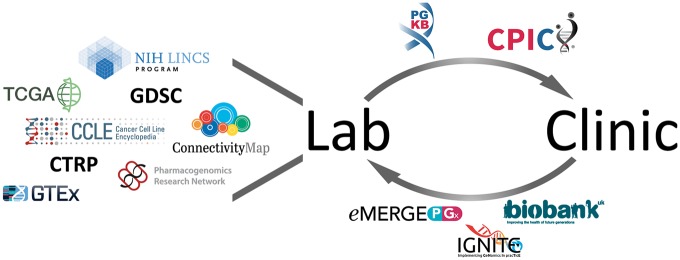
Figure 1.
General overview of the various resources that contain or utilize genomic data within pharmacogenomics (PGx), with arrows representing information flow. Much initial PGx discovery is carried out in the laboratory setting, through cell-line resources such as ConnectivityMap (4), LINCS (5), Genomics of Drug Sensitivity in Cancer (GDSC) (6), the Cancer Cell Line Encyclopedia (CCLE) (7) and Cancer Therapeutics Response Portal (CTRP) (8). The Cancer Genome Atlas (TCGA) (9) and the Genotype-Tissue Expression (GTEx) project (10) were not created for PGx research but have been used by PGx investigators. Additionally, the PGRN has sought to bring U.S. PGx research efforts together and has conducted numerous PGx studies. Published PGx findings are reviewed and aggregated by PharmGKB (11) in the form of variant and clinical annotations. The Clinical Pharmacogenetics Implementation Consortium (CPIC) brings together clinical and basic research scientists to evaluate the evidence and develop consensus for dosing guidelines in the clinical setting. Recent efforts, such as eMERGE-PGx (12), Implementing Genomics in Practice (IGNITE) (13) and the UKB (14), have sought to generate genomic data for individuals within clinical electronic health record systems and make that data available to PGx researchers for further discovery.
There are ongoing efforts to translate these laboratory findings to the clinic through the annotation, aggregation and integration of PGx information and the creation of reviewed clinical PGx guidelines (Fig. 1, top arrow). The widespread adoption of electronic medical records (EMR) systems within the United States over the last decade creates a large repository of information of phenotypes of potential interest to PGx researchers, such as clinical notes regarding drug efficacy and adverse drug events (ADE). Recent efforts undertaken to sequence large numbers of individuals with EMR data will enable large amounts of genomic data to flow from the clinic to the lab (Fig. 1, bottom arrow).
The field of PGx research has a long-standing tradition of laboratory and clinical collaboration (3), and the development of large genomic data sets will only further connect these two areas of research. This review covers recent PGx research efforts that have utilized large genomic data in the laboratory, the facilitation of those findings for clinical use and the future potential of PGx research with the recent integrations of clinical and genomic data.
PGx Research Efforts in the Laboratory
Initially, PGx discoveries were primarily the result of clinical observations of variation in drug response between patients (3). These clinical observations of systematic differences, often between patients of different ethnicities, were then investigated through in vitro laboratory to identify the genetic or environmental mechanisms behind the observed differences. Notable examples of early PGx discoveries are the role of G6PD in primaquine metabolism (15,16), differences in isoniazid response (17–19) later shown to be the result of NAT2 mutations (20), and the effect of TMPT alleles on the ability to metabolize 6-mercaptopurine (21–23). These early PGx discoveries were not the result of large systematic scientific efforts, but rather observant clinicians and small laboratory experiments.
As sequencing costs declined disease consortiums started to generate large cohorts of genotyped and/or sequenced individuals (24–26). The advantage of these large cohort studies is their ability to systematize genomic discovery via sufficient statistical power to detect real genetic differences associated with the phenotype of interest. PGx-related research has lagged behind these efforts as no similar resource currently exists. Genomic profiling of drug response has been conducted by other means, such as through cell lines, which have long been a mainstay of laboratory research. In vitro studies have several advantages over human studies, as cells can be easily cultured in large numbers, there are fewer ethical dilemmas surrounding the experimental use of cells, and cells have proven useful for studying many aspects of fundamental biology. The prominence of cell lines within biological research has led to the creation of resources with detailed information on their molecular profiles (6–8). These resources characterize cells at the genomic, transcriptomic and occasionally proteomic level. In aggregate, these profiled cell lines represent a big genomic data set for PGx discovery.
The relative ease of conducting cell line experiments has led to large studies that focus on the cellular response to chemical perturbations. Many studies have been conducted within the differential gene expression framework (4,5,8,27), notable among them are the Connectivity Map (CMap) (4) and the larger follow-up study the Library of Integrated Cellular Signatures (LINCS) (5) conducted by the National Institute of Health (NIH). The LINCS Consortium recently released 1.3 million new molecular profiles of compounds. These profiles capture changes in cellular gene expression in response to 19 811 different compounds in 3–77 different cell lines. Previous drug molecular profiles have been used to perform a variety of pharmacogenomic outcome predictions (28–31). The utility of cell line data has led to the development of resources that support access to and aggregation from the many different cell line data resources (32–35).
A study by Iorio et al. (28) investigated the ability to map tumor-level phenotypes for drug response onto human cancer cell lines using somatic mutations, copy number alterations, DNA methylation and gene expression data. They matched 11 289 tumors from 29 different tissues in the Cancer Genome Atlas (TCGA) (9) to 1001 profiled cancer cell lines from the Genomics of Drug Sensitivity in Cancer resource (6) based on the tissue of origin and ‘cancer functional events’ (CFEs). The CFEs, key molecular aberrations, were identified from the tumor and somatic data from COSMIC (36) and TCGA. Drug response information from cell lines with similar CFEs to the tumor of interest were used to predict the tumor’s response to a given drug therapy. Driver mutations and copy number alterations were the most predictive of drug response in specific tissue types, with methylation data incrementally improving the prediction models. Gene expression was most informative for pan-cancer predictions, while contributing little to tissue-specific models. Using their models, it could be possible to use cell line-derived data to prioritize treatment regimens for cancer patients based on their tumor profiles.
Another effort predicted drug–response for TCGA patients using a regression model trained on cell line gene expression and drug response data. The imputed drug–response was then used to investigate associations between the imputed drug response and somatic mutations in the TCGA patients (29). The authors replicated findings of known somatic biomarkers for a few drugs currently in use and report more than 140 novel gene–drug associations. They attribute the large number of novel associations to the increased statistical power of their TCGA-based approach compared with previous association studies. Imputation of drug–response may be one possible path for PGx researchers to fully utilize existing genomic data sets that lack drug–response information.
The above studies used gene expression data derived from array-based platforms, which has limited resolution for identifying transcriptional isoforms. An effort by the Pharmacogenomics Research Network (PGRN) used RNA-Seq to analyze the transcriptome profiles of 389 pharmacogenes across liver, kidney, heart and adipose tissues (37). Subsets of pharmacogenes that shared similar expression patterns across the tissue types were discovered, as well as pharmacogenes that were expressed across all tissues at similar rates. Novel splice events and high levels of person to person variability in expression for a subset of pharmacogenes were found.
To understand how different human populations might respond to different compounds and how useful existing PGx markers might be for a given population, efforts have sought to characterize global PGx variation. These studies focused on actionable PGx variants, as the unit of analysis, drawing from existing PGx knowledge resources at the Food and Drug Administration (FDA), the European Medicines Agency (EMA) and the Pharmacogenomics Knowledgebase (PharmGKB) (11). Wright et al. focused on 1000 Genomes Phase 3 (38) data and 120 pharmacogenes, for which they assessed the level of variation within these genes across 26 global populations (39). A median of three variants from PharmGKB with level 1 A/B evidence were found per individual, with most (97%) of individuals having at least one of these variants. Most of the variation (>90% of variants) within the pharmacogenes were rare with allele frequencies less than 0.5%. The work of Wright et al. demonstrates the potential impact of rare variants on the function of pharmacogenes on the individual basis, as well as the efficacy of sequencing data for assessing PGx variation. Further characterization of the impact of rare variants on drug–response is needed, as little is currently known. Rare variants present a unique challenge in that their low levels of prevalence require very large cohorts for there to be sufficient statistical power to distinguish their effects.
Mizzi et al. (40) conducted a similar study using genotype data and focused only on European populations and FDA/EMA-approved clinical PGx variants. Their findings showed significant differences in prevalence of seven PGx variants among ethnic groups within Europe. Differences in population prevalence identified in these studies could be used to help prioritize individuals for PGx testing based on their ethnic background.
Cell line and population level studies, among other studies conducted in the laboratory setting, aim to characterize the response of different genomic profiles to various therapeutics. Ideally, the knowledge created in the laboratory can be utilized in the clinical setting to inform prescribing practices and to improve patient outcomes.
Translation of Laboratory PGx Findings to the Clinic
In the hospitalized setting, studies have shown up to 6.7% of patients experience a serious ADE (41). The translation of pharmacogenomic discoveries in the laboratory to the clinic aims to tackle this problem by aiding clinicians in drug selection and drug dosing (42). The translation process requires that research findings be aggregated, reviewed and formed into clinical guidelines. Healthcare systems can then incorporate these guidelines into the workflows of their physicians, as long as they have genetic information about their patients.
PGx findings can be implemented in the clinic through the use of the Clinical Pharmacogenetic Implementation Consortium (CPIC) guideline recommendations and high level clinical annotations found on PharmGKB. In addition to CPIC, there are other efforts such as the Dutch Pharmacogenetics Work Group (DPWG) (43,44) to develop clinical dosing guidelines.
In order to implement these guidelines, it is necessary to first have patient genotype, haplotype and diplotype information in hand. The Pharmacogenomics Clinical Annotation Tool (PharmCAT) is a tool designed to aid researchers and clinicians in the translation of patient DNA sequence, exomes and SNP-chip genotypes into patient-based drug-prescribing recommendations from CPIC (and other dosing guidelines) using PharmGKB annotations and PGx gene haplotype definitions. These resources and their relationships are described below (Fig. 2).
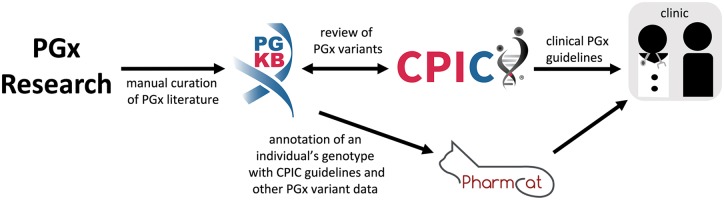
Figure 2.
Relationships between different pharmacogenomics (PGx) variant resources. PharmGKB manually curates PGx literature and acts as a repository of information about PGx variants. CPIC uses PharmGKB literature and drug label annotations to help rank gene–drug pairs for genotype-based drug prescribing guideline development, and incorporates PhamGKB annotations with independent literature reviews to create the guideline recommendations, that can then be used to support clinical decision making. PharmCAT facilitates genome annotation with actionable PGx information, starting with CPIC guideline recommendations.
PharmGKB is an online knowledge resource for pharmacogenomics (11). It focuses on the annotation, aggregation, integration and dissemination of pharmacogenomics peer-reviewed literature, drug labels with pharmacogenomics content and drug-prescribing guidelines based on patient genotypes. Staff scientists manually curate gene–drug–phenotype and variant–drug–phenotype relationships from these sources using standardized vocabularies and structured web forms.
The variant–drug–phenotype relationships from the literature are referred to as variant annotations, which are aggregated to create clinical annotations—summaries of all of the variant annotations of PGx literature found in PharmGKB. These clinical annotations are assigned a ‘level of evidence’ (LOE) based on the number and content from the aggregated annotations. LOE range from (1A) PGx associations known to be implemented in the clinic; (1B) individually replicated associations with high odds ratios, preferably in large populations; (2A) replicated associations for variants in known pharmacogenes; (2B) other individually replicated; (3) associations with little or conflicting evidence; (4) associations with weak evidence, including only in vitro evidence or case studies. The LOE for a clinical annotation can change, moving higher as more published studies regarding an association are annotated or moving lower if conflicting or contradictory published studies are annotated.
CPIC is focused on publishing that are evidence-based and genotype-dependent drug-dosing guidelines (45). These guidelines provide specific guidance to clinicians who have patient genotypes in hand and want to leverage it when prescribing medication, essentially implementing PGx. Each guideline is authored by a group of clinician and research experts in that field who conduct a comprehensive literature search and evidence review in combination with their own expertise to provide recommendations. Each recommendation is given a ‘strength’ based on the amount and quality of evidence found to support it. It is important to note that CPIC does not provide recommendations on who or when to test. Outside of the CPIC and PharmGKB ecosystem, other aggregations of PGx information exist. Among them, PharmacoGenomic Mutation Database is the most comprehensive, with a variety of data on more than 1300 approved drugs (46).
Given the effort that has been put in to create clinical PGx guidelines, several recent studies have focused on the logistics of translation to the clinic. The aim of these studies was understanding how pharmacogenetics can integrate into the clinical setting on a large scale. A study conducted at Vanderbilt through the Pharmacogenomics Resource for Enhanced Decisions in Care and Treatment (PREDICT) program has seen >10 000 patients undergo preemptive pharmacogenetic testing (47). The Vanderbilt effort focused on five drug–genome interactions and found 91% of the genotyped patients to have at least one actionable variant. Notably, they found the preemptive approach lowered the required number of genetic tests when compared to a reactive prescription-triggered approach. Another pilot project performed by the Right Drug, Right Dose, Right Time–Using Genomic Data to Individualize Treatment Protocol (RIGHT) genotyped over 1000 individuals across five major pharmacogenes—SLCO1B1, CYP2C19, CYP2C9, VKORC1 and CYP2D6 (48). The patients’ genotypes are integrated into their EMR and a decision support tool alerts clinicians of relevant genetic information when ordering drugs through the EMR. The RIGHT pilot project found that 99% of patients genotyped had a potentially actionable variant that could affect future drug prescribing. Similarly, the NIH Undiagnosed Disease Network has been genotyping important pharmacogenetic variants in their cohort. In a recent study, the NIH Undiagnosed Disease Network found 9 of their 359 patients with available medication records and pharmacogenetic genotyping were currently on a medication with altered efficacy based on PharmGKB pharmacogenetic guidelines (49). This study’s anecdotal cases highlight the potential impact of pharmacogenetics, – for example one patient whose genotype indicated decreased morphine efficacy had clinically reported better pain control with ibuprofen than with morphine, which is considered a more potent pain medication.
There are many different models for delivering pharmacogenomic information to providers and patients. Some healthcare systems are experimenting with preemptive genotyping of a large fraction of their population so that the information is in the medical record at the time of prescribing (e.g. Vanderbilt, U. Florida) (50,51). Others set up outpatient consultative clinics where experts review the results of genotyping and provide advice to primary care physicians about potential changes in medication use or dose (e.g. Stanford, U. Chicago) (52). Others are considering point-of-care testing in the inpatient setting for critical decisions when the genotyping can be done quickly enough to inform prescribing in real time.
There is a growing industry of companies that will check for common variation and provide reports for providers. The most comprehensive resource for finding PGx testing is the NCBI Genetic Testing Registry that ask companies to providing PGx testing or reporting to submit their data. Additionally, CPIC provides a list of implementers at https://cpicpgx.org/implementation/. In general, these companies rely on published data, such as CPIC guidelines to genotype common variants whose effects are well understood, at least in the populations in which they were discovered.
These genome–drug interactions must be viewed in the context of drug–drug interactions, and so the triad of drug–drug–genome can sometimes illuminate interactions that are more likely in a particular patient because of the genomic background associated with that drug–drug interaction. Although there are many research projects attempting to understand the effects of rare variants on drug response (particularly in genes that are known to be important for drug response based on their common variants, such as CYP2D6 and others), there is not yet routine use of exome or full genome sequencing in clinical settings.
In addition to trials investigating the effect of wide-scale preemptive pharmacogenetic testing, there are also clinical trials examining the impact of implementing individual drug–gene interactions. For example, a randomized control trial of warfarin pharmacogenetics found that using genotype-guided warfarin dosing compared to clinically guided dosing reduced the risk of adverse events in an older population initiating warfarin after elective knee or hip surgery (53).
The addition of PGx guidelines to patients’ EMRs adds an additional layer of information to the vast amount of data already being captured within the EMR. Further leveraging of EMR data could enable novel PGx discoveries if genomic information was also available.
Clinical Data to Big Genomic Data
Recent efforts to sequence large cohorts of individuals present numerous pharmacogenomic opportunities. Biobanks, such as the UK Biobank (UKB) (14) and others offer genetic data linked to a vast amount of phenotypic data including drug prescription information and clinical notes. Other initiatives such as eMERGE-PGX (12) have performed targeted sequencing of known pharmacogenetic genes and collected drug response phenotypes. These resources, though relatively new, present rich opportunity for discovery.
The UKB has made genetic and phenotypic data for ∼500 000 individuals available to researchers worldwide and presents opportunities to identify novel drug targets and pharmacogenes. Currently, the UKB provides clinical data in the form of ICD-10 coding and self-reported diagnoses. The only drug-related phenotypes currently available are the prescriptions each subject was taking at the time of enrollment with limited dosage information. This minimal drug data presents a challenge for conducting meaningful pharmacogenomic research, but the UKB has announced that clinical notes for each subject will be released in 2018. Clinical notes provide the opportunity for text mining to ascertain the specifics of a drug regimen and adverse event occurrences (54,55), which could then be used to build or refine a PGx cohort.
Additionally, an EMR-linked cohort at the scale of the UKB enables the use of phenome-wide association studies, termed pheWAS, for drug target discovery. The now famous example of protein-truncating variants (PTVs) in PCSK9 (56,57) that led to the development of LDL cholesterol lowering drugs exemplifies methods that can now be applied across the genome and across many phenotypes. By looking at the effect of PTVs, novel drug targets have been suggested for diseases such as asthma and hyperthyroidism, among other diseases (58).
A large-scale sequencing study, eMERGE-PGx, focused on pharmacogenomic variation has recently been made sequencing data publically available through dbGAP. eMERGE-PGx published targeted pharmacogenomic sequencing on 82 genes in 9010 individuals linked with and plans to release drug response phenotypes as well (59). These data will no doubt lead to the generation of new PGx hypotheses and clinical guidelines.
Conclusion
The broad use of pharmaceuticals in the healthcare setting speaks to the potential value of using PGx guidelines to enable safer and more efficacious drug-prescribing practices. Thus, PGx may be a primary way in which genomics can have a nearer term impact on patient care worldwide. The global recognition of the value of PGx information is exemplified by the fact that various agencies around the world already annotate drug labels with PGx information. Additionally, CPIC guidelines that take into account differences in risk due to population level differences in allele frequencies aid global efforts for PGx guideline adoption and implementation. Indeed, some countries have already adopted PGx into routine clinical care practices, such as the pharmacogenetic ID card system in Thailand and Taiwan (60).
Unfortunately, in the United States, there are challenges in getting PGx testing reimbursed, which may represent the biggest barrier to widespread use in the United States. A recent draft of national coverage determination for next-generation sequencing (NGS) diagnostic tests by the Centers for Medicare & Medicaid Services (CMS), the agency that regulates government funded healthcare programs, offered the possibility of CMS coverage/reimbursement for NGS cancer tests (61). A ruling from the CMS in favor of reimbursement for PGx clinical testing would be a major incentive for researchers in both academia and industry to pursue PGx test development in the United States.
Overall, PGx dosing guidelines have had positive results when evaluated for accuracy and efficacy (62), but there is still limited understanding of how other types of individual variability (e.g. rare variants, epigenetics, microbiome, etc.) impact drug response. Pursuing this opportunity will require the creation of new genomic data sets and creative us of existing resources. Hopefully, future studies will be of sufficient size and genetic resolution to measure the effects of rare variants on phenotypes of interest. Additionally, for them to have maximum impact on PGx research, they will need to be annotated with rich drug response phenotypes, such as the prescribed dosages of medications, occurrence of adverse drug reactions and the sequence in which medications were prescribed. This review highlighted some of the ways in which big genomic data has already made an impact on the field of pharmacogenomics, but the new efforts to link genetic information with large EMR repositories represents a potentially unprecedented opportunity for pharmacogenomic research.
Acknowledgements
A.L. and G.M. are supported by the Big Data to Knowledge (BD2K) from the National Institutes of Health (T32 LM012409). R.B.A, T.E.K. and M.W.C. are supported by the National Institutes of Health (R24 GM61374 and R24 GM115264). Additionally, R.B.A is supported by the National Institutes of Health (GM102365, LM05652) and the Food and Drug Administration (U01FD004979).
Conflict of Interest statement. None declared.
Funding
This work was supported by the National Institutes of Health [T32LM012409, R24GM61374, R24GM115264, GM102365, LM05652] and the Food and Drug Administration [U01FD004979].
Footnotes
PGx is used interchangeably for pharmacogenetics and pharmacogenomics throughout this publication.
References
- 1.Center for Disease Control National Center for Health Statistics: ‘Therapeutic drug use’. https://www.cdc.gov/nchs/fastats/drug-use-therapeutic.htm; January 18, 2018, date last accessed.
- 2. Kantor E.D., Rehm C.D., Haas J.S., Chan A.T., Giovannucci E.L. (2015) Trends in prescription drug use among adults in the United States from 1999–2012. JAMA, 314, 1818–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meyer U.A. (2004) Pharmacogenetics – five decades of therapeutic lessons from genetic diversity. Nat. Rev. Genet., 5, 669. [DOI] [PubMed] [Google Scholar]
- 4. Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.-P., Subramanian A., Ross K.N. (2006) The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science, 313, 1929–1935. [DOI] [PubMed] [Google Scholar]
- 5. Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K. (2017) A next generation Connectivity Map: L1000 platform and the first 1,000,000 profiles. Cell, 171, 1437–1452.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson I.R.. et al. (2013) Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res., 41, D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D.. et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seashore-Ludlow B., Rees M.G., Cheah J.H., Cokol M., Price E.V., Coletti M.E., Jones V., Bodycombe N.E., Soule C.K., Gould J.. et al. (2015) Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov., 5, 1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.M., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M., Network C.G.A.R. (2013) The Cancer Genome Atlas pan-cancer analysis project. Nat. Genet., 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. GTEx Consortium (2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whirl-Carrillo M., McDonagh E.M., Hebert J.M., Gong L., Sangkuhl K., Thorn C.F., Altman R.B., Klein T.E. (2012) Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther., 92, 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rasmussen-Torvik L.J., Stallings S.C., Gordon A.S., Almoguera B., Basford M.A., Bielinski S.J., Brautbar A., Brilliant M.H., Carrell D.S., Connolly J.J.. et al. (2014) Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin. Pharmacol. Ther., 96, 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weitzel K.W., Alexander M., Bernhardt B.A., Calman N., Carey D.J., Cavallari L.H., Field J.R., Hauser D., Junkins H.A., Levin P.A.. et al. (2016) The IGNITE network: a model for genomic medicine implementation and research. BMC Med. Genomics, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M.. et al. (2015) UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med., 12, e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clayman C., Arnold J., Hockwald R., Yount E.J., Edgcomb J., Alving A. (1952) 3. Toxicity of primaquine in caucasians. J. Am. Med. Assoc., 149, 1563–1568. [DOI] [PubMed] [Google Scholar]
- 16. Alving A.S., Carson P.E., Flanagan C.L., Ickes C.E. (1956) Enzymatic deficiency in primaquine-sensitive erythrocytes. Science, 124, 484–485. [DOI] [PubMed] [Google Scholar]
- 17. Hughes H.B. (1953) On the metabolic fate of isoniazid. J. Pharmacol. Exp. Ther., 109, 444–452. [PubMed] [Google Scholar]
- 18. Hughes H.B., Biehl J.P., Jones A.P., Schmidt L.H. (1954) Metabolism of isoniazid in man as related to the occurrence of peripheral neuritis. Am. Rev. Tuberc., 70, 266–273. [DOI] [PubMed] [Google Scholar]
- 19. Evans D.A.P., Manley K.A., McKusick V.A. (1960) Genetic control of isoniazid metabolism in man. Br. Med. J., 2, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vatsis K.P., Martell K.J., Weber W.W. (1991) Diverse point mutations in the human gene for polymorphic N-acetyltransferase. Proc. Natl. Acad. Sci. U. S. A., 88, 6333–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weinshilboum R.M., Sladek S.L. (1980) Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am. J. Hum. Genet., 32, 651–662. [PMC free article] [PubMed] [Google Scholar]
- 22. Lennard L., Van Loon J.A., Weinshilboum R.M. (1989) Pharmacogenetics of acute azathioprine toxicity: relationship to thiopurine methyltransferase genetic polymorphism. Clin. Pharmacol. Ther., 46, 149–154. [DOI] [PubMed] [Google Scholar]
- 23. Lennard L., Lilleyman J.S., Van Loon J., Weinshilboum R.M. (1990) Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia. Lancet (London, England), 336, 225–229. [DOI] [PubMed] [Google Scholar]
- 24. Wellcome Trust Case Control Consortium. (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature, 447, 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mahajan A., Go M.J., Zhang W., Below J.E., Gaulton K.J., Ferreira T., Horikoshi M., Johnson A.D., Ng M.C.Y., Prokopenko I.. et al. (2014) Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet., 46, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K.J., Ma C., Fontanillas P., Moutsianas L., McCarthy D.J.. et al. (2016) The genetic architecture of type 2 diabetes. Nature, 536, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keenan A.B., Jenkins S.L., Jagodnik K.M., Koplev S., He E., Torre D., Wang Z., Dohlman A.B., Silverstein M.C., Lachmann A.. et al. (2017) The library of integrated network-based cellular signatures NIH program: system-level cataloging of human cells response to perturbations. Cell Syst., 27, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H.. et al. (2016) A landscape of pharmacogenomic interactions in cancer. Cell, 166, 740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geeleher P., Zhang Z., Wang F., Gruener R.F., Nath A., Morrison G., Bhutra S., Grossman R.L., Huang R.S. (2017) Discovering novel pharmacogenomic biomarkers by imputing drug response in cancer patients from large genomics studies. Genome Res., 27, 1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Z., Clark N.R., Ma'ayan A. (2016) Drug-induced adverse events prediction with the LINCS L1000 data. Bioinformatics, 32, 2338–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ruffalo M., Stojanov P., Pillutla V.K., Varma R., Bar-Joseph Z. (2017) Reconstructing cancer drug response networks using multitask learning. BMC Syst. Biol., 11, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smirnov P., Safikhani Z., El-Hachem N., Wang D., She A., Olsen C., Freeman M., Selby H., Gendoo D.M.A., Grossmann P.. et al. (2016) PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics, 32, 1244–1246. [DOI] [PubMed] [Google Scholar]
- 33. Smirnov P., Kofia V., Maru A., Freeman M., Ho C., El-Hachem N., Adam G.-A., Ba-alawi W., Safikhani Z., Haibe-Kains B. (2018) PharmacoDB: an integrative database for mining in vitro anticancer drug screening studies. Nucleic Acids Res., 46, D994–D1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luna A., Rajapakse V.N., Sousa F.G., Gao J., Schultz N., Varma S., Reinhold W., Sander C., Pommier Y. (2016) rcellminer: exploring molecular profiles and drug response of the NCI-60 cell lines in R. Bioinformatics, 32, 1272–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Enache O.M., Lahr D.L., Natoli T.E., Litichevskiy L., Wadden D., Flynn C., Gould J., Asiedu J.K., Narayan R., Subramanian A. (2018) The GCTx format and cmap{Py, R, M} packages: resources for the optimized storage and integrated traversal of dense matrices of data and annotations. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L.. et al. (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res., 45, D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chhibber A., French C.E., Yee S.W., Gamazon E.R., Theusch E., Qin X., Webb A., Papp A.C., Wang A., Simmons C.Q.. et al. (2017) Transcriptomic variation of pharmacogenes in multiple human tissues and lymphoblastoid cell lines. Pharmacogenomics J., 17, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E.. et al. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wright G.E.B., Carleton B., Hayden M.R., Ross C.J.D. (2016) The global spectrum of protein-coding pharmacogenomic diversity. Pharmacogenomics J., 10.1038/tpj.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mizzi C., Dalabira E., Kumuthini J., Dzimiri N., Balogh I., Başak N., Böhm R., Borg J., Borgiani P., Bozina N.. et al. (2016) A European spectrum of pharmacogenomic biomarkers: implications for clinical pharmacogenomics. PLoS One, 11, e0162866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lazarou J., Pomeranz B.H., Corey P.N. (1998) Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA, 279, 1200–1205. [DOI] [PubMed] [Google Scholar]
- 42. Karczewski K.J., Daneshjou R., Altman R.B. (2012) Chapter 7: pharmacogenomics. PLoS Comput. Biol., 8, e1002817.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swen J.J., Wilting I., de Goede A.L., Grandia L., Mulder H., Touw D.J., de Boer A., Conemans J.M.H., Egberts T.C.G., Klungel O.H.. et al. (2008) Pharmacogenetics: from bench to byte. Clin. Pharmacol. Ther., 83, 781–787. [DOI] [PubMed] [Google Scholar]
- 44. Swen J.J., Nijenhuis M., de Boer A., Grandia L., Maitland-van der Zee A.H., Mulder H., Rongen G.A.P.J.M., van Schaik R.H.N., Schalekamp T., Touw D.J.. et al. (2011) Pharmacogenetics: from bench to byte–an update of guidelines. Clin. Pharmacol. Ther., 89, 662–673. [DOI] [PubMed] [Google Scholar]
- 45. Relling M.V., Klein T.E. (2011) CPIC: clinical pharmacogenetics implementation consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther., 89, 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaplun A., Hogan J.D., Schacherer F., Peter A.P., Krishna S., Braun B.R., Nambudiry R., Nitu M.G., Mallelwar R., Albayrak A. (2016) PGMD: a comprehensive manually curated pharmacogenomic database. Pharmacogenomics J., 16, 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Van Driest S.L., Shi Y., Bowton E.A., Schildcrout J.S., Peterson J.F., Pulley J., Denny J.C., Roden D.M. (2014) Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin. Pharmacol. Ther., 95, 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ji Y., Skierka J.M., Blommel J.H., Moore B.E., VanCuyk D.L., Bruflat J.K., Peterson L.M., Veldhuizen T.L., Fadra N., Peterson S.E.. et al. (2016) Preemptive pharmacogenomic testing for precision medicine: a comprehensive analysis of five actionable pharmacogenomic genes using next-generation DNA sequencing and a customized CYP2D6 genotyping cascade. J. Mol. Diagn., 18, 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee E.M.J., Xu K., Mosbrook E., Links A., Guzman J., Adams D.R., Flynn E., Valkanas E., Toro C., Tifft C.J.. et al. (2016) Pharmacogenomic incidental findings in 308 families: the NIH Undiagnosed Disease Program experience. Genet. Med., 18, 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoffman J.M., Dunnenberger H.M., Kevin Hicks J., Caudle K.E., Whirl Carrillo M., Freimuth R.R., Williams M.S., Klein T.E., Peterson J.F. (2016) Developing knowledge resources to support precision medicine: principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC). J. Am. Med. Inform. Assoc., 23, 796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weitzel K.W., Elsey A.R., Langaee T.Y., Burkley B., Nessl D.R., Obeng A.O., Staley B.J., Dong H.-J., Allan R.W., Liu J.F.. et al. (2014) Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am. J. Med. Genet. C. Semin. Med. Genet., 166, 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O’Donnell P.H., Wadhwa N., Danahey K., Borden B.A., Lee S.M., Hall J.P., Klammer C., Hussain S., Siegler M., Sorrentino M.J.. et al. (2017) Pharmacogenomics-based point-of-care clinical decision support significantly alters drug prescribing. Clin. Pharmacol. Ther., 10.1002/cpt.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gage B.F., Bass A.R., Lin H., Woller S.C., Stevens S.M., Al-Hammadi N., Li J., Rodriguez T.J., Miller J.P., McMillin G.A.. et al. (2017) Effect of genotype-guided warfarin dosing on clinical events and anticoagulation control among patients undergoing hip or knee arthroplasty: the GIFT randomized clinical trial. JAMA, 318, 1115–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harpaz R., Callahan A., Tamang S., Low Y., Odgers D., Finlayson S., Jung K., LePendu P., Shah N.H. (2014) Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Saf., 37, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Leeper N.J., Bauer-Mehren A., Iyer S.V., LePendu P., Olson C., Shah N.H. (2013) Practice-based evidence: profiling the safety of cilostazol by text-mining of clinical notes. PLoS One, 8, e63499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cohen J., Pertsemlidis A., Kotowski I.K., Graham R., Garcia C.K., Hobbs H.H. (2005) Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet., 37, 161–165. [DOI] [PubMed] [Google Scholar]
- 57. Dadu R.T., Ballantyne C.M. (2014) Lipid lowering with PCSK9 inhibitors. Nat. Rev. Cardiol., 11, 563–575. [DOI] [PubMed] [Google Scholar]
- 58. DeBoever C., Tanigawa Y., McInnes G., Lavertu A., Chang C., Bustamante C.D., Daly M.J., Rivas M.A. (2017) Medical relevance of protein-truncating variants across 337,208 individuals in the UK Biobank study. 10.1101/179762. [DOI] [PMC free article] [PubMed]
- 59. Bush W.S., Crosslin D.R., Owusu-Obeng A., Wallace J., Almoguera B., Basford M.A., Bielinski S.J., Carrell D.S., Connolly J.J., Crawford D.. et al. (2016) Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin. Pharmacol. Ther., 100, 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sukasem C., Chantratita W. (2016) A success story in pharmacogenomics: genetic ID card for SJS/TEN. Pharmacogenomics, 17, 455–458. [DOI] [PubMed] [Google Scholar]
- 61. Jensen T.S., Chin J., Baldwin J., Rollins J., Ashby L., Li C., Szarama K. (2018) Proposed decision memo for next generation sequencing (NGS) for medicare beneficiaries with advanced cancer (CAG-00450N).
- 62. Klein T.E., Altman R.B., Eriksson N., Gage B.F., Kimmel S.E., Lee M.-T.M., Limdi N.A., Page D., Roden D.M., Wagner M.J.. et al. (2009) Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med., 360, 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]